Abstract
Aerobic glycolysis is a metabolic pathway utilized by human cancer cells and also by yeast cells when they ferment glucose to ethanol. Both cancer cells and yeast cells are inhibited by the presence of low concentrations of 2-deoxyglucose (2DG). Genetic screens in yeast used resistance to 2-deoxyglucose to identify a small set of genes that function in regulating glucose metabolism. A recent high throughput screen for 2-deoxyglucose resistance identified a much larger set of seemingly unrelated genes. Here, we demonstrate that these newly identified genes do not in fact confer significant resistance to 2-deoxyglucose. Further, we show that the relative toxicity of 2-deoxyglucose is carbon source dependent, as is the resistance conferred by gene deletions. Snf1 kinase, the AMP-activated protein kinase of yeast, is required for 2-deoxyglucose resistance in cells growing on glucose. Mutations in the SNF1 gene that reduce kinase activity render cells hypersensitive to 2-deoxyglucose, while an activating mutation in SNF1 confers 2-deoxyglucose resistance. Snf1 kinase activated by 2-deoxyglucose does not phosphorylate the Mig1 protein, a known Snf1 substrate during glucose limitation. Thus, different stimuli elicit distinct responses from the Snf1 kinase.
Keywords: Snf1 kinase, AMP-activated protein kinase, 2-deoxyglucose, hexokinase, PP1 phosphatase
GLUCOSE is the preferred carbon source for most organisms and the yeast Saccharomyces cerevisiae is no exception. Glucose is avidly taken up by yeast and fermented to ethanol. In contrast, 2-deoxyglucose (2DG) is taken up by yeast cells but the missing hydroxyl group prevents fermentation at the isomerization step that would convert 2-deoxyglucose-6-phosphate to fructose-6-phosphate. Beyond being unavailable as a metabolic fuel, 2DG is a potent inhibitor of yeast cell growth even when other carbon sources are available. Early studies reported that 2DG was incorporated into and weakened cell walls (Johnson 1968; Biely et al. 1971). Additional research found that 2DG exposure promoted glucose repression of gene expression, a response that was severely maladaptive for cells growing on alternative carbon sources such as galactose and raffinose (Zimmermann and Scheel 1977). Addition of as little as 0.03% (g/100 ml) 2DG to yeast cells growing on 2% raffinose caused rapid and complete repression of invertase mRNA expression (Randez-Gil et al. 1995). Since invertase expression is required for growth on raffinose, the repression of invertase expression specifically and promotion of glucose repression in general provided an explanation for the inhibitory effects of 2DG.
2DG has been used in genetic studies to select for mutations that affect glucose repression. In one screen, spontaneous mutants able to grow on raffinose in the presence of 2DG identified three complementation groups now known to comprise the HXK2, REG1, and GRR1 genes (Zimmermann and Scheel 1977; Entian and Zimmermann 1980). In a second screen, cells were subjected to random mutagenesis followed by selection for growth on sucrose in the presence of 2DG (Neigeborn and Carlson 1987). This screen identified mutations in the REG1, HXK2, and GLC7 genes. Reg1 protein is a regulatory subunit of the Glc7 phosphatase (Dombek et al. 1999); Hxk2 is one of three hexokinase genes in yeast (De Winde et al. 1996); and Grr1 is an F-box containing subunit of a ubiquitin ligase complex (Li and Johnston 1997). Deletions of these genes confer resistance to 2DG, release cells from glucose repression, and produce very similar changes in global gene expression (Apweiler et al. 2012). Another key player in the glucose repression pathway is the Snf1 kinase, the AMP-activated protein kinase of S. cerevisiae. Release from glucose repression, sometimes referred to as derepression, requires the activation of the Snf1 kinase (Hedbacker and Carlson 2008). Deletion or loss of function mutations in REG1, GRR1, and HXK2 as well as specific missense alleles of GLC7 cause constitutive activation of Snf1 kinase, escape from glucose repression, and resistance to 2DG (Tu and Carlson 1995; Treitel et al. 1998; McCartney and Schmidt 2001).
In addition to its use for the study of glucose repression in yeast, 2DG is also being tested in humans as an anticancer agent (Raez et al. 2013). 2DG is thought to inhibit hexokinase and thereby reduce glycolysis, an energy-generating pathway of particular importance to cancer cells (Pelicano et al. 2006). Therefore, understanding the mechanism of 2DG action is of interest to more than those who study glucose repression in yeast. The idea that 2DG inhibits yeast cell metabolism by promoting glucose repression has been challenged recently by the study of Ralser et al. (2008). First, they showed that 2DG inhibited yeast growth on media containing glucose as the carbon source. If the mechanism of 2DG growth inhibition is the inappropriate promotion of glucose repression, then one would not necessarily expect to find growth inhibition on glucose where glucose repression is expected and should not be maladaptive at all. Second, Ralser et al. (2008) screened the yeast MATa knockout collection (Winzeler et al. 1999) and identified 19 genes whose deletion conferred resistance to 2DG. Their list of 2DG-resistant knockouts included REG1 and HXK2, genes known to regulate glucose repression and confer 2DG resistance, adding some confidence to the validity of their screen. They identified 17 additional genes that had not previously been linked to glucose repression or 2DG resistance, and these findings suggested that our understanding of the mechanism of growth inhibition by 2DG was incomplete. Finally, their demonstration that 2DG inhibited growth on glucose opened the possibility of performing genetic analysis of SNF1 and 2DG resistance. SNF1 is required for growth on sucrose and raffinose media, making it impossible to study 2DG resistance in snf1 mutants growing on these alternative carbon sources. However, SNF1 is not required for growth on glucose, thus allowing the determination of the epistatic relationships between SNF1 and genes that confer resistance to 2DG. While it is known that Snf1 kinase is activated by glucose starvation, we demonstrate that Snf1 kinase is active on high glucose and its activity is required for resistance to 2DG.
Materials and Methods
Yeast strains and genetic methods
The yeast strains used in this study were all derivatives of S228C. Yeast strains with specific gene deletions were generated in our laboratory or by the Saccharomyces Genome Deletion Project (Winzeler et al. 1999) and purchased from Thermo Scientific (Table 1). Cells were grown at 30° using standard media lacking nutrients needed for plasmid selection (Rose et al. 1990). Oligonucleotide-directed mutagenesis was performed with Pfu polymerase followed by DpnI digestion of the plasmid template (Fisher and Pei 1997). All of the mutations were confirmed by DNA sequencing. The wild-type SNF1 allele and all derivatives used in this study contained three copies of the hemagglutinin tag (3HA) at the C terminus (McCartney and Schmidt 2001).
Table 1. S. cerevisiae strains.
| Strain | Genotype | Source |
|---|---|---|
| MSY1212 | MATa ura3-52 leu2∆1 his3∆200 | This laboratory |
| MSY1217 | MATa ura3-52 leu2∆1 his3∆200 snf1∆10 | This laboratory |
| BY4741 | MATa ura3∆0 leu2∆0 his3∆1 met15∆0 | Thermo Scientific |
| MSY543 | MATa ura3-52 leu2∆1 trp1∆63 his3∆200 sip2∆::HIS3 gal83∆::HIS3 | This laboratory |
| MSY544 | MATα ura3-52 leu2∆1 trp1∆63 his3∆200 sip2∆::HIS3 sip1∆::HIS3 | This laboratory |
| MSY552 | MATa ura3-52 leu2∆1 trp1∆63 his3∆200 sip1∆::HIS3 gal83∆::HIS3 | This laboratory |
| MSY557 | MATα ura3-52 leu2∆1 trp1∆63 his3∆200 sip1∆::HIS3 sip2∆::HIS3 gal83∆::HIS3 | This laboratory |
| MSY676 | MATα ura3∆0 leu2∆0 his3∆1 lys2∆0 sak1∆::KAN | This laboratory |
| MSY682 | MATa ura3∆0 leu2∆0 his3∆1 met15∆0 tos3∆::KAN | This laboratory |
| MSY691 | MATα ura3∆0 leu2∆0 his3∆1 met15∆0 elm1∆::KAN | This laboratory |
| MSY1226 | MATα ura3-52 leu2∆1 his3∆200 reg1∆::HIS3 | This laboratory |
| MSY1227 | MATa ura3-52 leu2∆1 his3∆200 reg1∆::HIS3 snf1∆10 | This laboratory |
| MSY1254 | MATα ura3∆0 leu2∆0 his3∆1 hxk2∆::KAN | This laboratory |
| MSY1260 | MATa ura3 leu2 his3 hxk2∆::KAN snf1∆10 | This laboratory |
| RG7198 | MATa ura3∆0 leu2∆0 his3∆1 met15∆0 tup1∆::KAN | Thermo Scientific |
| RG6902 | MATa ura3∆0 leu2∆0 his3∆1 met15∆0 grr1∆::KAN | Thermo Scientific |
| RG4214 | MATa ura3∆0, leu2∆0, his3∆1, met15∆0, lsm6∆::KAN | Thermo Scientific |
2-Deoxyglucose resistance assays
Resistance to 2DG was measured in liquid culture growth assays. Fresh overnight cultures were diluted in fresh media to an A600 of 0.1 in the absence or presence of increasing concentrations of 2DG. Typically, 2DG was added to final concentrations of 0.01, 0.02, 0.05, 0.1, and 0.2% (g/100 ml). Other carbon sources were included at 2% (glucose and sucrose). When glycerol and ethanol were used as the carbon source, they were included at 3 and 2% (vol/vol), respectively. Cells were grown for 18 hr and the A600 was measured. Cell growth was plotted relative to growth in the absence of 2DG, defined as 100%.
Western blotting
The Mig1 and Snf1 proteins tagged with three copies of the HA epitope was detected with a 1:3000 dilution of HA probe (Santa Cruz). Goat antimouse IgG DyLight 680 (Thermo) diluted 1:10,000 was used as the secondary antibody. For detection of phosphorylated Snf1, Phospho-AMPKalpha (Thr172) antibody (Cell Signaling) diluted 1:1000 was used with goat antirabbit IRDye 800CW (Li-Cor) (1:10,000 dilution) as the secondary antibody. Blots were scanned by using an Odyssey scanner (Li-Cor). Integrated intensity values of bands were quantified by using Odyssey scanning software. Yeast extracts were prepared using the boiling method described by Kuchin and colleagues (Orlova et al. 2008).
Canavanine ploidy test
Genome copy number was assayed with the canavanine ploidy test (Schild et al. 1981). Patches of yeast cells replica plated to synthetic complete medium lacking arginine and supplemented with 60 mg/liter canavanine sulfate. Cells were mutagenized with UV light and allowed to grow at 30° for 3 days. Spontaneous mutants capable of growth indicated haploid cells.
Generation of dhh1 strains
The dhh1∆ strain from the MATa knockout collection was found to be a MATa/a diploid. To test the cosegregation of 2DG resistance with the dhh1∆ allele, the dhh1∆::KAN allele was PCR amplified and used to transform a wild-type diploid strain to kanamycin resistance. Accurate replacement of the DHH1 gene was confirmed by PCR analysis. The dhh1∆::KAN heterozygous diploid strain was subjected to sporulation and haploid progeny were analyzed for kanamycin and 2DG resistance.
Statistical analysis
For all bar plots, mean values using a minimum of three independent measurements are plotted with error bars representing one standard error. Statistical significance was determined using the Student t-test for unpaired variables with equal variance. Statistical significance is indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ns, P > 0.05.
Results
2DG toxicity is carbon source dependent
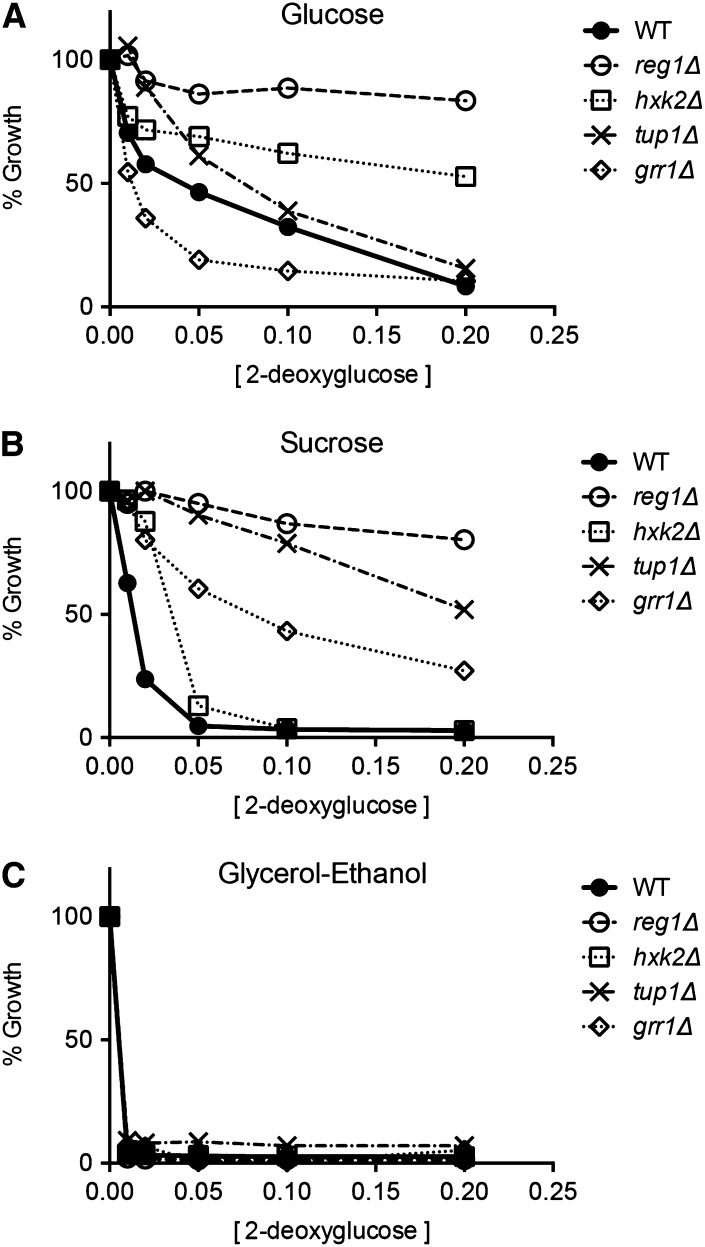
To study the effect of 2DG on yeast growth, we have used an assay in which liquid cultures diluted to an A600 of 0.1 are grown for 18 hr in the presence of increasing concentrations of 2DG. Since many mutants have different growth rates, we plot cell growth as a percentage of growth relative to cultures with no 2DG. This assay yields a reproducible measurement of 2DG toxicity and resistance (Tabba et al. 2010). Here, we examined the ability of 2DG to inhibit yeast cell growth in liquid cultures as a function of carbon source. When cells are grown in media with 2% glucose as the carbon source, addition of 2DG causes significant inhibition of cell growth (Figure 1). At 2DG concentrations of 0.1 and 0.2%, growth is almost completely blocked. When the alternative, fermentable carbon source sucrose was used, cells display a greater sensitivity to the presence of 2DG. Finally, when cells grow aerobically in media with glycerol and ethanol as the carbon source, they become hypersensitive to the presence of 2DG. Addition of only 0.01% 2DG is sufficient to completely inhibit cell growth. We conclude that 2DG inhibits yeast cell growth on all carbon sources tested with the greatest 2DG resistance observed during fermentation of glucose and the least observed during aerobic growth on a mixture of glycerol and ethanol.
Figure 1.
Effect of carbon source on the inhibition of yeast growth by 2DG. Wild-type yeast cells were grown in synthetic complete media with either glucose, sucrose, or a mixture of glycerol and ethanol as the carbon source. Overnight cultures were diluted in fresh media to an A600 of 0.1 with the indicated concentration of 2-deoxyglucose. Cultures were grown for 18 hr and the A600 was measured. Growth is plotted relative to the cell density of the culture without 2DG.
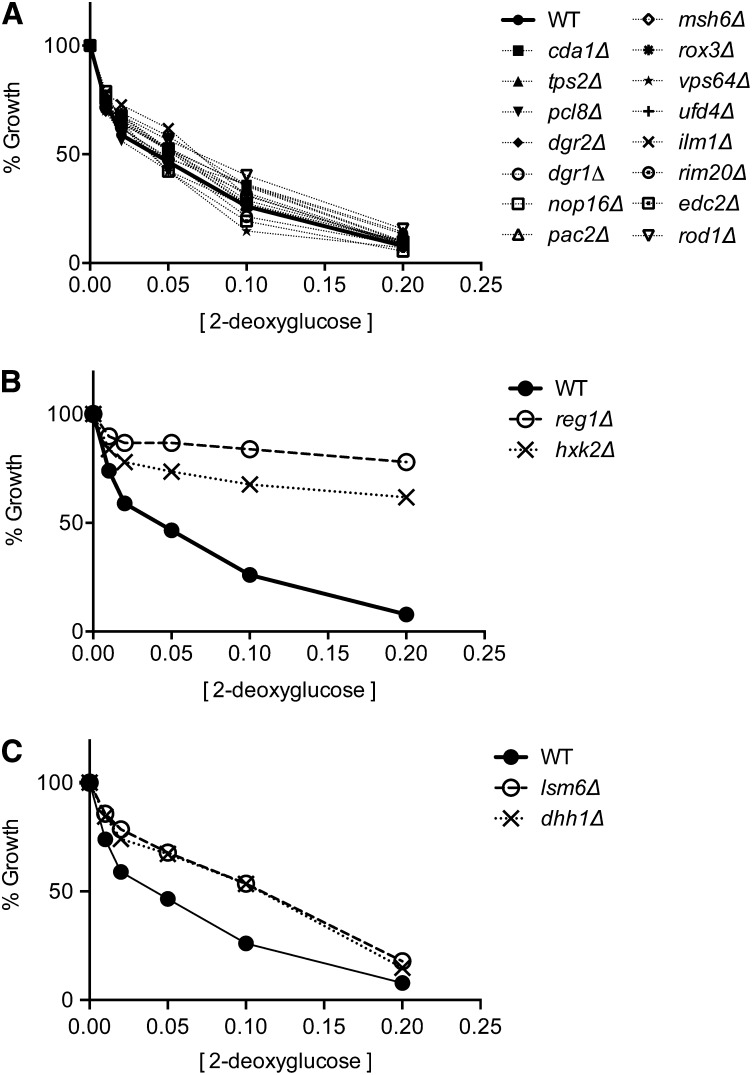
Gene deletions that confer resistance to 2DG
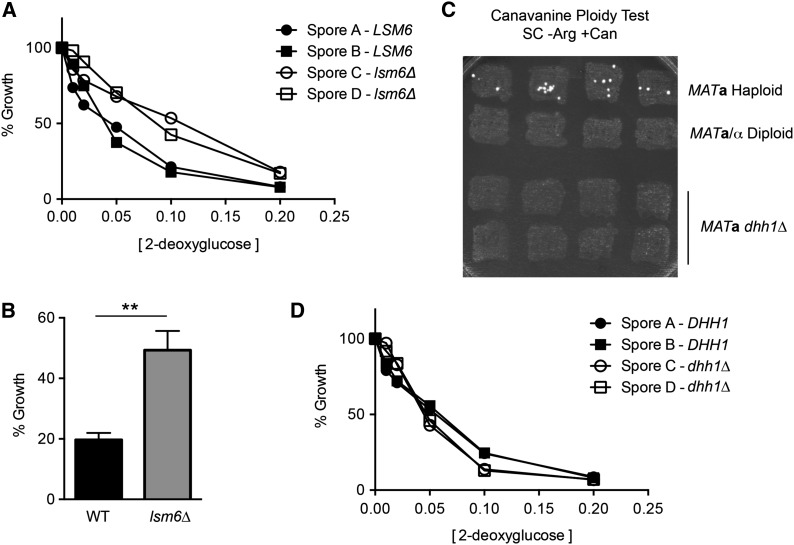
We next examined which genes when deleted could confer resistance to 2DG. Random mutagenesis studies had identified multiple complementation groups that conferred 2DG resistance to cells growing by fermentation of sucrose or raffinose (Zimmermann and Scheel 1977; Neigeborn and Carlson 1987). These complementation groups mapped to the REG1, GRR1, GLC7, and HXK2 genes. A recent high throughput screen of the yeast MATa knockout collection (>5000 strains) identified 19 genes whose deletion conferred significant 2DG resistance to cells growing by fermentation of glucose (Ralser et al. 2008). We sought to measure the 2DG resistance on glucose media of cells lacking each of the 19 genes identified by Ralser and colleagues. These strains were taken from the MATa knockout collection, grown in rich media without selection, and each gene knockout was confirmed by PCR. Each strain was tested for 2DG resistance on glucose. We found that 15 of the 19 strains identified by Ralser et al. (2008) were not significantly different from the wild-type strain with regard to growth in the presence of 2DG (Figure 2A). Two strains that showed significant resistance to 2DG were those with deletions REG1 and HXK2, genes known to confer 2DG resistance on sucrose (Figure 2B). Backcrosses to a wild-type strain confirmed that the reg1∆ and hxk2∆ deletions cosegregated with 2DG resistance (not shown). Finally, the strains bearing the dhh1∆ and lsm6∆ alleles showed a weak but reproducible 2DG resistance most evident when 2DG is present at 0.1% (Figure 2C). The lsm6∆ strain was backcrossed to wild type, and the haploid progeny were analyzed for 2DG resistance. The lsm6∆ allele did cosegregate with weak 2DG resistance (Figure 3A) that was reproducible and statistically significant (Figure 3B). We were unable to backcross the dhh1∆ strain taken from our MATa knockout collection. Upon analyzing the dhh1∆ strain, we found that this strain was not haploid as judged by its inability to form colonies in the canavanine ploidy test (Figure 3C). To test cosegregation of the dhh1∆ with 2DG resistance, we used the genomic DNA of the dhh1∆ strain from the MATa collection as a PCR template to amplify the dhh1∆::KAN cassette. The amplified DNA was used to transform a diploid strain, which was then subjected to sporulation. We found that the dhh1∆ allele did not cosegregate with 2DG resistance (Figure 3D). In summary, we tested all 19 of the knockout strains identified by Ralser et al. (2008) as being 2DG resistant. We found that the two knockouts that conferred strong 2DG resistance were in genes long known to confer 2DG resistance (REG1 and HXK2). Deletion of the LSM6 gene conferred weak but reproducible resistance. However, 16 of the 19 knockout strains identified by Ralser and colleagues do not confer any significant resistance to 2DG. Thus we are unable to reproduce the results reported by Ralser et al. (2008). Possible reasons for this discrepancy are discussed below.
Figure 2.
Effect of gene deletions on 2DG resistance. Wild-type yeast cells and the 19 knockouts strains identified by Ralser et al. (2008) were grown in the absence or presence of increasing concentrations of 2DG in rich media with glucose as the carbon source. Deletion of 15 genes provide no significant resistance to 2DG relative to wild-type yeast (A). Deletion of the REG1 and HXK2 genes provides strong resistance to 2DG relative to a wild-type strain (B). Deletion of the LSM6 and DHH1 genes provides weak 2DG resistance relative to a wild-type strain (C).
Figure 3.
Genetic analysis of 2DG resistance in lsm6∆ and dhh1∆ strains. (A) The lsm6∆ strain from the MATa collection was backcrossed to a wild-type strain. Haploid progeny were analyzed for cosegregation of the lsm6∆ and resistance to 2DG. (B) Multiple wild-type and lsm6∆ strains were grown overnight in YEPD medium with and without 0.1% 2DG. Mean values for relative growth were plotted ± SE. (C) Patches of yeast cells derived from single colonies of a MATa haploid strain (MSY1212), a MATa/α diploid strain (MSY210), or the dhh1∆ strain from the MATa knockout collection were subjected to the canavanine ploidy test. (D) Haploid progeny from a diploid strain heterozygous for the dhh1∆ allele were analyzed for cosegregation of the dhh1∆ and resistance to 2DG.
Snf1 kinase is required for 2DG resistance on glucose media
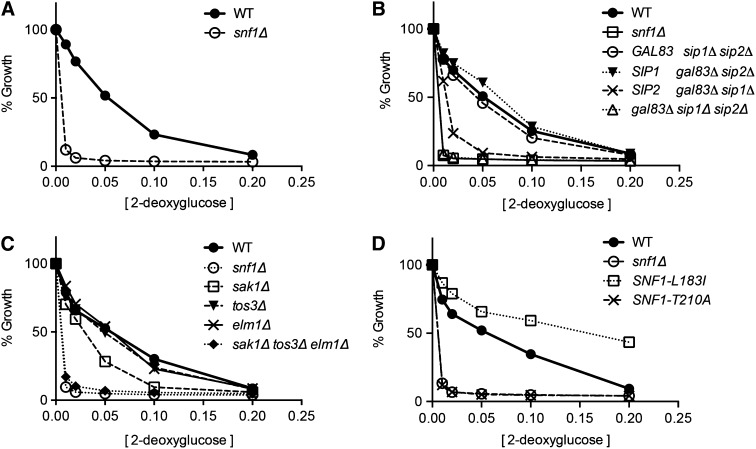
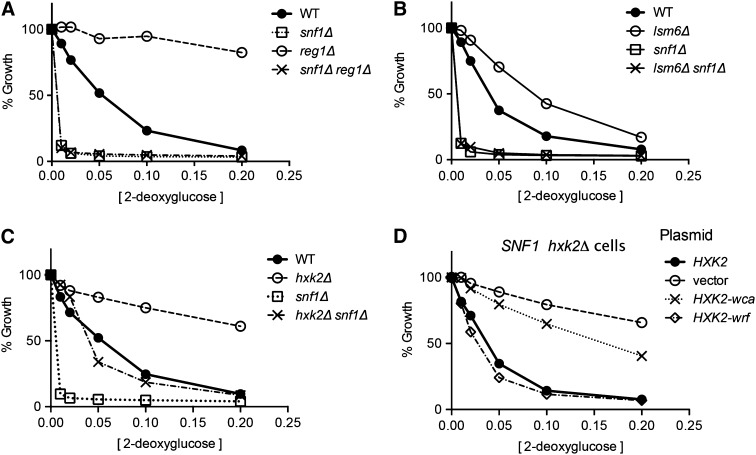
Since 2DG has been implicated as a compound that promotes glucose repression, we wanted to determine the role, if any, played by the Snf1 kinase. Snf1 is known to be a key signal transduction kinase in the glucose repression pathway (Hedbacker and Carlson 2008). We tested whether the Snf1 kinase is required for 2DG resistance in media with glucose as the carbon source. When wild-type and snf1∆ cells were subjected to our 2DG resistance assay, we found that the snf1∆ cells were hypersensitive to 2DG (Figure 4A). The lowest concentrations of 2DG used in this assay (0.01 and 0.02%) completely inhibit cell growth in the snf1∆ strain, while only causing a modest inhibition of growth in the wild-type strain. Therefore, SNF1 is required for resistance to low concentrations of 2DG on glucose media.
Figure 4.
Snf1 kinase is required for resistance to 2DG on glucose. 2DG resistance assays were conducted comparing wild-type cells to snf1∆ cells (A); to cells expressing one or none of the Snf1 β subunits, Gal83, Sip1, or Sip2 (B); to cells lacking one or all of the Snf1 activating kinases, Sak1, Tos3, or Elm1 (C); or to cells expressing Snf1 kinase with single-amino-acid substitutions (D).
The Snf1 kinase complex is an αβγ-heterotrimer and is present in vivo as three distinct isoforms that differ by the identity of the β-subunit (Schmidt and McCartney 2000). We tested whether each of the Snf1 isoforms was capable of conferring 2DG resistance in glucose media. Wild-type cells and cells deleted for different combinations of β-subunit genes were subjected to the 2DG resistance assay (Figure 4B). Cells lacking all three β-subunits (sip1∆ sip2∆ gal83∆) were hypersensitive to 2DG and indistinguishable from the snf1∆ strain. Interestingly, cells expressing Gal83 or Sip1 as the only β-subunit showed 2DG resistance curves similar to the wild-type cell, while cells expressing only Sip2 were clearly more sensitive to 2DG. Therefore, we conclude that the Sip2 isoform of Snf1 is not fully competent for providing 2DG resistance on glucose.
Activation of Snf1 is mediated by three distinct Snf1-activating kinases, Sak1, Tos3, and Elm1 (Hong et al. 2003; Sutherland et al. 2003). In the absence of all three Snf1-activating kinases, cells were hypersensitive to 2DG and indistinguishable from cells lacking the Snf1 kinase entirely (Figure 4C). We tested whether any one of the Snf1-activating kinases was required for 2DG resistance and found that cells lacking the Elm1 or Tos3 kinases showed 2DG resistance curves indistinguishable from the wild-type cell. Cells lacking the Sak1 kinase exhibited an increased sensitivity to 2DG relative to wild-type cells but did not display the hypersensitivity observed in the snf1∆ cell or the sak1∆ tos3∆ elm1∆ cell. These data indicate that a Snf1-activating kinase is required for 2DG resistance and that the Sak1 kinase plays a larger role than Elm1 or Tos3 but not an exclusive role in the activation of Snf1 in the presence of high glucose.
Snf1 kinase activity correlates with 2DG resistance
While the Snf1 kinase is activated under conditions of glucose limitation, our results with 2DG suggest that the Snf1 kinase plays a role in 2DG resistance in cells growing in abundant glucose. We have previously shown that acute inhibition of Snf1 kinase activity in cells growing on high glucose results in altered patterns of gene expression clearly detected by microarray analysis (Shirra et al. 2008). Here we asked whether Snf1 kinase activity is required for 2DG resistance by testing whether mutants of Snf1 that affect its catalytic potential also affect 2DG resistance. A mutation of threonine 210 to alanine completely blocks activation loop phosphorylation and is phenotypically similar to the snf1∆ allele (McCartney and Schmidt 2001). A mutation in the kinase domain hydrophobic core, leucine 183 to isoleucine (Leech et al. 2003), results in partial and constitutive activation of Snf1 kinase (Chandrashekarappa et al. 2013). When these alleles were tested for 2DG resistance, we found that the Snf1-T210A allele was hypersensitive to 2DG, indistinguishable from the snf1∆ strain (Figure 4D). Therefore activation loop phosphorylation is required for resistance to 2DG on glucose media. Cells expressing Snf1-L183I protein were more resistant to 2DG than wild-type cells. Since this mutation causes increased activity of the Snf1 kinase, we conclude that Snf1 kinase activity promotes 2DG resistance in cells growing on high glucose.
Effect of carbon source on gene deletions that confer resistance to 2DG
Earlier studies of 2DG resistance using alternative fermentable carbon sources (sucrose and raffinose) identified REG1, HXK2, as well as GLC7, GRR1, and TUP1 genes (Zimmermann and Scheel 1977; Neigeborn and Carlson 1987; Schuller and Entian 1991). Since GLC7 is an essential gene, its deletion is not present in the MATa deletion collection and thus could not have been detected by Ralser et al. (2008). However, the grr1∆ and tup1∆ strains are present in the MATa collection but were not identified as 2DG-resistant strains on glucose (Ralser et al. 2008). Here, we tested whether the carbon source affects 2DG resistance conferred by different gene deletions. We tested strains with complete deletions of the REG1, HXK2, GRR1, and TUP1 genes for 2DG resistance on glucose, sucrose, and glycerol/ethanol (Figure 5). The reg1∆ and hxk2∆ strains exhibited 2DG resistance on both glucose and sucrose media while the grr1∆ and tup1∆ strains exhibited significant 2DG resistance only on sucrose media. This result could explain why the grr1∆ and tup1∆ strains were not identified in the screen conducted by Ralser et al. (2008), as this was performed on glucose media. None of the deletions tested were able to confer any 2DG resistance to cells during aerobic growth on glycerol/ethanol media even with the lowest concentrations of 2DG (0.01%).
Figure 5.
Carbon source dependence of 2DG resistance conferred by mutations in the REG1, HXK2, TUP1, and GRR1 genes. Resistance to 2DG was assayed in media with glucose (A), sucrose (B), or glycerol–ethanol (C) as the carbon source. Wild-type cells were compared to cells with the following gene deletions: reg1∆, hxk2∆, tup1∆, and grr1∆.
Epistasis analysis of SNF1 and 2DG resistance genes
Since the snf1∆ cells are viable on glucose media, we decided to test the epistatic relationships between the snf1 mutation and the reg1, hxk2, and lsm6 mutations. These deletions were the only ones that confer 2DG resistance to cells growing on glucose. Strains with deletions in the SNF1, REG1, HXK2, and LSM6 genes were crossed and haploid progeny were tested for 2DG resistance. We found that the snf1∆ reg1∆ strain as well as the lsm6∆ snf1∆ strain were hypersensitive to 2DG (Figure 6, A and B). Thus snf1∆ is epistatic to the reg1∆ and lsm6∆ mutations. These data indicate that the 2DG resistance conferred by the reg1∆ and lsm6∆ alleles requires the presence of Snf1.
Figure 6.
Epistasis analysis of the snf1∆ mutation with the reg1∆, lsm6∆, and hxk2∆ mutations. 2DG resistance assays were conducted with glucose as the carbon source comparing wild-type (WT) and snf1∆ strains to strains with deletions in the REG1 (A), LSM6 (B), and HXK2 genes (C). Genotypes are indicated on the right of each plot. (D) The role of Hxk2 regulatory and catalytic functions in 2-deoxyglucose resistance was investigated with hxk2∆ cells transformed with plasmids that express either wild-type Hxk2, no Hxk2 (vector), Hxk2 without catalytic activity (Hxk2-wca), or Hxk2 without regulatory function (Hxk2-wrf).
The genetic relationship between the snf1∆ and hxk2∆ mutations is more complex. The snf1∆ hxk2∆ double deletion exhibits a 2DG resistance that is intermediate between either single mutant and similar to that observed in wild-type cells (Figure 6C). Thus deletion of HXK2 suppresses the hypersensitive phenotype observed in the snf1∆ strain, while deletion of SNF1 suppresses the 2DG resistance observed in the hxk2∆ strain. Therefore Hxk2 acts in part through Snf1 and in part independently of Snf1. The Hxk2 protein has two functional domains, a catalytic domain responsible for the phosphorylation of glucose and a regulatory domain that mediates regulation of gene expression (Pelaez et al. 2010). Specific mutations in the HXK2 gene that separate these two functions were tested for 2DG resistance. The mutation that eliminates the catalytic function of Hxk2 (without catalytic activity) (HXK2-wca) confers 2DG resistance, while the mutation that eliminates the regulatory function of Hxk2 (without regulatory function) (HXK2-wrf) does not confer any 2DG resistance (Figure 6D). Therefore the 2DG resistance that is conferred by the deletion of the HXK2 gene is due to the loss of the catalytic activity of yeast hexokinase II.
Persistence of 2DG growth inhibition
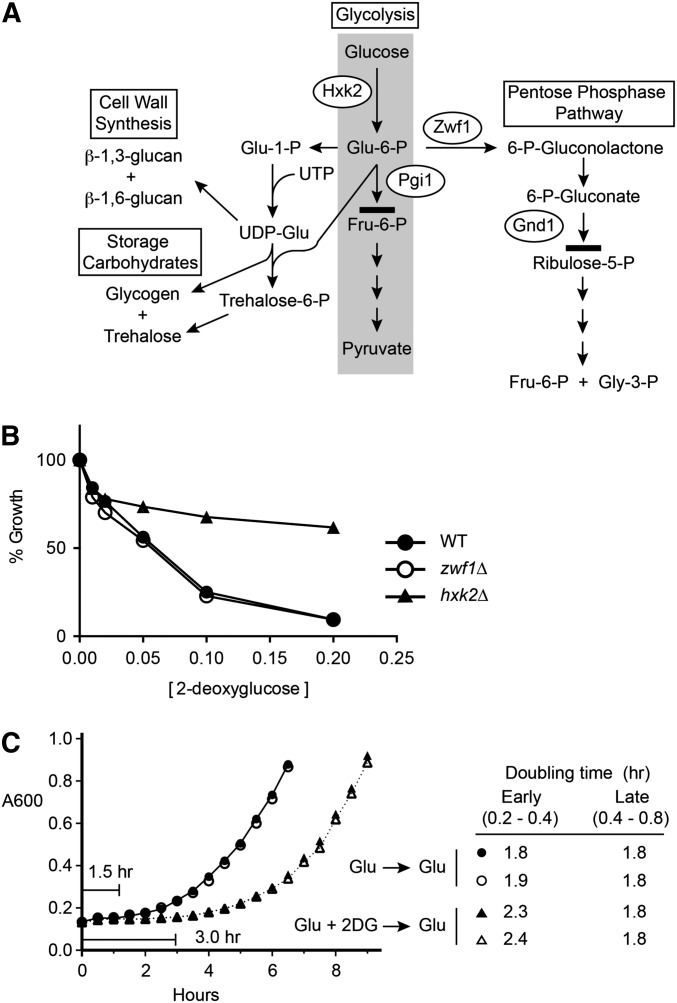
The finding that the loss of Hxk2 catalytic activity confers 2DG resistance suggests that the conversion of 2DG to 2-deoxyglucose-6-phosphate contributes to the toxicity of 2DG. Once 2DG is phosphorylated, it becomes trapped in the cell, since phosphorylated hexoses cannot be transported out of cells by the hexose transporter proteins. One possibility is that 2-deoxyglucose-6-phosphate accumulates within cells, since it cannot proceed in glycolysis beyond the phosphoglucose isomerase step (Figure 7A). However, 2-deoxyglucose-6-phosphate can be incorporated into the cell wall (Biely et al. 1971), into storage carbohydrates (Tran-Dinh et al. 1995) or undergo the first two steps of the pentose phosphate pathway. Once converted to 6-phospho-2-deoxygluconate, it is unlikely that 2DG can proceed beyond the step catalyzed by 6-phosphogluconate dehydrogenase, since the catalytic mechanism of that enzyme utilizes the C2 hydroxyl group missing in 2DG (Hanau et al. 2010). Ralser et al. (2008) reported that deletion of the ZWF1 gene, which encodes the enzyme catalyzing the first step in the pentose phosphate pathway, provides strong resistance to 2DG. When we endeavored to repeat this finding, we discovered that the zwf1∆ did not in fact confer any resistance to 2DG (Figure 7B). The Zwf1 protein is the only glucose-6-phosphate dehydrogenase in yeast. Deletion of the ZWF1 gene completely blocks entry of 2-deoxyglucose-6-phosphate into the pentose phosphate pathway and has no effect on 2DG sensitivity or resistance. Therefore entry of 2-deoxyglucose-6-phosphate into the pentose phosphate pathway is not required for 2DG toxicity. Here, we sought evidence to support the idea that the accumulation of a toxic intermediate in the metabolism of 2DG was responsible for its growth inhibition. If an intermediate accumulates in cells and it cannot be easily disposed of, then one would expect that 2DG-mediated growth inhibition would persist even after cells were transferred to media lacking 2DG. To test this, we grew wild-type cells overnight in the presence or absence of 2DG. After 18 hr, cells were washed to remove 2DG and then diluted into fresh media (Figure 7C). Cells that were grown overnight in 0.2% 2DG displayed a greater lag time (3 hr vs. 1.5 hr) and initially a reduced growth rate (doubling time of 2.3 hr vs. 1.8 hr). After two doublings and 7 hr of growth in the absence of 2DG, the growth rate of 2DG-treated cells was equivalent to cells that never experienced 2DG. The persistent inhibition of growth mediated by 2DG supports that idea that some inhibitory metabolic intermediate accumulates in cells exposed to 2DG and that this intermediate cannot be rapidly metabolized.
Figure 7.
Persistent inhibition by 2DG. (A) Four pathways of glucose metabolism in yeast are shown. 2-Deoxyglucose can be converted to 2-deoxyglucose-6-phosphate by hexokinase II (Hxk2) and used for cell wall synthesis and storage carbohydrate metabolism. 2-Deoxyglucose-6-phosphate cannot proceed further in glycolysis than the step catalyzed by phosphoglucose isomerase (Pgi1) but could enter the pentose phosphate pathway through oxidation by glucose-6-phosphate dehydrogenase (Zwf1). Further metabolism in the pentose phosphate pathway is blocked at the step catalyzed by 6-phosphogluconate dehydrogenase (Gnd1). (B) 2DG resistance assay of wild-type cells compared to zwf1∆ and hxk2∆ cells. (C) Wild-type cells were grown in duplicate cultures in YEPD media overnight in the absence (circles) or presence (triangles) of 0.2% 2DG. Cells were harvested, washed in YEPD, and diluted to an A600 of 0.15 in YEPD media. Cell growth was measured by reading absorbance at 600 nm. Lag times and doubling times for the cultures with and without 2DG treatment are indicated.
Effect of 2DG on Snf1 activation and signaling
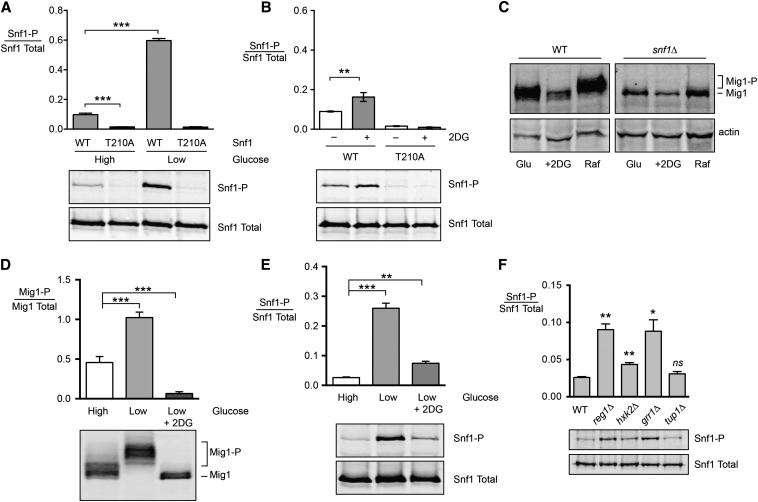
Our studies indicate that resistance to 2DG on glucose media requires the Snf1 kinase (Figure 4A) and that the Snf1 kinase must be phosphorylated on its activation loop to confer 2DG resistance (Figure 4D). These results are somewhat counterintuitive, since the Snf1 kinase is thought to be largely unphosphorylated and inactive on high glucose media. However, a more nuanced understanding would be that Snf1 kinase is less active on high glucose media. Indeed, several studies have documented Snf1 function in high glucose in response to sodium stress (McCartney and Schmidt 2001; Portillo et al. 2005), amino acid stress (Cherkasova et al. 2010), hydroxyurea stress (Dubacq et al. 2004), and alkaline stress (Hong and Carlson 2007). Phosphoproteomic studies have detected Snf1-dependent phosphorylation events in cells growing on high glucose (Bodenmiller et al. 2010; Braun et al. 2014), indicating that Snf1 kinase is active in high glucose. Furthermore, some studies suggest that the Snf1-T210A mutant, which cannot be phosphorylated on its activation loop, has detectable activity (Portillo et al. 2005; Braun et al. 2014). We analyzed the phosphorylation state of the Snf1 activation loop addition using quantitative Western blotting of triplicate samples grown in high and low glucose with or without a threonine residue in the activation loop at position 210. We found that the phosphorylation of the Snf1 activation loop is clearly detected in cells grown on high glucose (Figure 8A). The antibody reactivity is absolutely dependent on the threonine at position 210 and is lost upon treatment of samples with phosphatase (McCartney and Schmidt 2001). When cells were shifted to low glucose, the phosphorylation of the Snf1 activation loop is increased by approximately 10-fold.
Figure 8.
Effect of 2DG on Snf1 phosphorylation and signaling. (A) Snf1 activation loop phosphorylation was measured in triplicate in high and low glucose in cells expressing wild-type Snf1 (WT) or Snf1-T210A. The mean ratio of phosphorylated over total Snf1 is plotted ± SE. Representative Western blots are shown below. (B) Snf1 activation loop phosphorylation was measured in triplicate in cells expressing wild-type Snf1 or Snf1-T210A. Extracts were prepared from cells grown in high glucose or 1 hr after addition of 0.2% 2DG. (C) Mig1 phosphorylation was analyzed by Western blotting in wild-type and snf1∆ cells grown in high glucose or high glucose 1 hr after addition of 0.2% 2DG or 1 hr after shifting to raffinose. The SDS gel mobility of Mig1 and phosphorylated Mig1 (Mig1-P) is indicated. (D) Mig1 phosphorylation was measured by quantitative Western blotting from triplicate cultures grown in high glucose or 1 hr after shifting to low glucose with or without 0.2% 2DG. Representative Western blots are shown below. (E) Snf1 activation loop phosphorylation was measured in triplicate using the same growth conditions as in D. (F) Phosphorylation of Snf1 activation loop in glucose media was analyzed in triplicate cultures using wild-type cells or cells with the indicated gene deletion. Mean values statistically different from wild type are indicated.
We examined the effect of 2DG addition on the phosphorylation status of Snf1. If 2DG is added to cells growing on high glucose, we detected a small but statistically significant (P < 0.01) increase in Snf1 activation loop phosphorylation (Figure 8B). Interestingly, this increase in Snf1 activation loop phosphorylation did not translate into an increase in the phosphorylation of Mig1 (Figure 8C), a known Snf1 substrate in the response to glucose limitation (Treitel et al. 1998; Smith et al. 1999). We also analyzed the phosphorylation of Mig1 in cells grown in high glucose and then shifted to low glucose for 1 hr with or without 0.2% 2DG. Upon shifting to low glucose, the phosphorylation of the Mig1 protein causes its mobility to shift to a complex mixture of more slowly migrating species (Figure 8D). When cells are shifted to low glucose plus 0.2% 2DG, the Mig1 protein becomes hypophosphorylated with a mobility greater than that observed in high glucose. Analysis of Snf1 phosphorylation in cells grown in the same manner found that shifting to low glucose resulted in the well-known increase in Snf1 phosphorylation that was largely blunted by the presence of 2DG (Figure 8E). Finally, we tested whether there was a positive correlation between the gene deletions that confer 2DG resistance on glucose and the phosphorylation status of Snf1. Cells lacking the REG1, HXK2, GRR1, and TUP1 genes were grown in high glucose media. Triplicate extracts were prepared and the Snf1 protein was analyzed by Western blotting (Figure 8F). Cells lacking the Reg1 or Grr1 proteins showed the largest increase in Snf1 phosphorylation status, while deletion of HXK2 had only a minor effect on the phosphorylation of Snf1. Deletion of the GRR1 gene had a large effect on Snf1 phosphorylation status but does not confer 2DG resistance on glucose media (Figure 5A). Deletion of the HXK2 gene confers strong 2DG resistance on glucose media but has only a minimal effect on Snf1 phosphorylation. From these experiments we make the following conclusions: (1) a fraction of the Snf1 kinase complexes are phosphorylated and active in cells growing on high glucose; (2) addition of 2DG to cells growing on high glucose increases the fraction of Snf1 kinase complexes that are phosphorylated; (3) Snf1-dependent phosphorylation of Mig1 increases in response to glucose limitation but not in response to 2DG addition; and (4) gene deletions that increase Snf1 phosphorylation status do not necessarily confer 2DG resistance on glucose.
Discussion
We became interested in the genetics of 2DG resistance when it was reported that many additional and seemingly unrelated genes could confer resistance to 2DG (Ralser et al. 2008). However, when we tested the 19 gene deletion strains identified by Ralser and colleagues, we found that the vast majority (16 of 19) were not in fact any more resistant to 2DG than wild-type cells (Figure 2 and Figure 3). We believe that the source of this discrepancy is the high frequency with which yeast spontaneously acquire 2DG resistance, a finding reported many years ago (Heredia and Heredia 1988). Simple methods in yeast genetics, such as backcrossing a mutant with wild type (Figure 3), could have easily distinguished 2DG resistance due to spontaneous mutations from those linked to a specific gene deletion. Spontaneous 2DG resistant mutants could pose significant problems with false positives if one were to use 2DG resistant colonies as the source of an inoculum for subsequent experiments. Colonies of spontaneous 2DG resistant mutants growing on agar plates are clearly visible in many of the figures in Ralser et al. (2008). In this study, we have only used cells that had never been exposed to 2DG.
Carbon source has a large effect on 2DG toxicity and on the resistance conferred by gene deletions. Wild-type cells exhibit the greatest resistance to 2DG when growing on glucose and the least when growing aerobically on glycerol/ethanol. The ability of gene deletions to confer 2DG resistance was also found to be carbon source dependent. For instance, deletion of REG1, HXK2, TUP1, or GRR1 confers 2DG resistance to cells growing on alternative sugars but only deletion of REG1 or HXK2 could provide strong 2DG resistance on glucose (Figure 5). None of the genes tested here could provide any resistance to cells growing on a nonfermentable carbon source. The fact that the ability of these genes to confer 2DG resistance differs depending on the carbon source suggests that these gene deletions may be acting through distinct mechanisms.
The Snf1 kinase is a key regulatory kinase in the glucose repression pathway. However, its involvement in 2DG resistance had not been previously analyzed, since cells lacking SNF1 are not able to grow on alternative carbon sources. Here, we used glucose as the carbon source to test the requirement for SNF1 and the epistastic relationships between SNF1 and other genes that confer 2DG resistance. We found that SNF1 is required for resistance to 2DG on glucose. Cells lacking the Snf1 kinase are hypersensitive to 2DG (Figure 4). The SNF1 gene is epistatic to REG1 and LSM6 with regard to 2DG resistance. We interpret this result to mean that the Reg1 and Lsm6 proteins act through Snf1 kinase. The Reg1 protein is a regulatory subunit for the yeast PP1 phosphatase Glc7, which is needed to deactivate Snf1 kinase (McCartney and Schmidt 2001). Cells lacking Reg1 have constitutively active Snf1 kinase and are 2DG resistant. The Lsm6 protein promotes mRNA decapping as one component of the Lsm1-7–Pat1 complex (Chowdhury and Tharun 2009). An important role for Snf1 in the regulation of mRNA turnover has recently been uncovered in studies using conditional Snf1 kinase alleles (Young et al. 2012; Braun et al. 2014). Loss of Lsm6 may protect some Snf1-regulated mRNAs from decapping and thereby provide 2DG resistance.
Deletion of HXK2, the gene encoding yeast hexokinase II, confers strong 2DG resistance on glucose (Neigeborn and Carlson 1987; Schuller and Entian 1991; Ralser et al. 2008) (Figure 2), yet the mechanism of this effect is not fully understood. Hxk2 is thought to catalyze the phosphorylation of 2DG creating 2-deoxyglucose-6-phosphate, which cannot proceed in glycolysis but may enter into the cell wall, storage carbohydrate, or pentose phosphate pathways (Figure 7A). It seems plausible that deletion of the HXK2 gene would reduce the phosphorylation of 2DG. However, yeast express two hexokinases and one glucokinase enzyme, and while Hxk2 is the most highly expressed isozyme on glucose, transcriptome analyses comparing wild-type and hxk2∆ cells have shown that hxk2∆ cells compensate for the loss of the Hxk2 enzyme with greatly increased expression of its paralog HXK1 (Apweiler et al. 2012). Therefore, in the absence of direct measurement, it is not certain that deletion of HXK2 leads to reduced phosphorylation of 2DG. Our data support the idea that synthesis of 2-deoxyglucose-6-phosphate is required for 2DG toxicity. Hxk2 protein manifests two distinct activities, a catalytic activity and a regulatory activity (Pelaez et al. 2010). Earlier studies associated the catalytic activity of the hexose kinases with short-term glucose repression and the Hxk2-specific regulatory function with long-term glucose repression (De Winde et al. 1996). When mutations that specifically inactivate the catalytic activity or the regulatory function were tested in the 2DG resistance assay, we found that only disruption of the catalytic activity conferred 2DG resistance (Figure 6D). The pentose phosphate pathway is not likely to be involved in 2DG toxicity, since the initial oxidative steps of this pathway can be eliminated by deletion of the ZWF1 gene without any effect on 2DG toxicity or resistance (Figure 7B). This result directly contradicts the findings by Ralser et al. (2008) who reported that the zwf1∆ conferred 2DG resistance. We have measured 2DG resistance of the zwf1∆ strain in liquid assays (Figure 7B) as well as on agar plates (not shown) with multiple independent colonies, but have not been able to reproduce the 2DG resistance reported in the Ralser study. Our data indicate that the growth inhibition conferred by 2DG exposure is likely to be due to the accumulation of a long lasting toxic intermediate, since the inhibition persisted over several hours after the removal of 2DG from the media (Figure 7C). The toxic intermediate in 2DG-treated cells is most likely either 2-deoxyglucose-6-phosphate or some intermediate in the storage carbohydrate or cell wall pathways.
We also investigated the role played by the Snf1 kinase in resistance to 2DG. We found that Snf1 kinase is active when cells are growing on high glucose and that its kinase activity is required for 2DG resistance (Figure 4). The effect of 2DG addition on the activation status of Snf1 is complex and depends on the carbon source and the order of 2DG addition. When 2DG is added to cells growing on glucose, we find a small but statistically significant increase in the phosphorylation of the Snf1 activation loop (Figure 8B). While some studies have reported that the unphosphorylated Snf1-T210A protein is functional in vivo (Portillo et al. 2005; Braun et al. 2014), our data indicate that the phosphorylation of Snf1 is needed for 2DG resistance. First, the Snf1-T210A cannot provide any resistance to 2DG (Figure 4D). Second, deletion of all three Snf1-activating kinases produces a 2DG hypersensitivity indistinguishable from the snf1∆ strain (Figure 4C). The increase in Snf1 activation loop phosphorylation induced by 2DG does not translate into an increase in the phosphorylation of Mig1, a known Snf1 substrate in response to glucose limitation (Figure 8C). On the other hand, if 2DG is added at the same time that cells are shifted to low glucose media, the presence of 2DG blocks the phosphorylation and activation of Snf1 (Figure 8, D and E). Thus we find 2DG increasing the phosphorylation of Snf1 when added to cells growing on high glucose and decreasing phosphorylation of Snf1 when cells are shifted to low glucose plus 2DG. The observation that addition of 2DG to cells growing on glucose causes an increase in Snf1 phosphorylation without any detectable increase in the phosphorylation of Mig1 is reminiscent of earlier studies with regard to sodium stress (McCartney and Schmidt 2001). When cells growing in high glucose are exposed to high concentrations of sodium, phosphorylation of Snf1 increases without any increase in the phosphorylation of Mig1 (McCartney and Schmidt 2001; Ye et al. 2008). Thus different stimuli (glucose stress, sodium stress, alkali stress, and 2DG stress) all increase the phosphorylation status of Snf1, but only glucose stress results in the increased phosphorylation of the Mig1 protein. The targets of Snf1 following sodium stress or 2DG stress have not been identified. The mechanism(s) by which Snf1 kinase determines the appropriate substrates when activated by different stimuli is unknown and awaits further study.
Acknowledgments
We thank Karen Arndt, Karin Elbing, Sidney Morris, Axel Schmidt, and Allyson O’Donnell for helpful discussions. We are indebted to Peg Shirra for help with the canavanine ploidy test and Fernando Moreno for the gift of HXK2 plasmids. This work was supported by National Institutes of Health grant GM46443.
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Apweiler E., Sameith K., Margaritis T., Brabers N., Van De Pasch L., et al. , 2012. Yeast glucose pathways converge on the transcriptional regulation of trehalose biosynthesis. BMC Genomics 13: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biely P., Kratky Z., Kovarik J., Bauer S., 1971. Effect of 2-deoxyglucose on cell wall formation in Saccharomyces cerevisiae and its relation to cell growth inhibition. J. Bacteriol. 107: 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., et al. , 2010. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3: rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun K. A., Vaga S., Dombek K. M., Fang F., Palmisano S., et al. , 2014. Phosphoproteomic analysis identifies proteins involved in transcription-coupled mRNA decay as targets of Snf1 signaling. Sci. Signal. 7: ra64. [DOI] [PubMed] [Google Scholar]
- Chandrashekarappa D. G., McCartney R. R., Schmidt M. C., 2013. Ligand binding to the AMP-activated protein kinase active site mediates protection of the activation loop from dephosphorylation. J. Biol. Chem. 288: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V., Qiu H., Hinnebusch A. G., 2010. Snf1 promotes phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell. Biol. 30: 2862–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Tharun S., 2009. Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1–7-Pat1 complex. RNA 15: 1837–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winde J. H., Crauwels M., Hohmann S., Thevelein J. M., Winderickx J., 1996. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur. J. Biochem. 241: 633–643 [DOI] [PubMed] [Google Scholar]
- Dombek K. M., Voronkova V., Raney A., Young E. T., 1999. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol. Cell. Biol. 19: 6029–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C., Chevalier A., Mann C., 2004. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol. Cell. Biol. 24: 2560–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K., 1980. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 177: 345–350 [DOI] [PubMed] [Google Scholar]
- Fisher C. L., Pei G. K., 1997. Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23: 570–574 [DOI] [PubMed] [Google Scholar]
- Hanau S., Montin K., Cervellati C., Magnani M., Dallocchio F., 2010. 6-Phosphogluconate dehydrogenase mechanism: evidence for allosteric modulation by substrate. J. Biol. Chem. 285: 21366–21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M., 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia M. F., Heredia C. F., 1988. Saccharomyces cerevisiae acquires resistance to 2-deoxyglucose at a very high frequency. J. Bacteriol. 170: 2870–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. P., Carlson M., 2007. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282: 16838–16845 [DOI] [PubMed] [Google Scholar]
- Hong S. P., Leiper F. C., Woods A., Carling D., Carlson M., 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. F., 1968. Lysis of yeast cell walls induced by 2-deoxyglucose at their sites of glucan synthesis. J. Bacteriol. 95: 1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech A., Nath N., McCartney R. R., Schmidt M. C., 2003. Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot. Cell 2: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. N., Johnston M., 1997. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16: 5629–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney R. R., Schmidt M. C., 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276: 36460–36466 [DOI] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M., 1987. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115: 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M., Barrett L., Kuchin S., 2008. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25: 745–754 [DOI] [PubMed] [Google Scholar]
- Pelaez R., Herrero P., Moreno F., 2010. Functional domains of yeast hexokinase 2. Biochem. J. 432: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H., Martin D. S., Xu R. H., Huang P., 2006. Glycolysis inhibition for anticancer treatment. Oncogene 25: 4633–4646 [DOI] [PubMed] [Google Scholar]
- Portillo F., Mulet J. M., Serrano R., 2005. A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett. 579: 512–516 [DOI] [PubMed] [Google Scholar]
- Raez L. E., Papadopoulos K., Ricart A. D., Chiorean E. G., Dipaola R. S., et al. , 2013. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 71: 523–530 [DOI] [PubMed] [Google Scholar]
- Ralser M., Wamelink M. M., Struys E. A., Joppich C., Krobitsch S., et al. , 2008. A catabolic block does not sufficiently explain how 2-deoxy-D-glucose inhibits cell growth. Proc. Natl. Acad. Sci. USA 105: 17807–17811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randez-Gil F., Prieto J. A., Sanz P., 1995. The expression of a specific 2-deoxyglucose-6P phosphatase prevents catabolite repression mediated by 2-deoxyglucose in yeast. Curr. Genet. 28: 101–107 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. (Editors), 1990. Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schild D., Ananthaswamy H. N., Mortimer R. K., 1981. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics 97: 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. C., McCartney R. R., 2000. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19: 4936–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller H. J., Entian K. D., 1991. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J. Bacteriol. 173: 2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra M. K., McCartney R. R., Zhang C., Shokat K. M., Schmidt M. C., et al. , 2008. A chemical genomics study identifies Snf1 as a repressor of GCN4 translation. J. Biol. Chem. 283: 35889–35898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. C., Davies S. P., Wilson W. A., Carling D., Hardie D. G., 1999. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 453: 219–223 [DOI] [PubMed] [Google Scholar]
- Sutherland C. M., Hawley S. A., McCartney R. R., Leech A., Stark M. J., et al. , 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Tabba S., Mangat S., Mccartney R., Schmidt M. C., 2010. PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase. Cell. Signal. 22: 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Dinh S., Wietzerbin J., Courtois A., Herve M., 1995. A novel approach for investigating reaction mechanisms in cells. Mechanism of deoxy-trehalose synthesis in Saccharomyces cerevisiae studied by 1H-NMR spectroscopy. Eur. J. Biochem. 228: 727–731 [DOI] [PubMed] [Google Scholar]
- Treitel M. A., Kuchin S., Carlson M., 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J., Carlson M., 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Ye T., Elbing K., Hohmann S., 2008. The pathway by which the yeast protein kinase Snf1p controls acquisition of sodium tolerance is different from that mediating glucose regulation. Microbiology 154: 2814–2826 [DOI] [PubMed] [Google Scholar]
- Young E. T., Zhang C., Shokat K. M., Parua P. K., Braun K. A., 2012. The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity, and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae. J. Biol. Chem. 287: 29021–29034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I., 1977. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol. Gen. Genet. 154: 75–82 [DOI] [PubMed] [Google Scholar]