Abstract
In eukaryotic cells, the production of cellular energy requires close interplay between nuclear and mitochondrial genomes. The mitochondrial genome is essential in that it encodes several genes involved in oxidative phosphorylation. Each cell contains several mitochondrial genome copies and mitochondrial DNA recombination is a widespread process occurring in plants, fungi, protists, and invertebrates. Saccharomyces cerevisiae has proved to be an excellent model to dissect mitochondrial biology. Several studies have focused on DNA recombination in this organelle, yet mostly relied on reporter genes or artificial systems. However, no complete mitochondrial recombination map has been released for any eukaryote so far. In the present work, we sequenced pools of diploids originating from a cross between two different S. cerevisiae strains to detect recombination events. This strategy allowed us to generate the first genome-wide map of recombination for yeast mitochondrial DNA. We demonstrated that recombination events are enriched in specific hotspots preferentially localized in non-protein-coding regions. Additionally, comparison of the recombination profiles of two different crosses showed that the genetic background affects hotspot localization and recombination rates. Finally, to gain insights into the mechanisms involved in mitochondrial recombination, we assessed the impact of individual depletion of four genes previously associated with this process. Deletion of NTG1 and MGT1 did not substantially influence the recombination landscape, alluding to the potential presence of additional regulatory factors. Our findings also revealed the loss of large mitochondrial DNA regions in the absence of MHR1, suggesting a pivotal role for Mhr1 in mitochondrial genome maintenance during mating. This study provides a comprehensive overview of mitochondrial DNA recombination in yeast and thus paves the way for future mechanistic studies of mitochondrial recombination and genome maintenance.
Keywords: mitochondrial genome, recombination, yeast, intraspecies variability, genome maintenance
MITOCHONDRIA (mt) are central organelles for various cellular functions including respiration, apoptosis, ion homeostasis, and metabolite biosynthesis. Cellular energy production through oxidative phosphorylation requires the involvement of factors encoded by two different genomes: one located in the nucleus and the other in mitochondria. The interplay between these two genomes is crucial to ensure functionality and these interactions have been associated with phenotypic diversity (Zeyl et al. 2005; Solieri et al. 2008) as well as speciation due to genetic incompatibilities (Lee et al. 2008). The mt genome is a remnant of an ancestral α-proteobacterial symbiont and has undergone an important size reduction by gene transfer to the nucleus (Wallace 2007; Gray 2012). Nevertheless, the cell requires mtDNA to ensure respiration, as it encodes essential subunits of the oxidative phosphorylation pathway. Unlike nuclear genomic DNA, multiple copies of mtDNA are contained in each cell, the number of which varies depending on the species, the tissue (Preuten et al. 2010), or the culture conditions (Shay et al. 1990; Hori et al. 2009). High mtDNA copy number provides a pool of templates for intermolecular recombination, and mt recombination is a widespread phenomenon described in plants (Arrieta-Montiel et al. 2009; Galtier 2011), fungi (Dujon et al. 1974; Fourie et al. 2013), protists (Gray et al. 1999), and invertebrates (Ladoukakis and Zouros 2001). In animal mtDNA, although the recombination machinery has been described (Lakshmipathy and Campbell 1999), the common belief was that mtDNA is clonally inherited and devoid of recombination. However, some recent studies have challenged this dogma and evidence of mt recombination has been found in the animal kingdom, from statistical analyses of mtDNA sequences (Eyre-Walker and Awadalla 2001) and direct measurements (D’aurelio 2004; Kraytsberg et al. 2004).
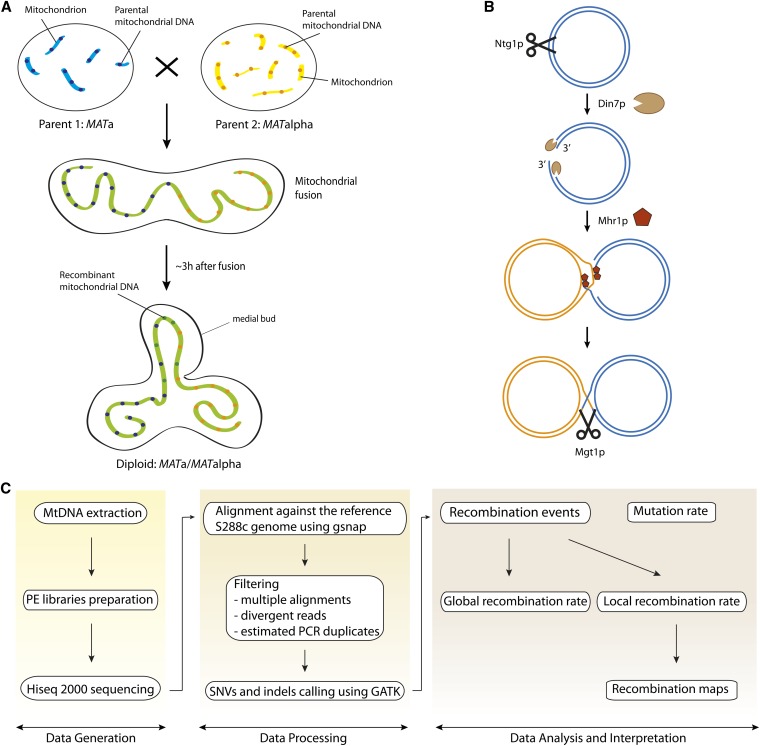
Saccharomyces cerevisiae has proved to be a relevant model for mitochondrial genetics due to its functional similarity with the human mtDNA and the ability to directly manipulate the mt genome. Yeast is also able to grow in the absence of functional mitochondria and to spontaneously produce respiratory-deficient mutants called petite mutants, in which the whole mtDNA or part of it is deleted. Additionally, the first mitochondrial genes to be sequenced originated from yeast and a full mtDNA genome sequence has been available since 1998 (Foury et al. 1998), a few years after the release of the nuclear genome (Goffeau et al. 1996). The major fraction of the yeast mt genome is organized as linear concatemers of 85-kb-long molecules suggested to originate from circular monomers (reviewed in Solieri 2010). Concatemers are predominantly present in mother cells and nondividing cells, while the monomers have been described to be located in the growing buds (Ling and Shibata 2002; Solieri 2010; Shibata and Ling 2007). The mt genome is composed of seven genes involved in oxidative phosphorylation (COX1, COX2, COX3, ATP6, ATP8, ATP9, and COB) and a gene coding for a ribosomal protein (VAR1). It also contains the mt small and large ribosomal RNA (rRNA) genes and 24 transfer RNA (tRNA) genes (Foury et al. 1998). Its copy number per cell ranges from 50 to 200 (Solieri 2010) and contrary to the mammalian mtDNA it is biparentally inherited after mating, allowing for recombination, although this heteroplasmic state is transient for only ∼20 generations. Yeast mt recombination has been described since the 1970s (Dujon et al. 1974). After mating, the parental mitochondria fuse, forming a single organelle throughout the newly formed zygote, with a rapid mixing of the mt proteins (Figure 1A). Subsequently, the parental mtDNAs are in the same compartment but they cannot diffuse freely through the mt reticulum, and the recombinants’ genome can thus be found only at the medial bud where both genomes are in direct physical contact (Figure 1A) (Strausberg and Perlman 1978; Zinn et al. 1987).
Figure 1.
Schematic representations of the mitochondrial recombination process and of the experimental and analysis workflow. (A) Mitochondrial recombination during mating (adapted from Chen and Butow 2005). When crossing two yeast cells of opposite mating type, mitochondria fuse to form a long reticulum, spreading within the zygote. MtDNA molecules do not move freely within this reticulum and the two parental mtDNA molecules are in contact only at the medial bud, where recombinant molecules can thus be found. (B) Mechanism of mtDNA recombination. Homologous recombination is initiated by a DSB generated by Ntg1. The 5′ ends of the molecules are then resected by Din7, leading to 3′ overhangs, which can then invade the homologous template under the action of Mhr1. This leads to Holliday junctions that can then be resolved by Mgt1. (C) MtDNA of the diploids pool was extracted and paired-end libraries were prepared and sequenced using a Hiseq 2000. The reads obtained were aligned against the reference S288c genome and subjected to several filtering rounds as well as SNPs and SVs calling, using GATK. The resulting reads were then scanned to detect recombination events, allowing us to determine the global recombination rate and to generate recombination maps of the local recombination rate. In parallel, the mutation rate was also estimated to compare it with the calculated recombination rates.
It has been proposed that mt homologous recombination is initiated by a double-strand break (DSB) generated by the repair enzyme Ntg1 (Figure 1B) (Ling et al. 2007). The 5′ ends at the DSB are then resected by the mt nuclease Din7, thereby producing 3′ overhangs (Mieczkowski et al. 1997; Fikus et al. 2000; Ling et al. 2013). These tails may then be bound by the single-strand binding protein Mhr1, which promotes strand invasion and homologous pairing (Ling and Shibata 2002). This step results in the formation of a Holliday intermediate that can be resolved by the cruciform-cutting endonuclease Mgt1 (also called Cce1) (Kleff et al. 1992; Lockshon et al. 1995). Despite the common steps to meiotic recombination, no proteins seem to be shared between the two processes. Several studies have focused on a better characterization of the mt recombination pathways over the years, but only very few proteins have been found and many factors still remain unknown. Moreover, they have mostly relied on the analysis of specific loci such as the 21S rRNA, COB, and VAR1 (Zinn et al. 1987) or on synthetic model systems (Ling et al. 1995, 2000, 2013), such that a genome-wide overview of mitochondrial recombination is still missing.
In this study, we present the first genome-wide map of mtDNA recombination in yeast. To do so, we first generated pools of diploids originating from a cross between two S. cerevisiae strains, SaUH (S) and YJM789 (Y). We then performed high-throughput sequencing of the mtDNA extracted from these pools and inferred recombination events by analyzing genotype switches within read pairs. This approach also allowed us to assess the impact of intraspecies variability on the occurrence of mt recombination, by comparing this map with one generated for a cross between the S288c and SK1 strains. Finally, to further comprehend the mechanisms underlying this phenomenon, we determined to what extent deleting recombination-associated genes affected the recombination profiles.
Materials and Methods
Yeast strains
All the strains used in this study are listed in Supporting Information, Table S1. The SaUH, SaUK, KalphaUH, and YJM789 strains were chosen due to their additional auxotrophies compared to the original S288c, SK1, and YJM789 strains. Given our experimental setting, we required strains with complementary auxotrophies to be able to select for diploids directly on plates. The mutant strains were constructed by deletion of the gene of interest in the SaUH and YJM789 strains and its replacement with the CORE cassette KanMX-klURA3 (Storici et al. 2001) by standard yeast chemical transformation procedures (Gietz and Schiestl 2007a,b). The positive transformants were selected on YPD (10 g/liter yeast extract , 20 g/liter bactopeptone, 20 g/liter dextrose ) plus geneticin (G418, 400 µg/ml) and were confirmed by colony PCR.
Mating and selection of diploids
The two parental haploid strains were grown overnight in 5 ml YPD. One hundred microliters of each cell suspensions were plated on YPD agar. The cells were incubated at 30° for 24 hr to allow them to mate. The cell layer was then scraped from the plate and resuspended in 1 ml sterile water. Two hundred microliters of this suspension were plated on selective media to allow for the selection of diploid cells. For the SaUH × YJM789-derived crosses, the cells were grown on synthetic dextrose medium (1.7 g/liter YNB without amino acids, 20 g/liter dextrose) and on synthetic dextrose medium supplemented with uracil (100 µg/ml) for the SaUK × KalphaUH cross. The cells were grown for 48 hr at 30°, scraped from the plates, and resuspended in 1 ml water. The cells were harvested by centrifugation for 3 min at 6000 rpm and washed once with 1 ml water. The cell pellet was either processed directly for mitochondrial DNA extraction or stored at −20° for future use. Three independent crosses and diploid selections were performed for each background, allowing us to compare biological replicates.
Mitochondrial DNA extraction
The mitochondrial DNA extraction protocol was adapted from Defontaine et al. (1991). This protocol allowed us to enrich for mitochondrial DNA compared to genomic DNA (Table S2). The cell pellet was washed once in water and once in wash buffer [1.2 M Sorbitol, 50 mM Tris (pH 7.5), 50 mM EDTA, 2% β-mercaptoethanol]. The pellet was then resuspended in 1 ml Solution A [0.5 M Sorbitol, 50 mM Tris (pH 7.5), 10 mM EDTA, 2% β-mercaptoethanol], 100 units Zymolyase 100T (Amsbio) were added, and the cells were incubated at 37° for 45 min under gentle horizontal shaking (400 rpm). The suspension was then centrifuged at 4000 rpm for 10 min. The supernatant was transferred to a new 1.5-ml tube and centrifuged at 14,000 rpm for 15 min. The crude mitochondrial pellet was washed three times with 1 ml Solution A, resuspended in 400 µl Solution B [100 mM NaCl, 10 mM EDTA, 50 mM Tris (pH 8)], and incubated at room temperature for 30 min to allow for lysis. The mtDNA was then purified by phenol-chloroform extraction followed by ethanol precipitation. The mtDNA pellet was resuspended in 50 µl EB buffer (10 mM Tris, pH 8). RNAs were degraded by RNAse cocktail treatment for 1 hr at 37°. Concentrations were measured using Nanodrop (Thermo Scientific). Usually, 1–10 µg of mtDNA was extracted using this protocol.
Library preparation and sequencing
The mitochondrial DNA libraries were generated using a protocol adapted from Wilkening et al. (2013). One to 10 µg of mitochondrial DNA was sheared using the S220 Focused ultrasonicator (Covaris) with the following conditions: duty cycle, 20%; intensity, 5; cycles per burst, 200; duration, 45 sec; temperature, 4°. The 350-bp fragments were purified using 1 vol Agencourt AMPure XP beads (Beckman Coulter). The fragments were submitted to end repair, A addition, and adapter ligation with enzyme heat inactivation after each step. The samples were purified using 1 vol Agencourt AMPure XP beads and the libraries were amplified by PCR (98° for 45 sec; 12 cycles of 98° for 15 sec, 65° for 30 sec, and 72° for 30 sec; and 72° for 5 min). This step was performed using 2–10 ng of DNA, 2× Phusion High Fidelity PCR Master Mix (New England Biolabs, Beverly, MA), and 0.2 µM PE1.0 and PE2.0 primers (Illumina, San Diego, CA). The PCR products were then cleaned with 1 vol Agencourt AMPure XP beads and the yield was quantified with a Qubit Fluorometer (Life Technologies, Carlsbad, CA). Up to 13 samples were pooled and purified for 400- to 450-bp fragments on an E-gel (Life Technologies, Carlsbad, CA). The multiplexed libraries were sequenced on a Hiseq 2000, yielding between 2.8 and 42 million reads per sample (Table S2).
Sequence alignment and read filtering
For each sample, the sequencing reads were aligned to the S288c reference genome, using GSNAP version 2011-12-28 (Wu and Watanabe 2005), allowing for gapped alignments and up to four mismatches. Each alignment data set was then filtered for pairs of reads with only one unambiguous alignment and convergent orientation (inward-facing read pairs). Estimated PCR duplicates (pairs of reads defining fragments with identical start and stop mapping positions) were also filtered out.
Detection of single-nucleotide polymorphisms and small structural variants calling
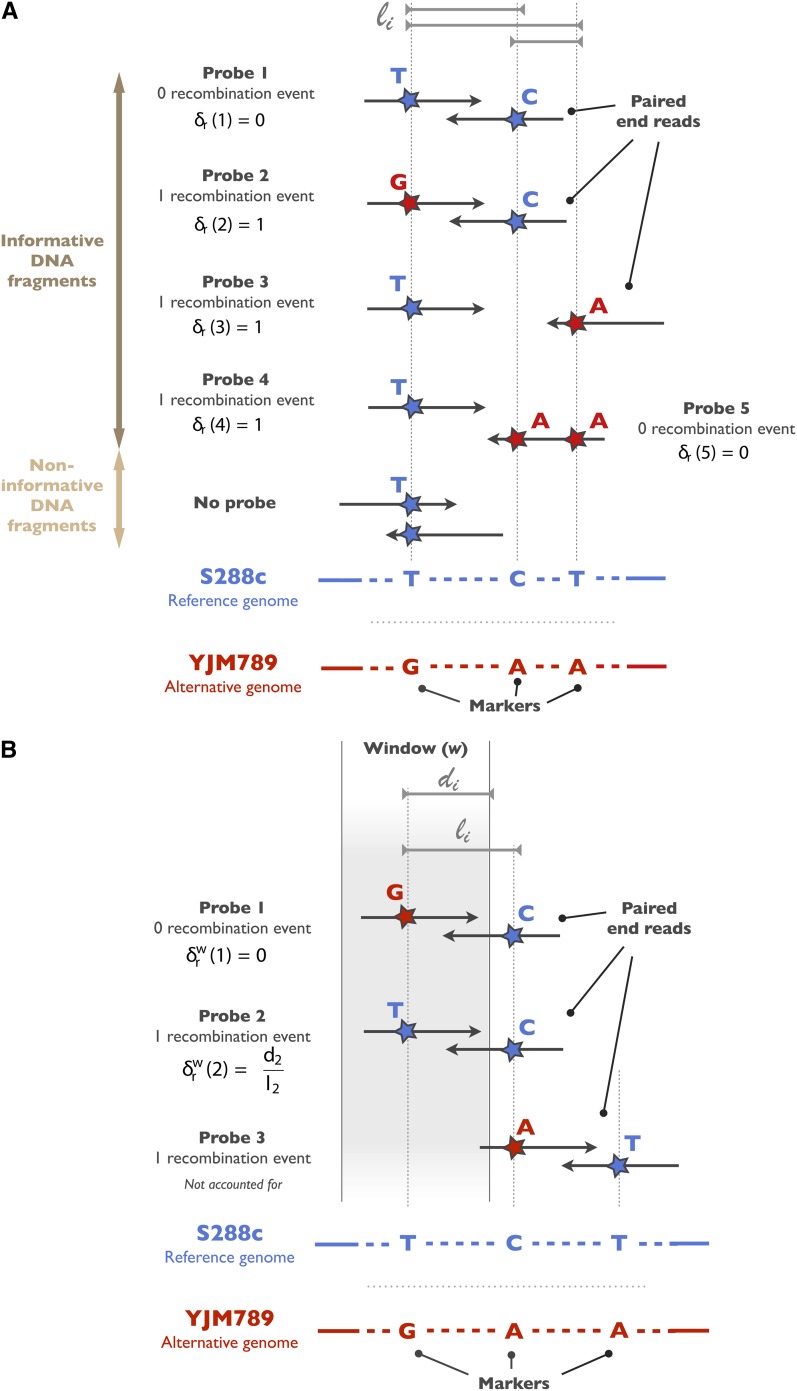
We identified single-nucleotide polymorphisms (SNPs) and small structural variants (SVs) in a genome-wide fashion, using the Unified Genotyper variant caller from GATK (McKenna et al. 2010). The vcf files obtained as output were then filtered out to keep only the SNPs and SVs that exhibited a coverage of >100×. In addition, for the parental genomes only the variants with an allele frequency of 1 were kept, as these strains are clonal and haploid and should therefore have only one single genotype for a given position. These SNPs and SVs are referred to as markers in the following sections (Figure 2).
Figure 2.
Determination of the global and local recombination rates. (A) Determination of the global recombination rate. Each read was scanned and a genotype was assigned at each marker position. Informative DNA fragments were defined as pairs of reads overlapping at least two markers. For each of these DNA fragments, we defined a probe i of length li as the region encompassed between two consecutive markers. A given fragment can thus contain several probes as represented for probes 4 and 5. If a genotype switch occurred at the extremities of the probe, as represented for probes 2–4, we concluded that a recombination event occurred within the probe. The recombination indicator δr(i) of probe i takes the value 1 if a recombination event occurred and 0 otherwise. The global recombination rate was then calculated as the ratio of the sum of the recombination indicators and the sum of probe lengths. (B) Determination of the local recombination rate. The local recombination rate was calculated for 1-kb windows shifted by 50 bp. Similarly to the global recombination rate, we determined probes, for which the window-recombination indicator δrw(i) was estimated. For each probe, the window-recombination indicator was then defined as 0 in the absence of a recombination event or as the ratio between di, the overlap length between the probe and the window, and li, the probe length, in the case of recombination. The local recombination rate was then calculated as the sum of the recombination indicators within a given window over the total overlapping probe length.
Determination of the coverage and variant density
To evaluate the coverage along the genome, we first binned the mitochondrial genome into adjacent 1-kb windows. We determined the boundaries of the genomic interval delimited by each pair of reads and evaluated the proportion of this interval overlapping with the aforementioned windows. These values were then summed over all the reads for each window and the final counts were plotted against the genomic coordinates to obtain a map of coverage (Figure S1).
To determine the variant density along the genome, the mitochondrial genome of the parental strains was binned into adjacent 1-kb windows. We counted how many variants fell in each window and plotted these values against the genomic coordinates to generate a map of variant density for the parental genomes (Figure S2). These maps were then used as a reference for the subsequent steps of genotype assignment of the reads originating from the different crosses.
Determination of the global recombination rates
Each read was scanned to determine its overlap with SVs and SNVs, thus allowing us to assign a genotype (reference or alternative) at each marker position in the read. Informative DNA fragments were defined as the pairs of reads spanning at least two markers (Figure 2). For a given fragment, we isolated the positions of consecutive markers; these two positions on the DNA fragment define a probe. It should be noted that a given fragment can contain several probes if it maps to a genomic locus containing more than two markers (Figure 2). To estimate whether a recombination event occurred within the probe, we determined the genotype at the probe extremities and concluded that a recombination event took place if a genotype switch was detected.
We denote by li the length of a probe i and δr(i) the recombination indicator for a probe i. δr(i) takes the value 1 when probe i contains a recombination event and 0 otherwise. We then define the global mitochondrial recombination rate as follows:
Determination of the local recombination rates
To generate a map of local recombination rates across the mitochondria, the mitochondrial genome was binned into 1-kb windows, shifted by 50 bp. To compute the recombination rate in a window, we examined the potential overlap between each probe and the considered window. When such an overlap existed, we denoted by di the length of that overlap. Using the same notations as introduced above for the global recombination rate, we defined a new recombination indicator δrw(i), which takes the value di/li if the probe delimits a recombination event within a window w and 0 otherwise (Figure 2). Such a definition was chosen to formalize the partial observation of a recombination event in a space-delimited window. Given the short size of the probes (usually <300 bp), we assumed that a unique recombination event occurs in fact within a probe and that its precise location is equiprobable along the probe. Therefore, we used the ratio of di over li to estimate the probability of a recombination event having occurred within the window w. The local recombination rate in the window was then computed as follows:
To mitigate the impact of lower coverage in some loci of the genome, we resampled (with replacement) 500,000 probes 1000 times and computed the local recombination rates for each of the probe sets. In each window, the median of all the recombination rates computed was used as a final value for the recombination map and all the subsequent analyses.
Determination of the mutation rate
We generated pileup files, which describe the base information at each chromosomal position, using the samtools mpileup command run on the final alignments. The mutation rate was then approximated as the ratio of the total number of bases that differ from the reference genome over the total number of bases that map perfectly to a genomic position. Bases mapping to positions of previously identified variants were excluded from that analysis. By processing only reads mapping to the nuclear or the mitochondrial genomes, we estimated the nuclear- and mitochondria-specific mutation rates (Table S3).
Determination of recombination hotspots
We used the MALDIquant package (Gibb and Strimmer 2012) to call recombination hotspot peaks. To use this package, we turned our data into mass objects, defining the positions as the masses and the local recombination rates as the intensities of these objects, analogous to mass spectrometry data. The global recombination rate was subtracted from the local recombination rates at each position, prior to a smoothing step performed with a Savitzky–Golay filter (using the MALDIquant defaults for the degree of the polynomial used in the filter, the parameter halfWindowSize was set to 15). Thus, the global recombination rates acted as a baseline in our analysis. Subtracting this baseline turned the recombination rates into hotspot rates. The detectPeaks function was then employed to detect peaks of the hotspot rates. Here the parameter SNR was set to 2 and the parameter halfWindowSize to 10. The boundaries of the hotspot regions were eventually defined as areas of consecutive non zero hotspot rates surrounding a peak.
Determination of differential hotspot regions
Using the reduce function from the IRanges package (http://www.bioconductor.org/packages/2.13/bioc/html/IRanges.html), we determined the common hotspot region boundaries between two selected samples of the two considered conditions (reference cross S×Y vs. either different background or mutants) as follows: we always selected the two samples displaying the highest number of peaks in the respective condition. The hotspot rates were further pretransformed using the Smithson and Verkuilen method (Smithson and Verkuilen 2006) to mitigate the influence of near zero rates. The rates of the three replicate samples per hotspot for a given cross were then pooled and considered as a single sample from a population. To apply the appropriate variance model for rate comparisons, we used a β-regression model to determine the hotspot regions showing statistically significant differences in hotspot rates between the two conditions. Using the “betareg” R package (Grün et al. 2011), we fitted an ANOVA model with “treatment contrast” parameterization and tested the coefficient presenting the fold change between the two conditions for significance, using a Wald test for each hotspot separately. The P-values were then adjusted using the Benjamini–Hochberg method to control the false discovery rate (FDR) (Benjamini and Hochberg 1995). We chose an FDR of 5% as a significance cutoff.
Results
Estimating the recombination rates of mitochondrial DNA
To achieve a detailed characterization of recombination events, we defined positions allowing us to distinguish between DNA fragments originating from the two parental strains, SaUH (S) and YJM789 (Y) (Table S1). Paired-end (PE) libraries from the parental mtDNAs were sequenced using next-generation technologies and the reads were aligned to the S288c mitochondrial reference genome, yielding 2,046,919 and 543,697 pairs of usable reads for S and Y, respectively (Figure 1C, Table S2). This extremely high genomic coverage for mitochondria (>1000×) allowed us to precisely call for variants corresponding to SNPs, as well as short deletions and insertions (<30 bp) (Figure S1A, Table S4, and Table S5). We thus defined 607 variants across the whole mitochondrial genome, which provided a marker every 140 bp on average. Some fluctuations in the marker distribution were observed, especially in the COX1 locus, which presented a lower density compared to the rest of the genome (Figure S2A). Despite these differences between loci, the number of markers was still sufficient to successfully detect recombination events across 1-kb sliding windows along the 85-kb genome.
MtDNA recombination occurs during mating of two yeast strains and can be detected in the resulting diploids (Figure 1A). In this study, we generated three independent pools of diploids from a cross between the S and Y strains. MtDNA of these pools was extracted and sequenced using deep-sequencing technology, yielding ∼650,000 usable pairs of sequencing reads after filtering the alignments against the S288c mitochondrial reference genome (Table S2). This corresponded to a sequencing coverage of >1000× on average, with few less-represented regions, yet covered by at least 100× (Figure S1C). The reads were then scanned to determine their overlap with the 607 markers and a genotype (S or Y) was assigned at each marker position (Figure 2). Pairs of reads covering at least two markers were defined as informative DNA fragments and each of these fragments was subdivided into probes corresponding to regions between two consecutive markers (Figure 2). We reasoned that a recombination event took place when a genotype switch was observed between two consecutive markers of a given probe. This allowed us to define the global recombination rate as the total number of recombination events over the total length of all examined probes. This recombination rate calculation differs from the standard nuclear meiotic recombination rate calculation in that it is expressed in number of events per sequenced kilobase and does not take into account the number of generations as the analysis is performed on a pool of diploid cells, with multiple mtDNA molecules present per cell.
For the S×Y crosses, the average global recombination rate was approximately three recombination events per kilobase (Table S3). To confirm that these events did not result from DNA template switching during the PCR amplification step, we prepared two independent PE libraries of the parental mtDNAs and pooled them prior to the final library amplification. This resulted in <0.01% of chimeric reads, indicating that the recombination events detected in the diploids were not artifactual, but of biological origin. As an additional control, we compared the recombination rates obtained with the mutation/sequencing error rate. We estimated this error rate as the ratio of the total number of bases differing from the reference genome over the total number of bases provided by the sequencing run (Table S3). As this rate was of the order of 10−5 mutations per kilobase, we considered it negligible in the calculation of the global recombination rate.
Hotspots are preferentially localized in intergenic and intronic regions
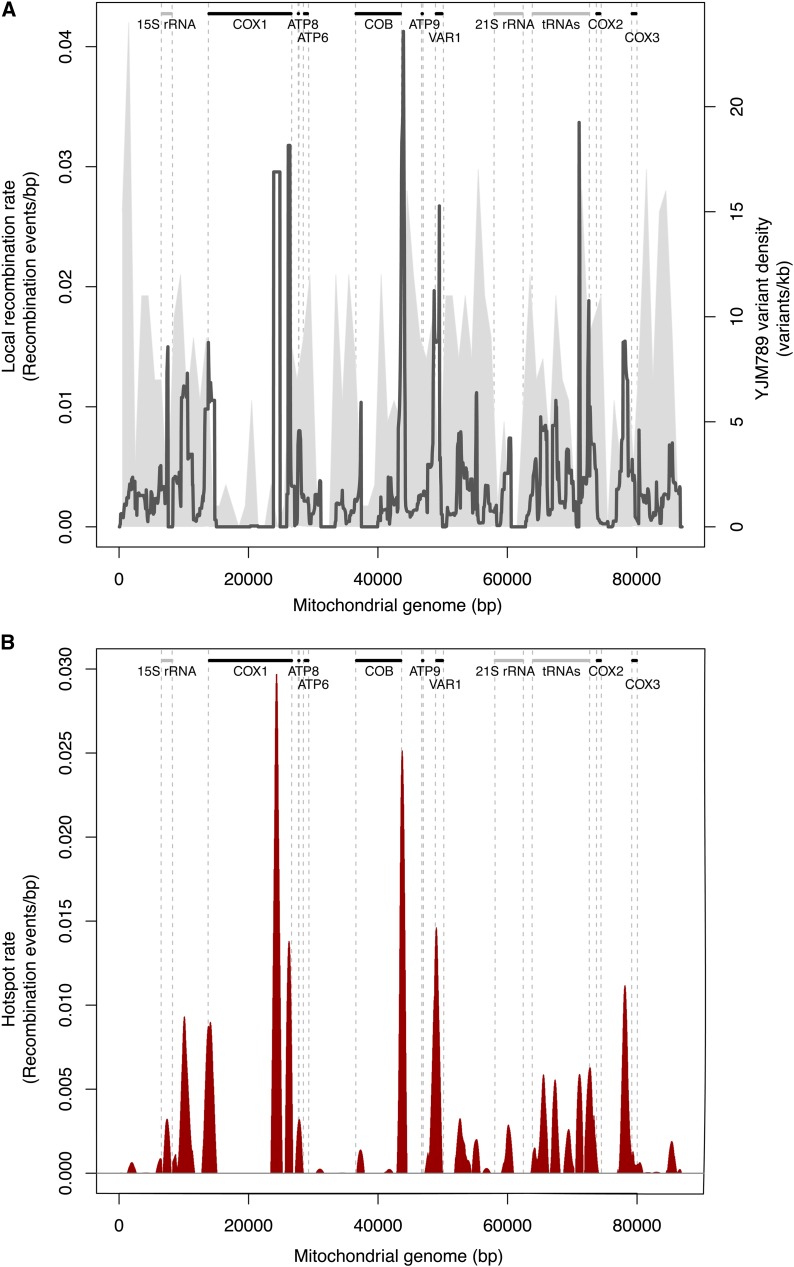
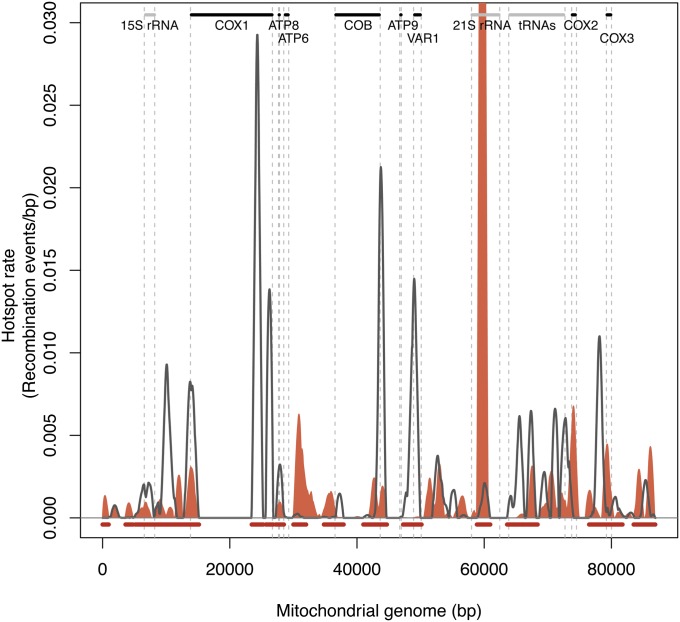
To evaluate the local recombination rates within 1-kb windows shifted by 50 bp, we took into account the partial overlap of the probes with the window intervals. The recombination rates were thus calculated as the total fraction of recombination events over the total probe length within each window (Figure 2). We generated a genome-wide map of mitochondrial DNA recombination in yeast by plotting the local recombination rates along the whole mitochondrial genome (Figure 3A). The maps obtained for the three S×Y biological replicates were highly reproducible, with a Spearman correlation coefficient >0.9, although some differences could still be observed in the profiles (Figure S3, A and B). The partially uneven distribution of the variants across the genome did not hinder the detection of recombination events as some regions with high variant density displayed very few recombination events, whereas some of the low-density regions were highly recombinogenic (Figure 3A). Although recombination events occurred throughout the entire genome, some regions were enriched, suggesting the existence of preferential recombinogenic sites. We therefore defined mitochondrial recombination hotspots as intervals involving more recombination events than predicted under a homogeneous genomic rate (approximately three recombination events per kilobase for the S×Y crosses). With this method, 23 hotspots were found, potentially encompassing multiple peaks (Figure 3B).
Figure 3.
Recombination profile for the S288c×YJM789 cross. (A) The local recombination rate was calculated for 1-kb windows shifted by 50 bp and is plotted as a black line. The variant density observed over the whole mitochondrial genome is represented in gray. The main yeast mitochondrial features are also indicated, with protein-coding genes represented as black bars and non-protein-coding genes as gray bars. (B) Recombination hotspot profile for the S×Y cross is plotted in red. The hotspot rate is calculated by subtracting the global recombination rate from the local one. The mean of the three replicates is plotted. Information concerning the standard deviation for the three replicates is indicated in Table S8. Mitochondrial protein-coding genes are indicated as black bars and non-protein-coding genes as gray bars.
We next analyzed in more detail the localization of hotspots with regard to annotated genetic features. Among the detected 23 hotspots, 15 (65%) lay within intergenic regions (Figure 3B) and one-third of these intergenic peaks occurred within the 8.8-kb tRNA gene cluster, suggesting that these clusters are highly recombinogenic compared to the rest of the genome. This result could partly be explained by the presence in these regions of repetitive and palindromic elements, which are sequences that can preferentially be used as templates for recombination. Additionally, we found one hotspot in each of the rRNA regions (15S and 21S). This observation is in agreement with previous studies reporting recombination between distinct yeast 21S rRNA alleles (Zinn and Butow 1985; Zinn et al. 1987).
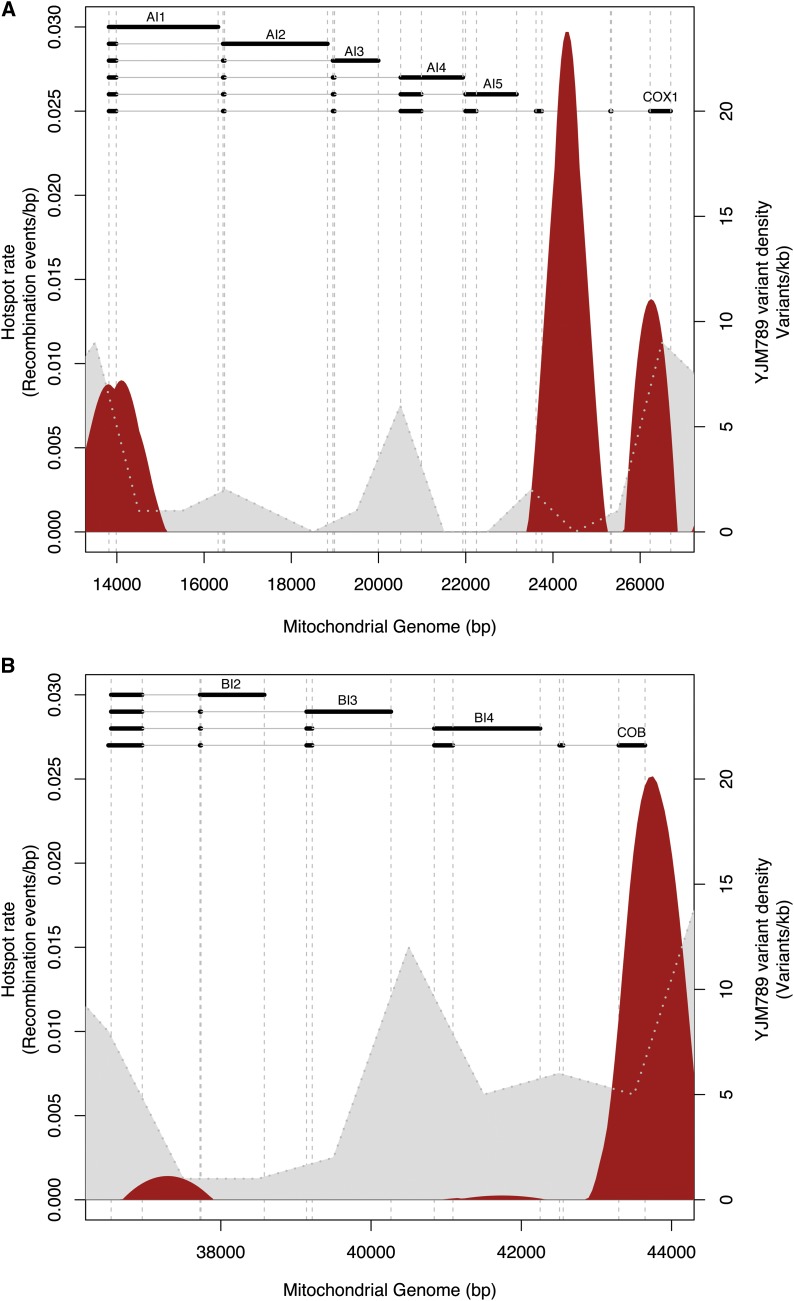
Recombination hotspots were also detected within three of the annotated genes: COX1, COB, and VAR1 (Figure 3B). For COX1, the major hotspot was located in the sixth intron of the gene (Figure 4). The second COX1-specific recombination peak was detected at the last intron–exon junction, while the third hotspot extends from the first COX1 exon until the first exon–intron junction, potentially disrupting this first exon. Three peaks could also be detected in COB; the major hotspot encompasses the last exon of COB but peaks in the intergenic region downstream of the gene (Figure 4). The two other peaks were situated in intronic regions of the gene. Recombination has previously been described for VAR1 and COB (Zinn et al. 1987) and homologous recombination within COX1 has already been performed to engineer strains with modified mitochondrial genomes (Pérez-Martínez et al. 2003). However, the present study is the first to report recombination hotspots in these loci. Altogether, these results suggest that recombination occurs preferentially in intronic and intergenic regions, thus avoiding potential gene disruption.
Figure 4.
Recombination profiles observed at the COX1 and COB genes for the S288c×YJM789 cross. The recombination hotspots are indicated in red, overlapping the variant density plotted in gray. This profile corresponds to the mean of the three replicates. Information concerning the standard deviation for the three replicates is indicated in Table S8. The different isoforms of COX1 and COB are represented with exons as black bars and introns as gray lines. (A) Recombination profile at the COX1 locus. (B) Recombination profile at the COB locus.
Intraspecies variability influences recombination hotspot profiles
To assess the impact of intraspecies variability on mtDNA recombination, we determined the recombination rates and profiles of a different cross of S. cerevisiae strains: SaUK isogenic to SaUH, differing from it only by its auxotrophies, and KalphaUH (K) (Table S1). Similarly to the analysis performed for the parental S and Y strains, we defined a set of genetic markers that could be used to detect recombination events. We sequenced the mtDNA of the SaUK and K strains and aligned the reads to the S288c genome, yielding >2,600,000 and 980,000 pairs of reads, respectively, after filtering, which corresponds to a coverage of >2000× (Table S2). Although the coverage for the K strain was high across most of the genome, two regions showed a much lower coverage (∼10×) (Figure S1B). However, this should not affect our estimations of local recombination rates, given the use of 1-kb sliding windows. We called for variants and defined 660 markers across the mitochondrial genome, providing one marker every 130 bp on average (Table S6 and Table S7). Similarly to the variant density in YJM789, the markers were not uniformly distributed over the genome but in sufficient numbers for an effective detection of the recombination events (Figure S2B). Although the variant density profiles appeared different between KalphaUH and YJM789, the same regions were covered and we therefore considered that the effect on recombination event detection was negligible (Figure S2, A and B).
We then generated three independent pools of diploids for an S×K cross, from which mtDNA was extracted and PE libraries were prepared. Sequencing these libraries yielded ∼692,000 and ∼960,000 pairs of reads for two of the replicates after filtering, corresponding to a coverage of >1600×. For the third replicate, only ∼59,000 pairs of reads were recovered after filtering, leading to a lower coverage of ∼138× (Table S2, Figure S1D). To detect recombination events, we applied the same analysis pipeline as described for S×Y. The global recombination rate was estimated to be ∼2.3 recombination events per kilobase and was thus lower than the S×Y rate (∼3 recombination events per kilobase) (t-test P = 0.006653; Figure S4, Table S3).
We determined the local recombination events for 1-kb sliding windows shifted by 50 bp and generated a recombination map for the S×K cross (Figure 5). The local recombination maps of the two replicates with the highest coverage were highly reproducible, with a Spearman correlation coefficient >0.9 and 26 common peaks (Figure 5 and Figure S3, A and B). The map of the third replicate showed less reproducibility (Spearman’s coefficient ∼0.7), probably due to the lower coverage (Table S2, Figure S1D).
Figure 5.
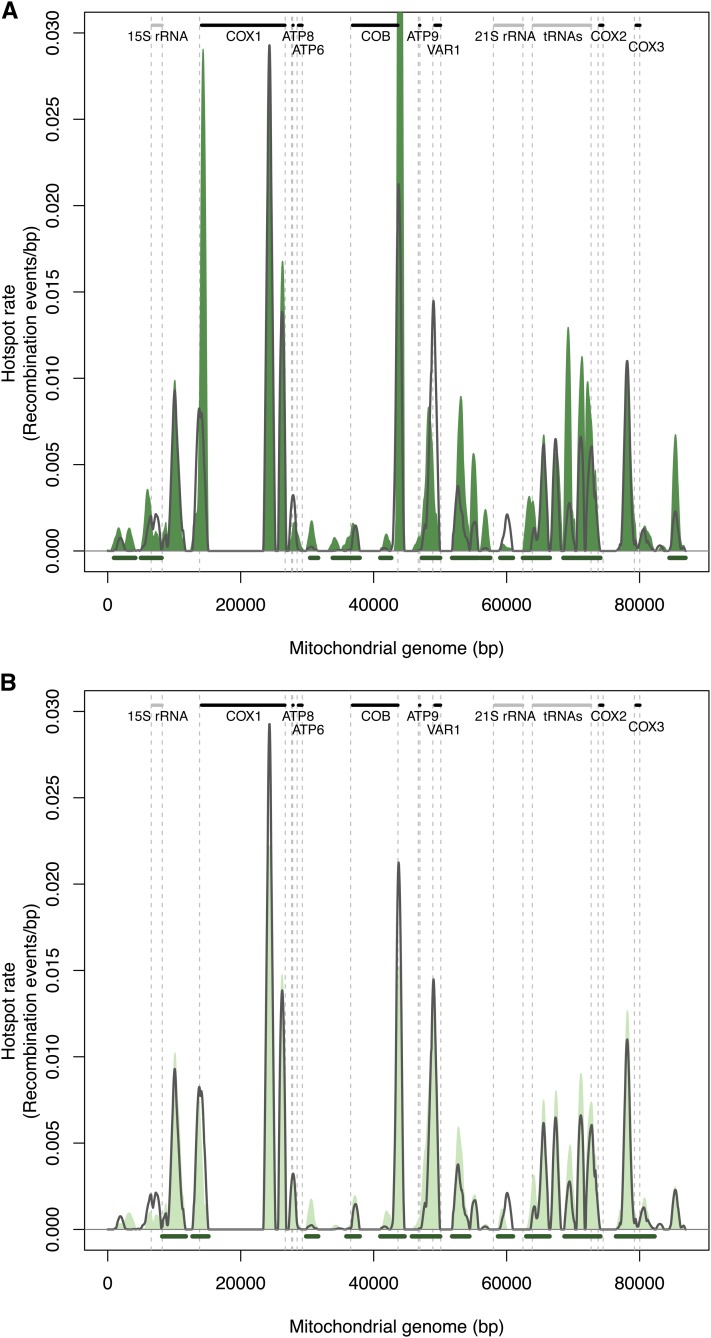
Recombination landscape comparison between the S288c×YJM789 and the S288c×SK1 crosses. The recombination hotspot profile obtained for the S×K cross is represented in orange and the S×Y profile as a black line. These profiles correspond to the mean of the three replicates. Information concerning the standard deviation for the three replicates is indicated in Table S8. Statistically significantly different hotspots at an FDR of 5% are underlined in red. Mitochondrial protein-coding genes are indicated as black bars and non-protein-coding genes as gray bars.
Comparison of S×K and S×Y recombination profiles showed that conserved peaks generally displayed a lower intensity, except for the hotspot located within the 21S rRNA locus. This is in concordance with the fact that the global recombination rate was slightly lower in S×K. Moreover, several hotspots were absent in S×K, notably the two major peaks in COX1 and the main peak at the exon–intron junction of COB as well as the one in VAR1. The intronic regions of COX1 have previously been described to be variable between different S. cerevisiae strains (Wei et al. 2007); however, in the case of SK1, these regions are highly similar (>90%) to the ones from S288c and thus the lack of recombination in these regions cannot be explained by sequence variations. One peak was strictly S×K specific and was located in the intergenic region between ATP6 and COB. We also observed that the recombination profiles in the tRNA gene cluster were different between the two crosses. Both regions were recombinogenic but different sequences seem to have been used as templates (Figure 5).
Our results provide evidence that the genetic background influences mitochondrial recombination, leading to variations in the global rates as well as affecting the hotspot localization observed throughout the mt genome.
Analysis of strains deleted for genes implicated in mtDNA recombination suggests the involvement of additional factors
To determine the impact of proteins involved in the main steps of the mtDNA recombination pathway on recombination profiles, we generated deletions of four genes, NTG1, DIN7, MHR1, and MGT1, in both S and Y backgrounds (Figure 1B, Table S1). NTG1 encodes a DNA glycosylase and apurinic/apyrimidic (AP) lyase. It has been described to create double-strand breaks (DSB) in the mitochondrial DNA, thus stimulating mtDNA recombination and replication (Ling et al. 2007; Hori et al. 2009). Din7 was identified as a mitochondrial nuclease involved in DNA repair and replication and has been proposed to be the exonuclease involved in the generation of 3′ single-strand tails from DSBs, required for homologous pairing in mitochondria (Mieczkowski et al. 1997; Fikus et al. 2000; Ling et al. 2013). MHR1 was identified to catalyze homologous pairing of single-strand DNA overhangs on the homologous template (Ling et al. 1995; Ling and Shibata 2002). Finally, MGT1 encodes a mitochondrial cruciform cutting endonuclease, which resolves the Holliday junctions formed during mtDNA recombination (Kleff et al. 1992; Ezekiel and Zassenhaus 1993; White and Lilley 1997). These deletion strains were crossed with the parental counterpart strain to generate three independent pools of diploids per background. As previously described, the mtDNA of these pools was extracted and sequenced. However, no libraries were generated from the S∆din7×Y∆din7 background, as most of the mtDNA was repeatedly lost after the fragmentation step, suggesting a potential degradation of the mtDNA .
For S∆mgt1×Y∆mgt1, between 102,586 and 261,198 pairs of reads were recovered after filtering, leading to a coverage ranging from 239× to 609× (Table S2, Figure S1E). The global recombination rate for this cross was estimated to be ∼3.2 recombination events per kilobase, showing no statistically significant difference with the global rate we calculated for S×Y (t-test, P = 0.7735; Figure S4). Sequencing the S∆ntg1×Y∆ntg1 diploid pools yielded between 821,782 and 1,378,186 pairs of reads and a genomic coverage of >1900× (Table S2, Figure S1F). The global recombination rate was ∼2.2 recombination events per kilobase, lower than the one observed for the S×Y background (3 recombination events per kilobase, t-test, P = 0.009462; Table S3, Figure S4). This observation is in agreement with the fact that deletion of NTG1, which creates DSBs necessary for recombination initiation, should lead to a decrease in recombination events. However, as some events are still detected, our results suggest that NTG1 contributes but is not solely responsible for initiating recombination in the S×Y background.
For both ∆mgt1 and ∆ntg1 diploid backgrounds, local recombination profiles showed high reproducibility between biological replicates (Spearman’s correlation coefficient >0.9, Figure S5, B, D, and E) and were highly similar to the wild-type profile obtained for S×Y (Spearman’s correlation coefficient >0.9, Figure S5, A and E). Most of the hotspot positions were conserved except for the 21S rRNA peak, which was hardly detected for the S∆mgt1×Y∆mgt1 background (Figure 6A) and was lower and shifted in the S∆ntg1×Y∆ntg1 recombination profile (Figure 6B). The hotspot intensities, notably at the tRNA gene cluster and in the intergenic region between VAR1 and the 21S rRNA, were slightly enriched in the mutant profiles compared to the wild type. Altogether, our results indicate that the MGT1 or NTG1 deletions have little effect on recombination rates and profiles, suggesting that either these genes are not key players in the occurrence of recombination events or other enzymes are redundant with these two proteins and complement for their loss.
Figure 6.
Recombination landscape observed in the ∆mgt1 and ∆ntg1 diploid backgrounds. The recombination hotspot profiles are plotted in green for the mutant backgrounds and as a black line for the S×Y cross. Each profile corresponds to the mean of the three replicates. Information concerning the standard deviations for the three replicates is indicated in Table S8. Statistically different hotspots are underlined in green. The mitochondrial map is represented at the top with protein-coding genes in black and non-protein-coding genes in gray. (A) Hotspot landscape for the ∆mgt1 diploid mutant. (B) Hotspot profile for the ∆ntg1 diploid mutant.
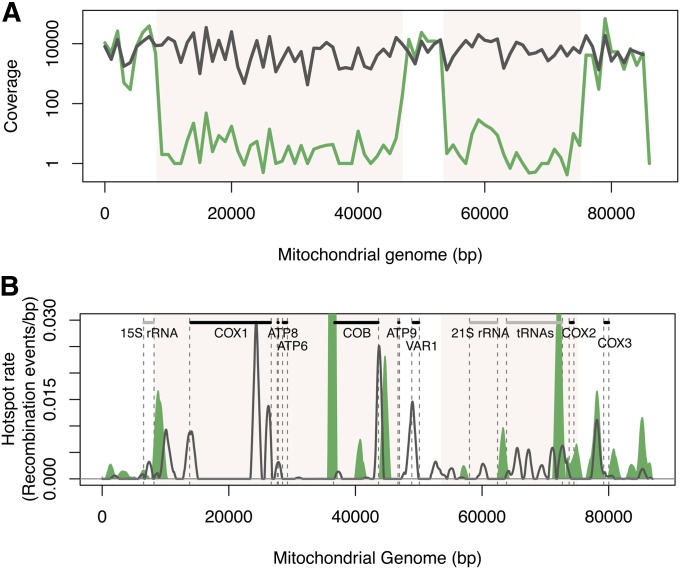
After sequencing the mtDNA of the S∆mhr1×Y∆mhr1 diploid pools, 102,714 and 312,264 pairs of usable reads were recovered, resulting in a coverage of >200× (Table S2). Interestingly, two large regions extending over >10 kb each displayed a coverage that was on average 1000-fold lower than in the rest of the genome, suggesting that these regions had been deleted (Figure 7A, Figure S1G). Analysis of coverage for parental deletion strains did not show any deletion, indicating that this event occurred during mating (Figure S1, H and I). Most of the mitochondrial gene features were absent with the exception of the 15S rRNA, VAR1, and COX3 (Figure 7B). As some of these preserved regions were recombinogenic in the S×Y wild-type background, the global recombination rate as well as the local recombination profiles were still determined for S∆mhr1×Y∆mhr1.
Figure 7.
Mitochondrial recombination in the ∆mhr1 diploid background. (A) Comparison of the coverage obtained after sequencing for the S×Y (gray) and the S∆mhr1×Y∆mhr1 (green) diploid pools. Regions of extremely low coverage are colored in beige and represent parts of the genome that were likely lost during the mating process. (B) Hotspot profile for the ∆mhr1 diploid mutant is plotted in green and the S×Y hotspot profile as a black line. Each profile represents the mean of the three replicates. Information concerning the standard deviation for the three replicates is indicated in Table S8. Regions of the genome that were lost in the S∆mhr1×Y∆mhr1 background according to the coverage analysis are colored in beige on the map. The mitochondrial map is indicated with protein-coding genes in black and non-protein-coding genes in gray.
The global recombination rates calculated for the three replicates were not consistent, ranging from 1.4 to 2.65 recombination events per kilobase (Table S3, Figure S4). Also, the local recombination profiles were poorly reproducible between replicates (Spearman’s correlation coefficient ∼0.6, Figure S5, D and F). Although some recombination peaks could be detected in the deleted regions, they do not represent real hotspots but arise from few reads that included recombination events in regions of very low coverage, which led to an overestimation of the recombination rates in those areas. Indeed, the coverage of the deleted regions is never equal to zero, suggesting that within the population, there might be a very small fraction of the cells that did not undergo the complete deletion event and therefore some potential recombination events can be detected. Nevertheless, we observed recombination peaks in the intergenic regions between COX2 and COX3, overlapping with a hotspot also present in the S×Y wild-type background. Two other recombination hotspots were also identified in the intergenic region downstream of COX3. Although both VAR1 and 15S regions were present in these mutants, no recombination hotspots were detected, in contrast to what was observed for S×Y. Our study demonstrates that deleting MHR1 has a strong impact on mtDNA maintenance, with the formation of partially deleted mitochondrial genomes due to large deletions. Nevertheless, in our experimental design, the cells are never isolated but always handled as a population and we could therefore not determine whether these deletions led to respiratory deficiencies or higher petite formation for the cells. Loss of MHR1 also affected the recombination profiles observed in the regions conserved between the mutant and the wild-type background. The invasion step is thus important for the determination of the recombination hotspots localization. Nevertheless, the fact that some hotspots were conserved suggests that this localization does not rely solely on the presence of MHR1.
Discussion
Mitochondrial recombination has been observed in many organisms, including several plants, fungi, and protists. In yeast mitochondria, recombination events have been described since the 1970s (Dujon et al. 1974; Strausberg et al. 1978; Zinn et al. 1987) and several mechanistic analyses have followed over the years (Ling et al. 1995, 2000, 2013; Phadnis et al. 2006). However, these studies focused on marker genes or relied on artificial systems. Here, we developed an analysis pipeline to generate the first genome-wide map of mitochondrial recombination. This approach also allowed us to assess the impact of intraspecies genetic variability on recombination profiles by comparing two different crosses of S. cerevisiae strains. Finally, we addressed the impact of recombination-associated genes on hotspot rate and localization.
Mitochondrial recombination occurs at a much higher frequency than mutations
Our calculation of recombination rate relies on the detection of markers along the genome to map genotype switches. Therefore, mutation and sequencing error rates can represent potential confounding factors. Particularly, if they are of the same order of magnitude as the recombination rate, it becomes difficult to disentangle the origin of events. In our study, we did not separate the mutation rate from the sequencing error rate and therefore we calculated an overall rate via an estimation of errors stemming from both sources. A previous study evaluated the yeast mitochondrial mutation rate to be 12.9 × 10−9 mutations per base pair per cell division (Clark-Walker 1991; Lynch and Blanchard 1998; Lynch et al. 2008). Due to our experimental setup, the number of generations undergone by the cells could not be measured precisely. However, we estimated this number to be between 40 and 50 generations, leading to a rate of ∼10−9 mutations and sequencing errors per base pair per cell division, in the same range as previously described but ∼100 times higher than the nuclear mutation rate. Several factors can explain this higher rate in mitochondria. First, mtDNA is subject to more replication rounds than the nuclear genome and may gather more mutations during this process. Second, mutations can accumulate due to DNA damage inflicted by the mitochondrial highly oxidative environment (Doudican et al. 2005). Finally, the mitochondrial DNA repair pathway has been shown to be less efficient than its nuclear counterpart (Song et al. 2005).
The analysis pipeline implemented in this study allowed us to evaluate the extent of recombination at the mitochondrial level in five different backgrounds. Globally, for the S×Y cross, this rate was approximately three recombination events per kilobase (Table S3). In previous studies, mitochondrial recombination has been described as a very efficient process, occurring more often than meiotic recombination, with up to 25% recombinants within the total population (reviewed in Dujon et al. 1974). However, these results are not directly comparable with ours as they rely on the analysis of marker genes and are not representative of a large population of diploids (millions in our case). Estimating the number of cells carrying recombinant mtDNA molecules in our study would have required microdissection and single-cell analysis of mother and daughter cells to obtain quantitative estimates of the segregation process. The recombination rate calculated in the present study was five orders of magnitude higher than the estimated mutation rate, and recombination should therefore be considered as a key player in genome reshuffling through the occurrence of rearrangements. These modifications can have drastic consequences, resulting in mitochondrial dysfunction (Bianchi et al. 2001; Lee et al. 2006), but may also drive evolution through the generation of novel DNA sequences (Shibata and Ling 2007; Arrieta-Montiel et al. 2009; Davila et al. 2011; Jung et al. 2012).
Mitochondrial recombination preferentially takes place in non-protein-coding regions
By determining the local recombination rate within 1-kb windows, we generated a genome-wide map for a cross between the SaUH and YJM789 strains. The distribution of the recombination events was uneven across the mitochondrial genome with enrichment in specific regions representing 23 hotspots. These hotspots were mostly localized in non-protein-coding regions, one-third of them being in the tRNA gene cluster (Figure 3). This observation could be explained by the presence of highly repetitive regions in these clusters. Although meiotic recombination between tRNA genes has also been reported (Munz et al. 1982; Pratt-Hyatt et al. 2006), genome-wide studies in yeast have not detected specific association of the tRNAs encoding regions with the presence of hotspots (Gerton et al. 2000; Mancera et al. 2008). Therefore, to our knowledge, it is the first time that a tRNA gene cluster is described as a recombination hotspot.
We also detected recombination in the rRNA regions (Figure 3). Particularly, in the 21S rRNA locus, a peak was detected at the exon–intron junction. This observation is in agreement with previous studies that have shown that different 21S rRNA alleles can recombine. This process was described to involve the SCE1 endonuclease gene encoded in the intron, which generates a double-strand break near the intron insertion site (Jacquier and Dujon 1985; Macreadie et al. 1985; Zinn and Butow 1985; Zinn et al. 1987). Hotspots were also detected in introns or in intron–exon junctions for COB and COX1 genes. Although recombination between different COB alleles and at COX1 has already been described (Zinn et al. 1987; Pérez-Martínez et al. 2003), this is the first time that recombination hotspots are reported in these loci.
The mitochondrial hotspots found in our study are prevalent in non-protein-coding regions. Similar observations have also been made for meiotic recombination, where hotspots occur more frequently in intergenic regions than within protein-coding genes (Baudat and Nicolas 1997; Gerton et al. 2000; Wahls et al. 2008; Buhler et al. 2009; Brick et al. 2012). Also, when they do occur within genes bodies, they are preferentially localized within introns (Kong et al. 2010). Our results thus indicate that, as observed for meiotic recombination, the localization of mitochondrial recombination hotspots is under selective pressure to preserve the sequence of the encoded proteins and to avoid the occurrence of potentially deleterious mutations or rearrangements, such as deletions, duplications, or inversions. A previous study reported that the RM11 yeast strain lacks the three last introns of COX1; recombination in these intronic regions is therefore unlikely to have a functional impact as they are not essential (Dimitrov et al. 2009). Additionally, as recombination can be a driving force for or in evolution, our observations agree with those of a previous genome-wide mitochondrial analysis of different Lachancea kluyveri strains, which indicated that intergenic regions evolve faster than genes (Jung et al. 2012).
We noted that the VAR1 hotspot is an exception to the observed protein-coding conservation, in agreement with previous reports of gene conversion at this locus (Strausberg et al. 1978; Strausberg and Butow 1981; Zinn et al. 1987). This gene encodes a mitochondrial protein of the small ribosomal subunit. It has previously been shown that the gene is missing in most of the hemiascomycete sequenced mitochondrial genomes (Jung et al. 2009) and is under reduced constraints in various L. kluyveri strains (Jung et al. 2012). Altogether, these observations indicate that the consequences of rearrangements occurring at this locus may be moderate as this gene is under decreased selective pressure compared to others such as COX1, which is conserved in all aerobic organisms (Wallace 2007).
The genetic background influences mitochondrial recombination
When comparing the recombination map obtained for the S×K cross with the S×Y profile, we observed that the global recombination rate was significantly lower in S×K, which also led to an overall lower local recombination rate. The sequences at the S×Y- and S×K-specific peaks were analyzed in more detail but did not show any striking variations that could explain the differential localization of hotspots. For instance, a previous study reported preferential recombination at GC clusters on the mitochondrial genome (Dieckmann and Gandy 1987); however, we did not detect any hotspot enrichment in these regions. Moreover, no allelic variation between the S, K, and Y strains was observed within the recombination-associated nuclear genes MGT1, MHR1, DIN7, and NTG1.
To further assess the impact of genomic variability on recombination profiles, additional crosses would be required. However, mtDNA and intergenic regions in particular are sometimes extremely variable between yeast strains and difficult to sequence due to their repetitive elements, thereby complicating the analysis (Jung et al. 2012). In the present study, our maps represent the regions common to the S288c genome, thus lacking YJM789- or SK1-specific sequences. For instance, an additional intron in COX1 has previously been reported in the YJM789 strain (Wei et al. 2007). Nevertheless, as these regions are very divergent from the S288c mtDNA, they do not represent a suitable template for homologous recombination between the different strains we selected for the study and should therefore not affect the observed recombination landscape. To address this limitation, complete assemblies of the mitochondrial genomes will be required in the future.
Although recombination pathways are distinct between mitochondrial and nuclear DNA, it is interesting to note that lower meiotic recombination has also been observed when analyzing the progeny of the S×K and S×Y crosses (Wilkening et al. 2013). Moreover, it has been suggested that meiotic hotspot activity relies on the accessibility of the DNA to the recombination machinery (Petes 2001) and that this accessibility may partially be due to chromatin structure (Székvölgyi and Nicolas 2010). Mitochondrial DNA does not contain histones but is compacted into 190-nm structures by Abf2p (Brewer et al. 2003; Kucej et al. 2008). MtDNA is also shaped through secondary structures. For instance, GC clusters have been described to fold into stem loops (De Zamaroczy and Bernardi 1986). Taken together, our results suggest that while no significant sequence variations have been detected within recombination hotspots, the topology of the genomes may differently influence DNA accessibility in the S, K, and Y genomes.
Several pathways shape the recombination landscape at the genome-wide level
To better understand the mechanisms underlying mitochondrial recombination, we generated deletion strains in the S and Y backgrounds for four genes previously associated with this process: MGT1, NTG1, MHR1, and DIN7 (Figure 1B).
MGT1 encodes a mitochondrial resolvase that cleaves the recombination intermediate structures (Kleff et al. 1992; Ezekiel and Zassenhaus 1993; White and Lilley 1997). Deleting this gene has been reported to result in the accumulation of unresolved Holliday junctions (Lockshon et al. 1995). However, in our study, neither the global recombination rate nor the local recombination profiles were strongly modified in the MGT1 deletion background, indicating that this protein is not essential in the recombination process and that an enzyme with a redundant function may exist in mitochondria. Ntg1p, a mitochondrial N-glycosylase, is required for repairing oxidative DNA damage but was also found to induce DSBs at the ori5 locus, thus initiating mtDNA recombination, using a synthetic system (Ling et al. 2007). We posited that NTG1 deletion should lead to a reduced number of DSBs and to a decreased recombination rate. A significantly lower recombination rate was indeed detected, although the deletion had very little effect on the localization of the hotspots. This result confirms previous findings, which showed that the mtDNA recombination rate was affected in the absence of NTG1, using a reporter assay (Phadnis et al. 2006). We did not detect any hotspot at ori5, suggesting that despite the evidence that DSBs occur at this position (Ling et al. 2011), it is not a preferential location at the genome-wide level. Our findings demonstrate that NTG1 contributes but is not essential to the initiation of mitochondrial recombination. The loss of NTG1 and MGT1 did not result in a strong defect in recombination, although these enzymes have been previously associated with the process. Complementary factors may therefore be involved. Previous studies have also pointed to additional pathways for homologous recombination and DSB repair in mitochondria (Ling et al. 1995, 2000) and nonhomologous end joining has been proposed as a mechanism for mtDSB repair as well (Fukui and Moraes 2009; Kalifa et al. 2012).
Our study revealed that deleting MHR1 had a very drastic effect on mt genome maintenance, as large regions of mitochondrial DNA were lost in the mutant diploid background during mating (Figure 7). Mhr1p promotes single-stranded DNA (ssDNA) pairing with DSBs (Ling and Shibata 2002) and a defective allele has been associated with higher mtDNA damage rate and petite production in the mutant strain (Ling et al. 1995, 2000). A previous study also suggested that it is involved in gene conversion but not in homologous crossing over (Ling et al. 1995). Our current method does not allow us to differentiate between these two types of events. However, we observed that the recombination profiles in the S∆mhr1×Y∆mhr1 background were modified compared to those in wild type. While the VAR1 hotspot was lost, demonstrating that pairing via MHR1 could be an important process for hotspot localization, some hotspots were maintained, thereby indicating the likely involvement of another pairing factor in an MHR1-independent mechanism.
Attempts to generate mtDNA libraries from pools of diploids originating from a cross between the S∆din7 and Y∆din7 strains resulted in repeated DNA degradation following the fragmentation step, although the parental genomes were intact before mating (Figure S1). DIN7 is a mitochondrial nuclease induced by DNA damage and involved in the resection of the 5′ ends after a DSB (Mieczkowski et al. 1997; Fikus et al. 2000; Ling et al. 2013). Our results suggest that mtDNA is sensitized to DNA fragmentation after mating in the ∆din7 diploid background. One explanation could be that petite formation occurred during the mating process, leading to very short fragments after shearing, thereby preventing subsequent library preparation. However, so far, only Din7p overexpression has been associated with increased petite formation (Fikus et al. 2000). Therefore, further analyses will be needed to decipher the mechanisms causing mtDNA degradation after fragmentation in this background.
Although, mt recombination in yeast has been described since the 1970s (Dujon et al. 1974), most of the factors affecting mtDNA recombination and DSBs remain unknown. Our approach paves the way for further mechanistic studies of recombination and genome maintenance. Several recombination-associated mutants and their combinations may now be tested to better characterize the functions as well as the interactions of the encoded proteins.
Supplementary Material
Acknowledgments
The authors thank Julien Gagneur for his input on the statistical analysis, Jonathan Landry and Tobias Rausch for their input on aligners and variant callers, and Sophie Adjalley and Aleksandra Pekowska for fruitful discussions. E.S.F., C.D.C., and L.M.S. conceived and designed the experiments. E.S.F. performed the experiments. E.S.F. and C.D.C. performed the data analysis. B.K. performed the statistical analysis. E.S.F., C.D.C., B.K., and L.M.S. drafted the manuscript. The authors declare that they have no competing financial interests. This study was technically supported by the European Molecular Biology Laboratory Genomics Core Facility, where the libraries were sequenced. This work was funded by the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013)/15 ERC agreement no. AdG-16 294542 (to L.M.S.). C.D.C. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds.
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.166637/-/DC1.
Raw sequence files from this article have been deposited at the Sequence Read Archive (SRA) under accession no. ERP005547 (http://www.ncbi.nlm.nih.gov/sra).
Communicating editor: N. M. Hollingsworth
References
- Arrieta-Montiel M. P., Shedge V., Davila J., Christensen A. C., Mackenzie S. A., 2009. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183: 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Nicolas A., 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94: 5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57(1): 289–300 [Google Scholar]
- Bianchi N. O., Bianchi M. S., Richard S. M., 2001. Mitochondrial genome instability in human cancers. Mutat. Res. 488: 9–23 [DOI] [PubMed] [Google Scholar]
- Brewer L. R., Friddle R., Noy A., Baldwin E., Martin S. S., et al. , 2003. Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p. Biophys. J. 85: 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C., Shroff R., Lichten M., 2009. Genome-wide mapping of meiotic DNA double-strand breaks in Saccharomyces cerevisiae. Methods Mol. Biol. 557: 143–164 [DOI] [PubMed] [Google Scholar]
- Chen X. J., Butow R. A., 2005. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6: 815–825 [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D., 1991. Contrasting mutation rates in mitochondrial and nuclear genes of yeasts vs. mammals. Curr. Genet. 20: 195–198 [DOI] [PubMed] [Google Scholar]
- D’aurelio M., 2004. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 13: 3171–3179 [DOI] [PubMed] [Google Scholar]
- Davila J. I., Arrieta-Montiel M. P., Wamboldt Y., Cao J., Hagmann J., et al. , 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defontaine A., Lecocq F. M., Hallet J. N., 1991. A rapid miniprep method for the preparation of yeast mitochondrial DNA. Nucleic Acids Res. 19: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G., 1986. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene 41: 1–22 [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Gandy B., 1987. Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 6: 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov L., R. Brem, L. Kruglyak, and D. Gottschling, 2009 Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383. [DOI] [PMC free article] [PubMed]
- Doudican N. A., Song B., Shadel G. S., Doetsch P. W., 2005. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 25: 5196–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B., Slonimski P. P., Weill L., 1974. Mitochondrial genetics IX: a model for recombination and segregation of mitochondrial genomes in Saccharomyces cerevisiae. Genetics 78: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Awadalla P., 2001. Does human mtDNA recombine? J. Mol. Evol. 53: 430–435 [DOI] [PubMed] [Google Scholar]
- Ezekiel U. R., Zassenhaus H. P., 1993. Localization of a cruciform cutting endonuclease to yeast mitochondria. Mol. Gen. Genet. 240: 414–418 [DOI] [PubMed] [Google Scholar]
- Fikus M. U., Mieczkowski P. A., Koprowski P., Rytka J., Sledziewska-Gójska E., et al. , 2000. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics 154: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourie G., van der Merwe N. A., Wingfield B. D., Bogale M., Tudzynski B., et al. , 2013. Evidence for inter-specific recombination among the mitochondrial genomes of Fusarium species in the Gibberella fujikuroi complex. BMC Genomics 14: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., Purnelle B., 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440: 325–331 [DOI] [PubMed] [Google Scholar]
- Fukui H., Moraes C. T., 2009. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18: 1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., 2011. The intriguing evolutionary dynamics of plant mitochondrial DNA. BMC Biol. 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton J. L., DeRisi J., Shroff R., Lichten M., Brown P. O., et al. , 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S., Strimmer K., 2012. MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics 28: 2270–2271 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007a Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 1–4 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007b High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31–34 [DOI] [PubMed] [Google Scholar]
- Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon et al, 1996 Life with 6000 genes. Science 274: 546, 563–567. [DOI] [PubMed]
- Gray M. W., 2012. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4: a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W., Burger G., Lang B. F., 1999. Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Grün, B., I. Kosmidis, and A. Zeileis, 2011 Extended beta regression in R: shaken, stirred, mixed, and partitioned. J. Stat. Softw. 48: 1–25. [Google Scholar]
- Hori A., Yoshida M., Shibata T., Ling F., 2009. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 37: 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Dujon B., 1985. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41: 383–394 [DOI] [PubMed] [Google Scholar]
- Jung P. P., Schacherer J., Souciet J.-L., Potier S., Wincker P., et al. , 2009. The complete mitochondrial genome of the yeast Pichia sorbitophila. FEMS Yeast Res. 9: 903–910 [DOI] [PubMed] [Google Scholar]
- Jung, P. P., A. Friedrich, C. Reisser, J. Hou, and J. Schacherer, 2012 Mitochondrial genome evolution in a single protoploid yeast species. G3 2: 1103–1111. [DOI] [PMC free article] [PubMed]
- Kalifa L., Quintana D. F., Schiraldi L. K., Phadnis N., Coles G. L., et al. , 2012. Mitochondrial genome maintenance: roles for nuclear nonhomologous end joining proteins in Saccharomyces cerevisiae. Genetics 190: 951–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S., Kemper B., Sternglanz R., 1992. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11: 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D. F., Masson G., Sigurdsson A., et al. , 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Schwartz M., Brown T. A., Ebralidse K., Kunz W. S., et al. , 2004. Recombination of human mitochondrial DNA. Science 304: 981. [DOI] [PubMed] [Google Scholar]
- Kucej M., Kucejova B., Subramanian R., Chen X. J., Butow R. A., 2008. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 121: 1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis E. D., Zouros E., 2001. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18: 1168–1175 [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U., Campbell C., 1999. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 19: 3869–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-C., Yin P.-H., Lin J.-C., Wu C.-C., Chen C.-Y., et al. , 2006. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann. N. Y. Acad. Sci. 1042: 109–122 [DOI] [PubMed] [Google Scholar]
- Lee H.-Y., Chou J.-Y., Cheong L., Chang N.-H., Yang S.-Y., et al. , 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Ling F., Shibata T., 2002. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 21: 4730–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Makishima F., Morishima N., Shibata T., 1995. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 14: 4090–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Morioka H., Ohtsuka E., Shibata T., 2000. A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA. Nucleic Acids Res. 28: 4956–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Hori A., Shibata T., 2007. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho-] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 27: 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Mikawa T., Shibata T., 2011. Enlightenment of yeast mitochondrial homoplasmy: diversified roles of gene conversion. Genes 2: 169–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Hori A., Yoshitani A., Niu R., Yoshida M., et al. , 2013. Din7 and Mhr1 expression levels regulate double-strand-break-induced replication and recombination of mtDNA at ori5 in yeast. Nucleic Acids Res. 41: 5799–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D., Zweifel S. G., Freeman-Cook L. L., Lorimer H. E., Brewer B. J., et al. , 1995. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81: 947–955 [DOI] [PubMed] [Google Scholar]
- Lynch M., Blanchard J. L., 1998. Deleterious mutation accumulation in organelle genomes. Genetica 102–103: 29–39 [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A., 1985. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell 41: 395–402 [DOI] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Fikus M. U., Ciesla Z., 1997. Characterization of a novel DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, which is a structural homolog of the RAD2 and RAD27 DNA repair genes. Mol. Gen. Genet. 253: 655–665 [DOI] [PubMed] [Google Scholar]
- Munz P., Amstutz H., Kohli J., Leupold U., 1982. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature 300: 225–231 [DOI] [PubMed] [Google Scholar]
- Petes T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369 [DOI] [PubMed] [Google Scholar]
- Pérez-Martínez X., Broadley S. A., Fox T. D., 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22: 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Mehta R., Meednu N., Sia E. A., 2006. Ntg1p, the base excision repair protein, generates mutagenic intermediates in yeast mitochondrial DNA. DNA Repair 5: 829–839 [DOI] [PubMed] [Google Scholar]
- Pratt-Hyatt M. J., Kapadia K. M., Wilson T. E., Engelke D. R., 2006. Increased recombination between active tRNA genes. DNA Cell Biol. 25: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T., Cincu E., Fuchs J., Zoschke R., Liere K., et al. , 2010. Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 64: 948–959 [DOI] [PubMed] [Google Scholar]
- Shay J. W., Pierce D. J., Werbin H., 1990. Mitochondrial DNA copy number is proportional to total cell DNA under a variety of growth conditions. J. Biol. Chem. 265: 14802–14807 [PubMed] [Google Scholar]
- Shibata T., Ling F., 2007. DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion 7: 17–23 [DOI] [PubMed] [Google Scholar]
- Smithson M., Verkuilen J., 2006. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol. Methods 11: 54–71 [DOI] [PubMed] [Google Scholar]
- Solieri L., 2010. Mitochondrial inheritance in budding yeasts: towards an integrated understanding. Trends Microbiol. 18: 521–530 [DOI] [PubMed] [Google Scholar]
- Solieri L., Antúnez O., Pérez Ortín J. E., Barrio E., Giudici P., 2008. Mitochondrial inheritance and fermentative : oxidative balance in hybrids between Saccharomyces cerevisiae and Saccharomyces uvarum. Yeast 25: 485–500 [DOI] [PubMed] [Google Scholar]
- Song S., Pursell Z. F., Copeland W. C., Longley M. J., Kunkel T. A., et al. , 2005. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc. Natl. Acad. Sci. USA 102: 4990–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A., 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776 [DOI] [PubMed] [Google Scholar]
- Strausberg R. L., Butow R. A., 1981. Gene conversion at the VAR1 locus on yeast mitochondrial DNA. Proc. Natl. Acad. Sci. USA 78: 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R. L., Perlman P. S., 1978. The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 163: 131–144 [DOI] [PubMed] [Google Scholar]
- Strausberg R. L., Vincent R. D., Perlman P. S., Butow R. A., 1978. Asymmetric gene conversion at inserted segments on yeast mitochondrial DNA. Nature 276: 577–583 [DOI] [PubMed] [Google Scholar]
- Székvölgyi L., Nicolas A., 2010. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 277: 571–589 [DOI] [PubMed] [Google Scholar]
- Wahls, W. P., E. R. Siegel, and M. K. Davidson, 2008 Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLoS ONE 3: e2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76: 781–821 [DOI] [PubMed] [Google Scholar]
- Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., et al. , 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104: 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. F., Lilley D. M., 1997. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 266: 122–134 [DOI] [PubMed] [Google Scholar]
- Wilkening S., Tekkedil M. M., Lin G., Fritsch E. S., Wei W., et al. , 2013. Genotyping 1000 yeast strains by next-generation sequencing. BMC Genomics 14: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Watanabe C. K., 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Zeyl C., Andreson B., Weninck E., 2005. Nuclear-mitochondrial epistasis for fitness in Saccharomyces cerevisiae. Evolution 59: 910–914 [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A., 1985. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell 40: 887–895 [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A., 1987. Kinetic and segregational analysis of mitochondrial DNA recombination in yeast. Plasmid 17: 248–256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.