Abstract
Based on his work with the Escherichia coli l-arabinose operon, Ellis Englesberg proposed in 1965 that the regulatory gene araC was an “activator gene” required for positive control of the ara operon. This challenged the widely held belief in a universal mechanism of negative regulation proposed earlier by Jacob and Monod. For years, Englesberg’s model was met with deep skepticism. Despite much frustration with complex ad hoc explanations used to challenge his model, Englesberg persisted until the evidence for positive control in ara and other systems became overwhelming. Englesberg’s pioneering work enriched the original operon model and had a lasting impact in opening new and exciting ways of thinking about transcriptional regulation.
Keywords: activation, operon, repression, transcription
ELLIS Englesberg was one of the pioneers in deciphering mechanisms of gene regulation (Figure 1). In 1965, Englesberg and colleagues Joseph Irr, Joseph Power, and Nancy Lee published an article now widely recognized as a landmark in understanding the mechanisms of transcriptional control (Englesberg et al. 1965). Examining regulation of the l-arabinose (ara) operon in Escherichia coli, their work led them to conclude that the regulatory gene araC is a new type of gene, termed an “activator gene,” that has a positive role in expression of the structural genes of the ara operon. They noted that their results were “in sharp contrast to the negative or repressor control” mechanism of Jacob and Monod as exemplified in the bacterial β-galactosidase (lac) operon and bacteriophage lambda. Today, the logic of Englesberg’s 1965 approach and the interpretation of his experiments demonstrating positive control seem elegant and clear-cut. However, the reaction of the molecular biology community at the time was one of deep skepticism, and resulted in both scientific and personal criticism of Englesberg until the early 1970s. Englesberg did not wither under this criticism, but persisted until evidence for positive control from ara and other unrelated systems was so overwhelming that it could not be dismissed.
Figure 1.

Ellis Englesberg in his laboratory at University of California Santa Barbara (UCSB), 1986. Photo from a UCSB publication courtesy of Barbara Englesberg.
Today we believe that positive control is probably the most widely used mechanism to regulate transcription in all forms of life and plays key roles in fundamental processes such as cell-type specificity, cell growth, development, and stress response. Significantly, when altered or misexpressed, transcription activator proteins can cause many human diseases. To understand why Englesberg’s pioneering work was so important in opening new ways of thinking about gene regulation, we first need to revisit the thinking of the field in 1965 and how his work overturned a key aspect of a widely accepted and highly influential paradigm.
After the discovery of DNA structure by Watson and Crick, arguably the next most influential work in 20th century molecular biology was the operon model of gene regulation proposed by Jacob and Monod. In 1961, they summarized their revolutionary work conducted over the preceding decade (Jacob and Monod 1961). Importantly, they also outlined the methodology and logic they developed for the study of gene regulation. They and others then rapidly applied these ideas and strategies to uncover the fundamental molecular mechanisms of how information flows from DNA to protein. The groundbreaking new concepts introduced included:
the concept of a “regulator gene,” a gene that controls the expression of other genes termed “structural genes” that encode enzymes involved in metabolism and other cellular processes
showing that gene induction involves new protein synthesis, in contrast, to previous theories where the inducer molecules mold the folding of enzyme active sites (Vogel 1957)
the deduction that protein synthesis must involve a short-lived intermediate between DNA and protein synthesis, which they concluded was an unstable RNA species termed messenger RNA
the operon concept, in which a single regulatory gene controls a set of linked genes using a common control region, and
the concept of allostery, whereby a small molecule can cause changes in the conformation and biological activity of proteins.
The revolutionary concept of regulator genes was developed from Jacob, Monod, and their colleagues’ studies on induction of the lac operon and induction of phage in cells lysogenic for bacteriophage lambda. They had the remarkable insight that repression and induction are different outcomes of fundamentally similar mechanisms and that repressors control gene expression in both systems. This led them to mistakenly propose that all regulatory genes act directly as repressors and that they act by inhibiting the expression of the structural genes (Monod and Jacob 1961).
Even though the new paradigm of regulatory genes and gene control mechanisms was a striking break from past thinking, it was rapidly accepted and many geneticists and molecular biologists rushed to apply this new logic and methodology to their own systems. One of the first to take up this approach was Ellis Englesberg. Englesberg, who had a great interest in carbohydrate metabolism, was no novice at bacterial genetics, having done his Ph.D. with Roger Stanier at Berkeley on inducible enzyme formation. In 1959, while at the Long Island Biological Laboratories in Cold Spring Harbor, New York, he collaborated with graduate student Julian Gross on the genetics of arabinose utilization (Gross and Englesberg 1959). According to Englesberg, “we picked the arabinose system to test the operon theory and negative control because it was completely different from the lactose operon, not because we had some insight into whether it would be under positive or negative control” (Fogle 1991, p. 68). At first, they tried to fit their genetic data into the negative control model. Mutations in the araC gene prevented expression of the araBAD genes, and they initially termed araC an operator gene, in a mistaken analogy to the Jacob and Monod regulation by repression model (Englesberg 1961). It soon became apparent, however, that araC behaves quite differently from that of a repressor since its presence is required for expression of the araBAD and unlinked arabinose permease genes (Helling and Weinberg 1963).
Soon after, in a stroke of good fortune, graduate student Joe Irr isolated araC constitutive mutations (araCc) that were instrumental in demonstrating positive control (Englesberg et al. 1965). Irr isolated these mutants by their resistance to fucose, an analog of arabinose that binds AraC and inhibits utilization of arabinose. Irr recounts that Englesberg had a drawer full of rare sugars and that he tried growing cells in mixtures of arabinose and these other sugars. The readily isolated fucose-resistant mutations all mapped to araC and caused constitutive expression of both araBAD and the unlinked araE low-affinity arabinose permease gene. The key findings that led Englesberg to propose that araC is an activator gene were:
araC+ acts in trans to activate expression of the araBAD and unlinked araE genes and is dominant to araC−.
araCC is dominant to araC−, showing that araC function is required for expression of the ara genes. This contrasts with regulation by a repressor, where constitutive expression is the consequence of eliminating repressor activity.
In an unexpected twist, araC+ was found to be dominant to araCC, suggesting that araC is both an activator and a repressor.
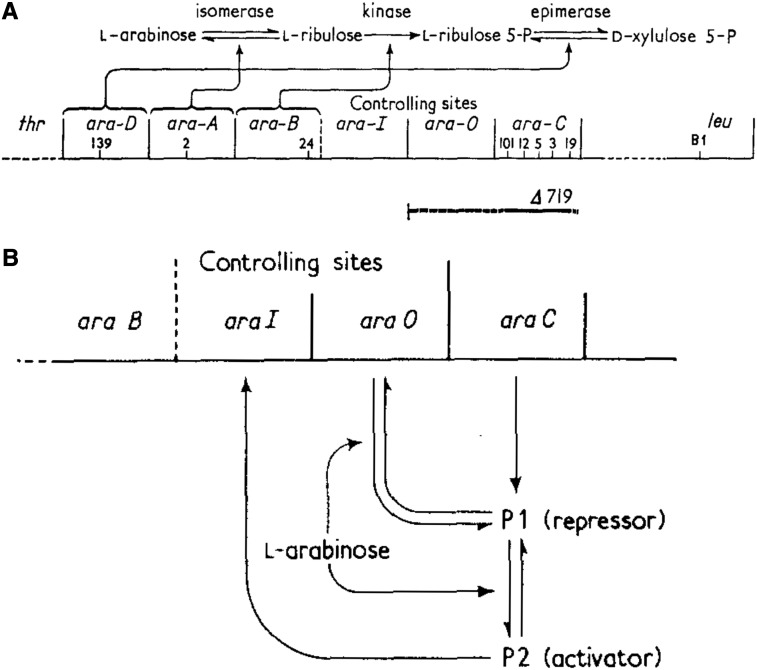
These findings led Englesberg to propose that AraC exists in two forms termed P1 (the repressor) and P2 (the activator). According to their model, in the absence of arabinose, AraC exists mostly in the P1 form that represses araBAD expression through an operator site termed araO. Arabinose was proposed to convert AraC to the P2 form that activates transcription via an inducing site termed araI (Figure 2).
Figure 2.
The l-arabinose operon. (A) Arrangement of genes and regulatory sites in the ara operon. The mapped position of araC deletion Δ719, which removes the araO site, is indicated. (B) The model for positive and negative control of the araBAD genes by AraC. Figure reprinted from Englesberg et al. (1969a).
Although there was great interest in Englesberg’s work, many people were skeptical of both the results and the model for positive control. Jacob and Monod had previously discussed the possibility of positive control, but as Jacob writes (Jacob 1979, p. 106): “Jacques...was not very fond of positive regulation, because he liked nature to provide unique solutions. Since a combination of two negatives were equivalent to one positive, he did not see the logical necessity of adding another, distinct mechanism.” Using only negative regulation, Monod developed schemes to explain gene regulation of complex phenomena such as development (Monod and Jacob 1961). He wrote that with only negative regulation “any number of systems may be interconnected into regulatory circuits endowed with virtually any desired property.” While today it is hard to understand why the concept of positive control met such resistance, this was an era when prominent discoveries revealed the great conservation of biological mechanisms, in organisms from bacteria to humans. Compared with the conservation of DNA structure, the universal genetic code, and the mechanism of protein synthesis, it seemed reasonable to many that negative regulation was another example of a universal biological mechanism (Beckwith 2011).
The most common alternative scientific explanation for Englesberg’s results was that AraC regulated the true repressor of the ara operon, perhaps by converting arabinose into a compound that was the true inducer of this hypothetical repressor. By this model, the araCc mutants converted a cellular metabolite into the true inducer. An alternative model was that AraC is a protein subunit required for activity of the other enzymes in the ara operon. Englesberg ruled out this model by showing that a deletion removing araC and putting the araA gene (arabinose isomerase) under leu regulation produced high levels of isomerase (Englesberg et al. 1965). Although Englesberg concedes that “by an elaborate series of ad hoc postulates, it may be possible to explain these results on the basis that the C gene is solely a repressor gene,” by far the simplest explanation was that araC is an activator gene. Nonscientific explanations were essentially that Englesberg was interpreting his results incorrectly and/or that he didn’t know how to do genetics.
At the time of Englesberg’s findings, the negative control paradigm dominated the field, and anything that contradicted this view was not viewed favorably. As Englesberg said (Fogle 1991, p. 71) “Any deviation from negative control was difficult to bear at the time. They would say to me, ‘I’ve just learned negative control. You cannot ask me to think about the possibility of positive control.” In defending the negative control model, Monod was especially persuasive and adamant. Regulation of alkaline phosphatase expression in E. coli was likely the first system where evidence for positive control was observed (Echols et al. 1961; Garen and Echols 1962a,b). Echols and Garen found that mutations in phoR could lead to either low level constitutive or complete loss of phoA expression. “Hatch” Echols recounts that when he presented his work on alkaline phosphatase regulation at the 1961 Cold Spring Harbor symposium and commented that regulation of the protein might be positive, “Monod ran to the front of the room, proclaiming, ‘No, no, we know that all regulation is negative—moreover, we have mutants like yours and know how to interpret them.’” (Beckwith 1996; Echols and Gross 2001). This interaction convinced Echols that he would be better off switching his research to a different subject. In Englesberg’s words (Fogle 1991, p. 71): “Sometimes I felt like I was holding a tiger by the tail. I’d ask myself ‘Who wants this?’ I didn’t want to have this battle on my hands. I just wanted to do my work. It was much easier to go along with the paradigm than it was to break out of it—easier, but boring.”
While in retrospect, the evidence in 1965 for positive control by AraC was as good or better than the data used to formulate the negative control model, Englesberg needed to accumulate much additional data to answer his critics. First, he isolated an extensive set of deletions ending in the araBAD control region that defined the location of the operator and initiator sites (Sheppard and Englesberg 1967; Englesberg et al. 1969b). Remarkably, the operator mapped upstream of the araI site. Consistent with the model that AraC was both an activator and repressor, deletions of araO showed a striking elevation of araBAD basal expression that was dependent on araC. Englesberg suggested that this was due to a small amount of the inducing form of AraC (P2) present in the absence of arabinose. Second, to counter the argument that he had “not looked hard enough” for the true ara repressor, Englesberg et al. (1969a) isolated many revertants of an araC deletion mutant. These revertants, which generated low but elevated levels of basal araBAD expression, were termed araI mutations. All the araI mutations mapped to the araBAD regulatory region and acted only in cis—again in contrast to results predicted by the repressor model. These and other genetic results led to a model for the organization of the araBAD operon and for the mechanism of AraC (Figure 2).
Although Englesberg’s results gradually convinced others, Monod remained unconvinced of positive control to the end. During this period, Englesberg gave several seminars at the Pasteur Institute. As told by Jacob (1979, p. 107): “After each seminar, however, [Englesberg] received a severe lesson in regulatory genetics from Monod, who always insisted on a notion ‘that even a schoolboy cannot ignore: negative × negative equals positive!’ Englesberg said that ‘whenever I spoke with Jacob and Monod, they would say that they were 33.3% convinced, and then 50% convinced, about positive control. When I gave a seminar at the Pasteur . . . in 1972, they said ‘Well, we are 66.6% convinced’” (Fogle 1991, p. 71). For others in the field, part of the difficulty accepting the positive control model for AraC was undoubtedly its complexity, with AraC acting as both an activator and a repressor. Another conceptual problem may have been that it was hard to imagine how a protein could stimulate transcription. We now know that, in many cases, activation in bacteria can be accomplished by simple protein–protein interactions that enhance polymerase recruitment to nonoptimal promoters (Benoff et al. 2002; Ptashne and Gann 2002; Jain et al. 2004).
Despite Englesberg’s elegant genetics, there wasn’t any single experiment that finally convinced the field of the existence and importance of positive control. An important early biochemical step in the study of AraC was the demonstration that it induced transcription in a cell-free system (Greenblatt and Schleif 1971). But was positive control confined only to the ara operon? Fortunately, evidence from other systems that could not be fit into the negative control model was rapidly accumulating (Englesberg and Wilcox 1974). Early examples were the lambda N and Q proteins (Skalka et al. 1967), regulation of the maltose operon (Hatfield et al. 1969), and the action of the catabolite activator protein, which, ironically, positively regulates lac, ara, and many other inducible enzymes involved in sugar metabolism (Emmer et al. 1970; Zubay et al. 1970). Even lambda repressor, one of the two original repressors studied by Jacob, Monod, and colleagues was later found to act as both an activator and a repressor (Ptashne 2004). All these new data that did not fit the original paradigm now left the field open to new possibilities and the discovery of other prokaryotic regulatory mechanisms such as attenuation, antitermination, and (much later) riboswitches, as well as the rich diversity of transcription regulatory mechanisms found in eukaryotes.
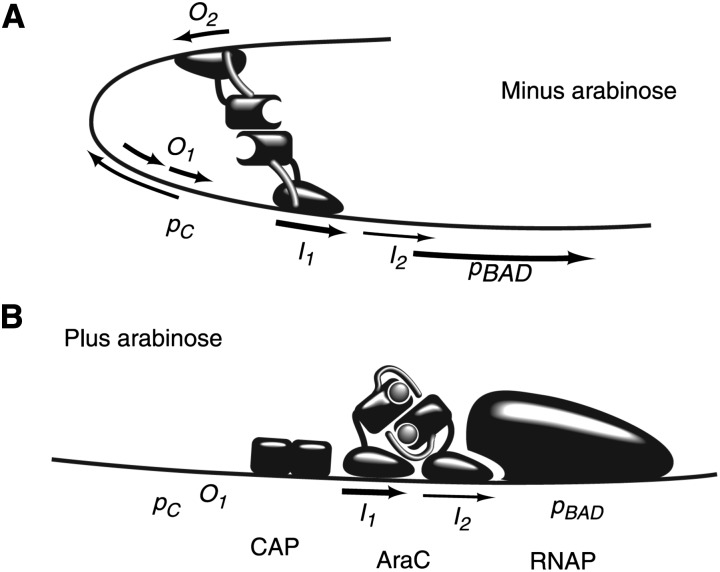
The molecular mechanisms of how AraC works were later pursued using purified proteins and modern methods of molecular biology, biochemistry, and structural biology (e.g., Horwitz et al. 1980; Dunn et al. 1984; Lee et al. 1987; Lobell and Schleif 1990). Comparing our current understanding of how AraC regulates transcription (Schleif 2010) (Figure 3) with Englesberg’s model shows a remarkable correspondence between the genetic and molecular models; nearly everything in Englesberg’s genetic model was correct. The biochemistry neatly explains, at a molecular level, all of Englesberg’s genetic evidence that AraC is both a repressor and an activator. In the absence of arabinose, the N-terminal arms of AraC are thought to interact weakly with the DNA-binding domains and hold these domains in an orientation that favors binding to distant binding sites via a DNA loop. The AraC binding site most distant from the araBAD promoter, termed araO2, is the upstream operator revealed by Englesberg’s original promoter deletion that caused an increase in basal expression of the araBAD operon. Arabinose binding to AraC causes a repositioning of the N-terminal arms, allowing the DNA binding domains to reorient and bind the adjacent half sites at araI, where AraC presumably activates transcription by direct contact with RNA polymerase. Fucose binds AraC, but presumably does not elicit the conformational change in the N-terminal arms. Most araC mutations are found in the N-terminal arms and likely allow the reorientation of the DNA-binding domains, leading to constitutive induction of the ara genes. The catabolite activator protein (CAP), binding upstream from araI, probably makes independent contacts with polymerase, assisting in its recruitment to the promoter.
Figure 3.
Present-day model for regulation of the ara operon. (A) In the absence of arabinose, the AraC N-terminal dimerization domain is constrained and favors binding of AraC to distant sites, forming a DNA loop between the araI1 and araO2 sites. (B) In the presence of arabinose, a reorientation of the dimerization domain favors AraC binding to two adjacent half sites at araI where it stimulates transcription from the araBAD promoter (pBAD). CAP (catabolite activator protein) bound to cAMP binds adjacent to AraC and likely makes separate contact with RNA polymerase (RNAP) to stimulate transcription. AraO1 is an operator where AraC binds to repress its own transcription from the araC promoter (pC). Figure reprinted from Schleif (2010).
The operon model had a profound impact on the study of gene regulation and its influence is still felt today. But the insistence that every aspect of the model be upheld inhibited progress in the field. Paradigms naturally influence the way scientists interpret their results but dogmatic adherence to a paradigm can lead to missing the obvious. As noted by Keith Yamamoto (Fogle 1991, p. 68): “as Englesberg discovered, such paradigms, once established, don’t ‘tip over’ easily. Thus, science moves forward by a ratchet-like process rather than in a smooth continuum.” In the final analysis, Englesberg’s discovery of positive control didn’t detract from the revolutionary concepts of the operon model. Rather his discovery added to the model, opening up new and exciting ways of thinking about gene regulatory mechanisms. Today we think that the only rule for gene regulation seems to be that there are no rules. Combining different mechanisms to regulate gene expression provides tremendous flexibility in control of spatial and temporal patterns of transcription and has undoubtedly led to the great diversity in organisms with highly related protein-coding DNA.
Ellis Englesberg passed away in May 2013. He was born in Brooklyn in 1921 and, after military service during WWII, earned his Ph.D. in bacteriology from University of California—Berkeley in 1952. During his time in Cold Spring Harbor, he worked on carbohydrate metabolism in both Yersinia pestis (plague bacteria) and E. coli. When he moved to the University of Pittsburgh in 1960 he was discouraged (for obvious reasons) from continuing to work with Y. pestis, so he concentrated his efforts on arabinose metabolism. In part because of academic freedom issues (Wofsy 1995), he moved in 1965 to chair the Department of Biological Sciences at University of California—Santa Barbara, where he turned the direction of the department toward modern molecular and genetic studies. Englesberg is fondly remembered by his students as a caring and encouraging mentor. He had a habit of pulling the most promising undergraduate students out of his advanced course on gene regulation, encouraging them to work in his laboratory as graduate students. Although he didn’t have an aggressive personality or promote himself, he was driven and expected the people in his group to have the same intensity. He was also an exceptional scholar and well read in art, philosophy, and history. And he had a passion for issues of social justice and human rights. After his retirement, he opened the Guernica Gallery of Graphic Arts in Santa Barbara, collecting artwork dealing with social and political issues.
Because Englesberg’s work was recognized as groundbreaking long after it was first published, he didn’t receive recognition until much later, being elected to the National Academy in 1986. Although there were long periods when he was frustrated, disappointed, even angry about the criticism of his model, he strongly believed he was right, and he didn’t shrink from defending his work. Today we admire Englesberg’s intellectual courage and his important contributions to our understanding of gene regulation.
Acknowledgments
Many thanks go to those who shared their reflections of Ellis and the work of the Pasteur group: Jon Beckwith, John Carbon, Dennis Clegg, Harvey Eisen, Jack Greenblatt, Joe Irr, Mark Ptashne, Bob Schleif, David Sheppard, and Gary Wilcox, with additional thanks to Barbara Englesberg for the photograph of Ellis and Ted Young for comments on the manuscript. S.H. is supported by National Institutes of Health grants GM053451 and 075114.
Footnotes
Communicating editor: A. S. Wilkins
Literature Cited
- Beckwith J., 1996. The operon: an historical account, pp. 1227–1231 in Eschericia coli and Salmonella, edited by Neidhardt F. C. Cellular and Molecular Biology, Washington, DC [Google Scholar]
- Beckwith J., 2011. The operon as paradigm: normal science and the beginning of biological complexity. J. Mol. Biol. 409: 7–13 [DOI] [PubMed] [Google Scholar]
- Benoff B., Yang H., Lawson C. L., Parkinson G., Liu J., et al. , 2002. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297: 1562–1566 [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F., 1984. An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. USA 81: 5017–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. G., Gross C. A., 2001. Operators and Promoters. Univ of California Press, Oakland, CA. [Google Scholar]
- Echols H., Garen A., Garen S., Torriani A., 1961. Genetic control of repression of alkaline phosphatase in E. coli. J. Mol. Biol. 3: 425–438 [DOI] [PubMed] [Google Scholar]
- Emmer M., deCrombrugghe B., Pastan I., Perlman R., 1970. Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc. Natl. Acad. Sci. USA 66: 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., 1961. Enzymatic characterization of 17 l-arabinose negative mutants in E. coli. J. Bacteriol. 81: 966–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Wilcox G., 1974. Regulation: positive control. Annu. Rev. Genet. 8: 219–242 [DOI] [PubMed] [Google Scholar]
- Englesberg E., Irr J., Power J., Lee N., 1965. Positive control of enzyme synthesis by gene C in the l-arabinose system. J. Bacteriol. 90: 946–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Sheppard D., Squires C., Meronk F., 1969a An analysis of “revertants” of a deletion mutant in the C gene of the l-arabinose gene complex in Escherichia coli B-r: isolation of initiator constitutive mutants (Ic). J. Mol. Biol. 43: 281–298 [DOI] [PubMed] [Google Scholar]
- Englesberg E., Squires C., Meronk F., 1969b The l-arabinose operon in Escherichia coli B-r: a genetic demonstration of two functional states of the product of a regulator gene. Proc. Natl. Acad. Sci. USA 62: 1100–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle S., 1991. Landmarks: positive control of enzyme synthesis by gene C in the l-arabinose system. J. NIH Res. 3: 63–72 [Google Scholar]
- Garen A., Echols H., 1962a Properties of two regulating genes for alkaline phosphatase. J. Bacteriol. 83: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen A., Echols H., 1962b Genetic control of induction of alkaline phosphatase synthesis in E. coli. Proc. Natl. Acad. Sci. USA 48: 1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Schleif R., 1971. Arabinose C protein: regulation of the arabinose operon in vitro. Nat. New Biol. 233: 166–170 [DOI] [PubMed] [Google Scholar]
- Gross J., Englesberg E., 1959. Determination of the order of mutational sites governing l -arabinose utilization in Escherichia coli B/r bv transduction with phage Plbt. Virology 9: 314–331 [DOI] [PubMed] [Google Scholar]
- Hatfield D., Hofnung M., Schwartz M., 1969. Genetic analysis of the maltose A region in Escherichia coli. J. Bacteriol. 98: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Weinberg R., 1963. Complementation studies of arabinose genes in Escherichia coli. Genetics 48: 1397–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz A. H., Morandi C., Wilcox G., 1980. Deoxyribonucleic acid sequence of araBAD promoter mutants of Escherichia coli. J. Bacteriol. 142: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., 1979. The Switch, pp. 95–108 in Origins of Molecular Biology. A Tribute to Jacques Monod, edited by Lwoff A., Ullmann A. Academic Press, New York [Google Scholar]
- Jacob F., Monod J., 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3: 318–356 [DOI] [PubMed] [Google Scholar]
- Jain D., Nickels B. E., Sun L., Hochschild A., Darst S. A., 2004. Structure of a ternary transcription activation complex. Mol. Cell 13: 45–53 [DOI] [PubMed] [Google Scholar]
- Lee N., Francklyn C., Hamilton E. P., 1987. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc. Natl. Acad. Sci. USA 84: 8814–8818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F., 1990. DNA looping and unlooping by AraC protein. Science 250: 528–532 [DOI] [PubMed] [Google Scholar]
- Monod J., Jacob F., 1961. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb. Symp. Quant. Biol. 26: 389–401 [DOI] [PubMed] [Google Scholar]
- Ptashne M., 2004. A Genetic Switch. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Ptashne M., Gann A., 2002. Genes and Signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schleif R., 2010. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34: 779–796 [DOI] [PubMed] [Google Scholar]
- Sheppard D. E., Englesberg E., 1967. Further evidence for positive control of the l-arabinose system by gene araC. J. Mol. Biol. 25: 443–454 [DOI] [PubMed] [Google Scholar]
- Skalka A., Butler B., Echols H., 1967. Genetic control of transcription during development of phage lambda. Proc. Natl. Acad. Sci. USA 58: 576–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1957. Repressed and induced enzyme formation: a unified hypothesis. Proc. Natl. Acad. Sci. USA 43: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wofsy L., 1995. Looking for the Future: A Personal Connection to Yesterday’s Great Expectations, Today‘s Reality, and Tomorrow’s Hope. I.W. Rose Press, Oakland, CA [Google Scholar]
- Zubay G., Schwartz D., Beckwith J., 1970. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. USA 66: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]