Abstract
In the yeast Saccharomyces cerevisiae, fermentation is the major pathway for energy production, even under aerobic conditions. However, when glucose becomes scarce, ethanol produced during fermentation is used as a carbon source, requiring a shift to respiration. This adaptation results in massive reprogramming of gene expression. Increased expression of genes for gluconeogenesis and the glyoxylate cycle is observed upon a shift to ethanol and, conversely, expression of some fermentation genes is reduced. The zinc cluster proteins Cat8, Sip4, and Rds2, as well as Adr1, have been shown to mediate this reprogramming of gene expression. In this study, we have characterized the gene YBR239C encoding a putative zinc cluster protein and it was named ERT1 (ethanol regulated transcription factor 1). ChIP-chip analysis showed that Ert1 binds to a limited number of targets in the presence of glucose. The strongest enrichment was observed at the promoter of PCK1 encoding an important gluconeogenic enzyme. With ethanol as the carbon source, enrichment was observed with many additional genes involved in gluconeogenesis and mitochondrial function. Use of lacZ reporters and quantitative RT-PCR analyses demonstrated that Ert1 regulates expression of its target genes in a manner that is highly redundant with other regulators of gluconeogenesis. Interestingly, in the presence of ethanol, Ert1 is a repressor of PDC1 encoding an important enzyme for fermentation. We also show that Ert1 binds directly to the PCK1 and PDC1 promoters. In summary, Ert1 is a novel factor involved in the regulation of gluconeogenesis as well as a key fermentation gene.
Keywords: gluconeogenesis, transcriptional regulator, Saccharomyces cerevisiae, fermentation, zinc cluster proteins
IN the yeast Saccharomyces cerevisiae, glucose is the preferred carbon source and fermentation is the major pathway for energy production, even under aerobic conditions. However, when glucose becomes scarce, ethanol produced during fermentation is used as a carbon source, a process requiring a shift to a respiration mode. Other nonfermentable carbon sources, such as lactate, acetate, or glycerol, can also be used by yeast (Turcotte et al. 2011). The shift from fermentative to nonfermentative metabolism results in massive reprogramming of gene expression for the use of ethanol (Derisi et al. 1997; Roberts and Hudson 2006). For example, increased expression of genes for gluconeogenesis and the glyoxylate cycle is observed upon a shift to ethanol and, conversely, expression of some fermentation genes is reduced under these conditions. An important player in the regulation of this process is the Snf1 kinase (Hedbacker and Carlson 2008; Zaman et al. 2008; Zhang et al. 2010; Broach 2012). Snf1 becomes activated under low glucose conditions resulting in the phosphorylation of a number of substrates that include DNA binding proteins such as Mig1, Cat8, Sip4, and Rds2 (for reviews see Schüller 2003; Turcotte et al. 2011).
Mig1 and Adr1 belong to the family of zinc finger proteins of the Cys2His2 type. Mig1 is a transcriptional repressor that, following phosphorylation by Snf1, undergoes nucleocytoplasmic shuffling, allowing derepression of target genes such as CAT8. Adr1 is a transcriptional activator of >100 genes involved in the utilization of ethanol, glycerol, and fatty acids, as well as genes involved in peroxisome biogenesis (Young et al. 2003). For example, Adr1 is a positive regulator of ADH2 encoding alcohol dehydrogenase, an enzyme involved in the conversion of ethanol to acetaldehyde. Cat8, Sip4, and Rds2 are members of the family of zinc cluster proteins (MacPherson et al. 2006). These transcriptional regulators are characterized by the presence of a well-conserved motif (CysX2CysX6CysX5-12CysX2CysX6-8Cys) where the cysteines bind to two zinc atoms that facilitate folding of the zinc cluster domain involved in DNA recognition (MacPherson et al. 2006). Zinc cluster proteins preferentially bind to CGG triplets oriented as direct (CGGNxCGG), inverted (CGGNxCCG), or everted repeats (CCGNxCGG) (MacPherson et al. 2006). These factors have been shown to bind DNA as monomers, homodimers, or heterodimers (MacPherson et al. 2006). Expression of CAT8 is increased under low glucose conditions and Cat8 regulates the expression of SIP4 (Hedges et al. 1995; Lesage et al. 1996). These factors control expression of various genes involved in the utilization of nonfermentable carbon sources. For example, expression of PCK1 encoding phosphoenolpyruvate carboxykinase (an essential enzyme for gluconeogenesis) is controlled by Cat8, Sip4, and Rds2. These factors mediate their effects by binding to carbon source response elements (CSREs) found in the promoters of target genes (Schüller 2003; Turcotte et al. 2011).
In this study, we focused on the gene YBR239C encoding a putative zinc cluster protein that we named ERT1 (ethanol regulated transcription factor 1). Ert1 shows high homology to Rds2 and data from a large-scale two-hybrid analysis suggests that the two proteins interact with each other (Ito et al. 2001). Importantly, AcuK, a homolog of Ert1 in Aspergillus nidulans, regulates expression of gluconeogenic genes (Suzuki et al. 2012). To unravel the role of Ert1, we have performed genome-wide location analysis (ChIP-chip). Results show that Ert1 binds to a limited number of promoters (e.g., PCK1) when cells were grown in the presence of glucose. However, enrichment of Ert1 with many genes involved in gluconeogenesis and other aspects of carbon metabolism was observed when ethanol was used as a carbon source. Finally, our results show that Ert1 acts redundantly with other regulators of gluconeogenesis, behaving as an activator of gluconeogenesis genes as well as a repressor of PDC1, a key fermentation gene. Taken together, our data identified Ert1 as a novel transcriptional regulator of the switch from fermentation to respiration.
Materials and Methods
Strains and media
Yeast strains used in this study are listed in Table 1. The wild-type strain is BY4741 (Brachmann et al. 1998). The open reading frame (ORF) of ERT1 was N-terminally tagged with a triple HA epitope using the approach of Schneider et al. (1995). Briefly, strain BY4741 was transformed with a PCR product generated using oligonucleotides PETYBR239-1 and PETYBR239-2 (Supporting Information, Table S2) and, as a template, plasmid pMPY-3XHA. Colonies were selected on plates lacking uracil. Transformants were grown in the absence of selection to allow loss of the URA3 marker by internal recombination, and Ura− cells were obtained using the 5-fluoroorotic acid selection (Schneider et al. 1995). Integrity of the DNA sequences encompassing the tag was verified by amplifying that region with oligonucleotides ERT1-Acheck and D2 and sequencing of the PCR product (Table S2). To generate the double deletion strains SC111A, SC113, and SC114 (Table 1), genomic DNA from Candida glabrata (strain ATCC 66032) was isolated and used as a template to amplify the HIS3 gene from −696 to +240 relative to the ORF using oligonucleotides ERT1-CgHIS3-1 and ERT1-CgHIS3-2 (Table S2). The PCR product containing sequences homologous to the 5′ and 3′ regions of the ERT1 ORF was transformed in single-deletion strains SC102A, SC103, and SC104 or wild-type W303-1A (Table 1) and transformants were selected on plates lacking histidine. Media were prepared according to Adams et al. (1997). Plasmid pPDC1(−850)-lacZ and mutants were linearized with StuI (site located in the URA3 gene) and integrated at the ura3-52 locus of strain YPH499 by selection on minimal plates lacking uracil and histidine. Quantitative PCR showed that two copies of the plasmids were integrated for selected clones.
Table 1. Strains used in this study.
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| BY4741 | N/A | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al. (1998) |
| SC12 | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [ERT1 tagged with three HA epitopes at the N-terminus] | This study |
| SC102A | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cat8∆::KanMX | Winzeler et al. (1999) |
| SC103 | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sip4∆::KanMX | Winzeler et al. (1999) |
| SC104 | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rds2∆::KanMX | Winzeler et al. (1999) |
| SC105A | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ert1∆::KanMX | Winzeler et al. (1999) |
| SC173 | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf1∆::KanMX | Winzeler et al. (1999) |
| SC111A | SC102A | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cat8∆::KanMX ert1∆::CgHIS3 | This study |
| SC113 | SC103 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sip4∆::KanMX ert1∆::CgHIS3 | This study |
| SC114 | SC104 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rds2∆::KanMX ert1∆::CgHIS3 | This study |
| W303-1A | N/A | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | Rothstein et al. (1977) |
| SC14 | W303-1A | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 ert1∆::CgHIS3 | This study |
| YPH499 | N/A | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | Sikorski and Hieter (1989) |
Plasmids
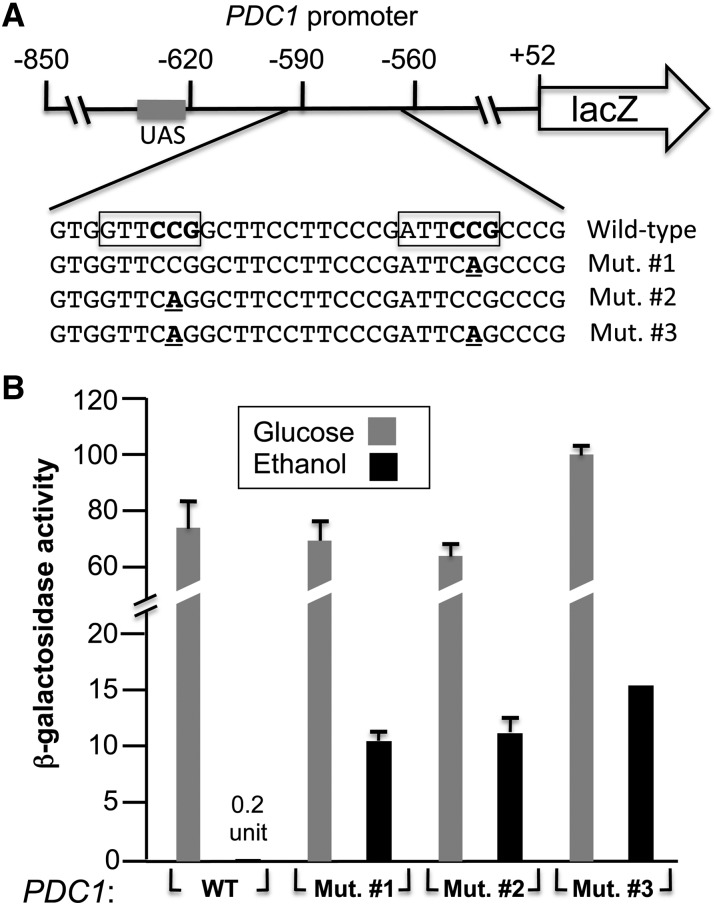
An integrative PDC1-lacZ reporter was constructed in multiple steps. Plasmid pRS303 (Sikorski and Hieter 1989) was cut with PvuII and religated to remove the multicloning sites yielding pRS303∆PvuII. Oligos LacZ-integr-1 and LacZ-integr-2 were used to amplify by PCR a region of plasmid YEP356 (Myers et al. 1986) containing the lacZ and the URA3 genes. The PCR product was cut with BglII and NheI and subcloned into pRS303∆PvuII cut with the same enzymes to yield pLacZ-integr. The PDC1 promoter (−850 to +52 bp relative to the ATG) was amplified with oligonucleotides PDC1-lacZ-3 and PDC1-lacZ-4 using genomic DNA isolated from strain BY4741. The PCR product was cut with EcoRI and SphI and subcloned into pLacZ-integr cut with the same enzymes to yield pPDC1(−850)-lacZ. Mutations (CGG to CTG, see Figure 8A) were introduced in pPDC1(−850)-lacZ by site-directed mutagenesis using oligonucleotides PDC1-CGG to CTG-1 and PDC1-CGG to CTG-2 (mutation at position −569) as well as PDC1-CGG to CTG-3 and PDC1-CGG to CTG-4 (mutation at position −587).
Figure 8.
Mutations that prevent binding of Ert1 to the PDC1 promoter result in increased activity of a PDC1-lacZ reporter. (A) Schematic view of the PDC1-lacZ reporter. The grey rectangle corresponds to the UAS (upstream activating sequence) present in the PDC1 promoter. The DNA sequence of the region bound by Ert1 is shown below. The key nucleotides for Ert1 binding are boxed and the CGG triplets (opposite strand) necessary for Ert1 binding are in boldface type. Mutations that were introduced in the reporter are underlined and in boldface type. (B) Strain YPH499 containing a wild-type PDC1-lacZ reporter (or mutants) integrated at the URA3 locus was grown in the presence of 2% glucose or 2% ethanol and β-galactosidase activity was measured (see Materials and Methods for more details). Results are the average of two experiments performed in duplicate and with two independent strains. In addition to histograms, one numerical value is given when a low β-galactosidase activity was measured. WT, wild\x{2010}type.
A GST-Ert1 (aa 1–152) bacterial expression vector was constructed by amplifying DNA sequences encoding the DNA binding domain of Ert1 by PCR using oligonucleotides D1 and D2 (Table S2) and genomic DNA isolated from strain BY4741 as a template. The PCR product was cut with BamHI and EcoRI and subcloned into pGEX-f (Hellauer et al. 1996) cut with the same enzymes to yield plasmid pGST-Ert1(1–152).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP)-chip assays were performed as described elsewhere (Larochelle et al. 2006). Cells from the wild-type (BY4741) and HA-ERT1 strains were grown in YPD to an approximate OD600 of 0.7, washed twice in water, and transferred to YPD or YEP media containing 2% ethanol as a sole carbon source and grown for 3 hr. Microarrays used for ChIP-chip (4× 44K) were obtained from Agilent. Significantly enriched regions from the ChIP-chip data were identified as described previously (Soontorngun et al. 2012) and tRNA genes were excluded from the analysis. Final assignments were then manually curated. Standard ChIP assays were performed as described (Sylvain et al. 2011).
Quantitative RT-PCR analysis
For quantitative RT-PCR (qRT-PCR), wild-type (BY4741 or YPH499) and deletion strains were grown as described above. RNAs were isolated using the hot-phenol extraction method and purified with an RNA clean-up kit (Qiagen). The cDNA was synthesized from 1 μg of total RNA according to the manufacturer’s protocol for the Maxima H minus Reverse Transcriptase (Thermo Scientific). The qRT-PCR was performed with an Applied Biosystems 7300 using Maxima SYBR Green/ROX qPCR Master Mix (2×) (Thermo Scientific). Quality control of the primers, listed in Table S2, was verified by efficiency and dimerization tests. The PCR parameters were 95° for 10 min, followed by 40 cycles at 95° for 10 sec, 60° for 15 sec, and 72° for 30 sec. The relative quantification of each transcript was calculated by the 2−ΔΔCT method (Livak and Schmittgen 2001) using the ACT1 gene (actin) as an internal control. Three independent cDNA preparations obtained from two independent cultures were used for analysis.
β-Galactosidase assays
Reporters used in this study are pJJ11b (FBP1-lacZ) (Vincent and Gancedo 1995), pPEPCK-lacZ (PCK1-lacZ) (Proft et al. 1995), and pPDC1(−850)-lacZ (this study). Following transformation and selection of the reporters, colonies were grown overnight in rich medium (YPD). Cell were then diluted into selective medium containing 0.67% yeast nitrogen base supplemented with 0.004% leucine, 0.004% histidine, 0.004% methionine, 0.2% glucose, and 2% ethanol and grown for ∼16 hr. β-Galactosidase assays were performed with permeabilized cells (Guarente 1983).
Electrophoretic mobility shift assay
Expression and purification of Ert1(aa 1–152), probe labeling and the electrophoretic mobility shift assay (EMSA) were performed as described (Hellauer et al. 1996; Larochelle et al. 2006). The binding reaction was performed in 4 mM Tris-HCl pH 8, 40 mM NaCl, 3 pmole/ml labeled probe, and 25 μg/ml sonicated salmon sperm DNA.
Results
Ert1 binds to the promoters of genes involved in the utilization of ethanol as a carbon source
Ert1 (Ybr239c) is a putative zinc cluster protein of unknown function and is a homolog of Rds2. To determine its function, ERT1 was tagged at its natural chromosomal location with a triple HA epitope, and ChIP-chip experiments were performed using tiling microarrays covering the entire yeast genome with probes located, on average, at every 290 bp. The initial ChIP-chip analysis was performed with cells grown in rich medium and with glucose as a carbon source. Under these conditions, Ert1 was bound to a limited number of genes (total of 11, see Table 2 and Table S1 for a complete list). The strongest enrichment (64-fold) was observed at the promoter of PCK1, a gene encoding phosphoenolpyruvate carboxykinase, an essential enzyme for gluconeogenesis. Ert1 was also detected at the promoter of FBP1 encoding fructose-1,6-bisphosphatase, another essential gluconeogenic enzyme. Other Ert1 targets include MAE1, encoding an additional gluconeogenic enzyme involved in the conversion of malate to pyruvate as well as PDC1, encoding a key enzyme for fermentation.
Table 2. Selected Ert1 target genes.
| Gene | Systematic name | Function | Enrichment (glucose) | Enrichment (ethanol) | Binding of Adr1, Cat8, or Rds2 |
|---|---|---|---|---|---|
| PCK1 | YKR097W | Phosphoenolpyruvate carboxykinase, key enzyme in the gluconeogenesis pathway | 64 | 47 | Adr1, Cat8, Rds2 |
| MAE1 | YKL029C | Mitochondrial malic enzyme, catalyzes the oxidative decarboxylation of malate to pyruvate | 11 | 35 | Rds2 |
| PDC1 | YLR044C | Major of three pyruvate decarboxylase isozymes, key enzyme in alcoholic fermentation | 4.8 | 26 | Rds2 |
| GAT1 | YFL021W | Transcriptional activator of genes involved in nitrogen catabolite repression | NS | 17 | — |
| AQR1 | YNL065W | Plasma membrane transporter of the major facilitator superfamily | 6.1 | 15 | Rds2 |
| ICY1 | YMR195W | Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA | 2.7 | 14 | Adr1, Cat8, Rds2 |
| LSC2 | YGR244C | β-Subunit of succinyl-CoA ligase | 3.0 | 14 | Adr1, Rds2 |
| VID24 | YBR105C | GID complex regulatory subunit; regulates fructose-1,6-bisphosphatase targeting to the vacuole | 4.4 | 9.4 | Rds2 |
| PET9 | YBL30C | Major ADP/ATP carrier of the mitochondrial inner membrane; exchanges cytosolic ADP for mitochondrially synthesized ATP; also imports heme and ATP | 2.2 | 7.1 | Rds2 |
| OPI1 | YHL020C | Negative regulator of phospholipid biosynthetic genes | 1.9 | 7.0 | Rds2 |
| FBP1 | YLR377C | Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway | 2.7 | NS | Adr1, Cat8, Rds2 |
| FMP48 | YGR052W | Putative protein of unknown function; the authentic, nontagged protein is detected in highly purified mitochondria in high-throughput studies | NS | 4.7 | Adr1, Rds2 |
| — | YGR050C | Unknown function | |||
| RPM2 | YML091C | Protein subunit of mitochondrial RNase P | NS | 4.6 | Rds2 |
| YPC1 | YBR183W | Alkaline ceramidase/ | NS | 4.5 | — |
| — | Putative protein of unknown function | ||||
| YBR182C-A | |||||
| — | YEL020C-B | Dubious open reading frame | NS | 4.4 | — |
| PTR2 | YKR093W | Integral membrane peptide transporter, mediates transport of di- and tri-peptides | NS | 4.2 | Adr1, Cat8, Rds2 |
| QDR3 | YBR043C | Multidrug transporter of the major facilitator superfamily | NS | 4.2 | Rds2 |
| PHD1 | YLR043W | Transcriptional activator that enhances pseudohyphal growth | NS | 3.3 | — |
| — | YLR044W | Protein of unknown function | |||
| HXT8 | YJL214W | Protein of unknown function with similarity to hexose transporter family members, expression is induced by low levels of glucose and repressed by high levels of glucose | NS | 3.3 | Rds2 |
| IMA5 | YJL216C | Alpha-glucosidase | |||
| PMA1 | YGL008C | Plasma membrane H+-ATPase | NS | 3.2 | — |
| TKL1 | YPR074C | Transketolase of the pentose phosphate pathway | NS | 2.9 | Rds2 |
| DIP5 | YPL265W | Dicarboxylic amino acid permease; mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate | NS | 2.7 | Adr1 |
| ATO2 | YNR002C | Putative transmembrane protein involved in export of ammonia | NS | 2.6 | Adr1, Rds2 |
| HAP4 | YKL109W | Limiting subunit of the heme-activated, glucose-repressed Hap2/3/4/5 complex involved in regulation of respiration genes | NS | 2.6 | Adr1, Rds2 |
| GID8 | YMR135C | Subunit of the GID complex involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase | NS | 2.3 | Adr1, Rds2 |
| FMP43 | YGR243W | Highly conserved subunit of the mitochondrial pyruvate carrier; a mitochondrial inner membrane complex comprised of Fmp37p/Mpc1p and either Mpc2p or Fmp43p/Mpc3p mediates mitochondrial pyruvate uptake; more highly expressed in glucose-containing minimal medium than in lactate-containing medium | NS | 2.1 | Adr1 |
| — | YPR036W-A | Protein of unknown function; transcription is regulated by Pdr1 | NS | 2.1 | Adr1, Cat8 |
| QCR6 | YFR033C | Subunit 6 of the ubiquinol cytochrome-c reductase complex | NS | 2.1 | — |
| ADR1 | YDR216W | Carbon source-responsive zinc-finger transcription factor, required for transcription of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization | NS | 1.9 | Adr1, Cat8 |
| PYC1 | YGL062W | Pyruvate carboxylase isoform involved in the conversion of pyruvate to oxaloacetate | NS | 1.9 | — |
| HXT2 | YMR011W | High-affinity glucose transporter of the major facilitator superfamily; expression is induced by low levels of glucose and repressed by high levels of glucose | NS | 1.8 | Adr1 |
| SFP1 | YLR403W | Regulates transcription of ribosomal protein and biogenesis genes; regulates response to nutrients and stress | NS | 1.8 | — |
| SFC1 | YJR095W | Mitochondrial succinate-fumarate transporter; required for ethanol and acetate utilization | NS | 1.8 | Adr1, Cat8, Rds2 |
Major targets of Ert1 are listed (P < 0.05 and with enrichment values >1.8). Gene functions were taken from the Saccharomyces cerevisiae database (http://www.yeastgenome.org/). Enrichment at a particular promoter is given for cells grown in glucose and ethanol; NS, not significant. ChIP-chip data for Adr1 and Cat8 (obtained under low glucose conditions) were taken from Tachibana et al. (2005) while data for Rds2 (obtained with ethanol) were taken from Soontorngun et al. (2007). For a complete list of targets, see Supporting Information, Table S1.
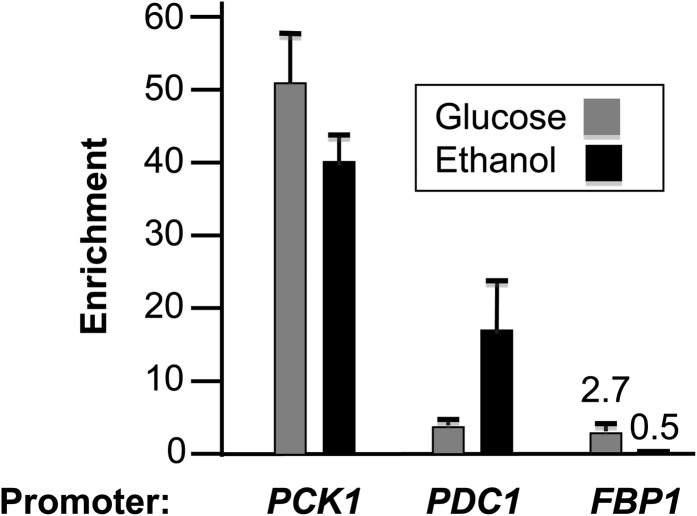
These results suggested that ERT1 encodes a factor involved in the utilization of nonfermentable carbon sources. To test this hypothesis, ChIP-chip assays were then performed with cells grown in the presence of ethanol as a sole carbon source. Results showed that Ert1 binds to the promoters of 70 different genes (P-value <0.05 with an enrichment >1.8). Gene Ontology (GO) term analysis of Ert1 targets showed a clear enrichment for processes related to glucose metabolism (Table 3). In agreement with this analysis, inspection of Ert1 targets revealed enrichment of Ert1 at genes involved in gluconeogenesis and related processes (Table 2). Strikingly, genes bound by Ert1 under glucose conditions showed increased enrichment with cells grown in the presence of ethanol (e.g., MAE1, AQR1, and VID24). A noticeable exception is FBP1, where no significant binding of Ert1 was observed with ethanol-grown cells as opposed to a 2.7-fold enrichment in glucose-grown cells. Additional targets of Ert1 include genes encoding transcription factors such as ADR1, HAP4 (an activator of respiration genes), and GAT1 (an activator of genes involved in the utilization of poor nitrogen sources). Interestingly, Ert1 was also detected at the promoter of PDC1 under both ethanol and glucose conditions. This gene encodes a major pyruvate carboxylase involved in fermentation via conversion of pyruvate to acetaldehyde. Standard ChIP assays for PCK1, PDC1, and FBP1 gave enrichment values highly similar to those obtained by ChIP-chip (Figure 1).
Table 3. Gene ontology enrichment of the Ert1 target genes identified by ChIP-chip analysis.
| GO term (process) | Fraction of the query | Fraction of the query/fraction of the genome (enrichment) | P value |
|---|---|---|---|
| Glucose metabolism | 8 of 75 genes | 9.7 | 4.8 × 10−4 |
| Hexose metabolism | 8 of 75 genes | 8.2 | 1.6 × 10−3 |
| Monosaccharide metabolism | 8 of 75 genes | 7.6 | 2.7 × 10−3 |
| Gluconeogenesis | 5 of 75 genes | 16.8 | 4.0 × 10−3 |
| Hexose biosynthetic process | 5 of 75 genes | 16.8 | 4.0 × 10−3 |
GO, Gene Ontology.
Figure 1.
Confirmation of ChIP-chip results for selected genes by standard ChIP. ChIP assays were performed with the PCK1, PDC1, and FBP1 genes using qPCR. Enrichment was calculated by normalization to signals obtained with an untagged strain and ARN1 as an internal control.
Ert1 is redundant with other factors involved in the utilization of ethanol as a carbon source
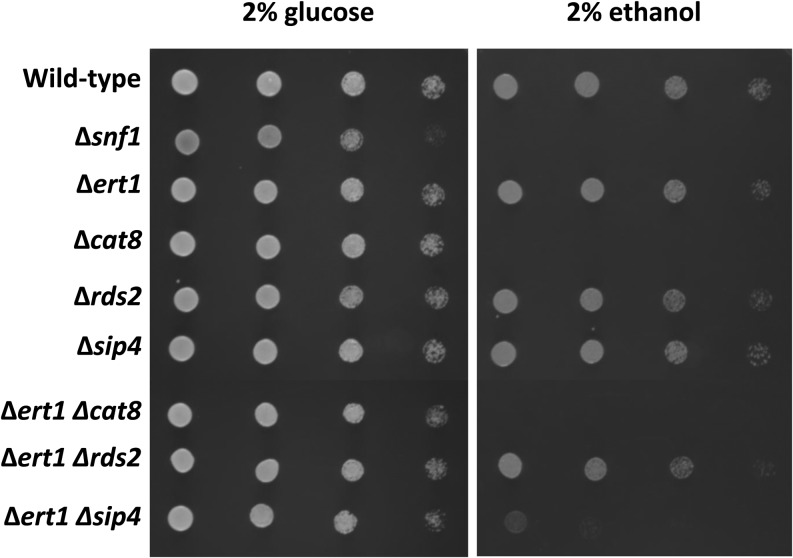
We were then interested to determine if Ert1 regulates the expression of the target genes identified by ChIP-chip analysis. We performed expression profiling experiments comparing mRNA levels of wild-type and ∆ert1 strains grown in the presence of glucose or ethanol as a carbon source. Minimal differences were observed between the wild-type and the ∆ert1 strains (data not shown). A control experiment showed that a shift from glucose to ethanol resulted in a massive effect on gene expression (data not shown). The lack of correlation between ChIP-chip data and expression profiling suggested that Ert1 is redundant with other factors involved in the utilization of nonfermentable carbon sources, namely ethanol. To test this hypothesis, we generated strains carrying double deletions of zinc cluster genes involved in utilization of nonfermentable carbon sources. Strains were grown in rich medium, washed in water, and diluted before spotting on plates containing glucose or ethanol as a carbon source (Figure 2). With plates containing glucose, growth was similar for all strains tested with the exception of ∆snf1, where slightly reduced growth was observed (Figure 2, left panel). With ethanol as a sole carbon source, no growth was observed with a strain carrying a deletion of SNF1 or CAT8, either alone or in combination with other mutants (Figure 2, right panel), a result expected from previous observations (Celenza and Carlson 1984; Hedges et al. 1995). While single deletions of ERT1, RDS2, or SIP4 had no effect on growth with ethanol as a carbon source, a strain lacking ERT1 and SIP4 showed greatly impaired growth on ethanol plates. Interestingly, no growth defect was observed with a strain carrying deletions of both ERT1 and RDS2. Collectively, these results strongly suggest that ERT1 is redundant with SIP4.
Figure 2.
Redundancy among transcriptional regulators of gluconeogenesis. Wild-type (BY4741) and deletion strains (Table 1) were grown overnight in YPD and washed twice in H2O. Cells were then serially diluted (from left to right: 25-, 100-, 400-, and 2000-fold). Ten microliters of diluted cells were spotted on minimal plates supplemented with 0.2% casamino acids and histidine, leucine, methionine, uracil (each at 0.004%), and 2% glucose or 2% ethanol, as indicated at the top. Glucose and ethanol plates were incubated at 30° for 1 and 5 days, respectively.
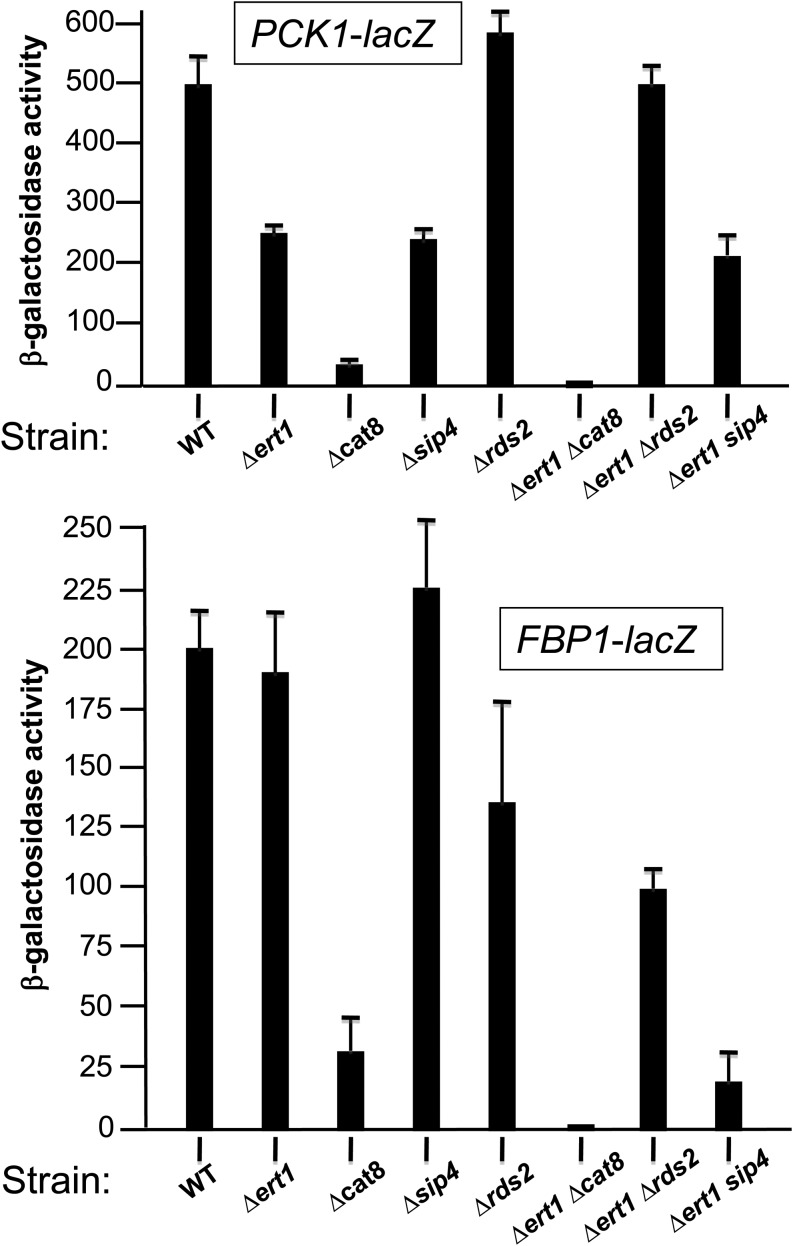
Ert1 controls the promoter activity of the gluconeogenic genes PCK1 and FBP1
Given these results, we were interested in determining if Ert1 controls the promoter activity of key gluconeogenic genes such as PCK1 and FBP1. Since regulation of these genes occurs at both the transcriptional and mRNA stability levels (Mercado et al. 1994), we assayed the activity of the PCK1 and FBP1 promoters using lacZ reporters transformed in the strains described above. For both reporters, background activity was measured when the wild-type strain was grown in the presence of glucose (data not shown). As expected (Proft et al. 1995), high activity of the PCK1-lacZ reporter was observed under inducing conditions (0.2% glucose and 2% ethanol) (Figure 3, top panel). Deletion of CAT8 reduced activity by 14-fold, while deletion of ERT1 or SIP4 reduced the activity by only 2-fold. In contrast, deletion of RDS2 had no significant effect on the activity of the PCK1 promoter. Similarly, with the double deletion strain ∆ert1∆sip4, no major change in activity was observed as compared to single deletion strains, while deletion of Rds2 did revert the downregulation observed in the ∆ert1 strain. Finally, deletion of both CAT8 and ERT1 resulted in background activity.
Figure 3.
Ert1 controls the activity of the PCK1 and FBP1 promoters. Wild\x{2010}type strain BY4741 and strains carrying deletion of genes encoding various zinc cluster proteins (as indicated at the bottom) were transformed with the lacZ reporters for PCK1 or FBP1. Cells were grown in the presence of 0.2% glucose and 2% ethanol to induce reporter activity (see Materials and Methods for more details).
With the FBP1-lacZ reporter, deletion of CAT8 strongly reduced β-galactosidase activity, while a slight increase was observed for a strain lacking SIP4 (Figure 3, bottom panel), in agreement with previous observations (Zaragoza et al. 2001). Deleting ERT1 gave β-galactosidase activity at levels comparable to a wild-type strain. Interestingly, the double mutant ∆ert1∆sip4 showed a 10-fold reduction in activity while deletion of both ERT1 and CAT8 resulted in background activity. Thus, the marked reduction in activity of the FBP1-lacZ reporter in strains ∆cat8, ∆ert1∆cat8, and ∆ert1∆sip4 correlates with their impaired growth, as assayed with ethanol as the sole carbon source. In summary, our results show that Ert1 controls the activity of the PCK1 and FBP1 promoters through a complex interplay with other zinc cluster transcription factors.
Ert1 acts as an activator and a repressor
We then focused on additional targets of Ert1 to determine if these genes are also regulated by this factor using qRT-PCR. Control experiments showed that no mRNA for ERT1, CAT8, RDS2, and SIP4 could be detected in the corresponding deletion strains (data not shown). We focused on five different genes: FBP1, SFC1 (encoding a mitochondrial transporter for succinate and fumarate), PDC1, ADR1, and GAT1. We determined changes in mRNA levels in a wild-type strain following a shift from glucose to ethanol (Figure 4A). As expected, the expression of FBP1 and SCF1 was greatly increased (FBP1: 380-fold; SFC1: 182-fold), while the expression of the fermentation gene PDC1 was strongly repressed (33-fold) upon a shift from glucose to ethanol (Schmitt et al. 1983; Bojunga et al. 1998). The expression of GAT1 and ADR1 was increased 3.2- and 5.9-fold, respectively. We then performed qRT-PCR with RNA prepared from wild-type and deletion strains that were grown under inducing conditions (0.2% glucose and 2% ethanol) (Figure 4B). Overall, results for FBP1 correlate with those obtained with a FBP1-lacZ reporter (Figure 3). For instance, reduced β-galactosidase activity and FBP1 mRNA levels were observed for the ∆cat8 and the ∆ert1∆sip4 strains. However, deletion of CAT8 reduced promoter activity by 8-fold, while no FBP1 mRNA could be detected in that strain. This difference could be due to changes in FBP1 mRNA stability (Mercado et al. 1994). As expected (Bojunga et al. 1998), deletion of CAT8 greatly diminished SFC1 mRNA levels, while a similar effect was observed upon deletion of SIP4 (Figure 4B). Single deletions of ERT1 or RDS2 had minor effects on expression of SFC1 but a ∆ert1∆rds2 strain showed reduction of SFC1 mRNA levels by almost 2-fold. For ADR1, single deletion strains had minor effects on expression of this gene and the strongest effect on mRNA levels was observed with the strain ∆ert1∆rds2 (∼2-fold reduction). Similarly for GAT1, the strongest effect (2-fold reduction) was observed with the double deletion strain ∆ert1∆sip4.
Figure 4.
Expression of Ert1 target genes in strains lacking various transcriptional regulators. (A) Wild-type strain BY4741 was grown in the presence of 2% glucose or 2% ethanol and RNA isolated for qRT-PCR analysis. Fold changes are given as ratios between mRNA levels measured in ethanol- and glucose-grown cells. (B) Wild-type strain BY4741 and deletion strains (Table 1) were grown in the presence of 2% ethanol and RNA was isolated for qRT-PCR analysis. mRNA levels were measured for various genes (as indicated at the bottom of each panel) and are given relative to the wild-type strain (100%). (C) Same as in B with the exception that strain W303-1A was used instead of BY4741.
As stated above, expression of PDC1 is repressed under nonfermentative conditions (Figure 4A). This gene encodes pyruvate decarboxylase, a key enzyme acting at the pyruvate branch point (Pronk et al. 1996). Pyruvate, the end product of glycolysis, can either enter the tricarboxylic acid cycle or be converted to acetaldehyde by pyruvate decarboxylase followed by conversion to ethanol. Thus, repression of PDC1 is one mechanism allowing cells to switch from fermentative to nonfermentative metabolism. Interestingly, deletion of ERT1 increased PDC1 mRNA levels by almost three-fold (Figure 4B). These results suggest that Ert1 is not only involved in activation of genes for nonfermentative metabolism but is also involved, in part, in shutting down the fermentative process. Deletion of RDS2 slightly increased PDC1 mRNA levels but no additive effect was observed with the double deletion ∆ert1∆rds2. Deletion of CAT8 or SIP4 had minor effects on PDC1 mRNA levels. However, strains ∆ert1∆cat8 and ∆ert1∆sip4 had reduced mRNA levels for PDC1. We also tested whether the effect of Ert1 on PDC1 expression is specific for the strain BT4741 or not. To this end, we generated a ∆ert1 strain in the W303-1A background. Deleting ERT1 resulted in increased (2.8-fold) PDC1 mRNA levels (Figure 4C), as observed in the BY4741 background. In summary, deletion of ERT1 minimally alters the expression of gluconeogenic genes but a more pronounced effect was observed in strains lacking ERT1 in combination with CAT8, SIP4, or RDS2. These results strongly suggest redundancy of function among these transcriptional regulators.
Ert1 binds directly to the PCK1 and PDC1 promoters
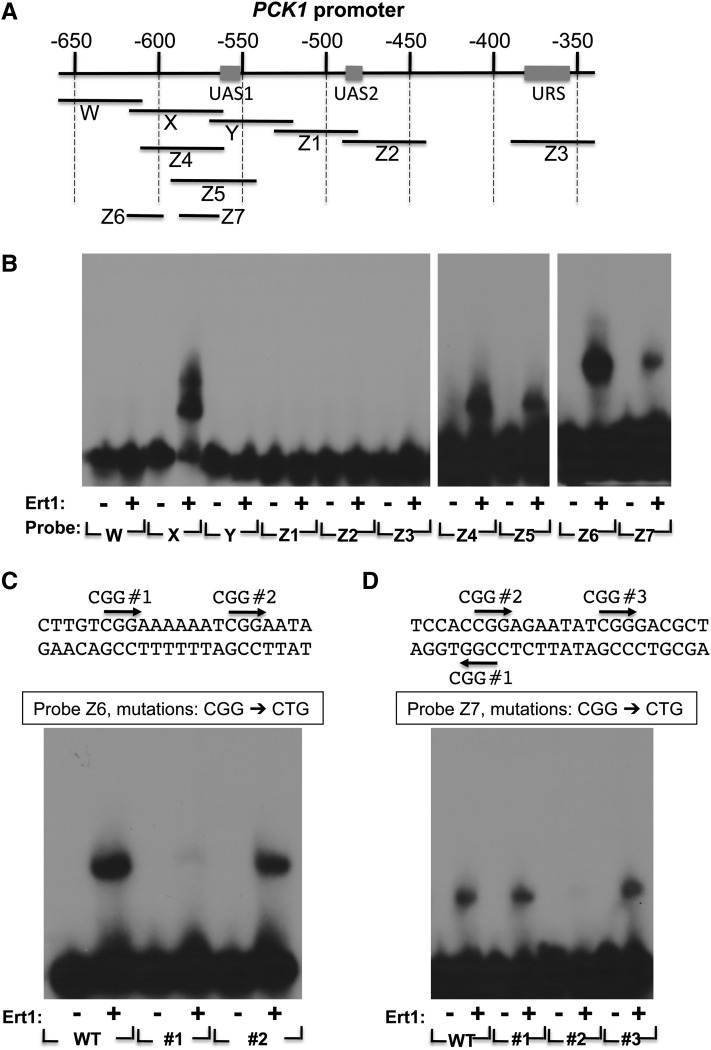
We were interested in determining if Ert1 directly interacts with DNA and, if so, what the DNA recognition site is. The DNA binding domain of Ert1 (aa 1–152) was expressed in bacteria as a GST fusion protein, purified, and the GST moiety removed by thrombin cleavage. The purified DNA binding domain of Ert1 was used in an EMSA. We initially focused on the PCK1 promoter. A series of DNA probes spanning the regulatory region of PCK1 (upstream activating sequences 1 and 2 (UAS1 and UAS2) and upstream repressing sequence (URS) was tested for Ert1(1–152) binding (Figure 5A). Binding was detected only when using probe X (Figure 5B, left panel). No Ert1(1–152) binding was observed when using probes located between probes Z2 and Z3 (data not shown). Thus, Ert1(1–152) binds to a region located upstream of UAS1 (a regulatory site recognized by the factors Cat8 and Sip4). With probe X, two complexes were observed. We compared the relative mobility of the Ert1(1–152)–DNA complex to zinc cluster proteins that bind DNA as homodimers (e.g., Gal4). Results suggest that the fast migrating complex is a monomer, while the slow migrating complex would be composed of two monomers bound to separate sites on probe X (data not shown). Use of the smaller probes Z4 and Z5 resulted in the formation of a single Ert1(1–152)–DNA complex (Figure 5B, middle panel). In addition, Ert1(1–152) bound to two smaller probes (Z6 and Z7) that do not overlap. Binding at Z7 was weaker when compared to site Z6. Thus, Ert1 binds to two distinct sites in the PCK1 promoter. As stated above, zinc cluster proteins preferentially bind to CGG triplets (MacPherson et al. 2006). As shown in Figure 5C, the Z6 probe contains two CGG triplets. Mutating CGG no. 2 to CTG had no effect on Ert1(1–152) binding. In contrast, a mutation in CGG no. 1 almost completely abolished binding of Ert1(1–152). Three CGG triplets are found in probe Z7; strikingly, only a mutation in CGG no. 2 reduced binding of Ert1(1–152).
Figure 5.
Ert1 binds to the PCK1 promoter. (A) Schematic view of the PCK1 promoter. Coordinates (in base pairs) are given relative to the ATG (+1 bp). Upstream activating sequences (UAS1 and UAS2) and an upstream repressing sequence (URS) (Proft et al. 1995) are shown as rectangles. Probes used for EMSA are shown as black lines. (B) Ert1 (aa 1–152) binds to two sites in the PCK1 promoter. Various probes were incubated (+) or not (−) with the purified DNA binding domain of Ert1 for EMSA analysis. (C) Mutational analysis of the binding site Z6 of Ert1. DNA sequence of probe Z6 (see A) is shown at the top as well as the two CGG triplets (CGG no. 1 and no. 2). The effect of mutating either CGG triplets to CTG on Ert1 binding was tested by EMSA. WT, wild\x{2010}type. (D) Mutational analysis of the binding site Z7 of Ert1. DNA sequence of probe Z7 (see A) is shown at the top as well as the three CGG triplets (labeled CGG nos. 1, 2, and 3). The effect of mutating either CGG triplets to CTG on Ert1 binding was tested by EMSA.
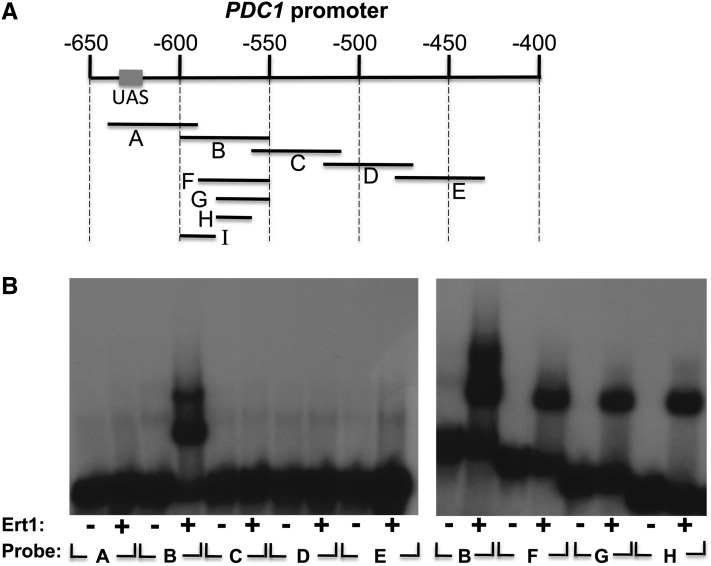
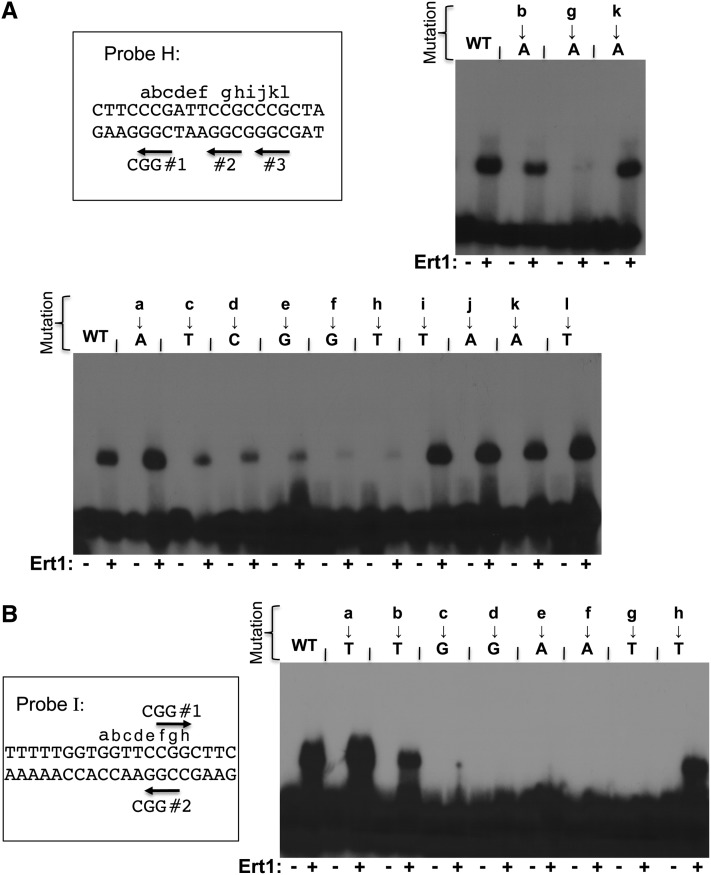
We used the same approach described above to identify Ert1 binding site(s) in the PDC1 promoter (Figure 6). Since ChIP-chip experiments suggested that Ert1 binds to a site located at ∼500 bp upstream of the ATG, we designed five probes (A–E) covering this region, including the UAS (Figure 6A). Binding of Ert1(1–152) was observed only with probe B (Figure 6B). In analogy to PCK1, two Ert1(1–152)–DNA complexes were observed with that probe. With smaller probes (F, G, and H), only a single complex of fast mobility was observed (Figure 6, right panel). Binding of Ert1(1–152) was also observed with probe I, which does not overlap with probes G or H (see below). Thus, the PDC1 promoter contains two binding sites for Ert1. We then performed mutational analysis of these two sites. Probe H contains three CGG triplets (Figure 7A). Slightly reduced binding of Ert1(1–152) was observed when mutating CGG no. 1 or no. 3. In contrast, mutation in the second CGG triplet completely abolished binding of Ert1(1–152) (Figure 7A, top panel). We tested additional mutations in the region encompassing CGG triplet no. 2 using transversions. No reduced binding of Ert1(1–152) was observed when mutating nucleotides labeled a, b, and i–l (Figure 7A). However, diminished binding was observed with mutations at positions c–f and h. A similar scan was performed with probe I (Figure 7B). EMSA analysis showed that nucleotides located at positions c–g and, to a lesser extent, nucleotide at position b, are critical for Ert1. Taken together, results show that the binding site of Ert1 at PDC1 is CGGAAY (where Y is C or T).
Figure 6.
Ert1 binds to the PDC1 promoter. (A) Schematic view of the PDC1 promoter. Coordinates (in base pairs) are given relative to the ATG (+1 bp). The upstream activating sequence (UAS) (Butler and McConnell 1988) is shown as a rectangle. Probes used for EMSA are shown as black lines. (B) Various probes were incubated (+) or not (−) with the purified DNA binding domain of Ert1 for EMSA analysis. Probe I was tested in Figure 7.
Figure 7.
Ert1 binds to two sites in the PDC1 promoter. (A) Mutational analysis of the binding site “H” of PDC1. DNA sequence of probe H (Figure 6A) is shown at the top as well as the three CGG triplets (labeled CGG nos. 1, 2, and 3). Letters a–l correspond to nucleotides that were mutated. The effect of mutating either CGG triplets to CTG on Ert1 binding was tested by EMSA using the purified DNA binding domain of Ert1 (aa 1–152), as shown in the top right panel. Additional nucleotides were also mutated (bottom panel). WT, wild\x{2010}type. (B) Mutational analysis of the binding site I of Ert1. DNA sequence of probe I (Figure 6A) is shown on the left as well as the two CGG triplets (labeled CGG nos. 1 and 2). Letters a–h correspond to nucleotides that were mutated. Mutants were analyzed by EMSA using the purified DNA binding domain of Ert1 (aa 1–152) (right panel).
Mutations that prevent Ert1 binding result in increased activity of a PDC1-lacZ reporter
We were interested in determining the effect of mutations that prevent binding of Ert1 on the PDC1 promoter activity. To this end, we constructed a PDC1-lacZ reporter that was integrated at the URA3 locus (Figure 8A). Three mutant versions were also tested. Mutant no. 1 has a mutation in the second CGG triplet while mutant no. 2 has a mutation in the first CGG triplet. These mutations abolished binding of Ert1(1–152) in vitro (Figure 7). A double mutant was also tested (Figure 8A). As expected with a wild-type PDC1 promoter (Butler and McConnell 1988), high β-galactosidase activity was observed with cells grown in the presence of glucose, while growth with ethanol as a carbon source gave background activity (Figure 8B). With glucose, mutants no. 1 and no. 2 gave β-galactosidase activity similar to the wild-type reporter. Interestingly, increased activity was observed for both mutants when assayed with ethanol. Finally, mutant no. 3 (carrying mutations in both CGG triplets) showed slightly increased β-galactosidase activity with glucose as compared to wild-type PDC1, while the activity was also slightly increased in the presence of ethanol when compared to mutants no. 1 and no. 2. In a ∆ert1 strain grown in the presence of ethanol, PDC1 mRNA levels are at ∼10% of the levels measured in a wild-type strain grown in the presence of glucose (Figure 4, A and B). Strikingly, a similar ratio (12%) was observed with lacZ reporters. Taken together, our results strongly suggest that Ert1 represses PDC1 promoter activity by binding directly to sites adjacent to the UAS.
Discussion
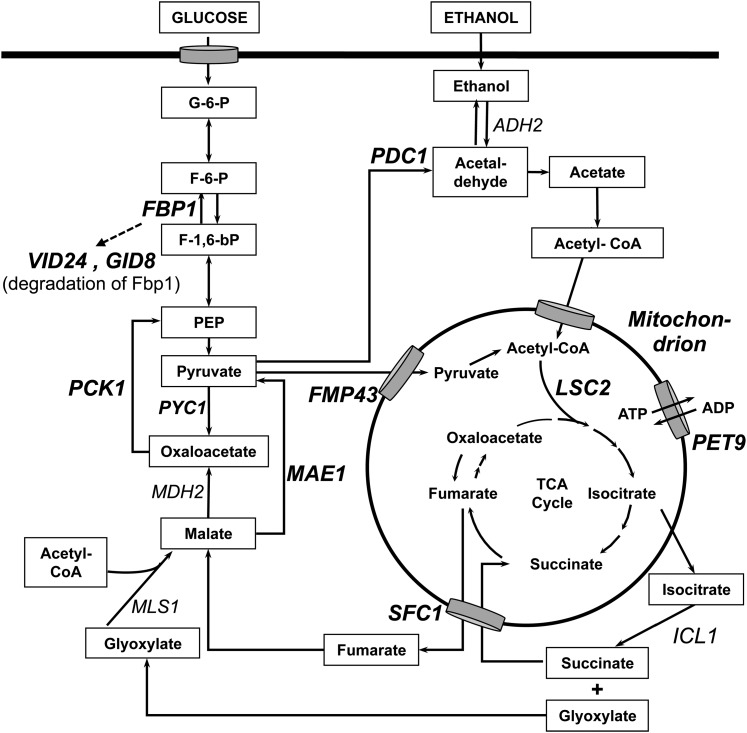
In this study, we have characterized the ORF YBR239C encoding a putative transcriptional regulator of the family of zinc cluster proteins and we named this gene ERT1 (ethanol regulated transcription factor 1). Figure 9 provides a schematic view of the pathways of fermentative and nonfermentative metabolism as well as some target genes of Ert1. ChIP-chip analysis showed that Ert1 is bound to a limited number of genes (total of 11), as assayed in rich medium supplemented with glucose. The strongest enrichment (64-fold) was observed at the promoter of the gluconeogenic gene PCK1 (Table 2). Given this result, we hypothesized that Ert1 is involved in regulating gluconeogenesis. When the ChIP-chip assay was repeated in the presence of ethanol as a carbon source, binding of Ert1 was detected at 70 promoters including the one of PYC1, another gluconeogenic gene. A similar phenomenon was observed for Rds2 where an increased number of targets was observed with nonfermentable carbon sources as compared to glucose (Soontorngun et al. 2007, 2012). A number of Ert1 target genes encode mitochondrial components. For example, SFC1 (ACR1) encodes a fumarate/succinate transporter, while the FMP43 gene product is a subunit of the pyruvate transporter (Palmieri et al. 1997; Bricker et al. 2012; Herzig et al. 2012). Some other Ert1 targets (PET9, FMP48, TAM41, RMP2, QCR6, and YHR080C) also encode mitochondrial proteins.
Figure 9.
Major Ert1 targets in the pathways of fermentative and nonfermentative metabolism. Some of the Ert1 target genes are indicated in boldface type. Only the mitochondrial compartment is shown. SFC1 encodes a fumarate/succinate transporter and FMP43 encodes a subunit of the pyruvate transporter. PET9 encodes an ATP/ADP carrier.
Ert1 is a DNA binding protein
EMSA analysis showed that Ert1(1–152) binds to specific DNA sequences in the PCK1 and the PDC1 promoters. Mutational analysis of the Ert1 binding sites at PDC1 suggests that the DNA recognition sequence of this factor is CGGAAY (where Y is C or T) (Figure 7). Strikingly, this matches exactly the consensus sequence obtained by protein binding microarray analysis (Badis et al. 2008). One site identified at PCK1 has the sequence CGGAAA, suggesting that the last position can tolerate some changes. In agreement with this observation, mutating the sequence CGGAAC found in PDC1 to CGGAAA reduced, but did not abolish, binding of Ert1(1–152) (Figure 7B). A similar effect was observed with the mutation CGGAAT to CGGAAG (Figure 7A). Finally, Ert1 bound weakly to probe Z7 located in the PCK1 promoter (Figure 6D), an observation that could be explained by the fact that this site (CGGAGAA) does not fit the consensus sequence. Our EMSA analysis suggests that Ert1(1–152) binds as a monomer. Although binding of a zinc cluster protein to DNA as monomer has been reported (Cahuzac et al. 2001), it is quite possible that Ert1 binds to DNA as a homo- or a heterodimer. For example, the DNA binding domain used in our studies may lack a dimerization domain. Alternatively, Ert1 may bind with a partner that remains to be identified. AcuM and AcuK are two A. nidulans homologs of Ert1/Rds2. AcuM and AcuK also are involved in the regulation of gluconeogenic genes and, interestingly, bind to DNA in an interdependent manner (Suzuki et al. 2012).
Redundancy of Ert1 with other transcriptional regulators
Our ChIP-chip analysis suggests that Ert1 is involved in controlling the expression of genes involved in the utilization of nonfermentable carbon sources such as ethanol. However, expression profiling using wild-type and ∆ert1 strains did not reveal any significant changes in expression of the Ert1 targets that were identified by ChIP-chip analysis (data not shown). These results suggest redundancy among transcriptional regulators. In agreement with these results, deletion of ERT1 did not impair growth on plates containing ethanol as a carbon source (Figure 2). However, a strain carrying deletions of both ERT1 and SIP4 showed greatly reduced growth when assayed with ethanol as a carbon source. These observations correlate with the minimal effect of a deletion of ERT1 on the activity of a FBP1-lacZ reporter. This contrasts with the marked reduction of promoter activity observed in a ∆ert1∆sip4 strain (Figure 3). These results were confirmed by qRT-PCR analysis (Figure 4B). Unexpectedly, Ert1 was detected at the FBP1 promoter under glucose conditions but not in the presence of ethanol as determined by ChIP-chip and standard ChIP analysis (Table 2, Figure 1). This lack of correlation between elevated expression of FBP1 and binding of Ert1 may be due to the possibility that, upon a shift to ethanol, binding of additional factors at the FBP1 promoter masks the epitope of HA-Ert1. Alternatively, the effect of Ert1 on expression of FBP1 may be indirect. For instance, in the presence of ethanol, Ert1 may increase the expression of a gene encoding positive regulator of FBP1.
Ert1 is a repressor of the fermentation gene PDC1
As stated above, Pdc1 is a key enzyme for fermentation, and its pyruvate decarboxylase activity is elevated under glucose conditions, promoting fueling of the glycolytic product pyruvate toward ethanol production. In contrast, low levels of pyruvate decarboxylase activity are observed with cells shifted to nonfermentative metabolism (Pronk et al. 1996). A major level of this regulation occurs via control of PDC1 transcription (Butler and McConnell 1988). With glucose as a carbon source, the transcription factor Pdc2 allows high levels of PDC1 expression (Hohmann 1993). However, it was not known how repression of PDC1 in the presence of ethanol is exerted. Our results show that Ert1 is responsible in part for this repression, since a 3-fold increase of PDC1 mRNA levels is observed in cells lacking Ert1, as assayed in ethanol (Figure 4B). In agreement with these results, mutations that prevent binding of Ert1(1–152) in vitro result in increased activity of a PDC1-lacZ reporter (Figure 8). These results suggest that Ert1 can act both as a transcriptional repressor and an activator, as observed for Rds2 (Soontorngun et al. 2007). Rds2 also binds to the PDC1 promoter (Soontorngun et al. 2007) and cells lacking RDS2 show a slight increase in PDC1 mRNA levels. However, a double mutant (∆ert1∆rds2) had PDC1 mRNA at lower levels as compared to a ∆ert1 strain. The effect of these two factors may be more apparent when assayed during the transition from fermentative to nonfermentative metabolism. It is also likely that additional transcriptional regulators control the expression of PDC1.
Transcriptional regulators of gluconeogenesis have common and distinct target genes
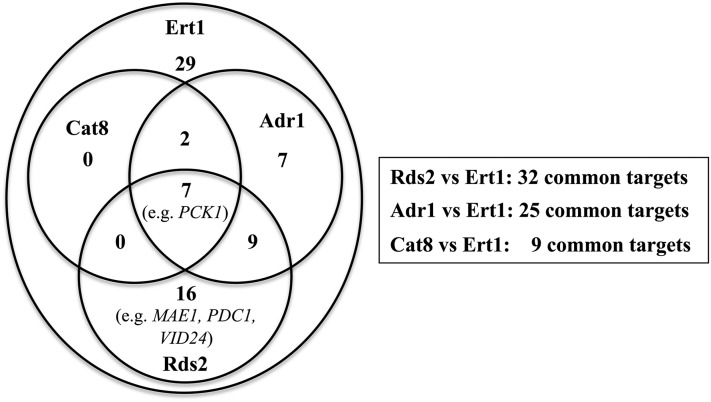
In analogy to the role played by PDC1 for fermentative metabolism, PCK1 encodes a key enzyme for committing cells toward gluconeogenesis. Gsm1 (glucose starvation modulator 1) is another zinc cluster protein that may control gluconeogenesis (Turcotte et al. 2011). This factor binds to the PCK1 promoter under glucose (van Bakel et al. 2008) and ethanol conditions (N. Gasmi, unpublished results). Remarkably, Ert1, Cat8, Sip4, Rds2, Gsm1, and Adr1 all bind to the PCK1 promoter, as determined by ChIP-chip analysis (Harbison et al. 2004; Tachibana et al. 2005; Soontorngun et al. 2007; van Bakel et al. 2008). Thus, the multiplicity of transcription factors bound to PCK1 may allow integration of multiple signals for a precise regulation of this key gluconeogenic gene. However, it is more common for these factors to have both common and distinct targets. For example, we have shown that approximately one-third of the Rds2 targets are bound by Adr1 (Soontorngun et al. 2012). Similarly, Adr1 and Cat8 have common and distinct target genes (Tachibana et al. 2005). With respect to Ert1, our results show that this regulator shares many targets with Rds2 and Adr1, while the overlap with Cat8 is minimal (Figure 10). These observations point to a complex combinatorial network of transcription factors involved in the utilization of nonfermentable carbon sources. Bmh proteins (also called 14-3-3 proteins) may also play an important role in this process. Adr1 and Cat8 are both required for maximal expression of the ADH2 gene (encoding alcohol dehydrogenase). Interestingly, inactivation of Bmh1 and 2 bypasses the requirement of Cat8 for expression of ADH2 (but not FBP1) (Braun et al. 2013). It has been proposed that Bmh1 and 2 are involved in combinatorial regulation of gene expression (Braun et al. 2013). It will be interesting to determine if Bmh proteins control the activity of additional transcriptional regulators of nonfermentative metabolism.
Figure 10.
Overlap between Ert1 targets and those of Cat8, Rds2, and Adr1. Values in the Venn diagram (left panel) refer to the number of genes that are common targets for specified regulators. The number of overlapping genes with Ert1 and a specific regulator is also indicated in the right panel. Forty-one percent (29/70) of the Ert1 target genes did not overlap with the Cat8, Rds2, and Adr1 targets. ChIP-chip data for Adr1 and Cat8 were taken from Tachibana et al. (2005), while data for Rds2 were from Soontorngun et al. (2007). ChIP-chip data for Cat8 and Adr1 were obtained with cells grown under low glucose conditions, while the assay for Rds2 was performed with ethanol as a carbon source.
Interplay among transcriptional regulators of gluconeogenesis
An additional level of regulation is exerted by the interplay among these transcriptional regulators. Expression of ERT1 and RDS2 does not vary much under fermentative and nonfermentative conditions (Soontorngun et al. 2007 and unpublished results). However, expression of CAT8, SIP4, GSM1, and ADR1 is increased under low glucose levels (Figure 4B) (Hedges et al. 1995; Lesage et al. 1996; Roberts and Hudson 2006). Similarly, expression of HAP4, encoding the limiting subunit of the Hap2/3/4/5 complex involved in the up-regulation of respiration genes, is increased upon a shift to respiration (Forsburg and Guarente 1989; Derisi et al. 1997; Soontorngun et al. 2007). Cat8 controls the expression of SIP4 (Lesage et al. 1996) while Ert1 and Rds2 control, in part, the expression of ADR1 (Figure 4B). Finally, Rds2 is a regulator of the expression of HAP4 (Soontorngun et al. 2007). These observations suggest an elaborate interplay among these factors involving complex dynamics. In summary, our studies on ERT1 have led to the identification of a novel factor involved in the regulation of gluconeogenesis as well as a key fermentation gene.
Supplementary Material
Acknowledgments
We thank Peter Kötter, Karl-Dieter Entian, Juana Maria Gancedo, and Hans-Joachim Schüller for the gift of reporter plasmids; Nitnipa Soontorngun, Xiao Bei Liang, and Piyasuda Thepnok for their input in this project; and Hugh Bennett for critical review of the manuscript. This work was supported by a grant to B.T. from the Natural Sciences and Engineering Research Council of Canada. F.R. was supported by a grant from the Canadian Institutes of Health Research (grant no. 82891).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.113.168609/-/DC1.
Communicating editor: M. Hampsey
Literature Cited
- Adams A., Gottschling D. E., Stearns T., 1997. Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Badis G., Chan E. T., van Bakel H., Pena-Castillo L., Tillo D., et al. , 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojunga N., Kotter P., Entian K. D., 1998. The succinate/fumarate transporter Acr1p of Saccharomyces cerevisiae is part of the gluconeogenic pathway and its expression is regulated by Cat8p. Mol. Gen. Genet. 260: 453–461 [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Braun K. A., Parua P. K., Dombek K. M., Miner G. E., Young E. T., 2013. 14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA Polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 33: 712–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker D. K., Taylor E. B., Schell J. C., Orsak T., Boutron A., et al. , 2012. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., 2012. Nutritional control of growth and development in yeast. Genetics 192: 73–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., McConnell D. J., 1988. Identification of an upstream activation site in the pyruvate decarboxylase structural gene (PDC1) of Saccharomyces cerevisiae. Curr. Genet. 14: 405–412 [DOI] [PubMed] [Google Scholar]
- Cahuzac B., Cerdan R., Felenbok B., Guittet E., 2001. The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein. Structure 9: 827–836 [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M., 1984. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 4: 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derisi J. L., Iyer V. R., Brown P. O., 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L., 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3: 1166–1178 [DOI] [PubMed] [Google Scholar]
- Guarente L., 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101: 181–191 [DOI] [PubMed] [Google Scholar]
- Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., et al. , 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M., 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges D., Proft M., Entian K. D., 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 1915–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellauer K., Rochon M.-H., Turcotte B., 1996. A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol. Cell. Biol. 16: 6096–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S., Raemy E., Montessuit S., Veuthey J. L., Zamboni N., et al. , 2012. Identification and functional expression of the mitochondrial pyruvate carrier. Science 337: 93–96 [DOI] [PubMed] [Google Scholar]
- Hohmann S., 1993. Characterisation of PDC2, a gene necessary for high level expression of pyruvate decarboxylase structural genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 241: 657–666 [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., et al. , 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M., Drouin S., Robert F., Turcotte B., 2006. Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol. Cell. Biol. 26: 6690–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P., Yang X. L., Carlson M., 1996. Yeast Snf1 protein kinase interacts with Sip4, a C-6 zinc cluster transcriptional activator: a new role for Snf1 in the glucose response. Mol. Cell. Biol. 16: 1921–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- MacPherson S., Larochelle M., Turcotte B., 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70: 583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado J. J., Smith R., Sagliocco F. A., Brown A. J. P., Gancedo J. M., 1994. The levels of yeast gluconeogenic mRNAs respond to environmental factors. Eur. J. Biochem. 224: 473–481 [DOI] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J., 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45: 299–310 [DOI] [PubMed] [Google Scholar]
- Palmieri L., Lasorsa F. M., Depalma A., Palmieri F., Runswick M. J., et al. , 1997. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417: 114–118 [DOI] [PubMed] [Google Scholar]
- Proft M., Grzesitza D., Entian K. D., 1995. Identification and characterization of regulatory elements in the phosphoenolpyruvate carboxykinase gene PCK1 of Saccharomyces cerevisiae. Mol. Gen. Genet. 246: 367–373 [DOI] [PubMed] [Google Scholar]
- Pronk J. T., Steensma H. Y., Vandijken J. P., 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12: 1607–1633 [DOI] [PubMed] [Google Scholar]
- Roberts G. G., Hudson A. P., 2006. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Genet. Genomics 276: 170–186 [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Esposito R. E., Esposito M. S., 1977. The effect of ochre suppression on meiosis and ascospore formation in Saccharomyces. Genetics 85: 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt H. D., Ciriacy M., Zimmermann F. K., 1983. The synthesis of yeast pyruvate decarboxylase is regulated by large variations in the messenger RNA level. Mol. Gen. Genet. 192: 247–252 [DOI] [PubMed] [Google Scholar]
- Schneider B. L., Seufert W., Steiner B., Yang Q. H., Futcher A. B., 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11: 1265–1274 [DOI] [PubMed] [Google Scholar]
- Schüller H. J., 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43: 139–160 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soontorngun N., Larochelle M., Drouin S., Robert F., Turcotte B., 2007. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol. Cell. Biol. 27: 7895–7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soontorngun N., Baramee S., Tangsombatvichit C., Thepnok P., Cheevadhanarak S., et al. , 2012. Genome-wide location analysis reveals an important overlap between the targets of the yeast transcriptional regulators Rds2 and Adr1. Biochem. Biophys. Res. Commun. 423: 632–637 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Murray S. L., Wong K. H., Davis M. A., Hynes M. J., 2012. Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans. Mol. Microbiol. 84: 942–964 [DOI] [PubMed] [Google Scholar]
- Sylvain M. A., Liang X. B., Hellauer K., Turcotte B., 2011. Yeast zinc cluster proteins Dal81 and Uga3 cooperate by targeting common coactivators for transcriptional activation of gamma-aminobutyrate responsive genes. Genetics 188: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., et al. , 2005. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 25: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B., Liang X. B., Robert F., Soontorngun N., 2011. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., van Werven F. J., M. Radonjic, M. O. Brok, D. van Leenen et al, 2008. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 36: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O., Gancedo J. M., 1995. Analysis of positive elements sensitive to glucose in the promoter of the FBP1 gene from yeast. J. Biol. Chem. 270: 12832–12838 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Young E. T., Dombek K. M., Tachibana C., Ideker T., 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278: 26146–26158 [DOI] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zaragoza O., Vincent O., Gancedo J. M., 2001. Regulatory elements in the FBP1 promoter respond differently to glucose-dependent signals in Saccharomyces cerevisiae. Biochem. J. 359: 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Vemuri G., Nielsen J., 2010. Systems biology of energy homeostasis in yeast. Curr. Opin. Microbiol. 13: 382–388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.