Abstract
In this commentary, Philip Hieter and Kym Boycott discuss the importance of model organisms for understanding pathogenesis of rare human genetic diseases, and highlight the work of Brooks et al., “Dysfunction of 60S ribosomal protein L10 (RPL10) disrupts neurodevelopment and causes X-linked microcephaly in humans,” published in this issue of GENETICS.
The application of next-generation DNA sequencing (NGS) technology has brought an unprecedented era of rare disease gene discovery (Boycott et al. 2013). The potential for clinical benefits is enormous, but how do we translate these gene discoveries into a mechanistic understanding of the disease gene’s function and identification of therapeutic targets?
Fortunately, we have model organisms that provide powerful tools for investigating the mechanistic basis of diseases, for identifying and developing potential therapeutic interventions, and for evaluating new treatments. The key to success in such endeavors will be to establish efficient mechanisms for catalyzing connections, collaboration, and cross-talk between basic and clinician scientists. In this month’s issue of GENETICS, Brooks et al. (2014) present the genetic, functional, and biochemical dissection of a multigenerational X-linked pedigree with syndromic microcephaly. This success story exemplifies the synergy that can exist between a clinical geneticist and a basic science research team to catalyze gene discovery and uncover novel disease mechanisms.
The Rare Disease Challenge Is Significant
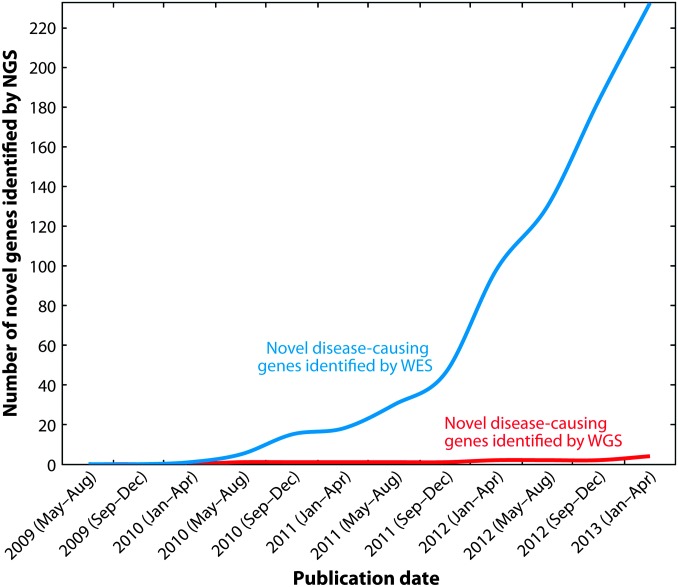
While each of the estimated 7000 single-gene inherited diseases is individually rare, they are collectively common, affecting as much as 3% of the population (Carter 1977; Baird et al. 1988). In aggregate, they present great healthcare challenges, contributing significantly to morbidity, mortality, and cost of care. Although the genes responsible for approximately 4000 distinct monogenic human diseases are known (Online Mendelian Inheritance in Man), recent estimates suggest that over 9000 single-gene phenotypes will ultimately be recognized and molecularly defined (Samuels 2010). Disease-causing genes that had eluded discovery because of their rarity, clinical heterogeneity, and the paucity of families with multiple individuals affected by a specific disease, are being increasingly identified by NGS approaches (Figure 1). Several large-scale initiatives are currently underway that are identifying new disease-associated genes at impressive rates, including the “Deciphering Developmental Disorders” (UK; http://www.ddduk.org/), “Neuromics” and “Eurenomics” (Europe; http://rd-neuromics.eu/, http://www.eurenomics.eu/) and, the “Centers for Mendelian Genomics” (USA; http://www.mendelian.org/). In Canada, the genetics community came together in 2010 to form the FORGE (Finding Of Rare Disease Genes in Canada) consortium. From 371 nationwide proposals, 264 rare diseases were selected for study, and at least one causative gene has now been identified for 146 diseases, yielding an impressive success rate of 55% over a 2-year period (Beaulieu et al. 2014). Sixty-seven of the 146 diseases solved resulted in the identification of a new disease gene, of which 41 have been genetically or functionally validated; 26 currently lack additional functional evidence. Despite the dramatic increase in the pace of rare disease gene discovery, the gap in our understanding of the molecular and cellular bases of rare diseases continues to grow. Thus, while we have at least a cursory understanding of perhaps 50% of human disease genes, our knowledge of the complete catalog of highly penetrant, disease-causing mutations is quite limited. This brings a real and immediate need for model organism research platforms to put disease-causing genes into a biological context.
Figure 1.
Number of novel genes discovered by whole-exome sequencing (WES) or whole-genome sequencing (WGS). This graph represents the results of PubMed searches using the terms “exome sequencing” and “whole genome sequencing,” sorted by date of publication and filtered for only single-gene diseases (references to complex diseases and cancer were excluded). Results were sorted to identify those that reported novel gene discovery, and duplicates were removed. The number of novel disease genes identified using WES is significantly greater than that by WGS. Reprinted with permission from: Boycott K. M., D. A. Dyment, S. L. Sawyer, M. R. Vanstone, and C. L. Beaulieu, 2014 Identification of genes for childhood heritable diseases. Annu. Rev. Med. 65: 19–31. PMID: 24422568.
Model Organisms Provide Valuable Insights Into Rare Disease Pathogenesis
The same building blocks are used to construct organisms as diverse as yeast, worms, flies, fish, mice, and humans. This allows researchers to determine biological mechanisms at the levels of genes, pathways, and networks through analysis of the equivalent (orthologous) genes in “model organisms” that can be readily manipulated in the laboratory. In the late 1980s, the architects of the Human Genome Project embraced this principle and had the foresight to include sequencing of the genomes of four key experimental organisms as part of the $3B international effort (Watson 1990). Comparing the genome sequences of yeast, worm, fly, mouse, and human confirmed the striking extent to which all organisms are built from the same set of genes (Dolinski and Botstein 2007) and highlighted the enormous potential of model experimental organisms for the study of gene function. These signal accomplishments showed that few, if any, biological processes are unique to humans at the gene level. Biomedical research, particularly since positional cloning of human disease genes became possible in 1987, has revealed that the principles of cross-species analysis of basic gene function extend to the study of all human disorders. Indeed, the need for model organism-based studies to provide functional insights into human disease and establish rational approaches to the development of novel therapeutics (Botstein 2012) has increased along with the pace of discovery of human disease mutations (Figure 1).
The identification of a human genetic variant that causes disease is an important breakthrough that provides a clear DNA-based diagnostic for a group of patients and their families. But the discovery of the human variant is a descriptive, hypothesis-generating milestone that necessitates further studies on the function of the gene and the biological consequences of specific gene mutations. This is where mechanistic studies in model organisms become so important (Spradling et al. 2006; Rine 2014). Following a disease gene discovery, key immediate goals are to understand the underlying molecular pathways and the reversibility of the phenotypes in genetic and pharmacologic rescue experiments. In some cases, phenotypes in a model experimental organism that clearly resemble hallmarks of the human disorder can be used. In other cases, a phenotype from a model organism may not ‘obviously’ be a hallmark of the human disease, and thereby be considered a Phenolog (McGary et al. 2010), but the molecular and cellular functions of the orthologue can still be investigated and shed essential light into the underpinnings of the human disorder. Importantly, phenologs, that do not mimic the human disease but conserve the regulation of a pathway and its defects, can be used to better understand disease mechanism and guide identification of candidate therapeutic targets. In either scenario, model organisms such as yeast, worms, and flies can offer powerful experimental approaches that facilitate and enhance experimental approaches in vertebrate models such as zebrafish and mice. In general, mouse models will be important for functional validation in a mammalian context to allow more comprehensive investigation at the tissue, organ, or whole-organism level. New technologies such as genome editing with CRISPR-Cas9 provide the ability to directly model specific human mutations (Sander and Joung 2014) including missense mutations that are nulls, hypomorphs, neomorphs (gain-of-function), or antimorphs (dominant negative) in nature.
Functional Analysis in Zebrafish Supports Identification of a Novel Disease Gene
Brooks et al. (2014) studied a rare disease characterized by microcephaly in a single family with suspected X-linked inheritance. The pedigree structure prompted the clinician to request a NGS X-linked Intellectual Disability diagnostic panel that simultaneously scrutinized 82 genes in an affected family member. The DNA sequence analysis revealed a single, novel, missense variant, K78E, in the 60S ribosomal protein L10 (RPL10), encoded by a gene previously reported as a rare autism susceptibility locus. The variant segregated with disease appropriately, and both carrier mothers demonstrated skewed X inactivation of the defective X chromosome. Although the combined genetic evidence was suggestive of a role for RPL10 in the disease in this family, the authors turned to zebrafish for functional studies of the gene. They showed that gene suppression with two independent morpholinos targeting rpl10 resulted in significantly reduced head size. Using in vivo complementation, they demonstrated that K78E is a loss-of-function variant. They monitored bulk translation and found a brain-specific decrease in protein synthesis. Finally, they explored the cellular processes that may contribute to microcephaly in their models, showing that although cell proliferation was normal in morphants, there was a sixfold increase in apoptosis localized to the forebrain of animals with reduced rpl10. Taken together, these results suggest that the loss-of-function mutation in RPL10 is driving the neurodevelopmental defects in this family, thereby defining a novel ribosomopathy.
Catalyzing Connections Between Clinician and Basic Scientists: Addressing the Next Grand Challenge
The history books will show that from 1980 to 2020 the “genetic anatomy of human diseases” was revealed (Altschuler 2012). Translating this genomic information into advances in the prevention and treatment of human disease will require elucidating gene function and understanding the ways in which biological mechanisms are affected by specific mutations. Over the next several decades, we must focus on a new “grand challenge”: determining the functions of genes, understanding how specific mutations cause disease, and creating practical protocols for using this information to prevent and treat disease. Fundamental aspects of most human disorders will be informed through analysis of orthologous genes and pathways in experimentally tractable model organisms using their sophisticated experimental toolboxes (Rine 2014). In addition, because of their complementary biology, model organisms in the aggregate offer much power for probing gene function.
Success in this endeavor will require increased connections, collaboration, and cross-talk between clinicians and basic scientists as early as possible after the discovery of new disease-causing mutations. One promising infrastructure development and large-scale collaboration is the International Rare Disease Research Consortium (IRDiRC; www.irdirc.org). Launched in 2011, IRDiRC is an umbrella organization of more than 30 global funding bodies and their funded research projects. With over $1B of investment, IRDiRC’s two main objectives are to develop diagnostic tools for most rare diseases and deliver 200 new therapies by 2020.
IRDiRC’s Model Organism Working Group is tasked with providing guidance and tools to increase collaboration. The way forward is to maintain an appropriate balance between fundamental research aimed at mechanistic understanding of gene, pathway, and network function, and translational research aimed at amelioration of human disease. The article by Brooks et al. (2014) in this issue of GENETICS embodies the synergy between model organism and human genetics research. Such coupling promises to establish the relevance to disease phenotype, reveal the pathogenic potential of private mutations, and elucidate mechanisms of pathology. We are in the midst of a golden age of disease gene discovery and understanding of the concomitant pathogenesis.
Footnotes
Communicating editor: M. Johnston
Literature Cited
- Altshuler D., 2012. 2011 Curt Stern Award Address. Am. J. Hum. Genet. 90(3): 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird P. A., Anderson T. W., Newcombe H. B., Lowry R. B., 1988. Genetic disorders in children and young adults: a population study. Am. J. Hum. Genet. 42(5): 677–693 [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C. L., Majewski J., Schwartzentruber J., Samuels M. E., Fernandez B. A., et al. , 2014. FORGE Canada Consortium: outcomes of a 2-year national rare disease gene discovery project. Am. J. Hum. Genet. 94: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., 2012. Why we need more basic biology research, not less. Mol. Biol. Cell 23(21): 4160–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott K. M., Vanstone M. R., Bulman D. E., Mackenzie A. E., 2013. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 14: 681–691 [DOI] [PubMed] [Google Scholar]
- Brooks S. S., Wall A. L., Golzio C., Reid D. W., et al. , 2014. Dysfunction of 60S ribosomal protein L10 (RPL10) disrupts neurodevelopment and causes X-linked microcephaly. Genetics 198: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. O., 1977. Monogenic disorders. J. Med. Genet. 14(5): 316–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K., Botstein D., 2007. Orthology and functional conservation in eukaryotes. Annu. Rev. Genet. 41: 465–507 [DOI] [PubMed] [Google Scholar]
- McGary K. L., Park T. J., Woods J. O., Cha H. J., Wallingford J. B., et al. , 2010. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl. Acad. Sci. USA 107(14): 6544–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., 2014. A future of the model organism model. Mol. Biol. Cell 25(5): 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels M. E., 2010. Saturation of the human phenome. Curr Genomics 11(7):482–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K., 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32(4): 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., Ganetsky B., Hieter P., Johnston M., Olson M., et al. , 2006. New roles for model genetic organisms in understanding and treating human disease: report from the 2006 Genetics Society of America meeting. Genetics 172(4): 2025–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., 1990. The human genome project: past, present, and future. Science 248(4951): 44–49 [DOI] [PubMed] [Google Scholar]