Abstract
In contrast to mice, in sheep no genome-wide demethylation of the paternal genome occurs within the first postfertilization cell cycle. This difference could be due either to an absence of a sheep demethylase activity that is present in mouse ooplasm or to an increased protection of methylated cytosine residues in sheep sperm. Here, we use interspecies intracytoplasmic sperm injection to demonstrate that sheep sperm DNA can be demethylated in mouse oocytes. Surprisingly, mouse sperm can also be demethylated to a limited extent in sheep oocytes. Our results suggest that the murine demethylation process is facilitated either by a sperm-derived factor or by male pronuclear chromatin composition.
Methylation of the DNA cytosine residues in CpG dinucleotides is part of the complex epigenetic mechanism that has evolved to silence genomic sequences where their transcription either is not required for development or may be detrimental to genomic stability (reviewed in refs. 1–3). DNA methylation also plays an important role in allele-specific repression at imprinted gene loci, regulating such processes as fetal growth and development as well as X inactivation (reviewed in refs. 4–6). Whereas the genome-wide methylation patterns and levels of differentiated somatic lineages remain largely constant, very dynamic changes have been reported to occur in the preimplantation embryo in association with the formation of pluripotent embryonic nuclei.
After fertilization, the sperm and egg genomes are remodeled into pronuclei, which appose within the oocyte cytoplasm before the first embryonic mitosis. However, the highly condensed sperm chromatin requires extensive nuclear remodeling and protamine-histone exchange, unlike nucleosomal maternal chromatin. By using an antibody against 5-methylcytosine (5mC), the presumptive male pronucleus of mouse, rat, pig, human, and, to a lesser extent, cow embryos have been shown to actively demethylate before syngamy, whereas the female pronucleus retains genome-wide methylation (7–12). In contrast, active demethylation of the paternal genome is not observed in early sheep or rabbit embryos (with an intermediate state in the cow) (12), which suggests that it is not an obligate requirement for mammalian development. The discovery that the dramatic changes in DNA methylation associated with early formative events in the mouse embryo are not conserved in the sheep allows a unique opportunity to investigate the regulatory mechanisms involved. Mouse ooplasm can fully demethylate multiple male pronuclei in polyspermic embryos, which raises the question of whether the demethylating activity resides in the fertilized oocyte or is intimately associated with the sperm (10). We have now used interspecies intracytoplasmic sperm injection (ICSI) to mimic the events of normal fertilization and investigate whether species differences in the oocyte environment or sperm composition determines the extent of male pronuclear demethylation.
Materials and Methods
All animal procedures were under strict accordance with U.K. Home Office regulations and within a project license issued under the Animal (Scientific Procedures) Act of 1986.
In Vitro and in Vivo Embryo Production. Ovine in vitro embryo production and in vivo surgical recovery were performed as described (13). Pronuclear stage embryos were then collected at appropriate times for immediate fixation. Superovulation in B6D2F1 mice (age 8–10 wk) was induced by injection of 5 international units of equine chorionic gonadotropin and 5 international units of human chorionic gonadotropin 48 h later. For in vivo fertilization, females were mated overnight. One-cell embryos were then collected and cultured in Chatot–Ziomek–Bavister (CZB) medium under 5% CO2 in air at 37°C until fixation. All embryos were fixed in freshly prepared 4% paraformaldehyde at 4°C.
ICSI. For ICSI into mouse metaphase II oocytes (17–19 h after human chorionic gonadotropin injection), frozen mouse, sheep, and bovine sperm were prepared by thawing briefly in a 37°C water bath, centrifuging for 2 min at 400 × g, and washing the pellet in 500 μl of PBS. After a final centrifugation of 5 min at 500 rpm, sperm in the supernatant were then immobilized to avoid oocyte lysis by diluting 1:5 in a 10% polyvinylpyrrolidone solution 30 min before injection. In the case of mouse sperm, tails were removed by the thawing/centrifugation process. Oocytes were placed in a drop of Hepes-buffered CZB medium, and single spermatozoa were injected with a piezo-driven micropipette. The reconstructed embryos cultured in CZB medium spontaneously activated.
For ICSI into sheep oocytes, abattoir-derived oocytes were matured in vitro as described (14). Fresh mouse sperm were allowed to disperse in Hepes/PBS for 2–5 min at room temperature. Bovine and ovine spermatozoa frozen in straws (kindly provided by Istituto Zooprofilattico Sperimentale, Teramo, Italy), were thawed in a water bath at 35°C and washed twice in Hepes/synthetic oviductal fluid (SOF). A small drop of sperm suspension from bovine, ovine, and murine samples was mixed thoroughly with 30 μl of injection medium (150 mM KCl/2 mM Pipes, pH 7.3) containing 12% polyvinylpyrrolidone immediately before ICSI. Sheep metaphase II oocytes where then injected with spermatozoa by using an inverted Nikon microscope fitted with a Narishighe manipulator. Injected oocytes were activated (ovine oocytes do not spontaneously activate after ICSI) by incubation in medium TCM 199 plus BSA containing 5 μM ionomycin (Sigma) for 5 min, followed by a 3-h incubation in SOFaaBSA medium (14) plus 10 μg/ml cycloheximide (Sigma). After activation, ICSI-fertilized oocytes were transferred to 20 μl of SOFaaBSA culture drops for 18–20 h until fixation.
For ICSI into bovine oocytes, abattoir-derived oocytes were matured as described (15). Motile spermatozoa (bull and ram) were obtained by centrifugation of frozen-thawed semen on a Percoll discontinuous density gradient for 40 min at 750 × g. Viable spermatozoa were washed in Ca2+-free Tyrode's solution containing albumin, lactate, and pyruvate (TALP) and pelleted by centrifugation for 10 min at 400 × g. Fresh mouse sperm was used. For ICSI, sperm was piezo-injected as described (16). In contrast to sheep, bovine oocytes do not require exogenous activation after ICSI. To investigate whether artificial activation had any effect on the methylation status of the pronuclei, both sheep and bovine sperm were injected into bovine oocytes either with (n = 11 and n = 9, respectively) or without (n = 12 and n = 13, respectively) subsequent 5 μM ionomycin (6 min), 10 μg/ml cycloheximide (5 h, 10 μg/ml) activation in SOF supplemented with essential and nonessential amino acids and 16 mg/ml BSA (SOFaaBSA). There was no observable difference in the methylation status of any group (results not shown). Bovine oocytes injected with mouse sperm were activated as above, and in all cases activated oocytes were cultured for 20–22 h in SOFaaBSA before fixation.
Immunodetection of 5mC. Immunodetection of 5mC, confocal microscopy, and image quantification were performed as in Beaujean et al. (13). Briefly, the three-dimensional confocal Z-series were summed with imagej software, version 1.30a. Total nuclear intensities were measured with simple pci imaging software by manually outlining each pronucleus. Pronuclear sizes were also determined on merged images and therefore represent the nuclear area at its greatest diameter. The three-dimensional Z-series confocal images were later merged with the confocal assistant software, version 4.02, to produce a two-dimensional image depicting the staining patterns and total intensities of all of the nuclei present. Processed images were assembled and pseudocolored with photoshop, version 6.0 (Adobe Systems, Mountain View, CA).
Results
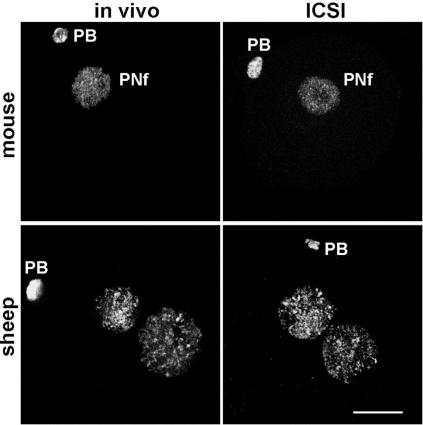
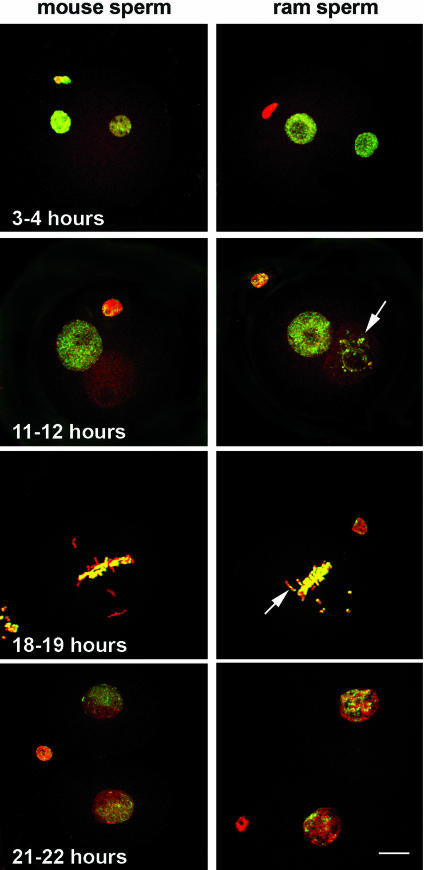
Interspecific ICSI Reveals Ooplasmic Regulation of Sperm Demethylation. We confirmed that intracytoplasmic microinjection of mature conspecific spermatozoa into both mouse and sheep oocytes recapitulates the pronuclear methylation status observed for in vitro and in vivo fertilized embryos (Fig. 1 and ref. 12). To test whether the absence of male pronuclear demethylation observed in the sheep embryo could be due to an increased protection of methylated cytosine residues in sheep sperm, we then compared mouse conspecific ICSI with interspecies ICSI performed by microinjecting sheep sperm into mouse oocytes. Fig. 2 shows that, for both types of sperm, demethylation correlates with sperm decondensation. It is noticeable that condensed regions of the sheep genome retain some methylation through mitosis in the mouse oocyte and possibly through to the two-cell stage. It is unclear why these regions in sheep sperm are refractory to the demethylation process, but it may be a consequence of their compact organization. Satellite sequences are relatively undermethylated in mouse sperm, so demethylation is normally not required at these regions (17, 18). Sheep/mouse hybrids also exhibit 5mC staining distributed more throughout the nucleus of the two-cell blastomeres, in contrast to the strong asymmetric distribution in the mouse–mouse combination. This experiment also demonstrates that the timings of pronuclear formation and first cleavage are similar in control and hybrid zygotes, suggesting cytoplasmic regulation of the cell-cycle progression regardless of the sperm origin.
Fig. 1.
Immunodetection of 5mC in mouse or sheep embryos produced either in vivo or by ICSI. Independent of the protocol, we detected methylation in both pronuclei for the sheep and only in the female pronucleus (PNf) for the mouse. PB, polar body. (Bar, 20 μm.)
Fig. 2.
Confocal images of hybrid zygotes obtained by either mouse sperm ICSI or ram sperm ICSI into mouse oocytes. Zygotes were stained with propidium iodide for chromatin (red) and FITC-conjugated secondary antibody for 5mC (green). Fixation was performed at various times after fertilization: 3–4, 11–12, 18–19 h, at mitosis; 21–22 h at the two-cell stage. For both sperm types, genome-wide DNA methylation starts to decrease with sperm decondensation to finally become undetectable by the end of the first cell cycle, except for some condensed regions in the sheep-derived pronucleus (arrows). At the two-cell stage, maternal and paternal compartments are still separated as shown by the differential green staining. However, in sheep/mouse hybrids, the distinction is less striking. (Bar, 10 μm.)
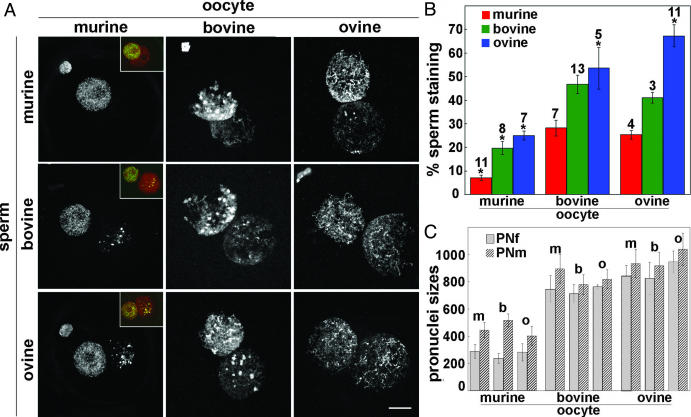
Because the sheep oocyte cytoplasm does not appear able to demethylate the paternal pronucleus after fertilization, in contrast to mouse ooplasm (with an intermediate scenario in the cow), we examined the methylation status of murine, bovine, or ovine sperm injected into murine, bovine, and ovine metaphase II oocytes (Fig. 3A). Quantification of relative male-versusfemale pronuclear 5mC intensity is shown in Fig. 3B. In contrast to sheep, mouse sperm injected into ovine oocytes (mouse/ovine hybrids) showed significant demethylation compared with the heavily demethylated mouse sperm in murine oocytes (Fig. 3A). The level of demethylation in the mouse/ovine hybrid seems to be the same as that seen in the mouse/bovine hybrid (Fig. 3). Ram sperm, which does not normally demethylate in the sheep oocyte, was also partially demethylated in the bovine oocyte (Fig. 3A). Bull sperm injected into mouse oocytes was also demethylated more heavily than in either bovine or ovine oocytes (Fig. 3A). The differential intensity of 5mC immunostaining between the male and female pronuclei is partially dependent on the source of the oocyte cytoplasm (Fig. 3B). Regardless of sperm species origin, murine oocytes have significantly higher demethylating activity than those of the sheep, with bovine oocytes demonstrating an activity between that of the mouse and sheep. This correlates very well with the observed degree of pronuclear demethylation observed within conspecific zygotes. Thus, the degree of demethylation of the male pronucleus relates primarily to the composition of the oocyte environment. However, in all types of ooplasm, the degree of mouse sperm demethylation is higher than for bull or ram sperm, suggesting that an undefined difference in the properties of the sperm also contributes to the demethylation process.
Fig. 3.
(A) Differential demethylating process in ICSI hybrid zygotes depends on oocyte source. Murine, bovine, or ovine sperm were microinjected into murine, bovine, and ovine oocytes. Hybrid zygotes were then processed for 5mC immunodetection at the end of the first cell cycle, after replication had taken place, i.e., 11–12, 20–22, and 18–20 h for mouse, cow, and sheep oocyte recipients, respectively. (Insets) Merged images of DNA (red) and methylation staining (green), with yellow showing overlapping signal. ICSI into mouse oocytes clearly demonstrates high demethylation capacity of the mouse cytoplasm regardlessofthe sperm origin. In contrast, sperm demethylation was lower in bovine oocytes and almost nonexistent in sheep oocytes. (Bar, 10 μm.) (B) Relative amounts of the methylation quantified in the male versus the female pronuclei (the number of hybrids analyzed is indicated above each column). Error bars represent the SE between hybrids within one group. Statistical analyses were performed with Student's t test. The asterisks denote values that are statistically different for one sperm origin between the different oocyte sources (P < 0.01). This demonstrates that, independently of the oocyte source, the mouse paternal pronucleus is significantly less methylated than bull- or ram-derived pronuclei (P < 0.01). In contrast, mouse and bull sperm have similar methylation levels in cow and sheep oocytes, whereas sheep sperm methylation is even higher after injection into sheep oocytes (P < 0.01). (C) Sizes of the female (PNf) and male (PNm) pronuclei (arbitrary units) quantified after injection of murine (m), bovine (b), or ovine (o) sperm into various oocytes. Error bars represent the SE between hybrids within one group.
We also observed some differences in pronuclear size. The normal pattern of a larger male versus female pronucleus in the mouse, but equivalent sizes in sheep and cow zygotes, was recapitulated by ICSI. In the hybrid zygotes, however, sheep and bovine sperm-derived pronuclei were smaller after injection into the mouse cytoplasm (Fig. 3A). On the other hand, the mouse sperm-derived male pronucleus was larger after injection in cow or sheep oocytes than in the mouse/mouse hybrid (Fig. 3A). The size of the male pronucleus seems to be conditioned by the oocyte environment regardless of the sperm origin. Because the larger male pronucleus in normal mouse embryos is associated with transcriptional activity, sperm-derived factors may also be important to induce the rapid remodeling required to help transcriptional activation of mouse sperm chromatin within a few hours of fertilization. On the contrary, major transcriptional activity in sheep and bovine embryos does not start before the 8- to 16-cell stage. Whether the degree of nuclear decondensation is associated with differences in transcriptional activity in the interspecific hybrids remains to be established.
Discussion
Considerable interest has recently arisen in the factors present in oocyte cytoplasm that reprogram not only the sperm and egg genomes in normal development but also somatic cells by the process of somatic cell nuclear transfer (reviewed in refs. 19–21). We were very surprised to initially uncover dramatic differences in the epigenetic reprogramming events occurring between sheep and mouse during normal preimplantation development (19). These species differences were extended to the rabbit embryo, which like sheep retains methylation in both pronuclei (12). In contrast, human 1-cell embryos generated by ICSI also show an asymmetric labeling of pronuclei (12). Here, we demonstrate that the species differences in male pronuclear demethylation can be attributed primarily to variation in the demethylase activity of the oocyte cytoplasm. However, it is evident that sperm composition also contributes to this process. Mouse, bovine, and sheep sperm show essentially the same trend of demethylation in interspecies hybrids, being less demethylated in the sheep and ovine oocytes with maximum/total demethylation in the mouse oocyte. Because the degree of pronuclear demethylation correlates negatively with pronuclear size in both conspecific and interspecific hybrid zygotes, demethylation activity may be influenced by the degree of nuclear condensation. The species differences we observed might also reflect variation in the efficiency of the demethylating factor, the presence or absence of activators/inhibitors, or even distinct protein quantities of the putative demethylase.
An Oocyte Demethylation Activity? The discovery of demethylation of the murine paternal pronucleus 4 h after fertilization, before DNA replication had taken place, demonstrated the existence of an active demethylation process in mammalian cells (7, 9–10). Our present findings provide evidence that this process is mainly maternally driven in mouse oocytes, although the nature of the activity is not known. Previous reports of active demethylase activity in a human lung carcinoma cell line (22), putatively identified as the methyl-binding protein MBD2b (23), were not reproduced in other systems (24–26). It is noteworthy that mutant MBD2 mice that lack a functional methyl-CpG binding domain showed the same demethylation profile of the paternal pronucleus as in wild-type controls (10). Although a testis-specific truncated Mbd2 transcript has been identified, there are no reports of an oocyte-specific form of MBD2 as yet that could account for the demethylase activity (27). Of interest is the finding that in bovine somatic cell nuclear transfer embryos, the donor nucleus showed more efficient satellite demethylation in the presence of the oocyte nucleus than in enucleated oocytes, suggesting that much of the demethylating activity is associated with the maternal nucleus (28). Because enucleated sheep oocytes are also able to demethylate serum-starved fibroblast nuclei by ≈50% (13), either sheep ooplasm has considerable residual demethylating activity or the sheep fibroblast DNA is particularly amenable to demethylation.
Effect of Sperm Composition. Regardless of the source of ovine 1-cell embryos we have examined (in vivo-derived, in vitro fertilization, or ICSI), we have observed no evidence for global active demethylation in the first cell cycle (refs. 12–13 and the present study). Whereas the mechanism of such resistance remains to be determined, we did observe that mouse sperm is demethylated to a greater degree than for either sheep or bovine sperm in each respective species of oocyte donor tested. Mouse sperm may possess either factors facilitating demethylation or a particular chromatin structure important to this process. One explanation might reside in genomic composition or in the methylation levels of mature spermatozoa. Using an HPLC assay, Jabbari et al. (29) demonstrated that sheep, cow, and pig have ≈10% lower levels of 5mC in sperm than human and mouse, despite little difference in the frequency of CpG in the respective genomes. Thus, the sheep genome may simply not require such an extensive loss of methylation to initiate reprogramming in the early embryo (12). Alternatively, species differences in the distribution of methylated DNA may determine whether a major male pronuclear demethylation event is required. Whereas the mouse sperm is considered globally hypermethylated relative to the mature oocyte (18), satellite sequences are relatively undermethylated in sperm (17, 18), whereas lower copy repeat sequences (IAP and MUP) are highly methylated in both gametes before fertilization (30). Species variations also exist in heterochromatic satellite sequences; whereas in mouse (17) and cow (31) sperm they are under-methylated relative to the oocyte, in pig sperm they are hyper-methylated relative to the oocyte (32), and both sheep gametes exhibit similar levels (H. Moore, R.R.M., and L.E.Y., unpublished results). Bisulfite sequencing suggests that Line1 elements undergo active demethylation in fertilized mouse embryos, whereas IAP elements remain methylated during preimplantation development (33). Thus, it is clear that not all sequences are equally prone to demethylation in the male pronucleus. Indeed, Santos et al. (10), using the 5mC antibody, report residual methylation in paternally derived chromosomes before syngamy. Our observations of incomplete demethylation of sheep and bovine sperm in mouse oocytes are consistent with some sequence specificity of the oocyte demethylating activity. This specificity may underlie why methylated plasmid DNA is not demethylated in mouse oocytes.
Function of Pronuclear Demethylation? We first considered that the selective pressure for maternal ooplasm to modify growth-promoting paternal imprints [previously suggested to underlie paternal demethylation (34, 35)] may be less in the mono/ditocous sheep than in the litter-bearing mouse, obviating the need for paternal demethylation. However, the existence of several growth-regulating imprinted genes in the fetal sheep and cow (36–40), asymmetric pronuclear demethylation in the monotocous human, and lack of demethylation in the polytocous rabbit zygote (12) provide no substantiation for an imprint-modifying role of paternal pronuclear demethylation.
Another suggested function for paternal demethylation is to allow more rapid remodeling/activation of the paternal genome in species where zygotic genome activation occurs relatively early (41). Maternally derived molecules are essential to remodel the sperm genome from protamine packaging to a nucleosomal state and to regulate gene expression in the first cleavage stages (reviewed in ref. 42). Observations of almost total genome-wide demethylation in the mouse, human, and pig male pronucleus but not in the sheep, rabbit, or even cow indeed seem to correlate with the relative timing of zygotic genome activation. Mouse, human, and pig are species in which major zygotic genome activation starts as early as the 2/4-cell stage (43, 44), therefore requiring more rapid remodeling of the sperm genome to a transcriptionally active state than in the sheep, rabbit, or cow. Thus, the timing of demethylation in the male pronucleus relative to the onset of the zygotic transcription clearly requires unraveling.
A variety of recent reports have emphasized the importance of studying DNA methylation events in the preimplantation embryo, where infertility-related procedures including superovulation and embryo culture (as well as somatic cell nuclear transfer) have been shown to alter methylation either globally or at specific loci. These changes are associated with both early developmental failure (35, 45) or serious phenotypic defects (reviewed in refs. 46 and 47). As supported by the present study, identification of key oocyte reprogramming factors may be of prime importance both in improving infertility treatments and in cellular dedifferentiation/transdifferentiation.
Acknowledgments
We are grateful to Dr. Alain Niveleau for the kind gift of the 5mC antibody, the Roslin Large Animal Unit staff, and Ali Ainslie for assistance with embryo collection. This work was supported by Biotechnology and Biological Sciences Research Council Grant GTH14114/15.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ICSI, intracytoplasmic sperm injection; 5mC, 5-methylcytosine.
References
- 1.Jaenisch, R. & Bird, A. (2003) Nat. Genet. 33, 245-254. [DOI] [PubMed] [Google Scholar]
- 2.Bestor, T. H. (2003) Ann. N.Y. Acad. Sci. 983, 22-27. [DOI] [PubMed] [Google Scholar]
- 3.Robertson, K. D. (2002) Oncogene 21, 5361-5379. [DOI] [PubMed] [Google Scholar]
- 4.Reik, W., Santos, F. & Dean, W. (2003) Theriogenology 59, 21-32. [DOI] [PubMed] [Google Scholar]
- 5.Li, E. (2002) Nat. Rev. Genet. 3, 662-673. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson-Smith, A. C. & Surani, M. A. (2001) Science 293, 1086-1089. [DOI] [PubMed] [Google Scholar]
- 7.Mayer, W., Niveleau, A., Walter, J., Fundele, R. & Haaf, T. (2000) Nature 403, 501-502. [DOI] [PubMed] [Google Scholar]
- 8.Oswald, J., Engemann, S., Lane, N., Mayer, W., Olek, A., Fundele, R., Dean, W., Reik, W. & Walter, J. (2000) Curr. Biol. 10, 475-478. [DOI] [PubMed] [Google Scholar]
- 9.Dean, W., Santos, F., Stojkovic, M., Zakhartchenko, V., Walter, J., Wolf, E. & Reik, W. (2001) Proc. Natl. Acad. Sci. USA 98, 13734-13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos, F., Hendrich, B., Reik, W. & Dean, W. (2002) Dev. Biol. 241, 172-182. [DOI] [PubMed] [Google Scholar]
- 11.Bourc'his, D., Le Bourhis, D., Patin, D., Niveleau, A., Comizzoli, P., Renard, J.-P. & Viegas-Pequignot, E. (2001) Curr. Biol. 11, 1542-1546. [DOI] [PubMed] [Google Scholar]
- 12.Beaujean, N., Hartshorne, G. M., Cavilla, J. L., Taylor, J. E., Gardner, J. O., Wilmut, I., Meehan, R. R. & Young, L. E. (2004) Curr. Biol. 14, R266-R267. [DOI] [PubMed] [Google Scholar]
- 13.Beaujean, N., Taylor, J. E., Gardner, J. O., Wilmut, I., Meehan, R. R. & Young, L. E. (March 3, 2004) Biol. Reprod., 10.1095/biolreprod.103.026559. [DOI] [PubMed]
- 14.Ptak, G., Tischner, M., Bernabo, N. & Loi, P. (2003) Biol. Reprod. 69, 278-285. [DOI] [PubMed] [Google Scholar]
- 15.Galli, C. & Lazzari, G. (1996) Animal Reprod. Sci. 42, 371-379. [Google Scholar]
- 16.Galli, C., Vassiliev, I., Lagutina, I., Galli, A. & Lazzari, G. (2003) Theriogenology 60, 1467-1480. [DOI] [PubMed] [Google Scholar]
- 17.Sanford, J., Forrester, L., Chapman, V., Chandley, A. & Hastie N. (1984) Nucleic Acids Res. 12, 2823-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monk, M., Boubelik, M. & Lehnert, S. (1987) Development (Cambridge, U.K.) 99, 371-382. [DOI] [PubMed] [Google Scholar]
- 19.Wilmut, I., Beaujean, N., de Sousa, P. A., Dinnyes, A., King, T. J., Paterson, L. A., Wells, D. N. & Young, L. E. (2002) Nature 419, 583-586. [DOI] [PubMed] [Google Scholar]
- 20.Vignon, X., Zhou, Q. & Renard, J.-P. (2002) Cloning Stem Cells 4, 363-377. [DOI] [PubMed] [Google Scholar]
- 21.Rideout, W. M., III, Eggan, K. & Jaenisch, R. (2001) Science 293, 1093-1098. [DOI] [PubMed] [Google Scholar]
- 22.Ramchandani, S., Bhattacharya, S. K., Cervoni, N. & Szyf, M. (1999) Proc. Natl. Acad. Sci. USA 96, 6107-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya, S. K., Ramchandani, S., Cervoni, N. & Szyf, M. (1999) Nature 397, 579-583. [DOI] [PubMed] [Google Scholar]
- 24.Ng, H. H., Zhang, Y., Hendrich, B., Johnson, C. A., Turner, B. M., Erdjument-Bromage, H., Tempst, P., Reinberg, D. & Bird, A. (1999) Nat. Genet. 23, 58-61. [DOI] [PubMed] [Google Scholar]
- 25.Boeke, J., Ammerpohl, O., Kegel, S., Moehren, U. & Renkawitz, R. (2000) J. Biol. Chem. 275, 34963-34967. [DOI] [PubMed] [Google Scholar]
- 26.Wade, P. A., Gegonne, A., Jones, P. L., Ballestar, E., Aubry, F. & Wolffe, A. P. (1999) Nat. Genet. 23, 62-66. [DOI] [PubMed] [Google Scholar]
- 27.Hendrich, B. & Bird, A. (1998) Mol. Cell. Biol. 18, 6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, Y. K., Koo, D., Park, J. S., Choi, Y., Lee, K. & Han, Y. (2001) FEBS Lett. 499, 55-58. [DOI] [PubMed] [Google Scholar]
- 29.Jabbari, K., Caccio, S., Pais de Barros, J. P., Desgres, J. & Bernardi, G. (1997) Gene 205, 109-118. [DOI] [PubMed] [Google Scholar]
- 30.Howlett, S. K. & Reik, W. (1991) Development (Cambridge, U.K.) 113, 119-127. [DOI] [PubMed] [Google Scholar]
- 31.Kang, Y. K., Koo, D. B., Park, J. S., Choi, Y. H., Chung, A. S., Lee, K. K. & Han, Y. M. (2001) Nat. Genet. 28, 173-177. [DOI] [PubMed] [Google Scholar]
- 32.Kang, Y. K., Koo, D. B., Park, J. S., Choi, Y. H., Kim, H. N., Chang, W. K., Lee, K. K. & Han, Y. M. (2001) J. Biol. Chem. 276, 39980-39984. [DOI] [PubMed] [Google Scholar]
- 33.Lane, N., Dean, W., Erhardt, S., Hajkova, P., Surani, A., Walter, J. & Reik, W. (2003) Genesis 35, 88-93. [DOI] [PubMed] [Google Scholar]
- 34.Reik, W. & Walter, J. (2001) Nat. Genet. 27, 255-256. [DOI] [PubMed] [Google Scholar]
- 35.Barton, S. C., Arney, K. L., Shi, W., Niveleau, A., Fundele, R., Surani, M. A. & Haaf, T. (2001) Hum. Mol. Genet. 10, 2983-2987. [DOI] [PubMed] [Google Scholar]
- 36.Young, L. E., Schnieke, A. E., McCreath, K. J., Wieckowski, S., Konfortova, G., Fernandes, K., Ptak, G., Kind, A., Wilmut, I., Loi, P. & Feil, R. (2003) Mech. Dev. 120, 1433-1442. [DOI] [PubMed] [Google Scholar]
- 37.McLaren, R. J. & Montgomery, G. W. (1999) Mamm. Genome 10, 588-591. [DOI] [PubMed] [Google Scholar]
- 38.Hagemann, L. J., Peterson, A. J., Weilert, L. L., Lee, R. S. & Tervit, H. R. (1998) Mol. Reprod. Dev. 50, 154-162. [DOI] [PubMed] [Google Scholar]
- 39.Killian, J. K., Nolan, C. M., Wylie, A. A., Li, T., Vu, T. H., Hoffman, A. R. & Jirtle, R. L. (2001) Hum. Mol. Genet. 10, 1721-1728. [DOI] [PubMed] [Google Scholar]
- 40.Ruddock, N. T., Wilson, K. J., Cooney, M. A., Korfiatis, N. A., Tecirlioglu, R. T. & French, A. J. (2004) Biol. Reprod. 70, 1131-1135. [DOI] [PubMed] [Google Scholar]
- 41.Haaf, T. (2001) Chromosome Res. 9, 263-271. [DOI] [PubMed] [Google Scholar]
- 42.De Sousa, P. A., Caveney, A., Westhusin, M. E. & Watson, A. J. (1998) Theriogenology 49, 115-128. [DOI] [PubMed] [Google Scholar]
- 43.Kanka, J. (2003) Theriogenology 59, 3-19. [DOI] [PubMed] [Google Scholar]
- 44.Braude, P., Bolton, V. & Moore, S. (1988) Nature 332, 459-461. [DOI] [PubMed] [Google Scholar]
- 45.Shi, W. & Haaf, T. (2002) Mol. Reprod. Dev. 63, 329-334. [DOI] [PubMed] [Google Scholar]
- 46.Maher, E. R., Afnan, M. & Barratt, C. (2003) Hum. Reprod. 18, 2508-2511. [DOI] [PubMed] [Google Scholar]
- 47.Young, L. E. (2003) Hum. Fertil. (Cambridge, U.K.) 6, 59-63. [DOI] [PubMed] [Google Scholar]