Summary
Hepatitis C virus (HCV) is endemic in many countries due to its high propensity to establish persistence1. The presence of HCV–specific T cells in repeatedly HCV–exposed subjects who test for HCV RNA and antibodies and do not have any history of HCV infection has been interpreted as T cell–mediated protection2-5. Here, we show in nonhuman primates that repeated exposure to human plasma with trace amounts of HCV induced HCV–specific T cells without seroconversion and systemic viremia, but did not protect upon subsequent HCV challenge. Rather, HCV–specific recall and de novo T cell responses as well as intrahepatic T cell recruitment and IFN-γ production were suppressed upon HCV challenge, concomitant to quantitative and qualitative changes in regulatory T (Treg) cells that began after subinfectious HCV exposure and increased after HCV challenge. In vitro Treg cell depletion restored HCV–specific T cell responses. Thus, T cells primed by trace amounts of HCV do not generate effective recall responses upon subsequent HCV infection. Subinfectious HCV exposure predisposes to Treg cell expansion, which suppresses effector T cells during subsequent infection. Strategies to reverse this exposure–induced suppression should be examined to aid the development of T cell–based vaccines against HCV and other endemic pathogens.
Keywords: hepatitis C virus, regulatory T cells, T cell response
At least 170 million people worldwide are persistently infected with HCV, a leading cause of chronic liver inflammation, cirrhosis and cancer1. Spontaneous HCV clearance occurs in less than 25% of acute infections, and is typically associated with T cell rather than antibody responses1,6-9. HCV–specific T cell responses have also been described in subpopulations of injection drug users who test negative for HCV RNA and antibodies and do not past HCV infection despite frequent exposure2-5, and in aviremic seronegative family members of HCV–infected subjects10,11. Based on these findings it has been proposed that repeated subinfectious (low–dose) exposure primes and maintains HCV–specific T cells that confer protective immunity3-5,10,11.
We set out to test this hypothesis using chimpanzees that had participated in a study to assess the infectivity of plasma and PBMC samples from HCV–antibody–positive patients with trace amounts of HCV, detected by nested RT–PCR below the detection limit of the standard clinical assay at the NIH (qualitative COBAS Amplicor HCV Test 2.0, Roche)12. The sporadic re–appearance of HCV RNA in these patients conincided with HCV–specific T cell responses and did not result in high–level viremia12.
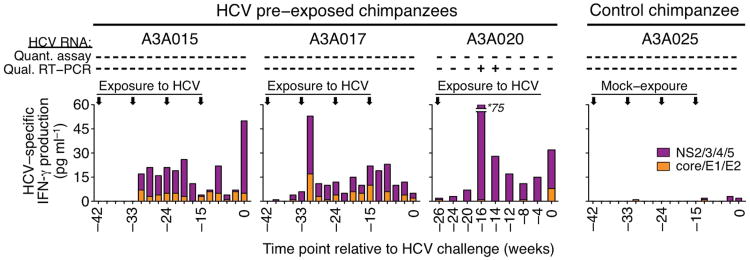
When chimpanzees A3A015 and A3A017 were infused at 9–week intervals with these plasma (infusions 1, 2, and 3) or PBMC samples (infusion 4) (Supplementary Table 1), they remained HCV RNA negative in blood and liver, did not mount antibody responses, but generated HCV–specific T cell responses as evidenced by IFN–γ secretion upon in vitro stimulation of PBMCs with HCV peptides (Fig. 1a). A third chimpanzee A3A020 transiently tested positive for HCV RNA in the blood by nested RT–PCR 10 and 12 weeks after plasma infusion, concomitant with increased HCV–specific T cell responses (Fig. 1a). Such responses were not observed in the control chimpanzee A3A025 after repeated exposure to blood products from HCV RNA–negative, HCV antibody–negative blood donors (Fig. 1a). Further characterization of the HCV–exposed chimpanzees revealed that both CD8+ and CD4+ T cells produced IFN-γ, TNF-α or MIP-1β in response to multiple HCV antigens (Supplementary Fig. 1a–c), but only a minority was polyfunctional (≤17% CD8+ T cells, ≤12% CD4+ T cells, Supplementary Fig. 1d). The majority of IFN-γ–producing CD8+ T cells were CD28 effector (61–88%) or effector memory cells (12–32%), and none were central memory cells (Supplementary Fig. 1e).
Figure 1.
Repeated exposure to blood samples from HCV–antibody–positive patients with trace amounts of HCV induces HCV–specific T cell responses. IFN-γ secretion by HCV–specific T cells as determined by cytometric bead array. Vertical arrows indicate the time points at which chimpanzees A3A015, A3A017 and A3A020 were intravenously infused with plasma or PBMC from HCV–antibody–positive patients with trace amounts of HCV, and chimpanzee A3A025 was infused with blood samples from HCV–antibody–negative HCV–RNA–negative healthy blood donors (Supplementary Table 1). “+” and “−” indicate “detection” and “no detection” of HCV RNA in the chimpanzee blood samples by quantitative COBAS Amplicor/COBAS Taqman HCV test (Roche) or by qualitative nested RT-PCR13, respectively. Numbers on the horizontal axis represent time points relative to challenge with 100 CID50 HCV genotype 1a at week 0.
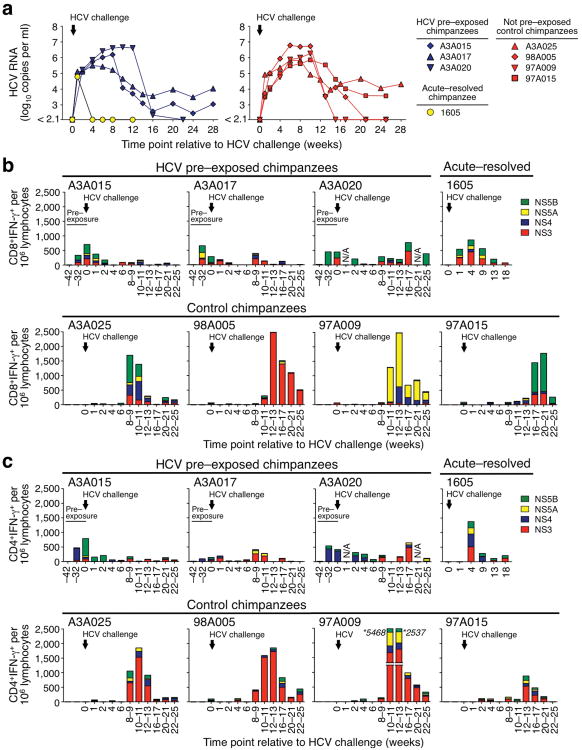
Chimpanzees that clear an acute HCV infection typically exhibit lower peak viremia levels and faster clearance of a secondary HCV challenge due to protective memory T cells8,9,13-16. However, when the HCV–pre–exposed chimpanzees A3A015, A3A017 and A3A020 with HCV–specific T cell responses were challenged with 100 CID50 HCV, they did not control viremia as rapidly as chimpanzee 1605 that had received the same HCV challenge after previous spontaneous clearance of acute HCV infection with high–titer viremia14 (Fig. 2a). Rather, they experienced the same prolonged high–titer viremia as four HCV–naïve control chimpanzees (A3A025, 98A005, 97A009 and 97A015) that had also been challenged with 100 CID50 17 (Fig. 2a). Two of three HCV–pre–exposed chimpanzees developed chronic infection (Supplementary Table 2).
Figure 2.
Repeated exposure to blood samples from HCV–antibody–positive patients with trace amounts of HCV suppresses T cell responses upon HCV challenge. (a) Serum HCV RNA titers after a 100 CID50 HCV genotype 1a challenge of three HCV pre–exposed chimpanzees (A3A015, A3A017 and A3A020), a chimpanzee that had spontaneously cleared HCV after acute infection with high–level viremia (1605)14, and four control chimpanzees (A3A025, 98A005, 97A009 and 97A015). Viral titers for chimpanzees 98A005, 97A009 and 97A015 and chimpanzee 1605 were previously reported14,17 and are shown for references purpose only. (b,c) frequencies of HCV–specific IFN-γ+CD8+ (b) and IFN-γ+CD4+ T cells (c) per 106 peripheral blood lymphocytes as determined by intracellular cytokine staining. The vertical arrow indicates the time point (week 0) at which all chimpanzees were challenged with 100 CID50 HCV genotype 1a. T cell data from week – 42 and week –32 (“pre-exposure”) are shown for those chimpanzees that had been infused with plasma or PBMC from HCV–antibody–positive patients with trace amounts of HCV.
Next, we investigated the reasons for the lack of immune protection in the three HCV–pre–exposed chimpanzees. At the time of HCV challenge (week 0), they displayed no HCV-E2-specific antibodies, but a higher frequency of HCV–specific IFN-γ–secreting CD8+ and CD4+ T cells than the HCV–recovered chimpanzee 1605 (Fig. 2b,c), which was supported by higher frequencies of TNF-α– and MIP-1β–secreting CD8+ and CD4+ T cells (Supplementary Fig. 2a,b). However, the T cell responses of the HCV–pre–exposed chimpanzees were not boosted after HCV challenge and rather decreased to minimum levels by week 4, the time point when HCV–specific CD8+ and CD4+ T cell recall responses of chimpanzee 1605 peaked (Fig. 2b,c for IFN-γ, Supplementary Fig. 2a,b for TNF-α and MIP-1β), consistent with other recovered and rechallenged chimpanzees8,16. Only ≤2% of the HCV–specific CD8+ T cells from HCV–pre–exposed chimpanzees, but 42% of those from chimpanzee 1605 were polyfunctional (Supplementary Fig. 2c).
Furthermore, new T cell responses were significantly suppressed in the three HCV–pre–exposed chimpanzees compared to the peak IFN-γ–response of HCV–specific CD8+ and CD4+ T cells in the four control chimpanzees (P < 0.05, Fig. 2b,c and Supplementary Fig. 2d). This was consistent with suppressed TNF-α+ and MIP-1β+ responses of HCV–specific T cells (Supplementary Fig. 2a,b), whereas CMV–specific T cell responses remained unchanged (Supplementary Fig. 3).
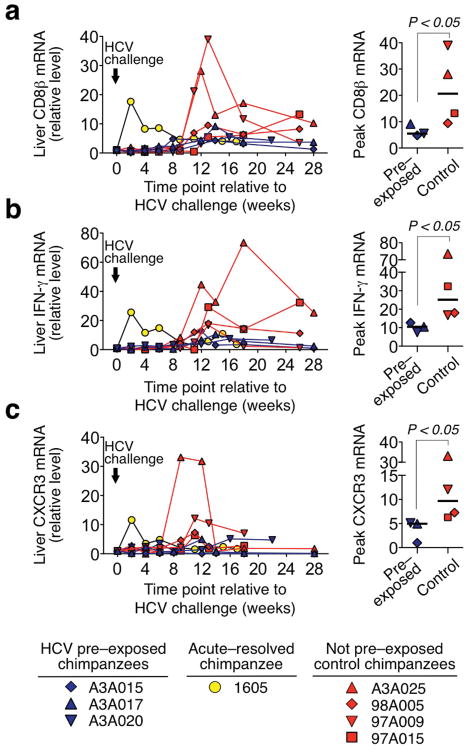
Suppression of immune responses occurred also in the liver, the site of HCV replication. Because liver biopsies yielded too few lymphocytes for ex vivo analysis, we used real–time PCR to monitor intrahepatic gene expression. There was almost no increase in the intrahepatic mRNA levels of CD8β, IFN-γ and CXCR3 during the first 6 weeks after HCV challenge in the three HCV–pre–exposed chimpanzees whereas they peaked early (week 2) in chimpanzee 1605, consistent with a rapid recall response (Fig. 3). The intrahepatic responses of the HCV–pre–exposed chimpanzees occurred at the same time (weeks 12-14) but with significantly lower magnitude than those of the four control chimpanzees (Fig. 3). Differences in CXCL11 mRNA levels were also observed but did not reach statistical significance (P = 0.114, not shown). Thus, prior exposures to trace amounts of HCV impede HCV–specific recall responses, induction of new T cells, and intrahepatic recruitment of CD8+ T cells and IFN–γ production after subsequent HCV challenge.
Figure 3.
Repeated exposure to blood samples from HCV–antibody–positive patients with trace amounts of HCV suppresses intrahepatic T cell recruitment and IFN–γ production upon subsequent HCV challenge. Intrahepatic (a) CD8β, (b) IFN-γ, and (c) CXCR3 mRNA levels as determined by real–time PCR, normalized to endogenous references (GAPDH and β7) and expressed as relative increase over week 0 expression, which was set to a value of 1. For chimpanzees 98A005, 97A009, 97A015 and Ch1605, the increase in CD8β and IFN-γ mRNA levels was calculated from previously reported data40. The vertical arrow indicates the time point (week 0) at which all chimpanzees were challenged with 100 CID50 HCV genotype 1a. Right panels represent the peak mRNA levels during acute HCV infection for each chimpanzee. The horizontal lines indicate the median. P values were calculated with the Mann–Whitney U test.
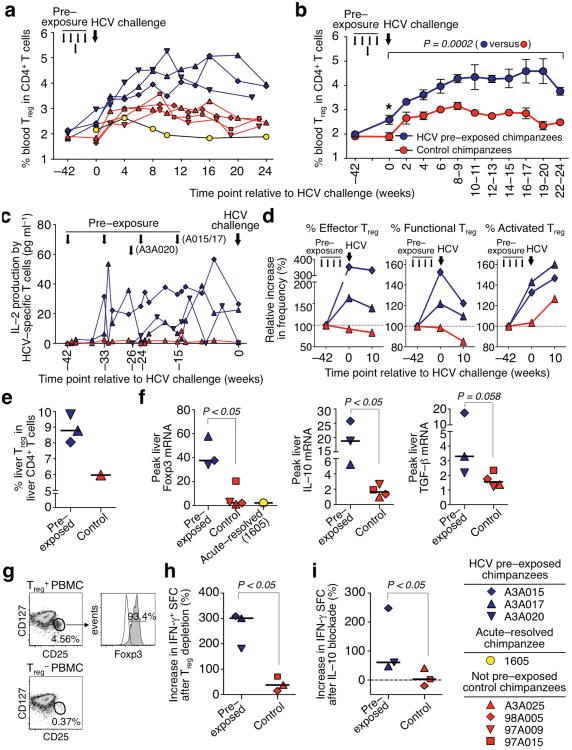
As a candidate mechanism we studied regulatory T (Treg) cells. Whereas the frequency of circulating CD4+CD25+Foxp3+ Treg cells did not differ among chimpanzees at the start of the study (week –42), it increased after repeated exposure to trace amounts of HCV (P < 0.05, week 0, Fig. 4a,b). This expansion may have been driven by IL–2 from HCV–specific effector T cells (Fig. 4c), because Treg cells require exogenous IL–2 for their maintenance and proliferation18. Challenge with 100 CID50 HCV further increased the Treg cell frequency in the three HCV–pre–exposed chimpanzees as compared to the previously HCV–recovered chimpanzee 1605 (Fig. 4a) and the four control chimpanzees (P = 0.0002, serial measures ANOVA, Fig. 4b). Furthermore, the Treg cell subset composition changed with an expansion of CD38+ activated, CD39+ functional19 and CD27– CD197–CD45RA– effector20 Treg cells (Fig. 4d).
Figure 4.
Repeated exposure to trace amounts of HCV predisposes to CD4+CD25+Foxp3+ Treg cell expansion upon subsequent HCV challenge. (a) CD4+CD25+Foxp3+ Treg cell frequency of three HCV pre–exposed chimpanzees, a chimpanzee that had spontaneously cleared HCV after acute infection with high–titer viremia and four HCV–naïve control chimpanzees, that were all challenged with 100 CID50 HCV genotype 1a (large arrow). Small arrows indicate HCV–pre–exposure of chimpanzees A3A015, A3A017 and A3A020 to trace amounts of HCV (Supplementary Table 1). (b) Mean CD4+CD25+Foxp3+ Treg cell frequency of HCV–pre–exposed and control chimpanzees after HCV challenge (mean ± s.e.m.; *, P < 0.05 at week 0, Mann–Whitney U test; comparison between both groups over time, serial measures ANOVA). (c) IL-2 produced by PBMC after HCV peptide stimulation. (d) Relative frequency of CD38+ activated, CD39+ functional19, and CD27–CD197–CD45RA– effector Treg cells20 at the study start (week –42), at HCV challenge (week 0), and 10 weeks thereafter. (e) Frequency of intrahepatic CD4+CD25+Foxp3+ Treg cells 6 weeks after HCV challenge. Horizontal lines indicate the median. (f) Intrahepatic Foxp3, IL–10 and TGF-β mRNA levels determined by real–time PCR, normalized to endogenous references and expressed as relative increase over pre-challenge levels. Horizontal lines indicate the median. (g) Efficacy of Treg cell isolation and depletion with magnetic beads. (h,i) Increase in IFN-γ–spot forming cells (SFC) after depletion of CD4+CD25+CD127dim/– Treg cells (h) or blockade of IL–10 signaling (i) as measured by IFN-γ ELISpot assay 8 weeks after HCV challenge. Statistical analyses in panels f–i, Mann–Whitney U test.
The three HCV–pre–exposed chimpanzees also displayed a higher intrahepatic Treg frequency (Fig. 4e) and higher peak intrahepatic Foxp3, IL–10 (P < 0.05, respectively) and TGF-β (P = 0.058) mRNA levels than chimpanzee 1605 and the four control chimpanzees after HCV challenge (Fig. 4f). In vitro depletion of Treg cells from PBMCs at 8 weeks and 19–37 weeks after HCV challenge enhanced HCV–specific T cell responses by a greater percentage in the HCV–pre–exposed chimpanzees than in the control chimpanzees (P < 0.05, Fig. 4g,h) with preserved immune hierarchy (Supplementary Fig. 4a). Since IL–10 secreted by Treg cells can directly suppress antigen–specific T cell responses, the suppressive function of Treg cells was confirmed by in vitro IL–10 neutralization combined with IL–10 receptor blockade that significantly enhanced HCV–specific T cell responses from HCV–pre–exposed but not control chimpanzees (P < 0.05, Fig. 4i, Supplementary Fig. 4b). The three HCV–pre–exposed chimpanzees also displayed a higher frequency of B7–H4+ circulating macrophages and higher intrahepatic B7–H4 mRNA levels than the four control chimpanzees after HCV challenge (Supplementary Fig. 5). Treg cells have been shown to suppress effector T cell responses by IL-10–mediated upregulation of B7-H4 on antigen–presenting cells21,22, and the timing of the intrahepatic B7–H4 mRNA peak coincided with the absence of HCV–specific T cell responses 8–12 weeks after HCV challenge in the HCV–exposed chimpanzees. The control chimpanzees mounted strong T cell responses at that time (Fig. 2b,c). These results suggest that HCV–specific T cells were induced after HCV challenge in all chimpanzees, but functionally suppressed in the HCV–pre–exposed chimpanzees. Of note, the sole HCV–pre–exposed chimpanzee (A3A020) that cleared the challenge inoculum did so after a decrease in Treg cell frequency (Fig. 4a) and a concomitant increase in HCV–specific CD8+ and CD4+ T cells between weeks 10 and 16 (Fig. 2b,c).
Summarily, we demonstrate that T cells primed by trace amounts of HCV do not generate effective recall responses upon subsequent HCV infection. Furthermore, exposure to trace amounts of HCV predisposes to Treg cell expansion, which suppresses HCV–specific effector T cells during subsequent acute infection. This scenario differs from allergen desensitization where exposure to increasing allergen doses suppresses IL–4 and IL–5 production via Treg cell induction and, contrary to our findings, increases IFN-γ responses23. Likewise, it differs from IL–10–dependent immunotolerance in beekeepers, which is induced by repeated high–dose antigen exposure24.
The absence of significant inflammation may be key to the observed Treg cell induction as suggested by the induction of Foxp3 in CD4+ T cells in mice that received small amounts of peptide25 without adjuvants25,26. Here we demonstrate that similar effects can be achieved by very small doses of live virus rather than inert antigen. A possible lack of signal 2 (costimulation) or, without sufficient viral replication, signal 3 (type I IFN) may impede the induction of optimal effector CD8+ T cells27 and instead favor Treg cell induction. The accelerated Treg cell expansion in the presence of high antigen levels after HCV challenge may imply HCV–specificity as suggested by in vitro studies28,29. IL–2 consumption by the expanding Treg cell population18 after HCV challenge may suppress new effector T cells, which depend on prolonged IL–2 signals30,31.
Our results were generated in the in vivo model that most closely represents HCV infection in humans, and warrant confirmation in human populations. At present, limited data is available to distinguish between subinfectious and full–dose HCV exposure in humans. Published studies on T cell responses in HCV RNA–negative and seronegative individuals are of cross–sectional design2-5,10,11 and thus do not exclude the possibility that T cell responses resulted from past acute HCV infection with systemic viremia and subsequent antibody loss32. However, we have recently shown in a prospective study that subinfectious HCV exposure of health care workers via accidental needlestick can indeed induce HCV–specific T cell responses in the absence of quantifiable viremia and HCV–specific antibodies33. Based on this study in humans and the HCV challenge data in the nonhuman primate model, we refute the notion that frequently low–dose HCV–exposed individuals who remain HCV–antibody–negative may have T cell mediated protective immunity upon HCV infection3,5,10,11. We propose that T cell–based protective immunity as described in re–challenged chimpanzees8 and suggested in some re–infected injection drug users34,35 requires clearance of a previous acute infection with systemic viremia.
The number of experimental chimpanzees in this study was too small to allow a statistical comparison of the outcome of HCV challenge in HCV–pre-exposed and naïve chimpanzees. However, it is conceivable that the observed Treg cell–mediated immune suppression also occurs in individuals who live in endemic areas rendering them less resistant upon subsequent HCV infection than individuals in non–endemic areas. This may extend to T cell responses to other endemic pathogens and would be consistent with the finding that the percentage and the number of Treg cells are higher among healthy individuals in a rural village with seasonal malaria than among individuals in an urban area where malaria is rare36.
Our findings may be relevant for vaccine research and for epidemiological studies because an increased Treg cell frequency reduces the response to vaccines37. They may, for example, offer a potential explanation for geographical variation in the efficacy of mycobacterium bovis bacilli Calmette–Guérin vaccination against tuberculosis, which is less effective in endemic areas38. Along this line, it would be of interest to examine whether an increased Treg cell frequency in HIV–negative individuals in HIV–endemic areas may have contributed to the vaccine failure in the HIV STEP 1 trial39. Considering that in vitro Treg cell–depletion restored the suppressed HCV–specific T cell responses in our study, strategies for in vivo blockade of these suppressive mechanisms may facilitate the development of T cell–based vaccines against endemic pathogens such as HCV, HIV, mycobacterium tuberculosis and plasmodium.
Online methods
Chimpanzees
Chimpanzees A3A015, A3A017, A3A020 and A3A025 were studied at New Iberia Research Center (NIRC), New Iberia, LA under protocols approved by the University of Louisiana Lafayette Animal Care and Use Committee (ACUC). Chimpanzees A3A015, A3A017 and A3A020 were intravenously infused with 17–31 ml plasma from HCV–antibody–positive patients who, after completion of interferon-based therapy, had tested HCV RNA–negative by qualitative COBAS Amplicor HCV Test 2.0 (Roche) but positive by nested RT–PCR12. Chimpanzees A3A015 and A3A017 were subsequently infused at 9–week intervals with two additional plasma samples and one PBMC sample (3.5 × 107) from additional patients with trace amounts of HCV RNA. The study was conducted to assess whether these samples were infectious. The control chimpanzee A3A025 was infused, at the same time intervals, with plasma and PBMC from blood donors without past HCV infection (Supplementary Table 1). Subjects were studied with informed consent under protocols approved by the NIDDK institutional review board of the US National Institutes of Health.
Fifteen weeks (for A3A015, A3A017 and A3A025) and 26 weeks (for A3A020) after the final infusion, all chimpanzees were challenged with 100 chimpanzee infectious doses 50 (CID50) HCV genotype 1a. At the indicated study time points, serum was isolated and cryopreserved on site. ACD–anticoagulated blood samples, a 10–20 mm liver biopsy piece in RPMI1640 with 5% fetal bovine serum and 2–5 mm snap–frozen liver biopsy piece were shipped overnight to NIH. PBMC and liver–infiltrating lymphocytes were isolated as described41.
Cryopreserved PBMC, serum and liver biopsies from (i) chimpanzees 98A005, 97A009 and 97A015 that had been challenged with 100 CID50 HCV genotype 1a in a previous study at NIRC17, and (ii) chimpanzee 1605 that had resolved an acute HCV infection prior to challenge with 100 CID50 HCV genotype 1a at the Food and Drug Administration (FDA), Rockville, Maryland in a previous study14 were studied for comparison (Supplementary Table 1) under protocols approved by the University of Louisiana Lafayette ACUC and the FDA ACUC, respectively.
Multicolor flow cytometry
(i) Analysis of HCV–specific T cell responses by cytometric bead array
Fresh PBMC (2 × 105 per well) were stimulated in RPMI1640 containing 5% fetal bovine serum, 100 IU mL1 penicillin, 100 μg mL−1 streptomycin and 2 mM L–glutamine (Mediatech) with 18 pools of overlapping fifteen–mer HCV genotype 1a (H77) peptides of the identical sequence as the challenge virus (1 μg ml−1 per peptide, 600 peptides total)12 or DMSO in 96–well plates. Culture supernatants were harvested after 42 hours to quantitate IFN-γ, TNF-α, MIP-1β and IL–2 by cytometric bead array (BD Biosciences). The HCV–specific response to each peptide pool was calculated by subtracting the background response (DMSO) from the response to each individual peptide pool. Cumulative responses to structural and nonstructural HCV antigens are shown.
(ii) Analysis of HCV–specific T cell responses by intracellular cytokine staining
Cryopreserved and thawed PBMC (2 × 106 per tube) were stimulated with or without 6 pools of fifteen-mer HCV genotype 1a peptides with an 11 amino acid overlap that spanned the HCV NS3, NS4, NS5A and NS5B sequences (4 μg ml−1 per peptides). Antibodies to CD28 (1 μg ml−1, clone CD28.2, BD Biosciences) and CD49d (1 μg ml−1, clone 9F10, BD Biosciences) were included in the peptide stimulation. After 1h incubation at 37°C and 5% CO2, 10 μg ml−1 brefeldin A (Sigma–Aldrich) was added. After 12h incubation, the cells were stained with antibodies to CD3AlexaFluor700 (clone UCHT1, 1:100) CD8APC H7 (clone RPA–T8, 1:33) and CD4BrilliantViolet421 (clone RPA–T4, 1:33) from BD Biosciences for 30 min at 4 °C, washed, fixed, permeabilized using FACS Perm 2 buffer (BD Biosciences) and stained with antibodies to IFN-γFITC (clone 1-D1K, 1:33, Mabtech), TNF-αAPC (clone MAb11, 1:20, BioLegend) and MIP-1βPE–Cy7 (clone D21-1351, 1:20, BD Biosciences) for 30 min at room temperature. After two washes, a minimum of 30,000 CD8+CD3+ T cell–gated events were acquired (Supplementary Fig. 6a). Data were expressed as number of cytokine–secreting CD3+CD8+ or CD3+CD4+ cells per 106 lymphocytes to allow comparison with published studies. The HCV–specific response to each pool was calculated by subtracting the background response (in the absence of HCV peptides) from the response to each individual peptide pool.
For phenotyping, PBMC were stimulated with HCV NS3 or NS5B peptide pools and antibodies to CD28 and CD49d as described above, stained with antibodies to CD3AlexaFluor700 (clone UCHT1, 1:100), CD28PE–Cy7 (clone CD28.2, 1:50) from BD Biosciences, CD8BrilliantViolet421 (clone RPA-T8, 1:33), CD45RABrilliantViolet605 (clone HI100, 1:33) from Biolegend and CCR7eFluor780 (clone 3D12, 1:50, eBioscience) for 30 min at 4 °C, washed, fixed, permeabilized, stained for IFN-γ (clone 1-D1K, 1:33, Mabtech) and analyzed as described in Supplementary Fig. 6a.
(iii) Analysis of Treg cells
PBMC or liver–infiltrating lymphocytes were incubated with 5 μg ml−1 ethidium bromide monoazide (Sigma–Aldrich) under light for 8 minutes, washed twice, stained with antibodies to CD3AlexaFluor700 (clone UCHT1, 1:100), CD4APC–Cy7 (clone RPA-T4, 1:33) and CD25PE (clone 2A3, 1:10) from BD Biosciences, permeabilized with eBioperm buffer (eBioscience) and stained with antibody to Foxp3FITC (clone PCH101, 1:20, eBioscience) (Supplementary Fig. 6b).
For phenotyping of Treg cell subpopulations, PBMC were incubated for 30 minutes with LIVE/DEAD Aqua fixable viability dye (Invitrogen), washed and stained with antibodies to CD3APC–Cy7 (clone SK7, 1:20), CD4V450 (clone RPA-T4, 1:20), CD38PerCP–Cy5.5 (clone HIT2, 1:20), CD25PE–Cy–7 (clone M-A251, 1:10), CD127APC (clone hIL-7R-M21, 1:10) from BD Biosciences, CD45Qdot800 (clone H130, 1:100), CD8Qdot605 (clone 3B5, 1:100), CD27Qdot655 (clone CLB–27/1, 1:50), HLA–DRPE–Cy5.5, (clone Tü36, 1:20), CD39FITC (clone A1, 1:10), CD103PE–Cy5 (clone LF61, 1:20) from Invitrogen, CD45RAPE–TR (clone 2H4LDH11LDB9, 1:20, Beckman Coulter) and CCR7Alexafluor700 (clone 150503, 1:20, R&D Systems), followed by permeabilization with eBioperm buffer (eBioscience), staining with antibody to Foxp3PE (clone PCH101, 1:20, eBioscience) and fixation in PBS containing 2% formaldehyde (Supplementary Fig. 6c).
All stained cells were analyzed on a LSRII flow cytometry (BD Biosciences) using FACS Diva version 6.1.2 (BD Biosciences) and FlowJo version 8.8.6 (Tree Star) software.
ELISpot assays
Three different ELISpot assays were performed:
Treg cells were isolated from PBMC with magnetic beads using the CD4+CD25+CD127dim/– regulatory T cell isolation kit II (Miltenyi) according the manufacturer's protocol. Treg cell–depleted or Treg cell–reconstituted PBMC (300,000 cells per well) were stimulated with 18 pools of overlapping HCV genotype 1a peptides (1 μg ml−1 per peptide, 600 peptides total).
300,000 PBMC per well were stimulated with HCV peptide pools with or without an IL–10 neutralizing (clone JES3–9D7, 10 μg ml−1, eBioscience) and IL–10Rβ-blocking (clone 90220, 10 μg ml−1, R&D Systems) antibodies.
300,000 PBMC per well were stimulated with pools of influenza A virus, CMV and EBV T cell epitopes (1 μg ml−1 per peptide, Supplementary Methods). All IFN-γ ELISpot assays were set up in duplicates and processed as described12. The specific response was calculated by subtracting the mean number of spots in the absence of peptides from the mean number of spots to each HCV peptide pool.
Quantitation of intrahepatic gene expression by real–time PCR
RNA was isolated from snap–frozen liver biopsies using Pico Pure RNA isolation kit (Arcturus) or RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Complementary DNA was synthesized using the MonsterScript 1st–Strand cDNA Synthesis Kit (Epicentre Biotechnologies). TaqMan Gene Expression Assays (Applied Biosystems) were performed in duplicates to determine CD8β, IFN-γ, Foxp3, IL–10, TGF-β, CXCR3, CXCL11 and B7–H4 mRNA levels. The amount of specific mRNA was calculated using comparative cycle threshold values and standard curves, normalized to internal controls (GAPDH and β7) and shown as increase over week 0 expression, which was set to 1. For chimpanzees 98A005, 97A009, 97A015 and Ch1605, previously reported data40 were used to calculate the increase in CD8β and IFN-γ mRNA levels over baseline, with baseline (week 0) expression set to 1.
Quantitative and qualitative detection of HCV RNA
Plasma HCV RNA was quantitated by COBAS Ampliprep/COBAS Taqman HCV test (Roche). The limit of quantitation was 116 copies per ml, the limit of detection was 28 copies per ml (95% probability rate). Viral titers for chimpanzees 98A005, 97A009 and 97A015 and chimpanzee 1605 were previously reported14,17 and are shown for references purpose only. For qualitative RT–PCR, RNA was extracted from 1 ml blood using the Roche High Pure Viral Nucleic Acid Large Volume Kit (Roche). Reverse transcription (RT) and qualitative nested RT–PCR were performed as previously described12 except that Superscript III (Invitrogen) and a primer amount of 2.5 pmol were used in the RT reaction. The RT-PCR sensitivity was 2 copies per ml as determined by testing serial dilutions of serum with known HCV titer (as assessed by COBAS Ampliprep/COBAS Taqman HCV test) in pre–infection serum samples from the same chimpanzees.
Statistical analysis
Non–parametric Mann–Whitney U test and serial measures ANOVA were performed with GraphPad Prism Version 5.0 (GraphPad Software) and JMP (SAS). A P value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank T. J. Rowell, J. Fontenot and the staff at New Iberia Research Center for the care of the chimpanzees and technical support, and L. Holz, NIDDK and W. Kastenmüller, NIAID for critical discussion and reading of the manuscript. This work was supported by the intramural research program of the US National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health.
Footnotes
Author contributions: SP, NSV and BR designed the study, analyzed the data and wrote the paper; SP, NSV and ECS processed blood and liver biopsy samples; SP and ECS characterized T cell responses; AB and JPM characterized Treg cell subsets by flow cytometry; NSV performed virological assays and some RT-PCRs; AF and SC challenged three additional control chimpanzees, and performed virological analyses. All authors discussed the results and commented on the manuscript.
Author Information: The authors declare no competing financial interests.
References
- 1.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizukoshi E, et al. Hepatitis C virus (HCV)-specific immune responses of long-term injection drug users frequently exposed to HCV. J Infect Dis. 2008;198:203–212. doi: 10.1086/589510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman AJ, et al. Prevalence of production of virus-specific interferon-gamma among seronegative hepatitis C-resistant subjects reporting injection drug use. J Infect Dis. 2004;190:1093–1097. doi: 10.1086/422605. [DOI] [PubMed] [Google Scholar]
- 4.Thurairajah PH, et al. Hepatitis C virus (HCV)-specific T cell responses in injection drug users with apparent resistance to HCV infection. J Infect Dis. 2008;198:1749–1755. doi: 10.1086/593337. [DOI] [PubMed] [Google Scholar]
- 5.Zeremski M, et al. Hepatitis C virus-specific T-cell immune responses in seronegative injection drug users. J Viral Hepat. 2009;16:10–20. doi: 10.1111/j.1365-2893.2008.01016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechner F, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thimme R, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoukry NH, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. The Journal of experimental medicine. 2003;197:1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grakoui A, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 10.Al-Sherbiny M, et al. Exposure to hepatitis C virus induces cellular immune responses without detectable viremia or seroconversion. Am J Trop Med Hyg. 2005;73:44–49. [PubMed] [Google Scholar]
- 11.Scognamiglio P, et al. Presence of effector CD8+ T cells in hepatitis C virus-exposed healthy seronegative donors. J Immunol. 1999;162:6681–6689. [PubMed] [Google Scholar]
- 12.Veerapu NS, Raghuraman S, Liang TJ, Heller T, Rehermann B. Sporadic reappearance of minute amounts of hepatitis C virus RNA after successful therapy stimulates cellular immune responses. Gastroenterology. 2011;140:676–685. e671. doi: 10.1053/j.gastro.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassett SE, et al. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology. 2001;33:1479–1487. doi: 10.1053/jhep.2001.24371. [DOI] [PubMed] [Google Scholar]
- 14.Major ME, et al. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. Journal of virology. 2002;76:6586–6595. doi: 10.1128/JVI.76.13.6586-6595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nascimbeni M, et al. Kinetics of CD4+ and CD8+ memory T-cell responses during hepatitis C virus rechallenge of previously recovered chimpanzees. Journal of virology. 2003;77:4781–4793. doi: 10.1128/JVI.77.8.4781-4793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanford RE, et al. Cross-genotype immunity to hepatitis C virus. Journal of virology. 2004;78:1575–1581. doi: 10.1128/JVI.78.3.1575-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folgori A, et al. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nature medicine. 2006;12:190–197. doi: 10.1038/nm1353. [DOI] [PubMed] [Google Scholar]
- 18.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biancotto A, Dagur PK, Fuchs JC, Langweiler M, McCoy JP., Jr OMIP-004: in-depth characterization of human T regulatory cells. Cytometry A. 2012;81:15–16. doi: 10.1002/cyto.a.21158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kryczek I, et al. Cutting edge: induction of B7-H4 on APCs through IL-10: novel suppressive mode for regulatory T cells. J Immunol. 2006;177:40–44. doi: 10.4049/jimmunol.177.1.40. [DOI] [PubMed] [Google Scholar]
- 22.Sica GL, et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;18:849–861. doi: 10.1016/s1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 23.Larche M, Akdis CA, Valenta R. Immunological mechanisms of allergen-specific immunotherapy. Nat Rev Immunol. 2006;6:761–771. doi: 10.1038/nri1934. [DOI] [PubMed] [Google Scholar]
- 24.Meiler F, et al. In vivo switch to IL-10-secreting T regulatory cells in high dose allergen exposure. J Exp Med. 2008;205:2887–2898. doi: 10.1084/jem.20080193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apostolou I, von Boehmer H. In vivo instruction of suppressor commitment in naive T cells. J Exp Med. 2004;199:1401–1408. doi: 10.1084/jem.20040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtsinger JM, Mescher MF. Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol. 2010;22:333–340. doi: 10.1016/j.coi.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebinuma H, et al. Identification and in vitro expansion of functional antigen-specific CD25+ FoxP3+ regulatory T cells in hepatitis C virus infection. J Virol. 2008;82:5043–5053. doi: 10.1128/JVI.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, et al. Analysis of FOXP3+ regulatory T cells that display apparent viral antigen specificity during chronic hepatitis C virus infection. PLoS pathogens. 2009;5:e1000707. doi: 10.1371/journal.ppat.1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalia V, et al. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 31.de Goer de Herve MG, Jaafoura S, Vallee M, Taoufik Y. FoxP3(+) regulatory CD4 T cells control the generation of functional CD8 memory. Nat Commun. 2012;3:986. doi: 10.1038/ncomms1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takaki A, et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nature medicine. 2000;6:578–582. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 33.Heller T, et al. Occupational Exposure to Hepatitis C Virus: Early T-Cell Responses in the Absence of Seroconversion in a Longitudinal Cohort Study. J Infect Dis. 2013;208:1020–1025. doi: 10.1093/infdis/jit270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osburn WO, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2010;138:315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta SH, et al. Protection against persistence of hepatitis C. Lancet. 2002;359:1478–1483. doi: 10.1016/S0140-6736(02)08435-0. [DOI] [PubMed] [Google Scholar]
- 36.Finney OC, Nwakanma D, Conway DJ, Walther M, Riley EM. Homeostatic regulation of T effector to Treg ratios in an area of seasonal malaria transmission. Eur J Immunol. 2009;39:1288–1300. doi: 10.1002/eji.200839112. [DOI] [PubMed] [Google Scholar]
- 37.Surls J, Nazarov-Stoica C, Kehl M, Casares S, Brumeanu TD. Differential effect of CD4+Foxp3+ T-regulatory cells on the B and T helper cell responses to influenza virus vaccination. Vaccine. 2010;28:7319–7330. doi: 10.1016/j.vaccine.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 38.Black GF, et al. Patterns and implications of naturally acquired immune responses to environmental and tuberculous mycobacterial antigens in northern Malawi. J Infect Dis. 2001;184:322–329. doi: 10.1086/322042. [DOI] [PubMed] [Google Scholar]
- 39.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin EC, et al. The kinetics of hepatitis C virus-specific CD8 T-cell responses in the blood mirror those in the liver in acute hepatitis C virus infection. J Virol. 2008;82:9782–9788. doi: 10.1128/JVI.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin EC, et al. Delayed induction, not impaired recruitment, of specific CD8(+) T cells causes the late onset of acute hepatitis C. Gastroenterology. 2011;141:686–695. doi: 10.1053/j.gastro.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.