Abstract
Although genome-wide association studies have successfully identified thousands of risk loci for complex traits, only a handful of the biologically causal variants, responsible for association at these loci, have been successfully identified. Current statistical methods for identifying causal variants at risk loci either use the strength of the association signal in an iterative conditioning framework or estimate probabilities for variants to be causal. A main drawback of existing methods is that they rely on the simplifying assumption of a single causal variant at each risk locus, which is typically invalid at many risk loci. In this work, we propose a new statistical framework that allows for the possibility of an arbitrary number of causal variants when estimating the posterior probability of a variant being causal. A direct benefit of our approach is that we predict a set of variants for each locus that under reasonable assumptions will contain all of the true causal variants with a high confidence level (e.g., 95%) even when the locus contains multiple causal variants. We use simulations to show that our approach provides 20–50% improvement in our ability to identify the causal variants compared to the existing methods at loci harboring multiple causal variants. We validate our approach using empirical data from an expression QTL study of CHI3L2 to identify new causal variants that affect gene expression at this locus. CAVIAR is publicly available online at http://genetics.cs.ucla.edu/caviar/.
Keywords: association studies, causal variants, fine mapping
ALTHOUGH genome-wide association studies (GWAS) reproducibly identified thousands of risk loci (Hakonarson et al. 2007; Sladek et al. 2007; Zeggini et al. 2007; Yang et al. 2011a,b; Kottgen et al. 2013; Lu et al. 2013; Ripke et al. 2013), only a handful of causal genetic variants (i.e., variants that biologically alter disease risk) have been found (Altshuler et al. 2008; Manolio et al. 2008; McCarthy et al. 2008), thus prohibiting the mechanistic understanding of the genetic basis of common diseases. The linkage disequilibrium (LD) (Pritchard and Przeworski 2001; Reich et al. 2001) structure of the human genome has greatly benefited GWAS in interrogating only a subset of all variants to assay common variation across the genome. Unfortunately, LD hinders the identification of causal variants at risk loci in fine-mapping studies as at each locus, there are often tens to hundreds of variants tightly linked to the reported associated single-nucleotide polymorphism (SNP) (Malo et al. 2008; Maller et al. 2012; Yang et al. 2012). In a continued effort to identify causal variants, many fine-mapping studies that assess genetic variation at known GWAS risk loci are currently underway (Bauer et al. 2013; Coram et al. 2013; Diogo et al. 2013; Gong et al. 2013; Marigorta and Navarro 2013; Peters et al. 2013; Wu et al. 2013).
Fine-mapping studies typically follow a two-step procedure. First, a statistical analysis of the association signal is performed to identify a minimum set of SNPs that can explain the signal. Second, the SNPs that are putatively causal are functionally tested using laborious and expensive functional assays. Therefore, the objective of the statistical component of fine mapping is to minimize the number of SNPs that need to be selected for follow-up studies while identifying the true causal SNPs. In this work, we focus on developing approaches for statistical refinement of the association signal with the goal of identifying the minimum set of variants to be tested to identify all the causal variants. Although in this work we primarily focus on common variants, our work can be extended to rare variants through careful regularization of normalized association scores (z-scores) (Navon et al. 2013).
The basic statistical fine-mapping approach is to select SNPs for functional validation based on the strength of the association signal. A standard statistical association test is performed, followed by the selection of the top k SNPs with the highest evidence of association for functional assays. The value of k depends on the budget and resources assigned for the follow-up study. This procedure is suboptimal as it does not properly account for the LD at a particular locus (Lawrence et al. 2005; Udler et al. 2009; Faye et al. 2013). For example, two SNPs in perfect LD will always show the same association statistic and it is unclear how to prioritize these SNPs for functional assays. In addition, the finite sampling of individuals in the fine-mapping study induces statistical noise in the association statistics that can result in higher association statistics at neighboring SNPs as opposed to the true causal SNP. Furthermore, even when the sample sizes are large enough such that the statistical noise can be ignored, the local LD structure can induce higher association statistics for neighboring SNPs rather than causal variants at loci with multiple causal variants (Udler et al. 2009). More fundamentally, this approach provides no guarantees that the actual causal SNPs are contained in the top k SNPs selected for functional assays.
In this article, as opposed to the basic top k approach, recent works (Maller et al. 2012; Beecham et al. 2013) have proposed to estimate the probability of each SNP to be causal at a given locus under the simplifying assumption that each GWAS associated locus harbors exactly one causal variant. Under this assumption the approximation of the posterior can be computed using only the marginal per-SNP association statistics. This induces a one-to-one relationship between marginal association statistics and the estimated posterior probabilities that yields the same ranking of SNPs within each locus. A major advantage of this approach is that confidence intervals (i.e., sets of SNPs that account for the 95% of all the posterior probability of causal variants in the locus) can be estimated and used to determine the number of SNPs for each locus to follow up in functional assays. A major drawback of this approach is that the confidence intervals rely on the assumption of a single causal variant per locus. As we show below, when applied to loci where there are more than one causal variant (Haiman et al. 2007; Allen et al. 2010; Galarneau et al. 2010; Chung et al. 2011; Trynka et al. 2011; Stahl et al. 2012; Flister et al. 2013), the confidence intervals may not contain any causal variants with a much higher than expected likelihood.
As opposed to the approaches above that yield the same ranking of SNPs, conditioning approaches to dissect the association signal that may change the ranking of variants have also been proposed (Allen et al. 2010; Galarneau et al. 2010; Chung et al. 2011; Trynka et al. 2011; Stahl et al. 2012; Flister et al. 2013). The conditional approach relies on an iterative selection of most associated SNPs followed by recomputation of the statistical score for the remaining SNPs conditional on the already selected SNPs. The iterations continue until no significant signal remains in the locus at a nominal or Bonferroni-corrected significance (Udler et al. 2009; Allen et al. 2010; Sklar et al. 2011; Yang et al. 2011a,b, 2012). Although conditioning is amenable for identifying the presence of multiple signals within the locus, it can also lead to the unfavorable situation of selection of no causal SNPs for follow-up assays. For example, in the case of two SNPs in perfect LD, where only one of the SNPs is the causal variant, the conditioning approach will drop one of the SNPs from the analysis, depending on the order in which the SNPs are selected in the iterative procedure. Since the statistics at these two SNPs are mathematically equal, the order can only be random (in the absence of other sources of information), leading to conditioning not finding any causal variants in 50% of the cases. This underlines a major drawback of the conditioning approach that can lead to highly suboptimal scenarios when searching for variants to test in functional assays.
Compared to previous work, we propose causal variants identification in associated regions (CAVIAR), a statistical framework that quantifies the probability of each variant to be causal while allowing an arbitrary number of causal variants. We accomplish this by jointly modeling the observed association statistics at all variants in the risk locus; posterior probabilities for sets of variants to be causal are then estimated using the conditional distribution of all association statistics in the locus conditional on the set of causal variants. The output of our approach is a set of variants that with a certain probability (e.g., 95%) contain all of the causal variants at that locus. Intuitively, the 95% causal confidence set is akin to a 95% confidence interval around an estimated parameter. Through extensive simulations we show that our method attains superior performance over all existing methods with comparable results at loci where there is a single causal variant. We validate our approach using empirical data from an expression QTL (eQTL) study of the CHI3L2 gene (Cheung et al. 2005), where the true causal variants are known. In this data, CAVIAR correctly identifies the true causal variant.
Results
Overview of statistical fine mapping
Our approach, CAVIAR, takes as input the association statistics for all of the SNPs (variants) at the locus together with the correlation structure between the variants obtained from a reference data set such as the HapMap (Gibbs et al. 2003; Frazer et al. 2007) or 1000 Genomes project (Abecasis et al. 2010) data. Using this information, our method predicts a subset of the variants that has the property that all the causal SNPs are contained in this set with the probability ρ (we term this set the “ρ causal set”). In practice we set ρ to values close to 100%, typically ≥95%, and let CAVIAR find the set with the fewest number of SNPs that contains the causal SNPs with probability at least ρ. The causal set can be viewed as a confidence interval. We use the causal set in the follow-up studies by validating only the SNPs that are present in the set. While in this article we discuss SNPs for simplicity, our approach can be applied to any type of genetic variants, including structural variants.
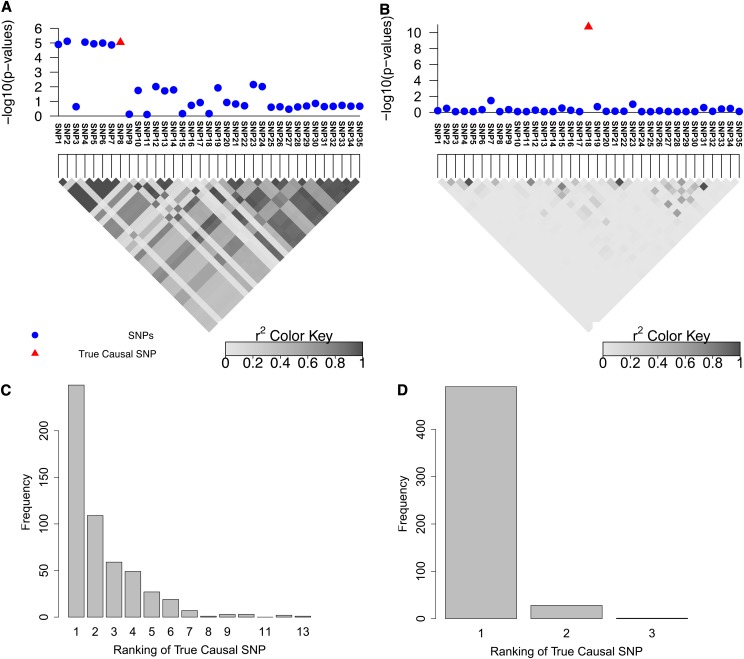
We used simulations to show the effect of LD on the resolution of fine mapping. We selected two risk loci (with large and small LD) to showcase the effect of LD on fine mapping (see Figure 1, A and B). The first region is obtained by considering 100 kbp upstream and downstream of the rs10962894 SNP from the coronary artery disease (CAD) case–control study. As shown in the Figure 1A, the correlation between the significant SNP and the neighboring SNPs is high. We simulated GWAS statistics for this region by taking advantage that the statistics follow a multivariate normal distribution, as shown in Han et al. (2009) and Zaitlen et al. (2010) (see Materials and Methods). CAVIAR selects the true causal SNP, which is SNP8, together with six additional variants (Figure 1A). Thus, when following up this locus, we have only to consider these SNPs to identify the true causal SNPs. The second region showcases loci with lower LD (see Figure 1B). In this region only the true causal SNP is selected by CAVIAR (SNP18). As expected, the size of the ρ causal set is a function of the LD pattern in the locus and the value of ρ, with higher values of ρ resulting in larger sets (see Table S1 and Table S2).
Figure 1.
(A and B) Simulated data for two regions with different LD patterns that contain 35 SNPs. A and B are obtained by considering the 100 kbp upstream and downstream of rs10962894 and rs4740698, respectively, from the Wellcome Trust Case–Control Consortium study for coronary artery disease (CAD). (C and D) The rank of the causal SNP in additional simulations for the regions in A and B, respectively. We obtain these histograms from simulation data by randomly generating GWAS statistics using multivariate normal distribution. We apply the simulation 1000 times.
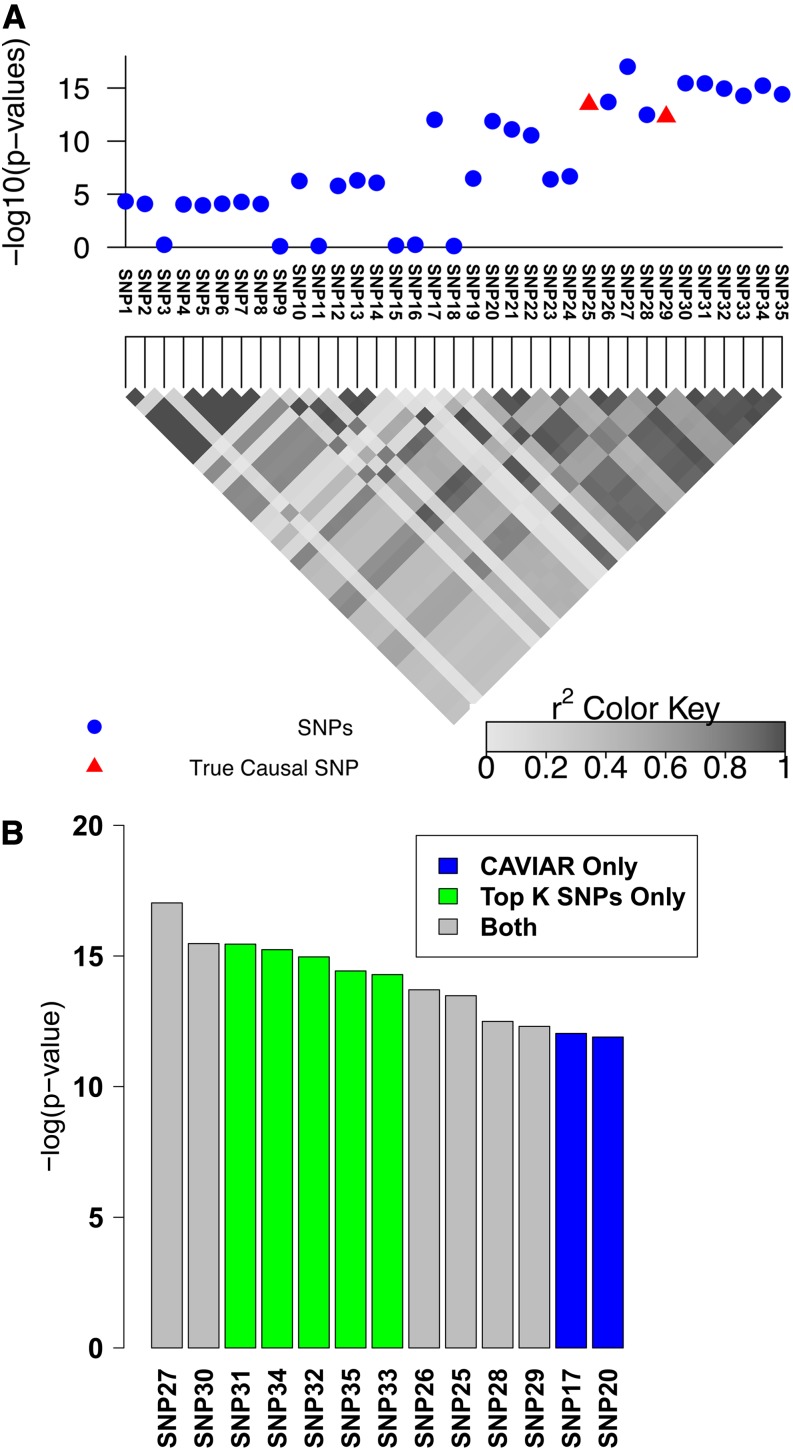
We also showcase the scenario of multiple causal variants (see Figure 2). We simulated data as before and considered SNP25 and SNP29 as the causal SNPs. Interestingly, the most significant SNP (SNP27, see Figure 2) tags the true causal variants but it is not itself causal, making the selection based on strength of association alone under the assumption of a single causal or iterative conditioning highly suboptimal. To capture both causal SNPs at least 11 SNPs must be selected in ranking based on P-values or probabilities estimated under a single causal variant assumption. As opposed to existing approaches, CAVIAR selects both SNPs in the 95% causal set together with five additional variants. The gain in accuracy of our approach comes from accurately disregarding SNP30–SNP35 from consideration since their effects can be captured by other SNPs.
Figure 2.
Simulated association with two causal SNPs. (A) The 100-kbp region around the rs10962894 SNP and simulated statistics at each SNP generated assuming two SNPs are causal. In this example SNP25 and SNP29 are considered as the causal SNPs. However, the most significant SNP is the SNP27. (B) The causal set selected by CAVIAR (our method) and the top k SNPs method. We ranked the selected SNPs based on the association statistics. The gray bars indicate the selected SNPs by both methods, the green bars indicate the selected SNPs by the top k SNPs method only, and the blue bars indicate the selected SNPs by CAVIAR only. The CAVIAR set consists of SNP17, SNP20, SNP21, SNP25, SNP26, SNP28, and SNP29. For the top k SNPs method to capture the two causal SNPs we have to set k to 11, as one of the causal SNPs is ranked 11th based on its significant score. Unfortunately, knowing the value of k beforehand is not possible. Even if the value of k is known, the causal set selected by our method excludes SNP30–SNP35 from the follow-up studies and reduces the cost of follow-up studies by 30% compared to the top k method.
Iterative conditioning is suboptimal in statistical fine mapping
We performed simulations to assess the performance of various approaches for identification of the causal variants in fine-mapping studies. In each simulation, we randomly selected one of the SNPs in this region as a causal SNP and generated association statistics for the 35 SNPs, using our data-generating model (see Materials and Methods). We set the statistical power at the causal SNP to be 50% at the genome-wide significance level of α = 10−8. This way, on average, the causal SNP statistic is significant in half of the simulation panels, and the causal SNP does not always attain the peak statistic in the region. Using this procedure, we generated 1000 simulation panels. Figure 1, C and D, indicates the ranking of the causal SNP for both regions, where the x-axis is the ranking of the true causal SNP and the y-axis is the number of simulations where the true causal SNP has that specific ranking. We observe the top k SNP where k is set to one and fails to find the true causal SNP 5–40% of the time, depending on how complex the LD pattern is in the region. Furthermore, this result illustrates that the first step of the conditional method, which selects the most significant SNP, will fail to select the right SNP 5–40% of the time.
CAVIAR outperforms existing approaches in fine mapping
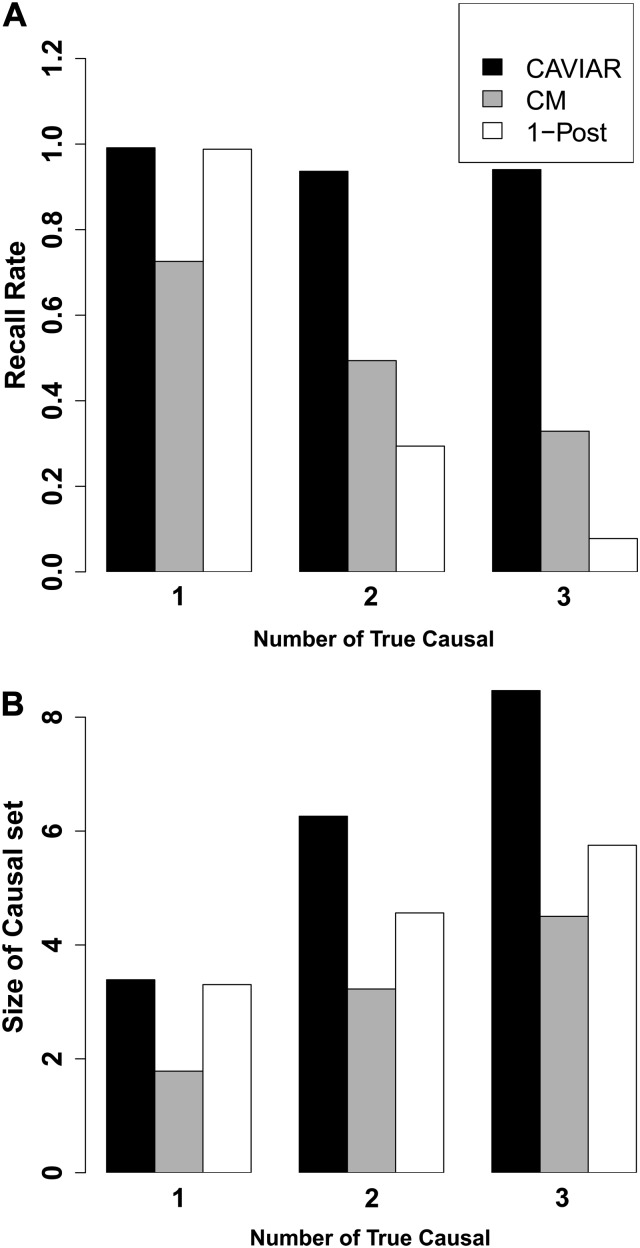
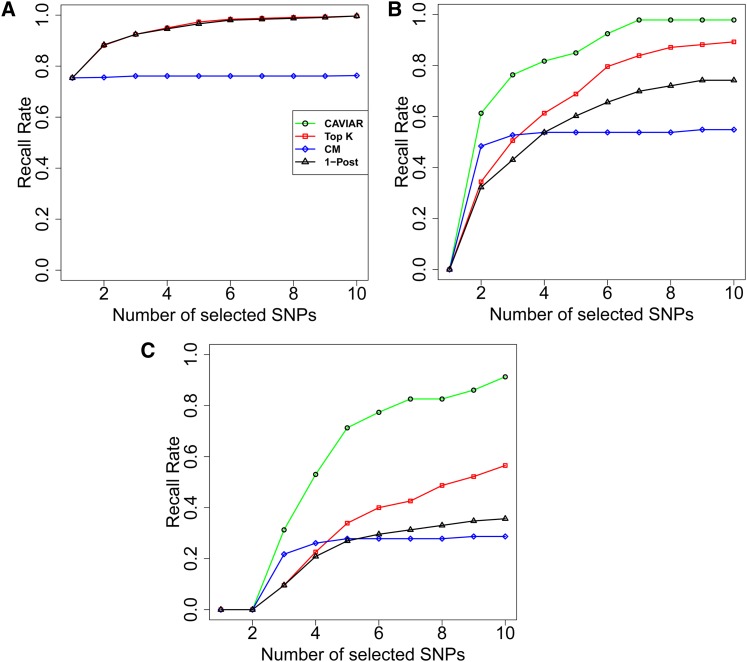
We used HapGen (Spencer et al. 2009) to simulate fine-mapping data across European populations in the 1000 Genomes project (Abecasis et al. 2010) across regions consisting of 50 SNPs. We randomly implanted one, two, or three causal SNPs in each region and then simulated case–control studies. We performed a t-test for each SNP to obtain the marginal statistical scores for each SNP. After obtaining the statistical scores and the LD correlation between each SNP, we applied our method. Figure 3 illustrates the recall rate and the size of the causal set for our method and the two competing methods (conditional and posterior methods). We define recall rate as the fraction of simulations where all the true causal SNPs are identified. The x-axis indicates the number of true causal SNPs implanted in each region. First we compared the recall rate of a probabilistic method that assumes a single causal variant [1-Post (Maller et al. 2012)] and CAVIAR. In simulations of a single causal variant both methods are well calibrated while in scenarios with multiple causals CAVIAR is the only approach that maintains a well-calibrated recall rate. Our simulations suggest that the approach that assumes a single causal variant will attain miscalibrated recall rates at loci with multiple causal variants.
Figure 3.
Comparison of each method’s performance on the simulated GWAS data. (A) The recall rate for each method. (B) The number of causal SNPs selected by each method. CM is the conditional method and 1-Post is the method proposed by Maller et al. (2012). In both panels the x-axis is the true number of causal SNPs that we have implanted in each region. In the scenario of one causal SNP both our method and 1-Post have similar results as both methods use the 95% confidence interval to select a SNP as causal. However, for scenarios in which we have more than one causal SNP, our method outperforms 1-Post.
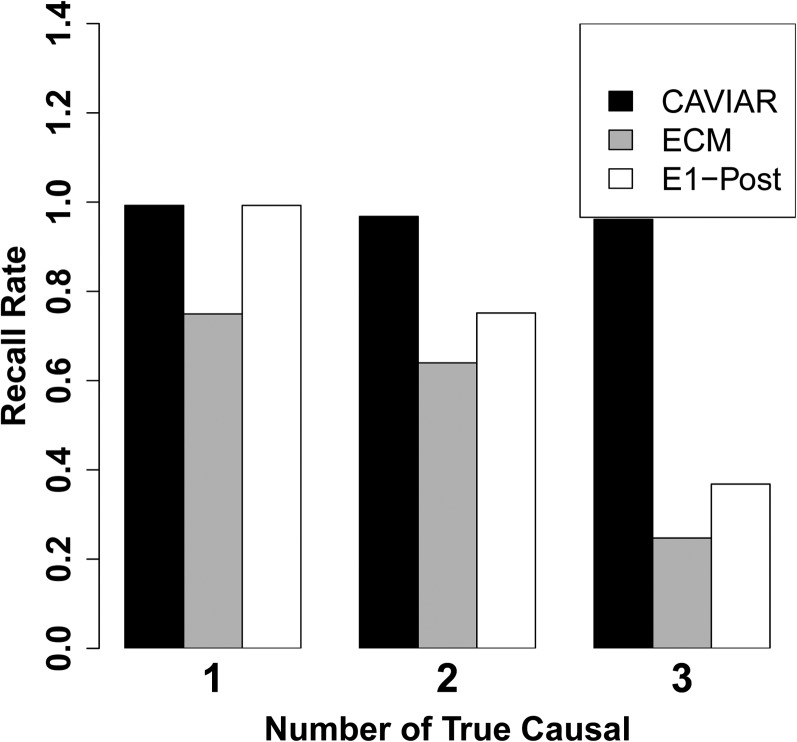
In the above experiments, CAVIAR shows the best recall rate compared to the competing methods. However, the number of SNPs selected by CAVIAR in the causal set is slightly higher than in those methods. To make the comparison among these methods fair, we extended the conditional method (CM) and the 1-Post method such that the number of SNPs selected by each method is equal to the number of SNPs selected by CAVIAR. The extensions of the CM and the 1-Post method are referred to as the ECM and the E1-Post method. As shown in Figure 4, our method has the highest recall rate among the competing methods for all the scenarios. Furthermore, we compared the ranking of the causal SNPs for each method. We vary the number of SNPs selected by each method from 1 SNP to 10 SNPs and compare the recall rate. The results are shown in Figure 5. The x-axis is the number of SNPs selected by each method and the y-axis is the recall rate for each method.
Figure 4.
Comparison of recall rates. ECM and E1-Post are our extension of the CM and the 1-Post method, respectively, where we allow them to select the same number of causal SNPs as CAVIAR.
Figure 5.
The recall rate compression for different methods while selecting the same number of causal SNPs. The x-axis is the number of SNPs selected by each method and the y-axis is the recall rate for each method. A, B, and C represent the scenarios where we have implanted one, two, and three causal SNPs, respectively. In the scenario of only one causal SNP CAVIAR, top k SNPs, and the 1-Post method obtain similar ranking for SNPs.
We also assessed the impact of the number of individuals in the fine-mapping study. As expected, we find that CAVIAR’s confidence set decreases with increased sample size (see Figure S1).
Fine mapping of the CHI3L2 locus
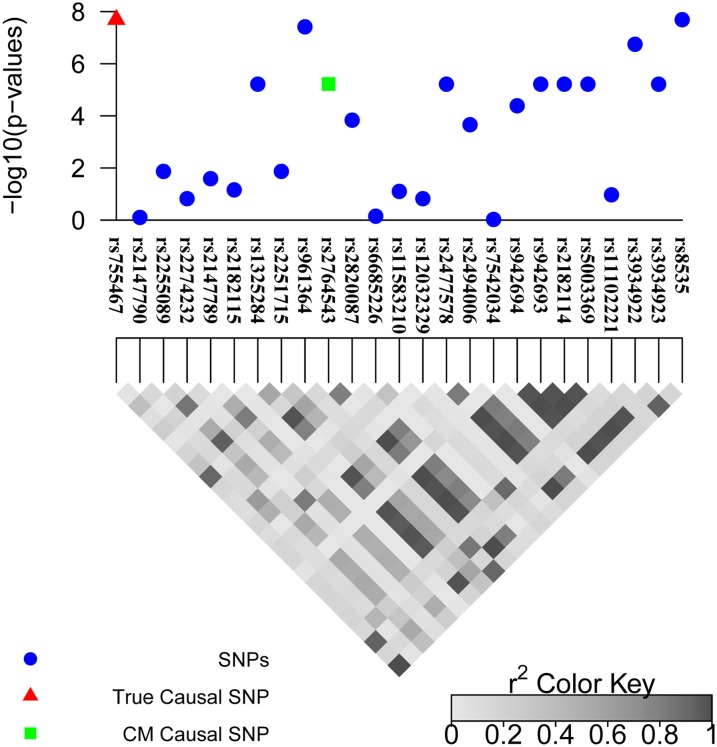
To validate simulation results, we applied CAVIAR to the CHI3L2 region, using the gene expression as a phenotype. This locus was extensively fine mapped with the true causal variant already identified (Cheung et al. 2005; Chen and Witte 2007; Malo et al. 2008). We obtained marginal statistical scores for each SNP from the Malo et al. (2008) study and inferred LD patterns from the HapMap data for 57 unrelated individuals of European ancestry (CEU), the same set of individuals used by previous studies. The result of our method and the LD pattern is shown in Figure 6. CAVIAR selects rs755467, rs961364, rs2764543, rs2477578, rs3934922, and rs8535 for the causal set. Cheung et al. (2005) illustrate the rs755467 SNP is the causal SNP through luciferase reporter and haplotype-specific chromatin immunoprecipitation assays. Furthermore, using the CM and conditioning on the known true causal SNP (rs755467), we obtain the secondary signal in the region, which is rs2764543. The E1-Post 95% causal set selected the same six SNPs as CAVIAR. The ECM selects rs755467, rs2274232, rs2182115, rs2764543, rs2820087, and rs11583210 for the causal set.
Figure 6.
The 95% causal set selected by CAVIAR for the CHI3L2 region. The red triangle represents the true causal SNP that is known using experimental methods (Cheung et al. 2005) and the green square represents the causal SNP detected using the CM conditional on the true causal SNP (rs755467).
Materials and Methods
The traditional fine-mapping study approach
A fine-mapping study is a procedure to identify, or predict, the disease causing SNPs from a given GWAS data set. It is assumed that the genotype data are dense enough, such that all the causal SNPs are genotyped, including the SNPs that are perfectly correlated to the causal variants other than SNPs. With the development of sequencing technologies, this assumption is becoming more realistic. Therefore, we assume that there exists a true label for each genotyped SNP on whether or not the SNP is causal in disease.
The traditional fine-mapping study approach performs the following iterative procedure to predict the causal SNPs within a genomic region. First, the association statistic of each SNP is computed and the most strongly associated SNP is chosen as a causal SNP. Intuitively, if the region contains a single causal SNP, then the most significantly associated SNP is likely to be the causal SNP itself (the assumption in the traditional fine-mapping approach). However, the region may contain multiple causal SNPs, and furthermore these SNPs may be correlated or in LD. In this scenario, the association statistic at a causal SNP may be contaminated by the presence of the causal SNPs that are in LD. To control for this contamination, at each iteration, the traditional approach recomputes the association statistic of the SNPs while conditioning on the presence of the causal SNPs that are identified in each iteration of the method. Given a statistic threshold, if the statistic of the most strongly associated SNP exceeds the threshold, the SNP is chosen as a causal SNP, or otherwise the procedure terminates.
We show through empirical and theoretical results that the traditional approach is underpowered to identify the causal SNP compared to our method. In the next section we present a data-generating model for fine-mapping studies.
Data-generating model for fine-mapping studies
We consider a GWAS on a quantitative trait where n individuals are genotyped on m SNPs. For individual k, we are given the phenotypic value yk and the genotype values at m SNPs, where for SNP i, gik ∈ {0, 1, 2} is the minor allele count. Let y denote the (n × 1) vector of the phenotypic values and xi denote the (n × 1) vector of normalized genotype values at SNP i such that 1Txi = 0 and
Let us assume that a SNP c is the only SNP involved in the disease. We assume the data-generating model follows a linear model,
where 1 denotes the (n × 1) vector of ones, μ is the intercept, βc is the effect-size of SNP c, and e is the (n × 1) vector of i.i.d. and normally distributed residual noise, where e ∼ (0, σ2I) with covariance scalar σ and (n × n) identity matrix I.
The estimates for μ and βc are obtained by maximizing the likelihood function,
The association statistic for SNP c, denoted by follows a noncentral t distribution, which is the ratio of a normally distributed random variable to the square root of an independent chi-square-distributed random variable,
with noncentrality parameter (NCP) and n d.f. Note that
where denotes the chi-square distribution with n d.f. and it can be shown that is independent of
For simplicity, we assume the sample size n is large enough, such that the association statistic Sc is well approximated by a normal distribution with NCP λc and unit variance
Furthermore, if SNP i is correlated with a disease-involved SNP c with coefficient r, i.e., the estimate of its effect size follows
The covariance between the two normal random variables reads
Therefore, the joint distribution of the association statistics of two SNPs in a region follows a multivariate normal distribution,
If we assume the ith SNP is causal, we have λj = rijλi, and if we assume the jth SNP is causal, we have λi = rijλj. Given the significance level α and the observed value of the test statistic the SNP is deemed significant, or statistically associated, if where Φ−1(.) is the quantile function of the standard normal distribution.
The equivalent derivation showing that the joint distribution of the association statistics in case/control studies follows the multivariate normal distribution has been shown in Han et al. (2009).
A new framework for computing the posterior probability of causal SNP statuses from GWAS data
Consider we are given a set of m SNPs ℳ, with their pairwise correlation coefficients Σ. We introduce a new parameter, c, an (m × 1) causal status indicator vector, with ci denoting an element for that vector. There are three possible causal statuses for each SNP: positive effect (ci = +1), negative effect (ci = −1), and no effect (ci = 0). The indicator vector c can take 3m possible causal statuses, denoted by the set , with 3m − 1 of them having at least one causal SNP.
We denote the association statistics of the SNPs by the (m × 1) vector S = [S1 … Sm]T, which follows a multivariate normal distribution,
| (1) |
where, for simplicity in presenting the model, we assume all causal SNPs have the same NCP, λc. Later, we relax this assumption by utilizing the standard Fisher’s polygenic model that effects size follows a normal distribution with mean zero. Although the above equation holds for common variants, we can extended it to rare variants through careful regularization of normalized association scores (z-scores) (Navon et al. 2013).
Let c∗ ∈ denote a particular causal status. We define a prior probability over the possible causal statuses, P(c), which assumes that each variant has a probability of being causal in either direction, γ,
Below, we extend the prior to allow for incorporating functional information into our approach.
Given the observed association statistics of the m SNPs, the posterior probability of the causal status can be expressed as
| (2) |
Given a set of SNPs ⊂ ℳ, we denote the set of causal SNP configurations rendered by with , which excludes all causal SNP configurations having a SNP outside of as causal. Note that our definition for includes the null configuration of having no causal SNPs as well. Using , we can compute the posterior probability of to include, or capture, all the causal SNPs,
We denote the value of this posterior probability with ρ, where and refer to it as the confidence level of in capturing the causal SNPs. Similarly, we refer to as a “ρ confidence set of causal SNPs” or a “ρ confidence set.”
Given a minimum confidence threshold ρ∗, there can be many confidence sets, each having a confidence level that is greater than the threshold. Among all these sets, the ones with a smaller number of SNPs are more informative, or have higher resolution, in locating the causal SNPs. Then, the problem we are interested in is to find the ρ∗ confidence set with the minimum size,
where has the minimum size.
Generalized framework for a locus with multiple causal SNPs with different NCP values
In the previous section we consider the case where all the causal SNPs in a locus have the same NCP. Thus, λcc indicates a point in a Rm space and the coordinates corresponding to the causal SNPs have value of ±λc and the coordinates corresponding to the noncausal SNPs have a value of zero. We relax this assumption to instead have the NCP for each causal SNP drawn from a distribution with mean 0 and variance σ2. This is the standard assumption of Fisher’s polygenic model.
We define the prior probability on the vector of NCP λc for a given causal status c, using the multivariate normal probability
where Σc is constructed as follows:
∈ is a small constant that ensures that the matrix Σc is of full rank. The final prior is then
| (3) |
where f(λc, 0, Σc) is the probability density function of the causal status (λc|c) ∼ (0, Σc). We use the above generalization as a prior on the mean of the distribution indicated in Equation 1. We know the LD between two SNPs is symmetric (ΣT = Σ) and the NCP λ = Σλc,
Thus, the association statistics of the SNPs follow a multivariate normal distribution,
Optimization
To compute the posterior probability for each set, which is shown in Equation 2, we calculate the summation over the likelihood of all the possible causal statuses. Unfortunately, computing this summation that is the denominator of the Equation 2 is computationally intractable in the general case (multiple causal SNPs with different NCP values). Thus, to simplify the calculation we assume the total number of causal SNPs in a region is bounded by at most six causal SNPs. Although this assumption simplifies the denominator in Equation 2, to detect the minimum causal set still we have to consider all the possible causal statuses. We utilize the following greedy algorithm to make the detection of the minimum causal set tractable. In each iteration of the greedy algorithm we select a SNP to be causal that increases the posterior probability the most. The process of selecting SNPs to be causal continues as long as the posterior probability of the causal set is at least a ρ fraction of the total posterior probability of the data.
Using simulated data, we show in Supporting Information, File S1, and Table S3 the proposed greedy method results are similar to the results obtained by solving Equation 2 exactly. In addition, for each causal status we define a prior. To compute the prior, we assume each SNP is independent and the probability of a SNP to be causal is equal to 10−2 (Eskin 2008).
To identify the causal SNP sets, we need to consider all possible subsets of the SNPs that number 2m (in the case of multiple causal SNPs with different NCP values, we consider two causal statuses for each SNP: have an effect or have no effect) when m is the number of SNPs in the region. In the process of computing the posterior probability for each of these possible subsets, we need to enumerate over each possible causal status for each SNP. There are two possible causal statuses for each SNP. The SNP has an effect or the SNP has no effect. Thus for each possible subset of SNPs, we need to consider 2m possible causal statuses for the SNPs. For each of these statuses, the multivariate normal distribution is utilized to compute the likelihood of the data given the causal statuses. Thus to identify the best causal SNP set, we must perform a significant amount of computation.
The computational burden is high because we need to consider every possible subset of SNPs to be in the causal set and for each subset we need to enumerate all of the possible causal SNP statuses. We propose two ideas to reduce the computational burden. The first idea only reduces the possible causal status that we need to consider for each subset. The second idea utilizes a greedy algorithm to identify the subset of SNPs in the causal set by eliminating our need to consider all possible subsets.
To reduce the computational burden, we assume in each region we have at most six causal SNPs. If we consider only causal statuses that have a total of i causal SNPs, there are possible different causal statuses. Thus, for the case where we consider only at most six causal SNPs we have possible causal statuses, which reduces the number of possible causal statuses. The intuition behind this assumption lies in the fact that causal variants are relatively rare. Using the simulated data we show (Table S3) the set obtained by considering only six causal SNPs in a region is highly similar to the set obtained by considering all the 2m causal statuses.
The assumption of at most six causal SNPs reduces the computational burden to compute the posterior probability for each subset of SNPs. However, to identify the causal SNP sets, we need to select the smallest subset of SNPs that has the desired posterior probability. This process can be extremely slow in some cases as we need to consider all the possible subsets of SNPs. We propose an efficient greedy method where in each iteration of the method we select a SNP that increases the posterior probability the most. We continue the process of adding SNPs to the causal set until we have the desired posterior probability for the causal set.
Incorporating functional data as a prior into CAVIAR
Although we consider a simple prior in our model, CAVIAR can easily be extended to incorporate external information such as functional data or knowledge from previous studies. This external information can be incorporated into CAVIAR as a prior. We allow the probability that a variant is part of a causal set to vary from variant to variant, depending on prior information. This variant-specific probability is denoted γi. We extend Equation 3 and instead of P(c) as the prior for each causal status, we compute P(c|γ = [γ1, γ2, …, γm]) as follows:
Conditional method for fine mapping
Here we show how to compute the statistics for the rest of the SNPs, given we have selected a SNP as the causal SNP. For simplicity we use only two SNPs to compute the conditional statistics. Thus, we have
Conditioning on one SNP is equivalent to making the statistics for that SNP equal to zero. Moreover, the variance of the remaining SNP is one. As a result,
We use the iterative method to obtain all the causal SNPs. In each iteration of the method we pick the SNP with the lowest P-value (the highest statistics) and recompute the statistics of the remaining SNP, using the formula mentioned above. We keep repeating this process until no significant SNP exists. In our experiment we set the significant threshold value to 0.001.
Discussion
Over the past few years, GWAS have identified hundreds of genetic loci harboring genetic variation affecting disease risk for hundreds of common diseases (Bauer et al. 2013; Coram et al. 2013; Diogo et al. 2013; Gong et al. 2013; Marigorta and Navarro 2013; Peters et al. 2013; Wu et al. 2013). Identifying the causal genetic variants affecting disease risk at these loci has the potential of providing clues to the mechanism of the disease, which can lead to identification of better targets for drug terrapins. Unfortunately, the pervasive LD and the uncertainty of data make the task of deconvoluting causal variants from tagging ones very challenging.
In this article, we present a novel framework for identifying the causal variants underlying GWAS risk loci. The key idea behind our framework is that instead of considering each variant one at a time, we instead analyze all of the variants in the entire locus simultaneously. The result of our method is a set of variants that with high probability contains (or captures) all the causal variants. Through extensive simulation results, we show that our approach is superior to existing methods in reducing the overall number of variants to be examined in functional follow-up to identify the causal variants.
In our method we make a series of assumptions to ease the computational burden and to simplify the model. We make the assumption that the number of causal SNPs in a region, in which we are interested to preform fine mapping, is at most six. Our method also makes the standard assumption of Fisher’s polygenic model that effects size follows a normal distribution with mean zero. This assumption is the basis of many recent approaches to estimate heritability (Yang et al. 2011a,b; Speed et al. 2012; Kostem and Eskin 2013) and to correct for population structure in GWAS (Kang et al. 2008; Lippert et al. 2011; Listgarten et al. 2012; Segura et al. 2012; Zhou and Stephens 2012).
Our method also assumes that we have genotyped each variant in the locus. With the increasing cost efficiency of high-throughput sequencing, this assumption is becoming more and more realistic. One future direction of research is to extend this approach to handle imputed association statistics. In this case, only a relatively small number of individuals in a GWAS must be fully sequenced at the locus while for the rest of the individuals the sequenced individuals can be used as an imputation reference panel.
Our method takes as input the association statistics and linkage disequilibrium patterns in the locus to identify the set of variants that are likely to contain the causal variants. The minor allele frequencies of the variants will affect the magnitude of the observed statistics as well as the linkage disequilibrium patterns. However, our approach is applied only to loci that harbor significant association signals at individuals’ variants. These types of signals are most likely driven by common variants. Most likely, additional rare variants in the locus that also have effects on the phenotype will not be selected because their association statistics are low. Extending our approach to discover additional rare variants in a locus is an interesting direction for future work.
CAVIAR can easily take into account data on putative function of variants either from functional genomic data (Bernstein et al. 2012) or from eQTL data that have been recently shown to help facilitate fine-mapping studies (Hoffman et al. 2012; Edwards et al. 2013). The way that this information can be incorporated is by assigning each variant a prior probability of affecting the trait (Eskin 2008; Jul et al. 2011; Darnell et al. 2012). In this framework, the functional genomic data are converted to a probability between 0 and 1 of that variant having an effect on the trait. These priors then affect the likelihood of each causal status and then ultimately are incorporated into the final causal set.
The method presented in this article has some conceptual similarities to methods for identifying associations in regions where there is more than one associated variant. These methods have become very popular in the context of rare variant association studies (Li and Leal 2008; Madsen and Browning 2009; Jul et al. 2011; Long et al. 2013; Navon et al. 2013). However, there are other methods that also consider common variants (Wu et al. 2011; Yi et al. 2011). Our method differs from these approaches in that our goal is to narrow down the possible set of variants in a locus that we suspect is associated while the previous approaches utilize multiple variants to attempt to identify an associated locus.
Compared to methods for association testing, methods for fine mapping, including the proposed method, are more complicated and make many implicit or explicit assumptions. For example, our method makes explicit assumptions about the effect size of causal variants while association methods make no such assumptions. In our view, this is inherent to the fact that fine-mapping methods attempt to control false negatives compared to association methods that attempt to control false positives. To control false negatives, fine-mapping methods must make explicit assumptions about the “alternate” distribution to understand how well the data fit the assumptions. Association methods on the other hand, to control false positives, need only to make assumptions about the null distribution, which in the case of association studies is the assumption that all of the variants at a locus have no effects. This asymmetry characterizes the fine-mapping problem and complicates attempts to merge fine mapping and association into a single framework.
Supplementary Material
Acknowledgments
F.H., E.K., E.Y.K., and E.E. are supported by National Science Foundation grants 0513612, 0731455, 0729049, 0916676, 1065276,1302448, and 1320589 and National Institutes of Health (NIH) grants K25-HL080079, U01-DA024417, P01-HL30568, P01-HL28481, R01-GM083198, R01-MH101782, and R01-ES022282. We acknowledge the support of the National Institute of Neurological Disorders and Stroke Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691). B.P. is supported in part by the NIH (R03 CA162200 and R01 GM053275). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167908/-/DC1.
Communicating editor: N. Yi
Literature Cited
- Abecasis G., Altshuler D., Auton A., Brooks L., Durbin R., et al. , 2010. A map of human genome variation from population-scale sequencing. Nature 467(7319): 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H. L., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467(7317): 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D., Daly M. J., Lander E. S., 2008. Genetic mapping in human disease. Science 322(5903): 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D. E., Kamran S. C., Lessard S., Xu J., Fujiwara Y., et al. , 2013. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342(6155): 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham A. H., Patsopoulos N. A., Xifara D. K., Davis M. F., Kemppinen A., et al. , 2013. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45(11): 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Birney E., Dunham I., Green E. D., Gunter C., et al. , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489(2): 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. K., Witte J. S., 2007. Enriching the analysis of genomewide association studies with hierarchical modeling. Am. J. Hum. Genet. 81(2): 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V. G., Spielman R. S., Ewens K. G., Weber T. M., Morley M., et al. , 2005. Mapping determinants of human gene expression by regional and genome-wide association. Nature 437(7063): 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. C., Ciampa J., Yeager M., Jacobs K. B., Berndt S. I., et al. , 2011. Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Hum. Mol. Genet. 20(14): 2869–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram M. A., Duan Q., Hoffmann T. J., Thornton T., Knowles J. W., et al. , 2013. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 92(6): 904–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell G., Duong D., Han B., Eskin E., 2012. Incorporating prior information into association studies. Bioinformatics 28(12): i147–i153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo D., Kurreeman F., Stahl E. A., Liao K. P., Gupta N., et al. , 2013. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am. J. Hum. Genet. 92(1): 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Beesley J., French J. D., Dunning A. M., 2013. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93(2): 779–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin E., 2008. Increasing power in association studies by using linkage disequilibrium structure and molecular function as prior information. Genome Res. 18(7319): 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye L. L., Machiela M. J., Kraft P., Bull S. B., Sun L., 2013. Re-ranking sequencing variants in the post-GWAS era for accurate causal variant identification. PLoS Genet. 9(8): e1003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flister J., Tsaih S., O’Meara C., B. Enders, M. J. Hoffmanet al. , 2013. Identifying multiple causative genes at a single GWAS locus. Genome Res. 467(7319): 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., Stuve L. L., et al. , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449(7164): 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau G., Palmer C. D., Sankaran V. G., Orkin S. H., Hirschhorn J. N., et al. , 2010. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet. 42(12): 1049–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. A., Belmont J. W., Hardenbol P., Willis T. D., Yu F., et al. , 2003. The international HapMap project. Nature 426(6968): 789–796 [DOI] [PubMed] [Google Scholar]
- Gong J., Schumacher F., Lim U., Hindorff L. A., Haessler J., et al. , 2013. Fine mapping and identification of BMI loci in African Americans. Am. J. Hum. Genet. 93(4): 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman C. A., Patterson N., Freedman M. L., Myers S. R., Pike M. C., et al. , 2007. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 39(5): 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H., Grant S. F. A., Bradfield J. P., Marchand L., Kim C. E., et al. , 2007. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 448(7153): 591–594 [DOI] [PubMed] [Google Scholar]
- Han B., Kang H. M., Eskin E., 2009. Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet. 5: e1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M. M., Ernst J., Wilder S. P., Kundaje A., Harris R. S., et al. , 2012. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 93(2): 779–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jul J. H., Han B., Eskin E., 2011. Increasing power of groupwise association test with likelihood ratio test. J. Comput. Biol. 18(11): 1611–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 5: e1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostem E., Eskin E., 2013. Improving the accuracy and efficiency of partitioning heritability into the contributions of genomic regions. Am. J. Hum. Genet. 92(2): 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottgen A., Albrecht E., Teumer A., Vitart V., Krumsiek J., et al. , 2013. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 45(2): 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R., Evans D. M., Morris A. P., Ke X., Hunt S., et al. , 2005. Genetically indistinguishable SNPs and their influence on inferring the location of disease-associated variants. Genome Res. 15(11): 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83(3): 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarte J., Liu Y., Kadie M., Davidson R. I., et al. , 2011. FaST linear mixed models for genome-wide association studies. Nat. Methods 8(7): 833–835 [DOI] [PubMed] [Google Scholar]
- Listgarten J., Lippert C., Kadie C. M., Davidson R. I., Eskin E., et al. , 2012. Improved linear mixed models for genome-wide association studies. Nat. Methods 9(7): 525–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long N., Dickson S. P., Maia J. M., Kim H. S., Zhu Q., et al. , 2013. Leveraging prior information to detect causal variants via multi-variant regression. PLoS Comput. Biol. 9(6): e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Vitart V., Burdon K. P., Khor C. C., Bykhovskaya Y., et al. , 2013. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet. 45(2): 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B. E., Browning S. R., 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5(2): e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller J. B., McVean G., Byrnes J., Vukcevic D., Palin K., et al. , 2012. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44(12): 1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo N., Libiger O., Schork N. J., 2008. Accommodating linkage disequilibrium in genetic-association analyses via ridge regression. Am. J. Hum. Genet. 82(2): 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Brooks L. D., Collins F. S., 2008. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118(5): 1590–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta U. M., Navarro A., 2013. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 9(6): e1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., et al. , 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9(5): 356–369 [DOI] [PubMed] [Google Scholar]
- Navon O., Sul J. H., Han B., Conde L., Bracci P. M., et al. , 2013. Rare variant association testing under low-coverage sequencing. Genetics 194: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters U., North K. E., Sethupathy P., Buyske S., Haessler J., et al. , 2013. A systematic mapping approach of 16q12.2/FTO and BMI in more than 20,000 African Americans narrows in on the underlying functional variation: results from the Population Architecture using Genomics and Epidemiology (PAGE) study. PLoS Genet. 9(1): e1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Przeworski M., 2001. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69(1): 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D. E., Cargill M., Bolk S., Ireland J., Sabeti P. C., et al. , 2001. Linkage disequilibrium in the human genome. Nature 411(6834): 199–204 [DOI] [PubMed] [Google Scholar]
- Ripke S., O’Dushlaine C., Chambert K., Moran J. L., Khler A. K., et al. , 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45(10): 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V., Vilhjlmsson B. J., Platt A., Korte A., Seren U., et al. , 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44(7): 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P., Ripke S., Scott L. J., Andreassen O. A., Cichon S., et al. , 2011. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43(10): 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R., Rocheleau G., Rung J., Dina C., Shen L., et al. , 2007. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445(7130): 881–885 [DOI] [PubMed] [Google Scholar]
- Speed D., Hemani G., Johnson M. R., Balding D. J., 2012. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 91(2): 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. C., Su Z., Donnelly P., Marchini J., 2009. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 5(5): e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E. A., Wegmann D., Trynka G., Gutierrez-Achury J., Do R., et al. , 2012. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 44(5): 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G., Hunt K. A., Bockett N. A., Romanos J., Mistry V., et al. , 2011. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43(12): 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler M. S., Meyer K. B., Pooley K. A., Karlins E., Struewing J. P., et al. , 2009. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum. Mol. Genet. 18(9): 1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare variant association testing for sequencing data with the sequence kernel association test (SKAT). Am. J. Hum. Genet. 89(2): 82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Waite L. L., Jackson A. U., Sheu W. H.-H., Buyske S., et al. , 2013. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet. 9(3): e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011a GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88(1): 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Manolio T. A., Pasquale L. R., Boerwinkle E., Caporaso N., et al. , 2011b Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43(6): 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ferreira T., Morris A. P., Medland S. E., Madden P. A., et al. , 2012. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44(4): 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Liu N., Li J., 2011. Hierarchical generalized linear models for multiple groups of rare and common variants: jointly estimating group and individual-variant effects. PLoS Genet. 12(7): e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N., Pasaniuc B., Gur T., Ziv E., Halperin E., 2010. Leveraging genetic variability across populations for the identification of causal variants. Am. J. Hum. Genet. 86: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E., Weedon M. N., Lindgren C. M., Frayling T. M., Elliott K. S., et al. , 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316(5829): 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed model analysis for association studies. Nat. Genet. 44(7): 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.