Abstract
P301L mutant tau transgenic mice develop neurofibrillary tangles, a histopathologic hallmark of Alzheimer's disease and frontotemporal dementia (FTDP-17). To identify differentially expressed genes and to gain insight into pathogenic mechanisms, we performed a stringent analysis of the microarray dataset obtained with RNA from whole brains of P301L mutant mice and identified a single up-regulated gene, glyoxalase I. This enzyme plays a critical role in the detoxification of dicarbonyl compounds and thereby reduces the formation of advanced glycation end products. In situ hybridization analysis revealed expression of glyoxalase I in all brain areas analyzed, both in transgenic and control mice. However, levels of glyoxalase I protein were significantly elevated in P301L brains, as shown by Western blot analysis and immunohistochemistry. Moreover, a glyoxalase I-specific antiserum revealed many intensely stained flame-shaped neurons in Alzheimer's disease brain compared with brains from nondemented controls. In addition, we examined a single nucleotide polymorphism predicting a nonconservative amino acid substitution at position 111 (E111A) in ethnically independent populations. We identified significant and consistent deviations from Hardy–Weinberg equilibrium, which points to the presence of selection forces. The E111A single nucleotide polymorphism was not associated with the risk for Alzheimer's disease in the overall population. Together, our data demonstrate the potential of transcriptomics applied to animal models of human diseases. They suggest a previously unidentified role for glyoxalase I in neurodegenerative disease.
Alzheimer's disease (AD) and frontotemporal dementia are common forms of age-related dementing diseases. They are characterized by proteinaceous aggregates, which are resistant to proteolysis due to conformational changes and posttranslational modifications such as hyperphosphorylation and glycation (1–4). In AD, these aggregates are β-amyloid plaques and neurofibrillary tangles (NFT). NFT formation, in the absence of overt amyloid plaques, is found in a group of neurodegenerative diseases, including frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17) (5, 6). In affected cells, the microtubule-associated protein tau is abnormally phosphorylated and relocalized from axonal to somatodendritic compartments, where it accumulates in aggregates that eventually assemble into NFT (7). The identification of mutations in the tau gene in FTDP-17 established that dysfunction of tau in itself can lead to dementia (6).
NFT formation has been reproduced in transgenic mice by expression of FTDP-17 mutant tau, both in neurons (8–12) and in glial cells (13–15). Moreover, intracerebral injection of β-amyloid fibrils caused significant increases of NFT in the amygdala of P301L (FTDP-17) mutant mice (16). A similar increase was achieved by crossing β-amyloid-producing amyloid precursor protein mutant mice with P301L mice (17).
The pathologic similarities between the P301L transgenic mice and AD suggest common pathogenic mechanisms. Therefore, we undertook a transcriptomic analysis of whole brains obtained from P301L transgenic mice and nontransgenic controls. By applying highly stringent criteria, we identified glyoxalase I (GLO) as the only up-regulated gene in P301L mice.
The glyoxalase system consists of the enzymes GLO and glyoxalase II, together with the cofactor glutathione, and detoxifies α-ketoaldehydes, especially methylglyoxal (MG), thereby preventing the formation of advanced glycation end products (AGEs) (18–20). AGE modification renders tau resistant to proteolysis, thus assisting in its aggregation (21).
To further address the role of GLO in AD, we assessed its mRNA and protein levels in P301L transgenic mice and its protein levels in AD brains. Finally, we observed that a nonsynonymous single nucleotide polymorphism (SNP) deviated from Hardy–Weinberg equilibrium in ethnically independent populations, suggesting the presence of yet unknown selection forces. This SNP has previously been shown to be associated with the risk for prostate cancer, demonstrating a potential role of GLO in a broad range of human pathogenic conditions. Whereas previous studies have shown a role for GLO in diabetes and cancer, our biochemical and immunohistochemical data suggest an additional role in AD.
Materials and Methods
Transgenic Mice. The transgenic mice used here express the longest human brain tau isoform with the human pathogenic mutation P301L under control of the neuron-specific mThy1.2 promoter (9). Line pR5–183 expressed tau in many brain areas, including hippocampus and cortex; however, NFT formation was mainly confined to the amygdala (16). The mice were caged in groups of up to five and kept on a 12-h light/dark cycle at 22°C. Food pellets and water were available ad libitum. This animal research has been approved by the local animal studies committee.
RNA Preparation, Labeling, and Hybridization of Affymetrix Microarrays and Northern Blot Analysis. At 12 months of age, three P301L transgenic mice and three nontransgenic littermates were killed by decapitation. The cerebrum was dissected and homogenized in TRIzol reagent (Life Technologies, Basel). The RNA pellet was dissolved in DEPC-H2O and stored at -80°C until use. The A260/A280 ratio was determined with the genequant pro RNA/DNA calculator software (Biochrom, Cambridge, U.K.) and was within a range of 1.9–2.1 for all samples. The high quality was further confirmed by agarose gel electrophoresis.
Double-stranded cDNA was synthesized from 20 μg of total RNA by reverse transcription using the SuperScript cDNA synthesis kit (Invitrogen) and a T7-(dT)24 primer (Microsynth, Balgach, Switzerland), and then cleaned with Phase Lock Gel Light (Vaudaux, Basel). Biotin-labeled cRNA was synthesized with T7 RNA polymerase using the BioArray RNA labeling kit (Loxo, Dossenheim, Germany), cleaned with the RNeasy kit (Qiagen, Valencia, CA), fragmented and hybridized to the Affymetrix microarray murine chip MG-U74Av2, following the manufacturer's recommendations (Affymetrix, Santa Clara, CA). Staining and scanning was carried out at the Microarray Suit workstation (Functional Genomics Center Zurich). The image data were converted into numeric data and analyzed with the Affymetrix software mas 5.0 and dmt 3.0. Each gene was accompanied by a measure of the expression (signal intensity) and by a detection flag (present, absent, and marginal).
To identify differentially expressed genes, the mean and median value of the signals was determined. Differences were considered significant when P ≤ 0.05 in the Student t test and/or the Mann–Whitney U test. We then applied a pairwise comparison using the dmt 3.0 program. Each of the three P301L mice was compared with all three wild-type mice, generating for each gene nine comparisons, which were classified as increase, marginal increase, no change, marginal decrease, and decrease. A gene was considered to be increased when it was detected as “increase” in all nine data sets. The same criterion was applied to decreased genes.
The RNAs used in the above microarray analysis were also used for Northern blot analysis as described (22). As a probe for GLO, a 346-bp probe was generated by PCR using the oligonucleotides 5′-CGGAACAAGGGGCAGCAGGCAGGAAACTTCG-3′ and 5′-TGAGTCAAGGGGGAGTCTGGCCTATGCTCTC-3′. For normalization control, a 1.9-kb β-actin cDNA was used (22).
Nonradioactive in Situ Hybridization. Oligonucleotide 5′-CTTGTCGAGGGAAATCTCTAGTGTCAACGATGACGGACAGAG-3′ was used as antisense probe for GLO. The complementary sequence was used as sense probe. For digoxigenin-dUTP labeling, the DIG oligonucleotide tailing kit (Roche, Mannheim, Germany) was used. It yielded ≈4 pmol/μl, as determined by dot blotting with an antidigoxigenin antibody (Roche). Paraffin sections (7 μm thick) were cut and incubated in 2× SSC, 1× Denhardt's, 10% dextran sulfate, 50 mM phosphate buffer, pH 7.0, 50 mM DTT, 5 μg/ml polydeoxyadenylic acid, 100 μg/ml polyadenylic acid, 250 μg/ml yeast tRNA, 500 μg/ml salmon sperm DNA, and 16% deionized formamide for 2 h at 37°C. Hybridization was carried out with 30 ng of labeled oligonucleotide in a volume of 30 μl per section for 20 h at 37°C. Negative controls included hybridization with the labeled sense probe, a 1:100 mixture of labeled and unlabeled antisense probe, and omission of the probe. After hybridization, sections were washed, blocked, and incubated with an anti-DIG-antibody (Roche, 1:200) for 4 h at room temperature. Finally, sections were incubated in a buffer containing 100 mM Tris·Cl, pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.3% nitroblue tetrazolium, 0.35% 5-bromo-4-chloro-3-indolyl phosphate, and 1 mM levamisol and protected from light for 20 h until the color developed.
Immunohistochemistry. Six-month-old P301L transgenic mice and nontransgenic littermates were perfused transcardially with 4% paraformaldehyde in phosphate buffer, pH 7.4. Stainings were done on 5-μm paraffin-embedded frontal brain sections as previously described (23). Human brain samples were from frontal and temporal cortex and the hippocampus of AD patients and nondemented controls aged 78–85 years. Sections were dehydrated and flat-embedded between glass slides and coverslips in Mowiol 4–88 (Roche). Antibody HT7 (Innogenetics, Ghent, Belgium; diluted 1:600) was used to detect human tau; AT100 to detect tau phosphorylated at Thr-212/Ser-214 (Innogenetics, 1:300); CP13 (P. Davies, Albert Einstein College of Medicine, Bronx, NY 1:500) to detect tau phosphorylated at Ser-202/Thr-205; and a monoclonal antibody (Innogenex, San Ramon, CA; 1:500) to detect glial fibrillar acidic protein. GLO-specific antisera were from S. Ranganathan and S. Staros (Fox Chase Cancer Center, Philadelphia) (1:50) and P. J. Thornalley (University of Essex, Wivenhoe Park, Colchester, U.K.) (1:10). For peroxidase/diaminobenzidine stainings, secondary antibodies were obtained from Vector Laboratories.
Western Blot Analysis. Total brain extracts were prepared from P301L transgenic and control brains. For human extracts, the gyrus inferior temporalis was obtained from three AD patients (69–85 years) and three nondemented controls (68–88 years). Extracts were normalized for protein content by using the DC Protein Assay (Bio-Rad) and separated by PAGE, followed by electrophoretic transfer onto a Hybond-ECL nitrocellulose membrane (Amersham Pharmacia). Ponceau stainings were included to confirm equal loading. Furthermore, signal intensities of GLO were normalized for β-actin and GAPDH levels. The membranes were blocked and reacted with primary and peroxidase-conjugated secondary antibodies as previously described (22). The following antibodies were used: HT7 (1:4,000), anti-GLO antiserum (S. Ranganathan and S. Staros; 1:500), anti-β-actin monoclonal antibody (Abcam, Cambridge, UK; 1:5,000), and anti-GAPDH monoclonal antibody (BioDesign, Kennebunk, ME; 1:6,000).
Human Studies: SNP Analysis and Statistics. Genetic studies on the association between GLO genotypes and AD were conducted on 926 subjects recruited from specialized memory clinics in Switzerland, Russia, and Greece. The clinical diagnoses of AD (n = 381; mean age of onset, 64.0 years; 165 women) were made according to the National Institute of Neurological Disorders and Stroke–Alzheimer's Disease and Related Disorders Association criteria (24). The control group (n = 545; mean age, 67.4 years; 224 women) consisted of elderly individuals who were either the spouses of patients with AD or subjects recruited from the outpatient memory clinics. Dementia and memory deficits in control subjects were excluded by neuropsychological testing, consisting of the Consortium to Establish a Registry for Alzheimer's Disease neuropsychological test battery (25) and the MiniMental State Examination. Studies on Hardy–Weinberg equilibrium of the GLO polymorphism in young individuals were done in 365 healthy subjects (mean age, 22.9 years; 248 women) recruited for neuropsychological testing. Information on polymorphic sites of GLO was derived from the SNP database (dbSNP) established by the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/SNP/index.html). SNP candidate rs2736654, predicting a nonconservative amino acid substitution at position 111 (E111A) was selected for genotyping by pyrosequencing on a PSQ 96 machine (www.pyrosequencing.com). Forward and reverse amplification primers for rs2736654 were 5′-GCAGGGGTTAGGCCAATTAT-3′ and 5′-CAGGCAAACTTACCGAATCC-3′, respectively. The reverse primer was biotinylated at the 5′ end. For the sequencing primer, 5′-GGGGCACTGAAGATG-3′ was used. Apolipoprotein E genotyping was done on a LightCycler as previously described (26). Informed consent was obtained from all participants, and the local human studies committee approved the study protocol. Genotype and allelic frequencies between patients with AD and control subjects were compared by Pearson χ2 tests and by logistic regression analysis. Statistical significance was assumed at P ≤ 0.05.
Results
Pairwise Comparison of Microarray Data Specifically Identifies Glyoxalase I as Up-Regulated Gene in P301L Mice. To identify differentially expressed genes in NFT-forming P301L tau transgenic mice, we used the Affymetrix microarray chip technology. Of the ≈6,000 known genes and 6,000 expressed sequence tags on the MG-U74Av2 chip, ≈40% were present in both P301L and wild-type mice, as defined by the mas 5.0 software. The general expression pattern was not affected by the transgene, as genes that were present in both groups showed a symmetrical pattern of alignment, ranging from very low signal intensities of around 100 to high intensities of up to 20,000 units, thus representing the entire spectrum of low to high abundant genes. Only a few genes were exclusively expressed by one group. To avoid false positives, these genes were not analyzed further as intensities were below 400 units.
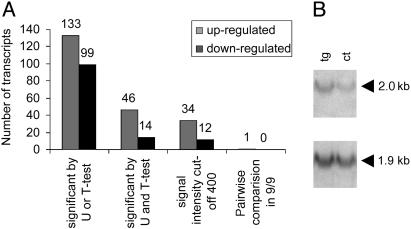
Based on absolute signal intensities of the present genes, we identified, by either the t test or the Mann–Whitney U test (P ≤ 0.05), 133 up-regulated and 99 down-regulated genes in P301L mouse brain (Fig. 1A). When both tests were applied simultaneously, 60 genes were differentially regulated. None of the genes displayed a >2-fold difference in signal intensities. These included low to high abundant genes with intensities ranging from 100 to 10,000 units. When a cutoff of 400 units was chosen, 34 up-regulated and 12 down-regulated genes remained (Fig. 1A).
Fig. 1.
Transcriptomic analysis of P301L and wild-type mice with Affymetrix chips. To identify differentially expressed genes in P301L tau transgenic mice, the Affymetrix microarray chip technology was applied (A). Of the ≈6,000 known genes and 6,000 expressed sequence tags on the MG-U74Av2 chip, 40% were present in both P301L and wild-type mice. Based on absolute signal intensities of the present genes, we identified, by either the t test or the Mann–Whitney U test (P ≤ 0.05), 133 up-regulated and 99 down-regulated genes in P301L brain. Of these, 46 genes were significantly up-regulated and 14 were down-regulated by both tests. To avoid false positives, a cutoff of 400 units was chosen. A further selection step was included by applying a highly stringent pairwise comparison, narrowing down the number of increased genes to just one, GLO, that was 1.6-fold up-regulated in P301L brains (P = 0.003, t test). Northern blotting of three P301L and three control RNAs followed by densitometric analysis revealed 1.5-fold increased GLO mRNA levels in P301L compared with control brains (P = 0.045, t test) (B). The GLO-(2.0 kb) and β-actin-specific (1.9 kb) bands are shown for one pair of P301L and control RNA samples.
When this group of genes was subjected to a further selection step, a highly stringent pairwise comparison (dmt 3.0 software), the number of differentially expressed genes was narrowed down to just one, GLO: probe 93269_at encoding GLO was increased in all nine datasets (Fig. 1A). A second probe for GLO, 93268_at, encoding a different portion of the gene, did not fulfill these criteria, possibly because it hybridizes to a second chromosomal locus. In terms of absolute intensities, GLO was 1.6-fold up-regulated in P301L compared with control brains (P = 0.003, t test).
Northern blotting followed by densitometric analysis revealed GLO mRNA levels, which were 1.52-fold increased in the P301L compared with control brains (P = 0.045, t test) (Fig. 1B).
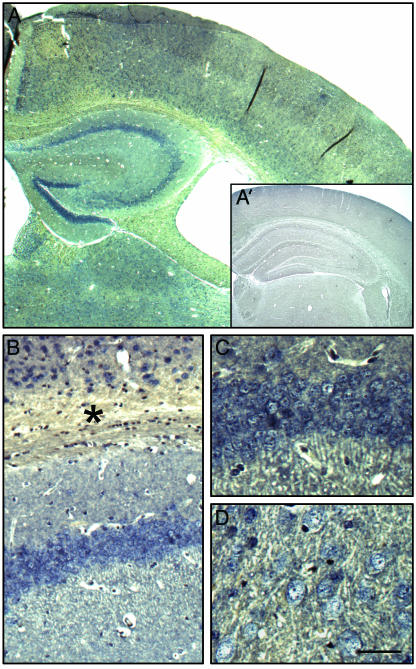
Distribution of Glyoxalase I mRNA in Wild-Type and P301L Transgenic Brain. To determine the mRNA distribution of GLO in mouse brain, coronal sections from wild-type and P301L mice were hybridized with a GLO antisense probe (Fig. 2). The sense probe was used as a negative control (Fig. 2 A Inset). GLO was strongly expressed in many brain areas, including cortex, hippocampus, and amygdala of all mice analyzed, irrespective of the genotype (Fig. 2). In the hippocampus, staining was most pronounced in CA1 pyramidal neurons and DG granule cells, whereas signal intensities were slightly lower for CA3 neurons (Fig. 2 A–C). The hybridization signal was weaker in additional brain areas such as cortex (Fig. 2 A and B) and amygdala (Fig. 2D). In addition to neurons, staining of cells in the corpus callosum demonstrated that glial cell types also transcribe the GLO gene at high levels (Fig. 2B). The in situ hybridization analysis did not reveal differences between P301L and control mice (data not shown). However, as this method is not sensitive enough to identify differences that are in the range of <3.5-fold (27), we analyzed brain extracts by Western blotting and stained brain sections with a GLO-specific antibody.
Fig. 2.
In situ hybridization analysis of glyoxalase I in murine brain. Digoxigenin-labeled oligonucleotide probes were hybridized to brain sections from wild-type mice (A–C). Digoxigenin-labeled sense probes were used as control (A′). The distribution pattern of the GLO mRNA is shown for cortex and hippocampus, where it was strongest (A–C), and the amygdala (D). GLO expression is not restricted to neurons, as shown by the presence of stained glial cells in the corpus callosum (*). The general pattern and relative intensities were identical in P301L mice (data not shown). Scale bar: 400 μm(A), 900 μm (A′), 115 μm (B), and 60 μm (C and D).
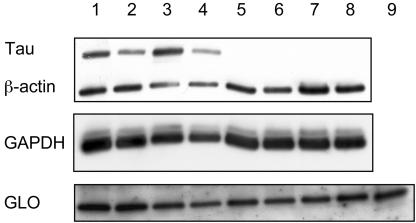
Increases in Glyoxylase I Protein in P301L Transgenic and Human AD Brain. Western blot analysis of whole brain extracts revealed 1.8-fold increased GLO levels in P301L mice when normalized for β-actin (P = 0.003, t test) and 1.4-fold when normalized for GAPDH (P = 0.037, t test) (Fig. 3A). A 2-fold dilution series of purified GLO protein and murine brain extracts was included as control (data not shown). When AD brain extracts were compared with those from healthy controls, normalization for β-actin revealed a 1.5-fold difference (P = 0.226, t test) and for GAPDH a 0.9-fold difference (P = 0.502, t test), possibly reflecting a more advanced stage of the disease with many dead neurons and likely lower GLO expression.
Fig. 3.
Western blot analysis of glyoxalase I. Western blot analyses of whole brain extracts reveals that GLO levels are 1.8-fold increased in P301L tauexpressing mice when normalized for β-actin (P = 0.003, t test) and 1.4-fold when normalized for GAPDH (P = 0.037, t test). Lanes 1–4, HT7 (human tau)-reactive P301L mice; lanes 5–8, wild-type littermate controls; lane 9, purified human GLO protein.
Next, we performed an immunohistochemical analysis of P301L transgenic brain. Many pyramidal neurons, as determined by costaining with a MAP2-specific antibody, were stained by the GLO-specific antibody (from Dr. Ranganathan) and at much higher intensities than in controls (Fig. 4 A and B and data not shown). To determine whether glial cells would also express GLO as suggested by the in situ hybridization analysis, we included costainings with a glial fibrillar acidic protein-specific antibody (data not shown). The GLO-specific antibody stained predominantly, but not exclusively, neurons.
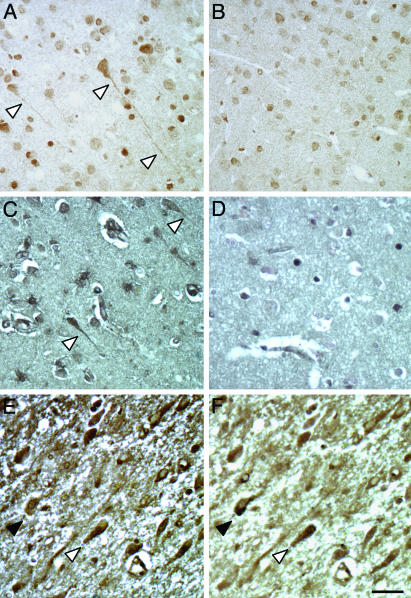
Fig. 4.
Distribution of glyoxalase I protein in transgenic and AD brain. An immunohistochemical analysis with a GLO-specific antiserum (from Dr. Ranganathan) identifies many flame-shaped pyramidal neurons in P301L transgenic (A) compared with control brain (B). When different brain areas are obtained from three AD brains, a prominent staining including flame-shaped, NFT-like neurons is found as shown for the temporal cortex (C), which is absent in nondemented controls (D). We first performed a GLO staining (E), stripped the sections, and restained them with the NFT-specific phosphorylation-dependent anti-tau antibody AT100 (F). AT100-tau is more condensed, whereas GLO shows a more uniform and mostly weak cellular distribution. In neurons with an intermediate AT100 reactivity, GLO and AT100-tau show a different subcellular distribution (open arrowhead). Those neurons that are strongly stained for AT100 do not display a strong GLO staining (black arrowhead). The reverse is also true. Scale bar, 50 μm.
To determine whether GLO was also differentially expressed in human AD brain, we stained sections taken from the frontal and temporal cortex and the hippocampus of human brains with confirmed AD pathology and corresponding sections of healthy controls. We found strong staining for GLO in all analyzed brain areas of AD but not control brains (Fig. 4 C and D). In particular, flame-shaped neurons were strongly stained by the antiserum. A similar staining pattern was obtained with a second antiserum (from P. J. Thornalley) but intensities were much lower (data not shown). To determine whether these flame-shaped neurons were bearing NFT, we did a GLO staining, stripped the sections, and restained them with the NFT-specific phosphorylation-dependent anti-tau antibody AT100 (Fig. 4 E and F).
Although GLO staining was mostly weak, those neurons that were strongly stained for GLO did not display a strong AT100 staining. The reverse was also true. When neurons showed an intermediate AT100 reactivity, GLO and AT100-tau showed a different subcellular distribution (Fig. 4 E and F, open arrowhead). Whereas AT100-tau was more condensed, GLO was more uniformly distributed. Together, our data show increased levels of GLO in P301L transgenic compared with wild-type brains and in AD brains compared with brains from nondemented controls.
Influence of a Coding SNP of Glyoxalase I on AD Risk and AD-Related Traits. In a first series of 677 unrelated AD cases and controls, genotype and allelic frequencies did not differ significantly (P > 0.2). However, sex-specific analysis revealed a significant association of the ala allele with AD in males (P = 0.01) but not in females (P = 0.4) There was no significant interaction between the apolipoprotein E and GLO genotypes with influence on AD risk. To confirm the sex-specific association of the GLO genotype with AD, we studied an additional sample of 249 male AD patients and control subjects and failed to detect a significant association both in this subsample and in the pooled sample (P = 0.3). Analysis of genotype frequency revealed significant deviations from Hardy–Weinberg equilibrium in the Swiss, the Russian, and the Greek samples independent of affection status (P < 0.04). To assess whether this deviation was confined to the elderly population, we additionally genotyped 365 young Swiss subjects between 18 and 35 years. Again, genotype frequencies deviated significantly from Hardy–Weinberg equilibrium (P < 0.001).
Discussion
The present study provides evidence for a role of GLO in AD. Selective up-regulation of GLO mRNA was found by a stringent microarray data analysis in P301L mutant tau transgenic mice, which model central aspects of the tau pathology of AD. Moreover, by analyzing murine and AD brains, we found that GLO protein accumulated in P301L transgenic mice and in human AD brain. Increased GLO levels have been reported for other human pathologic conditions such as cancer and diabetes (28, 29).
Of several organs studied, GLO expression has been shown in all tissues examined including brain (30). A detailed expression pattern, however, has so far not been performed. Here, we showed by in situ hybridization analysis that GLO is expressed in neurons and glial cells throughout the brain, with highest levels in the CA1 region and the dentate gyrus. The general expression pattern did not differ in the P301L mice compared with wild-type controls. Also, the staining did not reveal differences in signal intensities, possibly reflecting the insensitivity of this technique to detect 1.6-fold differences in gene expression. An at least 3.5-fold difference as detected by the Affymetrix chip is required to detect differences by in situ hybridization analysis (27). However, by Western blotting, we were able to demonstrate significantly increased protein levels of GLO in P301L mice. Similarly, by immunohistochemistry, we identified many flame-shaped pyramidal neurons that were strongly stained. When we analyzed human brain samples, we observed an even stronger staining in AD brains, whereas healthy control brains were only very weakly stained. Among the prominently stained cell types were many NFT-like flame-shaped neurons, suggesting that GLO is up-regulated in response to tau aggregation. When we used the NFT-specific phosphorylation-dependent anti-tau antibody AT100, we found that neurons which were strongly stained for AT100 did not display a strong GLO staining and vice versa. In neurons with an intermediate AT100 reactivity, AT100-tau was more condensed revealing tau filament (NFT) formation, whereas GLO was more uniformly distributed. This argues in favor of increases in GLO protein during earlier stages of the disease rather than for the inclusion or trapping of (hyperphosphorylated) GLO in NFT.
How the synthesis and enzymatic activation of GLO are regulated is not understood. The sequence of GLO contains several potential phosphorylation sites, and one study proposed that phosphorylation by PKA would affect its function (19, 28). As several kinases are up-regulated both in tau transgenic mice and in AD (31, 32), GLO may be differentially phosphorylated in P301L tau transgenic mice. However, in our Affymetrix dataset, we have no evidence for PKA up-regulation, although we cannot exclude slight increases that we would fail to detect by our method.
As GLO is up-regulated in the P301L tau transgenic mice with only modest numbers of NFT compared with AD (9, 16), this suggests that, in AD, aggregation of tau itself (and not neuronal cell death) may cause increased GLO levels, possibly as a response to cellular stress elicited by the accumulation of aggregated proteins. This notion is supported by the finding that GLO staining is not detectable in strongly AT100-positive neurons. Interestingly, the Alz50 conformational epitope of tau, which discriminates normal from fibrillar tau, is found in AD and P301L tau transgenic mice (5). This epitope is the result of the reaction of phosphorylated tau with the carbonyl hydroxy-nonenal, suggesting that an interplay between tau phosphorylation and oxidative stress is important for the formation of NFT (33).
Insight into the role of GLO in disease is also provided by the analysis of toxic substrates of GLO, such as MG and of AGEs. Under normal conditions, most MG is bound to cellular proteins as adducts formed with lysine, arginine, and cysteine residues. Elevated concentrations of MG can lead to irreversible modifications of lysine and arginine residues through formation of AGEs. Their formation is thought to contribute to several pathophysiological conditions, including tissue damage after ischemia, aging, and the development of blindness and vascular diseases in diabetic patients (34, 35). In AD, cross-linking by AGEs of lysine residues in the microtubule binding domain of tau results in the formation of detergent-insoluble and protease-resistant aggregates such as NFT (2, 3, 36, 37). In these, AGE determinants were localized in their fibrillar protein component, the tau filaments, by electron microscopy (38). NFT may interfere with axonal transport and intracellular protein traffic, as has been previously reported for tau transgenic mice (5). Finally, the finding that anti-AGE antibodies reacted predominantly with intracellular NFT in AD brain (an earlier stage) compared with extracellular NFT (a final stage) suggests that AGE modification may be involved in early stages of the disease and not be a secondary consequence of the deposits (3, 39).
This sequence of events may also hold true for GLO, as a relationship between AGE and GLO has been demonstrated. GLO inhibition is toxic to tumor cells (40), and overexpression of GLO in endothelial cells prevents hyperglycemia-induced AGE formation (41). In addition, a hemodialysis patient with unusual high levels of AGEs and a very low activity of GLO has been reported, demonstrating a possible causal relationship between low GLO levels and increased formation of AGEs (42). In the P301L transgenic mice and in AD, GLO may be up-regulated to reduce levels of MG and to decrease AGE formation. Alternatively, it may be up-regulated as a general stress response, independent of the levels of MG or reduced glutathione.
In the course of our study, we found that the ala allele of the coding SNP rs2736654 was moderately associated with the risk for AD in males. Because of the small effect size, we tried to confirm these results in additional samples and failed to replicate the initial association. Interestingly, the coding SNP deviated significantly from Hardy–Weinberg equilibrium in both young and elderly subjects. This deviation from Hardy–Weinberg equilibrium, which has been also reported in other populations (43), suggests the existence of yet unknown selection forces. Of note, this SNP has also been associated with diseases leading to decreased life expectancy such as prostate cancer and diabetes (44–47).
Together, our studies suggest a role for GLO in AD. Moreover, our approach demonstrates that transcriptomic approaches are suited to identify genes that are differentially expressed in human neurodegenerative disease. Crossing tau mutant mice with mice that either lack or overexpress GLO will provide additional insight into the role of GLO in AD.
Acknowledgments
We thank Drs. P. J. Thornalley, S. Ranganathan, and S. Staros for their generous gift of GLO-specific antisera, Dr. P. Davies for generously providing us with antibody CP13, Dr. de Quervain for human DNA samples, the Functional Genomics Center Zurich for support, K.-D. Huynh and E. Moritz for excellent technical assistance, and J. Tracy and Dr. M. Di Pietro for critical reading of the manuscript. This work was supported in part by grants from Interdisziplinäres Zentrum für Klinische Forschung Leipzig (01KS9504, Project N1), the A+D Fonds of the Swiss Academy of Medical Sciences, the Olga Mayenfisch Foundation, the Kurt and Senta Herrmann Foundation, the Hartmann Müller Foundation, the Zentrum für Neurobiologie Zürich, and the National Center of Competence in Research on Neural Plasticity and Repair.
Abbreviations: AD, Alzheimer's disease; AGE, advanced glycation end product; GLO, glyoxalase I; MG, methylglyoxal; NFT, neurofibrillary tangle; SNP, single nucleotide polymorphism.
References
- 1.Grundke-Iqbal, I., Iqbal, K., Tung, Y. C., Quinlan, M., Wisniewski, H. M. & Binder, L. I. (1986) Proc. Natl. Acad. Sci. USA 83, 4913-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ledesma, M. D., Medina, M. & Avila, J. (1996) Mol. Chem. Neuropathol. 27, 249-258. [DOI] [PubMed] [Google Scholar]
- 3.Buee, L., Bussiere, T., Buee-Scherrer, V., Delacourte, A. & Hof, P. R. (2000) Brain Res. Rev. 33, 95-130. [DOI] [PubMed] [Google Scholar]
- 4.Liu, F., Zaidi, T., Iqbal, K., Grundke-Iqbal, I., Merkle, R. K. & Gong, C. X. (2002) FEBS Lett. 512, 101-106. [DOI] [PubMed] [Google Scholar]
- 5.Götz, J. (2001) Brain Res. Rev. 35, 266-286. [DOI] [PubMed] [Google Scholar]
- 6.Lee, V. M., Goedert, M. & Trojanowski, J. Q. (2001) Annu. Rev. Neurosci. 24, 1121-1159. [DOI] [PubMed] [Google Scholar]
- 7.Goedert, M., Spillantini, M. G., Jakes, R., Crowther, R. A., Vanmechelen, E., Probst, A., Götz, J., Burki, K. & Cohen, P. (1995) Neurobiol. Aging 16, 325-334. [DOI] [PubMed] [Google Scholar]
- 8.Lewis, J., McGowan, E., Rockwood, J., Melrose, H., Nacharaju, P., Van Slegtenhorst, M., Gwinn-Hardy, K., Murphy, P. M., Baker, M., Yu, X., et al. (2000) Nat. Genet. 25, 402-405. [DOI] [PubMed] [Google Scholar]
- 9.Götz, J., Chen, F., Barmettler, R. & Nitsch, R. M. (2001) J. Biol. Chem. 276, 529-534. [DOI] [PubMed] [Google Scholar]
- 10.Tanemura, K., Akagi, T., Murayama, M., Kikuchi, N., Murayama, O., Hashikawa, T., Yoshiike, Y., Park, J. M., Matsuda, K., Nakao, S., et al. (2001) Neurobiol. Dis. 8, 1036-1045. [DOI] [PubMed] [Google Scholar]
- 11.Tatebayashi, Y., Miyasaka, T., Chui, D. H., Akagi, T., Mishima, K., Iwasaki, K., Fujiwara, M., Tanemura, K., Murayama, M., Ishiguro, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13896-13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen, B., Ingram, E., Takao, M., Smith, M. J., Jakes, R., Virdee, K., Yoshida, H., Holzer, M., Craxton, M., Emson, P. C., et al. (2002) J. Neurosci. 22, 9340-9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Götz, J., Tolnay, M., Barmettler, R., Chen, F., Probst, A. & Nitsch, R. M. (2001) Eur. J. Neurosci. 13, 2131-2140. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi, M., Ishihara, T., Zhang, B., Hong, M., Andreadis, A., Trojanowski, J. & Lee, V. M. (2002) Neuron 35, 433-446. [DOI] [PubMed] [Google Scholar]
- 15.Lin, W. L., Lewis, J., Yen, S. H., Hutton, M. & Dickson, D. W. (2003) Am. J. Pathol. 162, 213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Götz, J., Chen, F., van Dorpe, J. & Nitsch, R. M. (2001) Science 293, 1491-1495. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, J., Dickson, D. W., Lin, W.-L., Chisholm, L., Corral, A., Jones, G., Yen, S.-H., Sahara, N., Skipper, L., Yager, D., et al. (2001) Science 293, 1487-1491. [DOI] [PubMed] [Google Scholar]
- 18.Thornalley, P. J. (1990) Biochem. J. 269, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Herreweghe, F., Mao, J., Chaplen, F. W., Grooten, J., Gevaert, K., Vandekerckhove, J. & Vancompernolle, K. (2002) Proc. Natl. Acad. Sci. USA 99, 949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornalley, P. J. (1996) Gen. Pharmacol. 27, 565-573. [DOI] [PubMed] [Google Scholar]
- 21.Ledesma, M. D., Bonay, P., Colaco, C. & Avila, J. (1994) J. Biol. Chem. 269, 21614-21619. [PubMed] [Google Scholar]
- 22.Kins, S., Crameri, A., Evans, D. R., Hemmings, B. A., Nitsch, R. M. & Götz, J. (2001) J. Biol. Chem. 276, 38193-38200. [DOI] [PubMed] [Google Scholar]
- 23.Götz, J., Probst, A., Ehler, E., Hemmings, B. & Kues, W. (1998) Proc. Natl. Acad. Sci. USA 95, 12370-12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D. & Stadlan, E. M. (1984) Neurology 34, 939-944. [DOI] [PubMed] [Google Scholar]
- 25.Morris, J. C., Heyman, A., Mohs, R. C., Hughes, J. P., van Belle, G., Fillenbaum, G., Mellits, E. D. & Clark, C. (1989) Neurology 39, 1159-1165. [DOI] [PubMed] [Google Scholar]
- 26.Nauck, M., Hoffmann, M. M., Wieland, H. & Marz, W. (2000) Clin. Chem. 46, 722-724. [PubMed] [Google Scholar]
- 27.Zirlinger, M., Kreiman, G. & Anderson, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranganathan, S., Walsh, E. S., Godwin, A. K. & Tew, K. D. (1993) J. Biol. Chem. 268, 5661-5667. [PubMed] [Google Scholar]
- 29.Davidson, S. D., Cherry, J. P., Choudhury, M. S., Tazaki, H., Mallouh, C. & Konno, S. (1999) J. Urol. 161, 690-691. [PubMed] [Google Scholar]
- 30.Hayes, J. D., Milner, S. W. & Walker, S. W. (1989) Biochim. Biophys. Acta 994, 21-29. [DOI] [PubMed] [Google Scholar]
- 31.Götz, J. & Nitsch, R. M. (2001) NeuroReport 12, 2007-2016. [DOI] [PubMed] [Google Scholar]
- 32.Pei, J. J., Braak, H., Gong, C. X., Grundke-Iqbal, I., Iqbal, K., Winblad, B. & Cowburn, R. F. (2002) Acta Neuropathol. 104, 369-376. [DOI] [PubMed] [Google Scholar]
- 33.Takeda, A., Smith, M. A., Avila, J., Nunomura, A., Siedlak, S. L., Zhu, X., Perry, G. & Sayre, L. M. (2000) J. Neurochem. 75, 1234-1241. [DOI] [PubMed] [Google Scholar]
- 34.Corman, B., Duriez, M., Poitevin, P., Heudes, D., Bruneval, P., Tedgui, A. & Levy, B. I. (1998) Proc. Natl. Acad. Sci. USA 95, 1301-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oya, T., Hattori, N., Mizuno, Y., Miyata, S., Maeda, S., Osawa, T. & Uchida, K. (1999) J. Biol. Chem. 274, 18492-18502. [DOI] [PubMed] [Google Scholar]
- 36.Ko, L. W., Ko, E. C., Nacharaju, P., Liu, W. K., Chang, E., Kenessey, A. & Yen, S. H. (1999) Brain Res. 830, 301-313. [DOI] [PubMed] [Google Scholar]
- 37.Munch, G., Deuther-Conrad, W. & Gasic-Milenkovic, J. (2002) J. Neural Transm. Suppl. 62, 303-307. [DOI] [PubMed] [Google Scholar]
- 38.Yan, S. D., Chen, X., Schmidt, A. M., Brett, J., Godman, G., Zou, Y. S., Scott, C. W., Caputo, C., Frappier, T., Smith, M. A., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki, N., Fukatsu, R., Tsuzuki, K., Hayashi, Y., Yoshida, T., Fujii, N., Koike, T., Wakayama, I., Yanagihara, R., Garruto, R., et al. (1998) Am. J. Pathol. 153, 1149-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto, H., Mashima, T., Kizaki, A., Dan, S., Hashimoto, Y., Naito, M. & Tsuruo, T. (2000) Blood 95, 3214-3218. [PubMed] [Google Scholar]
- 41.Shinohara, M., Thornalley, P. J., Giardino, I., Beisswenger, P., Thorpe, S. R., Onorato, J. & Brownlee, M. (1998) J. Clin. Invest. 101, 1142-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyata, T., van Ypersele de Strihou, C., Imasawa, T., Yoshino, A., Ueda, Y., Ogura, H., Kominami, K., Onogi, H., Inagi, R., Nangaku, M. & Kurokawa, K. (2001) Kidney Int. 60, 2351-2359. [DOI] [PubMed] [Google Scholar]
- 43.Togan, I., Yilmaz, A., Erguven, A., Dereli, I. & Ozbas, F. (1996) Anthropol. Anz. 54, 121-124. [PubMed] [Google Scholar]
- 44.Kim, N. S., Sekine, S., Kiuchi, N. & Kato, S. (1995) J. Biochem. (Tokyo) 117, 359-361. [DOI] [PubMed] [Google Scholar]
- 45.Samadi, A. A., Fullerton, S. A., Tortorelis, D. G., Johnson, G. B., Davidson, S. D., Choudhury, M. S., Mallouh, C., Tazaki, H. & Konno, S. (2001) Urology 57, 183-187. [DOI] [PubMed] [Google Scholar]
- 46.Kirk, R. L., Theophilus, J., Whitehouse, S., Court, J. & Zimmet, P. (1979) Diabetes 28, 949-951. [DOI] [PubMed] [Google Scholar]
- 47.Subramanian, V. S., Krishnaswami, C. V. & Damodaran, C. (1994) Diabetes Res. Clin. Pract. 25, 51-59. [DOI] [PubMed] [Google Scholar]