Abstract
Apoptosis in activated T cells in vivo requires the proapoptotic Bcl-2 family member Bim. We show here that, despite its ability to bind LC8, a component of the microtubule dynein motor complex, most of the Bim in both healthy and apoptotic T cells is associated with mitochondria, not microtubules. In healthy resting T cells Bim is bound to the antiapoptotic proteins Bcl-2 and Bcl-xL. In activated T cells, levels of Bcl-2 fall, and Bim is associated more with Bcl-xL and less with Bcl-2. Our results indicate that, in T cells, Bim function is regulated by interaction with Bcl-2 family members on mitochondria rather than by sequestration to the microtubules.
During immune responses, antigen-specific T cells expand in large numbers and then die (1–3). Sometimes this death involves death receptors (4). However, under some circumstances, the death of activated T cells is driven by changes in the activity of Bcl-2-related proteins.
Bcl-2-related proteins are classified by the presence or absence in their sequences of BH1–4 domains. The antiapoptotic family members contain all four of these domains. Family members that are thought to be the executioners of cell death, Bax and Bak, contain only BH1–3. Many family members express only BH3 regions. These proteins are also proapoptotic.
It is not clear how Bcl-2 family proteins kill activated T cells. One hypothesis suggests that apoptotic stimuli activate BH3-region-only proteins and these proteins in turn activate Bax or Bak, either by direct interaction, or by neutralizing the antiapoptotic Bcl-2-like proteins (5–7). Alternatively, it has been suggested that antiapoptotic proteins such as Bcl-2 may inhibit the activity of death-dealing caspases. In this model, proapoptotic proteins act by binding to Bcl-2, thus interfering with its inhibition of the caspases.
Apoptosis of activated T cells requires the BH3-only Bcl-2 family member Bim (8); however, the processes that regulate Bim activity in T cells are unknown. Some cell types express low levels of Bim constitutively and rapidly increase its levels when they die (9, 10). However, healthy T cells contain appreciable levels of Bim that do not change very much when the cell is activated and undergoes Bim-dependent death (8). In other cell types, Bim activity is controlled by its location. Bim binds with high affinity to LC8, a component of the microtubule-associated dynein motor complex (11). Thus, in some cells, when healthy, Bim is sequestered to the microtubular dynein motor complex. In these cells, death-inducing signals cause release of Bim plus LC8 from the dynein motor complex, translocation of the complex to mitochondria, and Bim-induced death of the cell (11).
In this study, we tested the idea that Bim might move from microtubules to mitochondria during activation-induced T cell death. Surprisingly, we found that Bim was not bound to microtubules, even in healthy cells, but rather was associated with mitochondria. In both healthy and dying T cells, Bim was already bound to Bcl-2 and Bcl-xL although the ratio of binding to these two proteins changed in dying vs. healthy cells. Several regions of Bim contribute to its association with mitochondria. Our data favor a model in which T cell viability depends on the ability of mitochondrial antiapoptotic Bcl-2 family members, such as Bcl-2, to keep mitochondrial-associated Bim in check.
Materials and Methods
Reagents, Mice, T Cell Preparations, and Analysis. Reagents were as follows: FBS (Atlanta Biologicals, Norcross, GA); MEM and DMEM (GIBCO/BRL); staphylococcal enterotoxin B (SEB), paclitaxel, and saponin (Sigma); protease inhibitor mixture and Complete Mini (Roche Applied Science, Indianapolis, IN); Mitotracker Red CMXRos, Hoechst 33342, and Alexa Fluor 488 or 647 conjugated anti-rabbit IgG (Molecular Probes); Cy3, Cy5 or peroxidase-conjugated anti-mouse IgG, peroxidase-conjugated anti-goat IgG or anti-rabbit IgG or anti-rat IgM (Jackson ImmunoResearch). Rat mAb against Bim (clone 14A8) was a generous gift from A. Strasser (Walter and Eliza Hill Institute of Medical Research, Parkville, Victoria, Australia) (12).
Mice were maintained under specific pathogen-free conditions in the Biological Resource Center of the National Jewish Medical and Research Center. Mice transgenic for the Vβ8.2+ T cell antigen receptor (TCR) β-chain of the DO.11.10 TCR VβDO (13) were on the B10.D2 background (The Jackson Laboratory). Gp33 TCR transgenic (Tg) mice (14) were on the C57BL/6 background. Their CD8+ T cells recognize the lymphocytic choriomeningitis virus-derived gp33–41 peptide. Bim-/- mice were the generous gift of A. Strasser (15).
Naive and SEB-stimulated T cells were prepared from lymph nodes and spleen as described (2) from normal mice or mice given 150 μg of SEB 2–3 days previously. T cells used for retroviral infections were prepared according to a published protocol (16). Activated T cells from normal or SEB-injected mice were analyzed as described (1, 2). To assess cell viability, purified T cells were incubated with 0.65 μg/ml propidium iodide for 10 min at room temperature.
Immunofluorescent Microscopy. T cells were incubated with 50 nM Mitotracker Red, CMXRos, for 15 min at 37°C, washed with warm medium, loaded onto coverslips pretreated with 100 μg/ml poly-l-lysine and incubated at 37°C for 10 min. Immobilized cells were fixed with prewarmed 3% paraformaldehyde at 37°C and then permeabilized with 0.2% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) (Pierce). Permeabilized cells were blocked with 5% FBS for 30 min, then incubated with a mixture of anti-Bim primary antibody (pAb) (Stressgen Biotechnologies, Victoria, BC, Canada) and anti-tubulin mAb clone DM1α (Sigma) for 2 h, and finally with Alexa Fluor 488 conjugated anti-rabbit IgG and Cy5-conjugated anti-mouse IgG for 1 h. Six washes were performed after each antibody incubation. Finally, the cells were stained with 1 μM Hoechst 33342 for 5 min and mounted onto slides. Visual data were acquired by examining slides under a Leica (Deerfield, IL) DMXRA epifluorescence microscope equipped with a Sensi-Cam charge-coupled device camera (PCO CCD Imaging, Kelheim, Germany) at a final magnification of ×1900 and were analyzed with slidebook software (Intelligent Imaging Innovations, Denver).
T cells expressing various GFP fusion proteins were processed as described above, except that Mitotracker Red staining was omitted and different antibodies were used for staining. Primary antibodies were rabbit anti-TOM20 pAb and mouse anti-tubulin mAb. Secondary antibodies were Alexa Fluor 647 conjugated anti-rabbit IgG and Cy3-conjugated anti-mouse IgG.
Subcellular Fractionation and Western Blotting. Subcellular fractionation by sequential extraction was adapted from a published method (17). Forty million T cells were resuspended in 550 μl of saponin buffer (0.1 M Pipes, 5 mM MgSO4, 10 mM EGTA, 2 mM DTT, protease inhibitor mixture, 13% glycerol, 10 μM paclitaxel, and 0.02% saponin), incubated at 37°C for 10 min, and centrifuged at 1300 × g for 5 min. The pellets were washed with 200 μl of saponin buffer. Supernatants of the two centrifugations were pooled (cytosolic fraction). The pellets were resuspended in 550 μl of TX-100 buffer (saponin buffer plus 1% Triton X-100 without saponin), incubated at 37°C for 5 min, and then centrifuged at 13,500 × g for 7 min. The pellets were washed with 200 μl of TX-100 buffer and centrifuged again. Supernatants of these two centrifugations were pooled (membrane fraction). The pellets were resuspended in 750 μl of SDS buffer (saponin buffer plus 1% SDS without saponin, cytoskeleton fraction). Fractions were sonicated, and protein was recovered by chloroform extraction (18).
The heavy membrane fraction was prepared as follows: 4.5 × 107 T cells per ml were resuspended in ice-cold mitochondrial buffer (19). Cells were homogenized in a ball-bearing homogenizer [H&Y Enterprise, Redwood City, CA (20)] with 20 strokes. Cell lysates were centrifuged at 950 × g for 15 min twice to remove nuclei and intact cells. The supernatants were centrifuged at 10,000 × g for 25 min to acquire the mitochondria-enriched heavy membrane fraction. The heavy membrane pellets were then resuspended in either mitochondrial buffer (pH 7.5) or 0.2 M Na2CO3 (pH 11.5). After an incubation of 40 min, the “washed” heavy membranes were pelleted at 10,000 × g and solubilized in SDS sample buffer.
Proteins were separated by SDS/PAGE on Criterion 10–20% gradient gels (Bio-Rad) and transferred to Hybond ECL nitrocellulose membranes (Amersham Pharmacia) by a semidry method. Membranes were blocked at room temperature for 1–2 h with Blotto [5% milk/1% FBS in Tris-buffered saline (TBS) plus 0.05% Tween 20] and incubated with the primary antibody for 4 h, washed with several changes of TBS-T (TBS plus 0.05% Tween 20), and then incubated with secondary antibodies for 3 h. Bound secondary antibodies were visualized by ECL detection reagents (Amersham Pharmacia). The pAB used included anti-Bim pAb and anti-heat shock protein 60 (Hsp60) mAb (Stressgen); anti-LC8 mAb (Alexis Biochemicals, San Diego); anti-lactate dehydrogenase (LDH) pAb (Rockland Immunochemicals, Gilbertsville, PA), and anti-TOM40 pAb.
Immunoprecipitation. Purified T cells were lysed in ice-cold lysis buffer (150 mM NaCl/10 mM TRIZMA Base/2 mM sodium orthovanadate/50 mM sodium fluoride/1 mM PMSF, pH 7.5/Complete Mini protease inhibitor mixture) containing 2% CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate} for 30 min at 1.5 × 107 cells per ml. Lysates were centrifuged at 15,000 × g for 20 min to pellet nuclei and insoluble debris. Supernatants were incubated with Sepharose-conjugated anti-Bim mAb (clone 14A8) on an end-to-end rotor in the cold room for 4 h. Beads were pelleted at 800 × g, and the post-immunoprecipitation supernatant was saved (Fig. 4, sup). The beads were washed six times with lysis buffer, and the immunoprecipitated protein (Fig. 4, IP) released with SDS sample buffer and heating to 95°C. Primary blotting antibodies were anti-Bim pAb (Stressgen); anti-Bcl-2 mAb (clone 10C4, Zymed); anti-Bcl-x mAb (clone 4, Transduction Laboratories, San Jose, CA); anti-LC8 mAb (rat IgM, Alexis Biochemicals); anti-Bak pAb (Upstate Biotechnology, Lake Placid, NY); anti-Bax pAb (N-20, Santa Cruz Biotechnology). When necessary, probed membranes were stripped with Restore Western Blot Stripping Buffer (Pierce) to allow reprobing with another antibody.
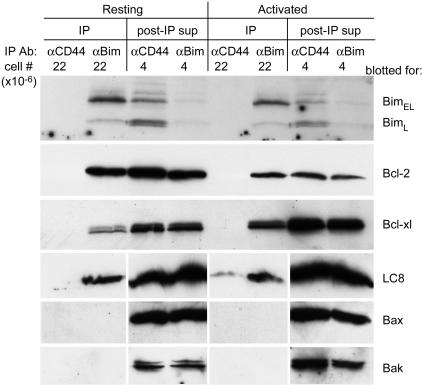
Fig. 4.
Protein binding pattern of Bim in resting and activated T cells. Lysates of resting or activated T cells were immunoprecipitated with rat anti-Bim (12) or with a control rat antibody that reacts with CD44. Immunoprecipitates (IP) with a cell equivalency of 22 × 106 per lane, together with post-IP supernatants (sup) with a cell equivalency of 4 × 106 per lane, were analyzed by Western blotting to evaluate the associations of Bim with Bcl-2, Bcl-x, LC8, Bax, or Bak.
Production and Transduction of Bim-Expressing Retroviruses. BimEL was amplified by PCR from the cDNA of C57BL/10 T cells. The PCR product and those of various Bim truncations were cloned into a plasmid downstream of an ORF encoding the humanized green fluorescent protein variant, EGFP. The stop codon of EGFP and the first codon (ATG) of Bim were removed to allow expression of the GFP-Bim fusion protein. A linker encoding GGAGGGGS was genetically inserted between EGFP and Bim. The constructs were cloned into the retroviral vector pEzeo (21). Sequences of all PCR primers are available upon request.
Maloney murine leukemia-based retroviruses expressing these constructs were produced in Phoenix cells and transduced into activated T cells as described (22, 23).
Results
Immunofluorescence Studies Reveal That Bim Is Localized to Mitochondria in T Cells. To study apoptosis in activated T cells, we used Vβ8+ T cells from Vβ8+ TCR β-chain transgenic (VβDO) mice, which had been activated in vivo by injection of the animals 48–72 h previously with SEB, a treatment that activates ≈50% of the CD4+ and 85% of the CD8+ T cells in the mice. Although healthy at the time of harvest, we have shown that these activated T cells, unlike resting T cells, die rapidly in vitro and in vivo (1, 2). The resting and activated T cells were fixed at 37°C, to avoid the disintegration of microtubules that occurs at cold temperatures, and stained with anti-Bim and anti-tubulin antibodies and Mitotracker to identify functional mitochondria. The specificity of the anti-Bim staining was checked by the lack of staining in resting Bim-/- T cells (Fig. 1A).
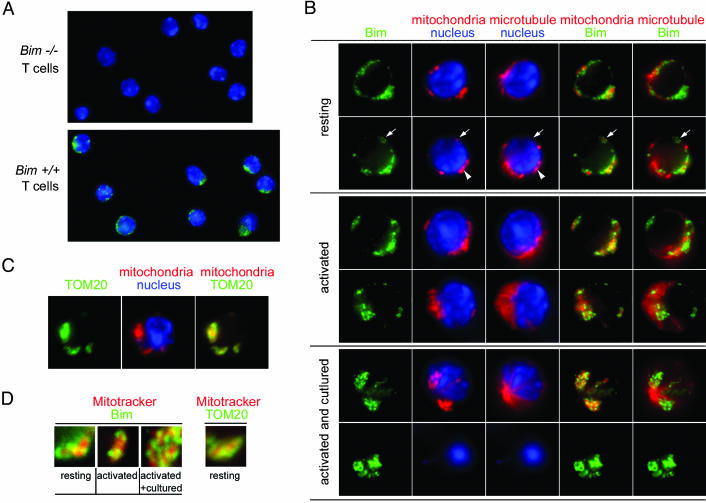
Fig. 1.
Immunofluorescence studies show that Bim is localized to mitochondria in T cells. (A) Two-color stainings of resting T cells from Bim+/+ and Bim-/- mice to reveal Bim (green) and nuclei (blue). (B) Resting, activated, and activated plus cultured T cells were stained for Bim (green), mitochondria (red), tubulin (red), and nuclei (blue). The arrow marks Bim staining associated with the mitochondria, not microtubules. The arrowhead indicates areas in T cells where mitochondria and microtubules overlap spatially. (C) Resting T cells were stained to show TOM20 (green), mitochondria (red), and nuclei (blue). (D) Equally enlarged views of areas from B and C showing Bim (green) or TOM20 (green) overlaid with mitochondrial staining (red).
Some of the Bim, in resting and, to some extent, activated T cells, was cytoplasmic, with a staining pattern that was in some places hazy and elsewhere focused (Fig. 1B). This staining was not particularly associated with microtubules. However, in both resting and activated T cells, a good deal of the Bim was associated with mitochondria. The Bim staining often appeared as a ring-like structure surrounding the area identified by Mitotracker (Fig. 1B, arrow, and enlarged views of such areas in Fig. 1D). Although very close, Bim and Mitotracker did not completely overlap (Fig. 1D). Staining of a known mitochondrial outer membrane protein [translocase of the outer mitochondrial membrane, 20 kDa (TOM20) (24)] showed the same pattern as that of Bim, surrounding, but not completely overlapping, the Mitotracker stain (Fig. 1 C and D). Unfortunately, we could not do the straightforward experiment of costaining with anti-TOM20 and anti-Bim antibodies because both antibodies were derived from rabbits.
Dead cells, identified by their condensed, featureless nuclei and absence of microtubule network, appeared among the cultured activated T cells. Mitotracker red did not stain these cells, because of their diminished mitochondrial membrane potential. (Fig. 1B, Bottom) In these cells, Bim seemed to be even more associated with condensed structures, many of which had the ring-like appearance characteristic of staining of outer mitochondrial membranes (compare Fig. 1 B and C).
Thus, some of the Bim in T cells is associated with mitochondria even if the cells are resting and alive, in contrast to some, but not all, previous reports on the subject (11, 25). In resting and activated T cells, the Bim that is not associated with mitochondria is not bound to microtubules.
Subcellular Fractionation Demonstrates That Bim Is Bound to Membranes in T Cells. Resting T cells are small. Thus, mitochondria and microtubules in these cells, visualized by microscopy, often overlap (see arrowhead in Fig. 1B). Therefore, we used biochemical methods to confirm the observation that Bim was on mitochondria in healthy T cells. Cytosolic, membrane/organelle, and cytoskeletal fractions were prepared from the cells (17). Because the microtubules were known to be fragile and to disintegrate at cold temperature, the fractionation process was carried out at 37°C in the presence of the microtubule-stabilizing agent paclitaxel (26).
The validity of the fractionation was demonstrated by various marker proteins (Fig. 2). The cytosolic protein lactate dehydrogenase (LDH) was found mainly in the cytosolic fraction; mitochondrial heat shock protein 60 (Hsp60) was found mainly in the membrane/organelle fraction, and tubulin was found in both the cytosolic and cytoskeletal fractions, representing the monomeric and polymerized pools of tubulin, respectively.
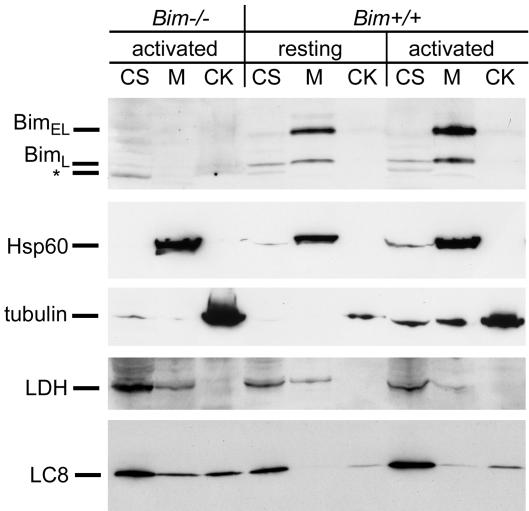
Fig. 2.
Subcellular fractionation demonstrates that Bim is membrane-associated in T cells. Cytosolic (CS), membrane (M), and cytoskeleton (CK) fractions from 8 × 106 per lane resting or activated Bim+/+ or activated Bim-/- T cells were analyzed by Western blots to detect Bim, heat shock protein 60 (Hsp60), tubulin, lactate dehydrogenase, and LC8. Bim-/- T cells were activated in vitro with anti-CD3 and anti-CD28 and then expanded by culturing with IL-2. Analysis of the Bim Western blot of Bim-/- T cells identified a nonspecific band, indicated with an asterisk.
Control fractionations using T cells from Bim-deficient mice showed that the anti-Bim was indeed specific for Bim (Fig. 2).
Averaged over three experiments, the fold increase of BimEL per cell in activated vs. resting cells was 1.34 ± 0.80 at day 2 and 0.9 ± 0.4 at day 3. Thus, although others have recently reported that Bim levels rise after T cell stimulation in vitro (27), this does not seem to be the case in vivo, as we have reported (8).
As in our immunofluorescence microscopy studies, most of the Bim appeared in the membrane/organelle fraction and not in the cytoskeletal fraction in both resting and activated T cells (Fig. 2).
We also analyzed these fractions for their content of the Bim-binding protein LC8. In resting and activated T cells, most of this protein was cytosolic, a smaller proportion coisolated with the cytoskeleton, and even less of the protein was identified in the membrane fraction. These results raise the question, why the LC8 is not completely associated with the cytoskeleton because LC8 is a dynein light chain? Perhaps T cells express LC8 in excess, as reported for neurons (28), such that the T cell dynein is saturated with LC8 and some of the LC8 is consequently free in the cytoplasm or bound by means of other proteins to intracellular membranes. Another light chain of the dynein motor complex has been shown to be present on mitochondria (29).
Alkali Wash Shows That Much of the BimEL Is Firmly Bound to Membranes. When depicted as an alpha helical wheel, one surface of the C-terminal domain of Bim is very hydrophobic in nature whereas another surface is positively charged (Fig. 3A). Such an amphipathic structure allows binding of proteins, as peripheral proteins, to membranes (30–33). To find out whether this idea applies to Bim, a heavy membrane fraction was prepared from resting T cells and washed with pH11.5 buffer, which releases peripheral membrane proteins from membranes or control, pH 7.5 buffer. Wash with the basic buffer had little effect on the amount of membrane-bound integral mitochondrial membrane protein, TOM40 (34), released ≈30% of the BimEL, and 99% of the BimL (Fig. 3B). Thus, BimL may indeed be bound as a peripheral membrane protein to mitochondria and other cellular membranes. Much of the BimEL, on the other hand, is either bound directly or indirectly, by means of another protein(s), as an integral membrane protein.
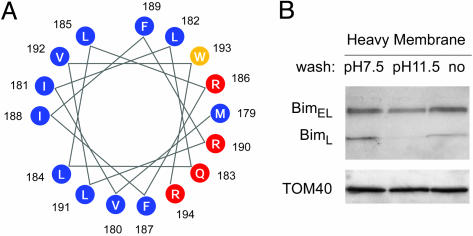
Fig. 3.
Strong association of BimEL with the mitochondria-enriched heavy membrane fraction of T cells. (A) A helical wheel representation of the C-terminal hydrophobic domain of Bim, showing the potential of this domain to form an amphipathic α-helical structure. Hydrophilic, hydrophobic, and neutral amino acids were drawn in red, blue, and yellow, respectively. (B) Heavy membrane fractions of healthy, resting T cells (14 × 106 per lane) were prepared and washed with either mitochondrial buffer (pH 7.5) or 0.2M Na2CO3 (pH 11.5). Washed fractions and unwashed controls were then Western blotted for Bim and TOM40.
Overall, these data show that the proapoptotic protein Bim is associated with membrane structures, probably mitochondria, given the results reported above, even in healthy T cells. Bim, especially in its “L” form, may bind to these membranes as a peripheral protein. However, BimEL is bound to these membranes either as an integral membrane protein or by means of engagement of some other protein(s).
Bim Binds to Bcl-2 and Bcl-xL in Healthy, Resting T Cells. Immunoprecipitation was used to study the proteins bound to endogenous Bim in T cell lysates. Resting and activated T cells were lysed in buffer containing CHAPS, a detergent that does not cause conformational changes in Bcl-2-related proteins such as Bax and Bak (35), and immunoprecipitated with anti-Bim or control, anti-CD44, antibody (Fig. 4).
Almost all of the Bim was present in the anti-Bim immunoprecipitates with very little remaining in the supernatants from the anti-Bim beads.
LC8 specifically coprecipitated with Bim so at least some of the Bim was bound to LC8 in both resting and activated T cells. As discussed above, perhaps T cells contain excess LC8, and therefore the fact that at least some Bim is bound to LC8 may not be contradictory to Bim's location in these cells on mitochondria and not on microtubules.
Bcl-2 coprecipitated with Bim from both healthy, resting, and activated T cells (Fig. 4). Bcl-2 did not coprecipitate with Bim lacking its BH3 region (data not shown) so the binding of Bim to Bcl-2 probably occurred by means of the engagement of Bim's BH3 to the BH3 binding groove of Bcl-2. This result and the fact that another mitochondrially located protein, Bak, did not detectably coprecipitate with Bim show that the isolation of Bcl-2 in Bim immunoprecipitates was not due to nonspecific isolation of mitochondrially bound proteins with Bim (Fig. 4). The fact that Bak did not coprecipitate with Bim, nor with Bcl-2 (data not shown), also showed that, as expected, CHAPS was not causing untoward unfolding of Bcl-2-related proteins, thus allowing post-cell lysis association of these proteins with Bim.
Estimates from the sum of the immunoprecipitates and supernatants showed that overall Bcl-2 levels fell in activated vs. resting T cells as we and others have reported (8, 36).
In agreement with findings that Bim binds Bcl-xL as well as Bcl-2, Bim also coprecipitated with Bcl-xL. The magnitude of Bcl-xL coisolated was greater in activated than in resting T cells. We could not, however, detect association of Bim, in resting or activated T cells, with either of the proteins thought to be executioners of cell death in T cells, Bax, or Bak.
The constitutive association of Bim with Bcl-2 and Bcl-xL in healthy resting T cells and our failure to detect Bim binding to Bax or Bak in activated cells have implications for hypotheses about the role of Bim in the deaths of activated cells.
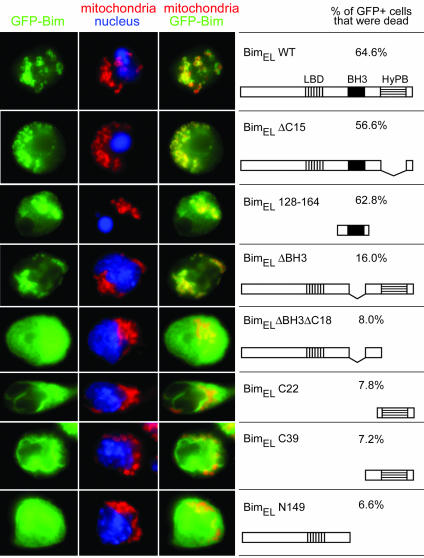
Several Regions of Bim Are Involved in Localization of Bim to Mitochondria. To identify the element(s) on Bim responsible for its localization, we made constructs coding for BimEL mutants fused to GFP (37). These constructs were cloned into a retroviral vector (21) and transduced into vigorously proliferating T cells (14). Fourteen hours later, the T cells were examined for localization of the GFP-Bim proteins and for death.
Overexpression of all of the variant BimEL proteins that retained the BH3 domain induced apoptosis, but Bim deleted in its BH3 domain did not kill (Fig. 5). Therefore, for BH3-containing constructs, we could examine Bim localization only in apoptotic cells. GFP-BimEL colocalized with the outer mitochondrial membrane protein TOM20 (24) in the apoptotic T cells. Therefore, the GFP tag did not interfere with the mitochondrial location of Bim or its ability to induce apoptosis. The Bim BH3 domain was sufficient for mitochondrial localization although the minimal construct, GFP-BimEL128–164, was not as exclusively localized to the mitochondria as the wild-type construct, GFP-BimEL. Because BH3 domains bind to BCl-2-related proteins (7, 38–40), it is likely that a mitochondrial Bcl-2 family member provides the anchor for BH3-mediated Bim localization to mitochondria.
Fig. 5.
Bim uses two different mechanisms to localize to mitochondria. Activated gp33 TCR transgenic T cells were infected with retroviruses encoding chimeric proteins of GFP fused to various Bim truncations, for which the schematic diagrams are shown to the right. After infection, cells were cultured with IL-2 for 14 h. The percentage of GFP+ cells in each preparation that were dead as assessed by propidium iodide after culture is shown. Cells were fixed and stained with Hoechst to reveal the nuclei and with anti-TOM20 to reveal the mitochondrial outer membrane. The fluorescence signals for GFP (green), TOM20 (red), and nuclei (blue) are shown.
A Bim construct lacking only the BH3 domain GFP-BimELΔBH3, however, also localized well to mitochondria in the healthy cells expressing it, suggesting that regions of Bim other than its BH3 domain could guide the protein to mitochondria. Constructs lacking both BH3 and the C-terminal stretch, GFP-BmELN149 and GFP-BimELΔBH3ΔC18, were diffusely cytoplasmic. Constructs containing only the C-terminal stretch of Bim, GFP-BimELC39 or GFP-BimELC22, were localized on membranes, but not specifically that of the mitochondria. Thus, in the absence of the BH3 domain, sequences from both ends of the Bim molecule are required for specific mitochondrial homing with the C-terminal region, perhaps guiding Bim to membranes and some portion of the N-terminal region responsible for mitochondrial specificity. Immunoprecipitation studies showed that this method of localization did not require binding to Bcl-2 as an intermediary (data not shown).
The simplest interpretation of these results is that Bim localizes to mitochondria by means of sequences in the N-terminal half of the protein and its C-terminal membrane-associating amphipathic helix. Once there, Bim binds Bcl-2 and Bcl-xL by means of its BH3 domain and is thus eventually held on the mitochondria by means of two distinct processes. After such localization, Bim is then available to participate together with other Bcl-2 family members in determination of the fate of the resting or activated T cell.
Discussion
Several publications have shown that Bim is crucial to the death of activated T cells (8, 15). Two mechanisms for controlling the killing activity of Bim have been described. The first mechanism, exemplified by removal of growth factors from neuronal cell lines, operates by means of changes in the level of Bim protein/cell (10). The second mechanism operates by means of movement of Bim. In some healthy cells, Bim is sequestered on microtubules by its very tight binding to the dynein light chain LC8. Under some circumstances, death is precipitated in these cells by movement of Bim and LC8 to mitochondria, where Bim participates in the death of the cell (11).
The work described here suggests that neither of these mechanisms applies to activated T cells. Microscopy and Western blots show that resting, relatively long-lived T cells contain Bim protein and the amounts of this protein/cell do not change very much when the cells are activated (Figs. 2 and 3). Moreover, three different types of experiments, using microscopy, cell fractionation, and transduction of genes coding for Bim covalently bound to GFP, show that, regardless of the state of the T cell, much of the Bim in the cell is localized to mitochondria and not to microtubules.
Others (41) have recently reported that Bim is located on microtubules in the T cell tumor line JURKAT. Perhaps this finding reflects part of the process leading to the transformation of JURKAT cells because, as shown here, this is clearly not the case in healthy, normal, resting T cells.
Bim may be located on mitochondria in T cells for several reasons. As demonstrated here by conjugates of GFP to various Bim constructs, the C-terminal region of Bim draws the protein to intracellular membranes. This finding, together with some other, more N-terminal region, locates the protein on mitochondria (Fig. 5). Similar functions for the C-terminal regions of other Bcl-2 family members have previously been reported (42–44). However, Bim is also drawn to mitochondria in both healthy and dying cells by the ability of its BH3 region to bind to two proteins, Bcl-2 and Bcl-xL. Both these proteins, particularly the former, are mitochondrially located.
The fact that most of the Bim is bound to Bcl-2 and Bcl-xL, even in healthy cells and that much of the protein is always located on mitochondria, suggests that the way in which Bim participates in the death of T cells is different from its modes of action in the deaths of other cells. Clearly, the events that trigger Bim's action must not depend on movement of, or dramatic increases in the amount of, Bim, unlike the situation in cell lines such as MCF-7, FDC-1, or in neuronal cells respectively. Mitochondrially located Bim must not be automatically lethal to T cells. T cell death cannot be driven by the mere existence of Bim/Bcl-2 or Bim/Bcl-xL complexes because the proteins are always to some extent bound to each other. Hypotheses about the role of Bim in activated T cell death must also include the finding that Bim is not detectably associated with either of the proteins thought to be the executioners of T cells, Bax and Bak.
Given these considerations, we prefer a model in which T cell death is initiated by the drop in levels of Bcl-2 that accompanies activation. Several hypotheses can then account for the death of the cell. The fall in Bcl-2 levels reduces the amount of Bim that is bound to Bcl-2 and therefore increased occupancy of Bcl-xL by Bim, leaving less Bcl-xL to perform other, perhaps crucial, antiapoptotic functions. Thus, death of the cell might be caused by reduced availability of Bcl-xL. Alternatively, the fall in Bcl-2 levels may lead to small increases in the amounts of free Bim. This small increase in free Bim may allow the protein to participate in what has been called “hit and run” activation of the proapoptotic proteins Bax and Bak (45).
Acknowledgments
We thank Drs. Philippe Bouillet and Lorraine O'Reilly of the Walter and Eliza Hill Institute of Medical Research (Parkville, Victoria, Australia), and Dr. Andreas Strasser for their generous gifts of the Bim-/- mice and anti-Bim mAb. We thank Bill Townend, Hannah Kupfer, and Dr. Abraham Kupfer for help with immunofluorescence microscopy. M.W. is a National Institute of Child Health and Human Development (NICHD) Fellow of the Pediatric Scientist Development Program (NICHD Grant Award K12-HD00850-17). This work was supported by U.S. Public Health Service Grants AI-17134, AI-18785, AI-22295, and AI-52225.
Abbreviations: SEB, staphylococcal enterotoxin B; TCR, T cell antigen receptor; pAb, primary antibody.
References
- 1.Mitchell, T., Kappler, J. & Marrack, P. (1999) J. Immunol. 162, 4527-4535. [PubMed] [Google Scholar]
- 2.Hildeman, D. A., Mitchell, T., Teague, T. K., Henson, P., Day, B. J., Kappler, J. & Marrack, P. C. (1999) Immunity 10, 735-744. [DOI] [PubMed] [Google Scholar]
- 3.Russell, J. H. (1995) Curr. Opin. Immunol. 7, 382-388. [DOI] [PubMed] [Google Scholar]
- 4.Green, D. R., Droin, N. & Pinkoski, M. (2003) Immunol. Rev. 193, 70-81. [DOI] [PubMed] [Google Scholar]
- 5.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B. & Korsmeyer, S. J. (2001) Science 292, 727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong, W. X., Lindsten, T., Ross, A. J., MacGregor, G. R. & Thompson, C. B. (2001) Genes Dev. 15, 1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letai, A., Bassik, M., Walensky, L., Sorcinelli, M., Weiler, S. & Korsmeyer, S. (2002) Cancer Cell 2, 183-192. [DOI] [PubMed] [Google Scholar]
- 8.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Bouillet, P., Strasser, A., Kappler, J. & Marrack, P. (2002) Immunity 16, 759-767. [DOI] [PubMed] [Google Scholar]
- 9.Putcha, G. V., Moulder, K. L., Golden, J. P., Bouillet, P., Adams, J. A., Strasser, A. & Johnson, E. M. (2001) Neuron 29, 615-628. [DOI] [PubMed] [Google Scholar]
- 10.Dijkers, P. F., Medema, R. H., Lammers, J. W., Koenderman, L. & Coffer, P. J. (2000) Curr. Biol. 10, 1201-1204. [DOI] [PubMed] [Google Scholar]
- 11.Puthalakath, H., Huang, D. C., O'Reilly, L. A., King, S. M. & Strasser, A. (1999) Mol. Cell 3, 287-296. [DOI] [PubMed] [Google Scholar]
- 12.O'Reilly, L. A., Cullen, L., Moriishi, K., O'Connor, L., Huang, D. C. & Strasser, A. (1998) BioTechniques 25, 824-830. [DOI] [PubMed] [Google Scholar]
- 13.Fenton, R. G., Marrack, P., Kappler, J. W., Kanagawa, O. & Seidman, J. G. (1988) Science 241, 1089-1092. [DOI] [PubMed] [Google Scholar]
- 14.Pircher, H., Burki, K., Lang, R., Hengartner, H. & Zinkernagel, R. M. (1989) Nature 342, 559-561. [DOI] [PubMed] [Google Scholar]
- 15.Bouillet, P., Metcalf, D., Huang, D. C., Tarlinton, D. M., Kay, T. W., Kontgen, F., Adams, J. M. & Strasser, A. (1999) Science 286, 1735-1738. [DOI] [PubMed] [Google Scholar]
- 16.Manjunath, N., Shankar, P., Wan, J., Weninger, W., Crowley, M. A., Hieshima, K., Springer, T. A., Fan, X., Shen, H., Lieberman, J. & von Andrian, U. H. (2001) J. Clin. Invest. 108, 871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollenbeck, P. J. (1989) J. Cell Biol. 108, 2335-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessel, D. & Flugge, U. I. (1984) Anal. Biochem. 138, 141-143. [DOI] [PubMed] [Google Scholar]
- 19.Gross, A., Jockel, J., Wei, M. C. & Korsmeyer, S. J. (1998) EMBO J. 17, 3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden, M. G., Chandel, N. S., Williamson, E. K., Schumacker, P. T. & Thompson, C. B. (1997) Cell 91, 627-637. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer, B. C., Mitchell, T. C., Kappler, J. W. & Marrack, P. (2001) Anal. Biochem. 297, 86-93. [DOI] [PubMed] [Google Scholar]
- 22.Naviaux, R. K., Costanzi, E., Haas, M. & Verma, I. M. (1996) J. Virol. 70, 5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan, M., Schallhorn, A. & Wurm, F. M. (1996) Nucleic Acids Res. 24, 596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanaji, S., Iwahashi, J., Kida, Y., Sakaguchi, M. & Mihara, K. (2000) J. Cell Biol. 151, 277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connor, L., Strasser, A., O'Reilly, L. A., Hausmann, G., Adams, J. M., Cory, S. & Huang, D. C. (1998) EMBO J. 17, 384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parekh, H. & Simpkins, H. (1997) Gen. Pharmacol. 29, 167-172. [DOI] [PubMed] [Google Scholar]
- 27.Sandalova, E., Wei, C. H., Masucci, M. G. & Levitsky, V. (2004) Proc. Natl. Acad. Sci. USA 101, 3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuhrmann, J. C., Kins, S., Rostaing, P., El Far, O., Kirsch, J., Sheng, M., Triller, A., Betz, H. & Kneussel, M. (2002) J. Neurosci. 22, 5393-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarzer, C., Barnikol-Watanabe, S., Thinnes, F. P. & Hilschmann, N. (2002) Int. J. Biochem. Cell Biol. 34, 1059-1070. [DOI] [PubMed] [Google Scholar]
- 30.Agasoster, A. V., Halskau, O., Fuglebakk, E., Froystein, N. A., Muga, A., Holmsen, H. & Martinez, A. (2003) J. Biol. Chem. 278, 21790-21797. [DOI] [PubMed] [Google Scholar]
- 31.Chen, C., Seow, K. T., Guo, K., Yaw, L. P. & Lin, S. C. (1999) J. Biol. Chem. 274, 19799-19806. [DOI] [PubMed] [Google Scholar]
- 32.Hoyt, D. W., Cyr, D. M., Gierasch, L. M. & Douglas, M. G. (1991) J. Biol. Chem. 266, 21693-21699. [PubMed] [Google Scholar]
- 33.Johnson, J. E. & Cornell, R. B. (1999) Mol. Membr. Biol. 16, 217-235. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, H., Okazawa, Y., Komiya, T., Saeki, K., Mekada, E., Kitada, S., Ito, A. & Mihara, K. (2000) J. Biol. Chem. 275, 37930-37936. [DOI] [PubMed] [Google Scholar]
- 35.Hsu, Y. T. & Youle, R. J. (1998) J. Biol. Chem. 273, 10777-10783. [DOI] [PubMed] [Google Scholar]
- 36.Grayson, J. M., Zajac, A. J., Altman, J. D. & Ahmed, R. (2000) J. Immunol. 164, 3950-3954. [DOI] [PubMed] [Google Scholar]
- 37.Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. (1996) Nat. Biotechnol. 14, 315-319. [DOI] [PubMed] [Google Scholar]
- 38.Kelekar, A., Chang, B. S., Harlan, J. E., Fesik, S. W. & Thompson, C. B. (1997) Mol. Cell. Biol. 17, 7040-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sattler, M., Liang, H., Nettesheim, D., Meadows, R. P., Harlan, J. E., Eberstadt, M., Yoon, H. S., Shuker, S. B., Chang, B. S., Minn, A. J., Thompson, C. B. & Fesik, S. W. (1997) Science 275, 983-986. [DOI] [PubMed] [Google Scholar]
- 40.Liu, X., Dai, S., Zhu, Y., Marrack, P. & Kappler, J. (2003) Immunity 19, 341-352. [DOI] [PubMed] [Google Scholar]
- 41.Chen, D. & Zhou, Q. (2004) Proc. Natl. Acad. Sci. USA 101, 1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cartron, P. F., Priault, M., Oliver, L., Meflah, K., Manon, S. & Vallette, F. M. (2003) J. Biol. Chem. 278, 11633-11641. [DOI] [PubMed] [Google Scholar]
- 43.Kaufmann, T., Schlipf, S., Sanz, J., Neubert, K., Stein, R. & Borner, C. (2003) J. Cell Biol. 160, 53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motz, C., Martin, H., Krimmer, T. & Rassow, J. (2002) J. Mol. Biol. 323, 729-738. [DOI] [PubMed] [Google Scholar]
- 45.Wei, M. C., Lindsten, T., Mootha, V. K., Weiler, S., Gross, A., Ashiya, M., Thompson, C. B. & Korsmeyer, S. J. (2000) Genes Dev. 14, 2060-2071. [PMC free article] [PubMed] [Google Scholar]