Abstract
Cystic fibrosis (CF) is a fatal, autosomal, recessive genetic disease that is characterized by profound lung inflammation. The inflammatory process is believed to be caused by massive overproduction of the proinflammatory protein IL-8, and the high levels of IL-8 in the CF lung are therefore believed to be the central mechanism behind CF lung pathophysiology. We show here that digitoxin, at sub nM concentrations, can suppress hypersecretion of IL-8 from cultured CF lung epithelial cells. Certain other cardiac glycosides are also active but with much less potency. The specific mechanism of digitoxin action is to block phosphorylation of the inhibitor of NF-κB (IκBα). IκBα phosphorylation is a required step in the activation of the NF-κB signaling pathway and the subsequent expression of IL-8. Digitoxin also has effects on global gene expression in CF cells. Of the informative genes expressed by the CF epithelial cell line IB-3, 58 are significantly (P < 0.05) affected by gene therapy with wild-type (CFTR CF transmembrane conductance regulator). Of these 58 genes, 36 (62%) are similarly affected by digitoxin and related active analogues. We interpret this result to suggest that digitoxin can also partially mimic the genomic consequences of gene therapy with CF transmembrane conductance regulator. We therefore suggest that digitoxin, with its lengthy history of human use, deserves consideration as a candidate drug for suppressing IL-8-dependent lung inflammation in CF.
Cystic fibrosis (CF) is a fatal, autosomal, recessive genetic disease that is characterized by an IL-8-related, proinflammatory environment in the lung (1). Morbidity and mortality from CF is commonly due to loss of lung function, which inexorably follows a chronic course of intrinsic inflammation, bacterial infection, and airway obstruction (1). CF is due to inactivating mutations in the chloride channel CF transmembrane conductance regulator (CFTR) gene (2–4). The most common CFTR mutation, ΔF508 CFTR, causes defective trafficking of the mutant protein (5–7) and compromises chloride channel function (8). The CFTR mutation and the process of intrinsic lung inflammation are presumably causally related. Consequently, both gene therapy (9–14) and pharmacotherapy (15–28) have been targeted toward correction of either the trafficking or channel defects, in anticipation that successful correction would suppress the proinflammatory phenotype of the CF airway (16, 29). The chemical basis for the inflammatory phenotype of the CF lung is believed to be the production of massively high levels of IL-8 by CF lung epithelial cells (30–41). We describe here a method for finding drugs that suppress baseline hypersecretion of IL-8 from CF lung epithelial cells and identify digitoxin and certain other cardiac glycosides as potential high-potency therapeutics for CF.
In this study we have applied modern proteomic and genomic methods to determine the mechanism of digitoxin action on CF lung epithelial cells. We find that the mechanism of action is the blockade of inhibitor of NF-κB(IκBα) activation. Blocking IκBα activation suppresses the NF-κB signaling pathway and, thus, IL-8 expression (16, 42). We also report the results of a genomic comparison between the effects of gene therapy with wild-type CFTR and digitoxin. Of the 58 genes significantly affected by gene therapy, 62% are also similarly and proportionately affected by digitoxin and related active analogues. This result may be of profound and fundamental importance because it indicates that a small molecule such as digitoxin can partially mimic gene therapy for CF. Digitoxin has a long history of being administered to humans (43), and we therefore conclude that this drug should be considered as a candidate for therapeutic use in CF.
Methods
Cells and Culture Methods. The CF lung epithelial cells IB3-1 and adeno-associated virus, wild-type CFTR-repaired IB3-1/S9 have been previously described (16, 44, 45). Both IB3-1 and IB3-1/S9 cells were grown in serum-free LHC-8 medium (Biofluids, Bethesda), formulated without gentimycin. To avoid potential problems with clonal drift, new batches of original frozen cells were thawed at least every three months during the course of the experiment.
Detection of IL-8. The details of this assay have been described in a previous publication (16). Briefly, IB-3 cells were grown in 96-well microtiter plates to 80% confluency. Drugs diluted in LHC-8 medium at the given concentrations were added, and the cells were allowed to incubate for an additional 24 h. To initiate the experiment, cells were washed with fresh LHC-8 medium and then incubated for an additional 16 h in the same medium supplemented with drug. At the end of the time period, the supernatant solutions were collected and assayed for IL-8 by an ELISA assay (R & D Systems). Data were normalized to cellular DNA by propidium iodide (kit from Boehringer Mannheim, which is now Roche Molecular Biochemicals).
Medicinal Chemistry and Organic Reagents. Oleandrin was obtained from Indofine Chemical (Hillsborough, NJ; >98% purity). Cardiac glycosides were purchased from Sigma (>95% purity). Oleandrin and other cardiac glycosides were prepared in ethanol and diluted to a final solvent concentration of 0.01%. Solubilized drugs were stored at 4°C. Crystalline drugs were stored in a desiccator at room temperature in the dark.
Pharmacogenomic Analysis with cDNA Microarrays. cDNA microarrays were purchased from Clontech (Atlas Human 1.2 Array, catalog no. 7850-1), and used as the pharmacogenomic platform. RNA was prepared from IB-3 or IB-3/S9 cells treated exactly as described for the screening paradigm with concentrations of the cardiac glycosides at 90% inhibiting dose (ID90) (see Fig. 2 Lower). These concentrations were as follows: I, 10 nM; II, 10 nM; III, 30 nM; IV, 30 nM; V, 200 nM; VI, 200 nM; and VII, 200 nM. The inactive cardiac glycoside, VIII, was formulated as for VII (namely, 200 nM). Cognate cDNAs were radiolabeled with 32P, allowed to bind to identified sites on the nitrocellulose-based arrays, imaged on a Typhoon PhosphorImager (Molecular Dynamics/Amersham Pharmacia Biosciences), and analyzed as described (46).
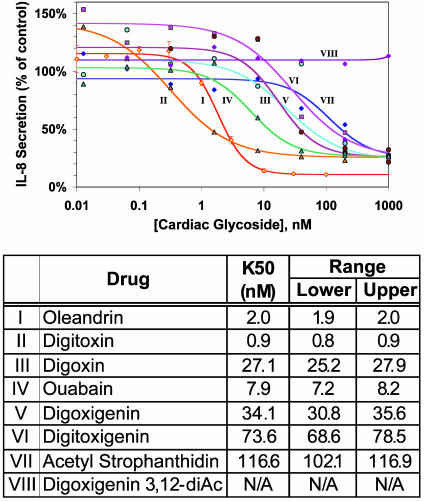
Fig. 2.
Concentration–response curves for different cardiac glycosides as a function of IL-8 secretion from IB-3 CF lung epithelial cells. (Upper) Roman numerals correspond to fitted curves and common names for the compounds in the table. Curves are prepared from at least three independent assays. IL-8 data are normalized to DNA content of each well, as defined by propidium iodide fluorescence. (Lower) Cardiac glycoside compounds are numbered as in Fig. 1. The lower and upper ranges are calculated from the same data used to compute the K50.
Pharmacoproteomic Analysis with Protein Microarrays. Reverse-phase protein microarrays were manufactured as described (47, 48). Briefly, 20 nl of denatured protein lysates (4 M urea/125 nM Tris, pH 6.8/2% Chaps/2% 2-mercaptoethanol) were immobilized onto nitrocellulose-coated glass slides by using a GMS470 Affymetrix microarrayer in an ordered array. After the incubation with the secondary antibody, detection and analysis were as described (47, 48).
Antibodies for Protein Microarrays. Anti-rabbit (T202,Y204)ERK (extracellular signal-regulated kinase), total ERK, (Ser-176,Ser-180)IκB kinase-β (IKKβ), cleaved caspase-3, total caspase-3 of p65, (Ser-576)p65, and anti-mouse (Ser-32,Ser-36)IκBα were purchased from Cell Signaling Technology (Beverly, MA). Anti-mouse total IκBα and total IKKβ were obtained from BD Transduction Laboratories (Franklin Lakes, NJ). Horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies were from Vector Laboratories (Gaithersburg, MD). Nitrocellulose-coated FAST slides were purchased from Schleicher and Schuell. Reagents used for catalyzed signal amplification were from Dako-Cytomation (Glostrup, Denmark).
Statistics. For compounds designated as authentic “hits” in the screen, detailed concentration/response curves were then performed in quadruplicate in at least three independent experiments. Titration data were analyzed by a curve-fitting routine to estimate the concentration at which 50% inhibition of IL-8 secretion is achieved (K50). Statistical significance was measured by ANOVA. Hierarchical clustering algorithm analyses were performed by using the cluster and treeview applications from Stanford University (http://rana.lbl.gov/EisenSoftware.htm), as described (16).
Results
Cardiac Glycosides Suppress IL-8 Secretion. The first cardiac glycoside tested for CF IL-8 suppression was oleandrin (Fig. 1, structure I). This hit proved to be profoundly active, with a K50 of ≈2 nM. Subsequently, digitoxin (Fig. 1, structure II) was found to be even more potent, with a K50 value of ≈0.9 nM. We therefore tested a variety of cardiac glycosides for capacity to suppress constitutive IL-8 secretion from IB-3 cells. The structures of these compounds are shown in Fig. 1. Complete pharmacodynamic titrations are shown in Fig. 2. The computed K50 values and the common names of the tested compounds are shown in Fig. 2 Lower. The least potent of these compounds is VIII, which is inactive even at 1 μM. The relationships among the activities in terms of progressively higher K50 values are as follows: II > I > IV > III, V > VI > VII >> VIII.
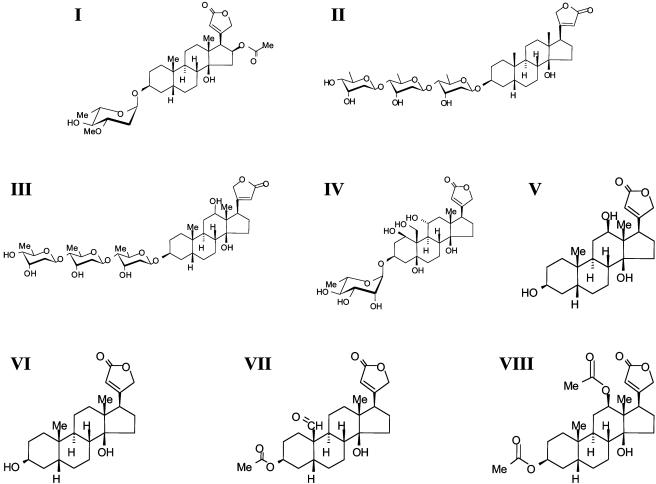
Fig. 1.
Structures of cardiac glycosides used to set the structure activity relation for suppression of constitutive IL-8 secretion from IB-3 CF lung epithelial cells. Roman numerals correspond to titrations and column names in Fig. 2.
Pharmacogenomic Analysis Distinguishes Potency of Cardiac Glycosides. We hypothesized that potent analogues and possible mechanistic indications could be distinguished among the cardiac glycosides by using the technique of pharmacogenomics. We therefore treated IB-3 cells for 24 h with all eight compounds at concentrations that inhibited constitutive hypersecretion of IL-8 by ≈90% (namely, ID90). Gene expression was measured on cDNA microarrays. Of 1,200 informative genes, 58 genes were determined to significantly distinguish IB-3 CF cells from the wild-type CFTR-repaired IB-3/S9 (P < 0.05). Expressions of these 58 significant genes were then examined as a function of the different drug treatments by using a hierarchical clustering algorithm for data display (see Fig. 3a).
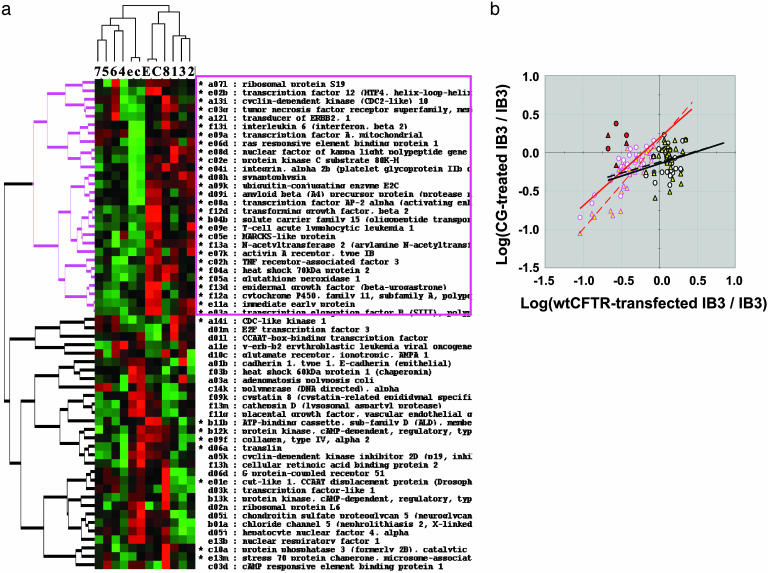
Fig. 3.
Pharmacogenomic analysis of cardiac glycoside drugs. (a) Pharmacogenomic analysis of cardiac glycosides with representation by a hierarchical clustering algorithm. Cardiac glycoside concentrations were chosen at ID90 points, as calculated from Fig. 1. The exact concentrations are given in Methods. Genes for the analysis were selected from those differing significantly (P < 0.05) between CF lung epithelial IB-3 cells and the wild-type CFTR-repaired daughter cells, IB-3/S9. Drugs 1, 2, and 3, corresponding to cells treated with drugs I, II, and III, respectively, cluster on the immediate right. The inactive cardiac glycoside (VIII) clusters with control cells, with (E) or without (C) 0.01% ethanol vehicle. A separate cluster contains a subcluster for drugs 4 and 6 (drugs IV and VI) and drugs 5 and 7 (drugs V and VII). IB-3/S9 cells corrected with wild-type CFTR are shown as controls with (e) or without (c) 0.01% ethanol vehicle. The diadic hierarchical cluster marked in red corresponds to most of the genes affected in common by both cardiac glycoside therapy and CFTR gene therapy. (b) Pharmacogenomic comparison between IB-3 cells treated with active cardiac glycosides or with wild-type CFTR. The effects of drug treatment or wild-type CFTR gene therapy are shown as the log of the ratio of the respective treatments to IB3-1 cells treated with vehicle only. Genes with relative expression in the range of 0.1–1.0 are relatively highly correlated (symbols with red borders) compared with those in the relative expression range of >1.0 (symbols with black borders). The symbols with red borders correspond to 27 genes marked by an asterisk in a and further denoted in a by a red-bordered gene cluster and text box. Eight genes marked by asterisks are also located in the lower diad. Red-bordered symbols are further differentiated by circles with white fill (drugs I, II, and III; y = 0.88x + 0.19, R2 = 0.75) or triangles with yellow fill (drugs IV, V, VI, and VII; y = 1.16x + 0.16, R2 = 0.76). Thus, regression analysis indicates that the slopes are close to 1 and the intercepts are close to 0. Three black-bordered “outlier” genes, somewhat more heavily expressed by drugs than by gene therapy, are marked by red fill (see text for gene identification). Symbols in black borders correspond principally to genes in the lower cluster in a. The symbols and fill colors code for the same drug categories as for the red-bordered symbols. However, for the latter genes, there is virtually no correlation between cardiac glycosides and gene therapy: drugs I, II, and III (y = 0.32x - 0.15, R2 = 0.05) and drugs IV, V, VI, and VII (y = 0.29x - 0.13, R2 = 0.03).
As shown in Fig. 3a, cardiac glycosides 1 (I, oleandrin), 2 (II, digitoxin), and 3 (III, digoxin) form one cluster (cluster 1) on the extreme right of the horizontal axis. Drugs I and II are among the most potent drugs tested by pharmacokinetics (see Fig. 2 Lower). As anticipated, the inactive drug (VIII, digitoxigenin 3,12-diacetyl) clusters separately (cluster no. 2) with untreated or vehicle-treated IB-3 cells. The other five drugs cluster separately from the others on the left side of the horizontal axis. Within the latter cluster we find two subclusters of drugs. Subcluster 3 includes cardiac glycosides 4 (IV, ouabain) and 6 (VI, digitoxigenin), which are more and less potent, respectively. The remaining subcluster 4 includes cardiac glycosides 5 (V, digoxigenin) and 7 (VII, acetyl strophanthidin), which are among the least potent. These data can be summarized symbolically as follows: {[I,II,III]1 ≠ [VIII]2} ≠ {[IV,VI]3 ≠ [V,VII]4}. Thus these pharmacogenomic clusters appear to correlate approximately with potency, as measured by pharmacodynamic data in Fig. 2 Lower.
Cardiac Glycosides Mimic CFTR Gene Therapy. We also wished to test whether the genomic changes induced in IB-3 cells by repair with wild-type CFTR paralleled in any way the genomic consequences of exposure to active cardiac glycosides. The repaired cell line is IB3-13/S9. As shown in Fig. 3b, there is a close correlation between the genomic effects of all active cardiac glycosides (I–III and IV–VII) and the subset of the genes significantly affected by wild-type CFTR. This correlation, principally in the range of relative expression of 0.1–1.0, is R2 = 0.76 for drugs I–III (red-bordered circles with white fill), and R2 = 0.75 for drugs IV–VII (red-bordered triangles with yellow fill). Three genes in the red cluster are clearly outliers in that they are more profoundly enhanced in expression by the more potent cardiac glycosides. These genes, marked by black borders with red fill for easier identification, are NF-κB,p105; IL-6 receptor; and transcription factor A, mitochondrial. For genes whose relative expression in wild-type CFTR-repaired CF cells are >1.0, the appearance in the plot suggests no correlation with drug effects. This conclusion is supported by the very low R2 values (R2 = 0.05 and R2 = 0.03, respectively).
The locations of these highly correlated genes in the hierarchical cluster diagram (Fig. 3a) are marked by an asterisk next to the gene names. Seventy-five percent of these genes are in the upper of the two major clusters on the vertical axis. The upper cluster is marked in red, to correlate with the outline of symbols in Fig. 3b. These genes identified by asterisks in the upper cluster are outlined with a red box. Eight other genes in this highly correlated set are located in the lower gene cluster and are also marked by asterisks. It is important to emphasize that the cDNA array used here does not include every gene in the genome and that other key or even yet unidentified genes may prove to be informative in the future.
Cardiac Glycosides Block Activation of IκBα. To further test the hypothesis that proteins from the NF-κB signaling pathway might also be changed in expression or activation by active cardiac glycosides (Fig. 2 Lower), we used quantitative reverse-phase protein microarray technology. For this test, we used antibodies to several of the kinases that are known to either control or impact on the NF-κB signaling pathway. As shown in Fig. 4a, microarrays of cell extracts treated with ID90 doses of different cardiac glycosides were probed with antibodies against ERK and phospho-ERK, IKKβ and phospho-IKKβ, IκBα and phospho-IκBα, NF-κB,p65, and phospho-NF-κB,p65. Inasmuch as the “divide or die” paradigm suggests the possibility of activation of apoptotic processes in the face of suppression of inflammation, we also tested for activation of caspase-3.
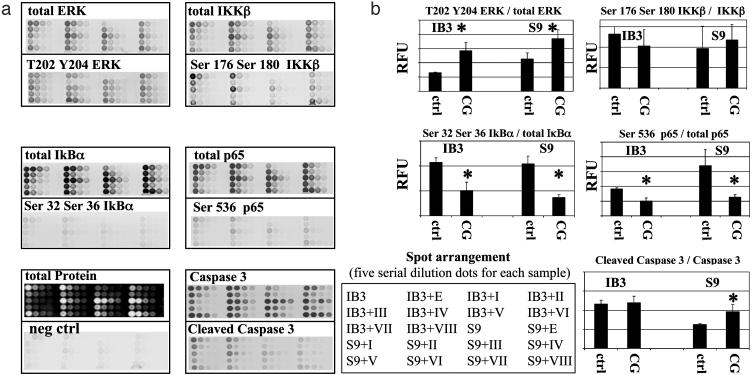
Fig. 4.
Cardiac glycosides block activation of IκBα and NFκB,p65 in CF lung epithelial IB-3 cells. (a) Cell extracts from IB3-1 and IB3-1/S9, repaired with wild-type CFTR, were robotically arrayed in serial 2-fold titremetric series on nitrocellulose-coated slides. Arrays were probed with antibodies against proteins known to be associated with activation of the NF-κB signaling pathway. Cells were incubated with ID90 concentrations of compounds I–VIII. Quantitative analyses are shown for total ERK and (T202,Y204)phospho-ERK; IKKβ and (Ser-176,Ser-180)phospho-IKKβ; total IκBα and (Ser-32,Ser-36)phospho-IκBα; and total p65 and (Ser-536)phospho-p65. Total caspase-3 and cleaved caspase-3 are included as a control, as is total protein concentration. The negative control (neg ctrl) is isotype-matched primary mouse IgG with common second antibody. An equivalent negative control is prepared for rabbit antibodies (data not shown). The data are representative of three independent experiments. (b) Fractional phosphorylation values of ERK, IKKβ, IκBα, and p65 are calculated as a function of cardiac glycoside treatment. The control (ctrl) value is calculated as the average of untreated cells, cells treated with ethanol vehicle (0.01%), and cells treated with the inactive compound VIII. The cardiac glycoside (CG) value is calculated as the average of effects for compounds I–VII. The asterisk represents differences between control and cardiac glycoside, with a significance of P < 0.05. RFU, relative fluorescence unit.
As shown in Fig. 4b, the active cardiac glycosides, as a class, activate upstream ERK. The cardiac glycosides also appear to somewhat affect activation of IKKβ. However, the active cardiac glycosides, as a class, significantly reduce the phosphorylation of IκBα. This result is of potentially fundamental mechanistic importance, because the cytoplasmically NF-κB complex only becomes active in the nucleus when the IκBα component is phosphorylated and proteosomally destroyed. As might therefore be expected, phosphorylation of the immediate downstream target, NF-κB,p65, is also significantly reduced. As further shown in Fig. 4b, caspase-3 activation is unaffected by treatment with active cardiac glycosides (Fig. 2 Lower) in the CF lung IB-3 cells. However, caspase-3 activation is significantly elevated by these compounds in the wild-type CFTR-repaired IB-3/S9 cells. Protein microarrays were found to correlate closely with cognate Western blots (e.g., phospho-ERK, R2 = 0.91; total ERK, R2 = 0.80). These data lend firm support to the concept that suppression of IκBα phosphorylation and consequent suppression of NF-κB,p65 activation are the mechanisms by which digitoxin and other active cardiac glycoside drugs suppress constitutive hypersecretion of IL-8 from CF lung epithelial cells.
Discussion
Taken together, our results show that that digitoxin and other active cardiac glycoside drugs can potently suppress constitutive hypersecretion of IL-8 from cultured CF lung epithelial cells. The mechanism of action of these drugs is to block the proinflammatory NF-κB pathway by inhibiting activation of IκBα. This result is in complete agreement with data from our and other laboratories identifying a dysfunctional NF-κB pathway as responsible for the proinflammatory character of the CF lung (16). Thus digitoxin and its related active analogues can be considered as potentially useful in addressing the central mechanism of CF lung pathophysiology. Cardiac glycoside pharmacodynamics indicate that the most active suppressor of IL-8 secretion is digitoxin (II). In this role, digitoxin is ≈30-fold more potent than the concentration classically used to treat cardiac failure (43). By contrast, digoxin (III), a highly potent cardiotonic drug, is 15-fold less potent than digitoxin (II) as a CF IL-8 suppressor. Considering the pharmacology of the entire series, it appears that the suppressive actions of cardiac glycosides on IL-8 secretion and the classical positive actions on cardiac contractility are mediated by completely separate mechanisms. Structurally, the most active species, I and II, are characterized by sugars of different structures and by the absence of oxygen-containing substitutions at or near the 12th position on the C ring. We therefore give the equatorial 12th position and its neighbors crucial negative pharmacophoric importance, and glycosidic substitution at the third position positive pharmacophoric importance for the control of IL-8 secretion from CF lung epithelial cells.
Mechanism of Cardiac Glycoside Suppression of IL-8. The mechanism of digitoxin action appears to involve inhibition of the NF-κB signaling pathway. Data in Fig. 4 indicate that phosphorylation of IκBα is greatly reduced, as is phosphorylation of the downstream NF-κB,p65 itself. Activated IKKβ is canonically responsible for phosphorylation of IkBα. When phosphorylated, IκBα loses its capacity to interact with and hold the otherwise inactive NF-κB,p65/NF-κB,p50 complex in the cytosol and is destroyed by the proteosome. The now unencumbered NF-κB complex is thus free to translocate into the nucleus and activate IL-8 expression by binding to the κB site on the IL-8 promoter. Separately, phosphorylation of NF-κB,p65 potentiates translocation of this transcription factor into the nucleus once it is free of IκBα (49). Thus, the cardiac glycoside-induced suppression of phospho-NF-κB,p65 can be interpreted as an internal reporter on primary drug-induced suppression of upstream IκBα activation. We conclude that the principal site of action of the active cardiac glycosides in CF epithelial cells appears to be at the interface of the reaction by which activated IKKβ or a related kinase activates IκBα.
Pharmacogenomics of Cardiac Glycosides. The pharmacogenomic analysis of the complete cardiac glycoside series shows that the high and low potency cardiac glycosides cluster separately when compared by using the hierarchical clustering algorithm. These data from Fig. 3a can be symbolized as follows: {[I,II, III]1 ≠ [VIII]2} ≠ {[IV,VI]3 ≠ [V,VII]4}. This equation shows that the more potent drugs in cluster 1 generally occur in a separate cluster from the less potent drugs. An important caveat to keep in mind is that these genomic data are collected on the basis of ID90 concentrations, meaning that there is much more of drug III in the assay than either drugs I or II. These observations are of particular importance because they constitute an explicit example of cross validation between pharmacodynamically and pharmacogenomically active and inactive species, respectively.
Cardiac Glycosides As CFTR Gene Therapy Mimetics. From a completely different perspective, a most remarkable and fundamentally important result is obtained when the genomic effects of digitoxin and other active cardiac glycosides are compared with the genomic effects of gene therapy with wild-type CFTR. This bioinformatics experiment was performed to test the hypothesis that small molecules, such as cardiac glycosides, could mimic CFTR gene therapy. Of the 58 genes that were significantly changed in IB-3 cells by gene therapy (P < 0.05; n = 1,200), 36 (62%) were found to be equivalently and proportionately changed by the active cardiac glycosides. The graphical comparison in Fig. 3b shows that the genes that are down-regulated by gene therapy of the IB-3 cell are also down-regulated by active cardiac glycosides. By contrast, the genes that are up-regulated by wild-type CFTR seem to be virtually unaffected by drug exposure. Thus, both wild-type CFTR and the active cardiac glycosides appear to act as tonic inhibitory “gatekeepers” for a specific group of genes in the CF epithelial cell. As shown in Fig. 3a, the hierarchical clustering algorithm also identifies these genes by clustering most of them separately from the others. The genes in this coregulated cluster, denoted by asterisks, contain many from the group of genes known to be associated with the proinflammatory tumor necrosis factor α/NF-κB signaling pathway and IL-8 expression (16). We interpret this result to be consistent with quantitative protein microarray data showing that active cardiac glycosides inhibit phosphorylation and activation of IκBα and NF-κB,p65.
Acknowledgments
We thank the Cystic Fibrosis Foundation for support of this work.
Abbreviations: CF, cystic fibrosis; CFTR, CF transmembrane conductance regulator; ERK, extracellular signal-regulated kinase; IκBα, inhibitor of NF-κB; IKK, IκB kinase; ID90, dose at which inhibition is 90%; K50, dose at which inhibition of IL-8 secretion is 50%.
References
- 1.Welsh, M. J., Ramsey, B. W., Accurso, F. & Cutting, G. R. (2001) Cystic Fibrosis, eds. Scriver, C. L., Beaudet, A. L., Valle, D., Sly, W. S. (McGraw–Hill, New York), 8th Ed., pp. 5121-5188
- 2.Riordan, J. R., Rommens, J. M., Karem, B-S., Alon, N., Rozmahel, R., Grzelczak, Z, Zielenski, J., Lok, S., Plavsic, N., Chou, J. L, et al. (1989) Science 245, 1066-1073. [DOI] [PubMed] [Google Scholar]
- 3.Rommens, J. M., lannuzzi, M. C., Karem, B-S., Drumm, M. L., Melmer, G., Dean, M., Rozmahel, R., Cole, J. L., Kennedy, D., Hidaka, N., et al. (1989) Science 245, 1059-1065. [DOI] [PubMed] [Google Scholar]
- 4.Kerem, B.-S., Rommens, J. M., Buchanan, J. A., Markiewicz, D., Cox, T. K., Chakravarti, A., Buchwald, M. & Tsui, L.-C. (1989) Science 245, 1073-1080. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, S. H., Gregory, R. J., Marshall, J., Paul, S., Souza, D. W., White, G. A., O'Riordan, C. R. & Smith, A. E. (1990) Cell 63, 827-834. [DOI] [PubMed] [Google Scholar]
- 6.Ward, C. L. & Kopito, R. R. (1994) J. Biol. Chem. 269, 25710-25718. [PubMed] [Google Scholar]
- 7.Ward, C. L., Omura, S. & Kopito, R. R. (1995) Cell 83, 121-127. [DOI] [PubMed] [Google Scholar]
- 8.Pasyk, E. A. & Foskett, J. K. (1995) J. Biol. Chem. 270, 12347-12350. [DOI] [PubMed] [Google Scholar]
- 9.Flotte, T. R. & Carter, B. J. (1998) Methods Enzymol. 292, 717-732. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz, F. E., Clancy, J. P., Perricone, M. A., Bebok, Z., Hong, J. S., Cheng, S. H., Meeker, D. P., Young, K. R., Schoumacher, R. A., Weatherly, M. R., et al. (2001) Hum. Gene Ther. 12, 751-761. [DOI] [PubMed] [Google Scholar]
- 11.Walters, R. W., van't Hof, W., Yi, S. M., Schroth, M. K., Zabner, J., Crystal, R. G. & Welsh, M. J. (2001) J. Virol. 75, 7703-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noone, P. G., Hohneker, K. W., Zhou, Z., Johnson, L. G., Foy, C., Gipson, C., Jones, K., Noah, T. L., Leigh, M. W., Schwartzbach, C., et al. (2000) Mol. Ther. 1, 105-114. [DOI] [PubMed] [Google Scholar]
- 13.Flotte, T. R., Zeitlin, P. L., Reynolds, T. C., Heald, A. E., Pedersen, P., Beck, S., Conrad, C. K., Brass-Ernst, L., Humphries, M., Sullivan, K., et al. (2003) Hum. Gene Ther. 14, 1079-1088. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari, S., Geddes, D. M. & Alton, E. W. (2002) Adv. Drug Delivery Rev. 54, 1373-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eidelman, O., Guay-Broder, C., van Galen, P. J. M., Jacobson, K. A., Fox, C., Turner, R. J., Cabantchik, Z. I. & Pollard, H. B. (1992) Proc. Natl. Acad. Sci. USA 89, 5562-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eidelman, O., Srivastava, M., Zhang, J., Murthy, J., Heldman, E., Jacobson, K. A., Metcalfe, E., Weinstein, D. & Pollard, H. B. (2001) Mol. Med. 7, 523-534. [PMC free article] [PubMed] [Google Scholar]
- 17.Guay-Broder, C., Jacobson, K. A., Sarnoy, S., Cabantchik, V., Guggino, W. B., Zaitlin, P. L., Turner, R. J., Vergara, L., Eidelman, O. & Pollard, H. B. (1995) Biochemistry 34, 9079-9087. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson, K. A., Guay-Broder, C., van Galen, P. J. M., Gallo-Rodriguez, C., Melman, N., Jacobson, M. A., Eidelman, O. & Pollard, H. B. (1995) Biochemistry 34, 9088-9094. [DOI] [PubMed] [Google Scholar]
- 19.Cohen, B. E., Lee, G., Jacobson, K. A., Kim, Y-C., Huang, Z., Sorscher, E. J. & Pollard, H. B. (1997) Biochemistry, 36, 6455-6461. [DOI] [PubMed] [Google Scholar]
- 20.Andersson, C. & Roomans, G. M. (2000) Eur. Respir. J. 15, 937-941. [DOI] [PubMed] [Google Scholar]
- 21.McCarty, N. A., Standaert, T. A., Teresi, M., Tuthill, C., Launspach, J., Kelley, T. J., Milgram, L. J., Hilliard, K. A., Regelmann, W. E., Weatherly, M. R., et al. (2002) Pediatr. Pulm. 33, 90-98. [DOI] [PubMed] [Google Scholar]
- 22.Arispe, N., Ma, J., Jacobson, K. A. & Pollard, H. B. (1998) J. Biol. Chem. 273, 5724-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubenstein, R. C. & Zeitlin, P. L. (2000) Am. J. Physiol. 278, C259-C267. [DOI] [PubMed] [Google Scholar]
- 24.Wang, F., Zeltwanger, S., Yang, I. C., Nairn, A. C. & Hwang, T. C. (1998) J. Gen. Physiol. 111, 477-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz, B. D., Singh, A. K., Devor, D. C. & Bridges, R. J. (1999) Physiol. Rev. 79, S109-S144. [DOI] [PubMed] [Google Scholar]
- 26.Knowles, M. R., Church, N. L., Waltner, W. E., Yankaskas, J. R., Gilligan, P. H., King, M., Edwards, L. J., Helms, R. W. & Boucher, R. C. (1990) N. Engl. J. Med. 322, 1189-1194. [DOI] [PubMed] [Google Scholar]
- 27.Burns, J. L., Van Dalfsen, J. M., Shawar, R. M., et al. (1999) J. Infect. Dis. 179, 1190-1196. [DOI] [PubMed] [Google Scholar]
- 28.Geller, D. E., Pitlick, W. H., Nardella, P. A., Tracewell, W. G. & Ramsey, B. W. (2002) Chest 122, 219-226. [DOI] [PubMed] [Google Scholar]
- 29.Accurso, F. J., Sagel, S. D., Sontag, M. K. & Wegener, J. S. (2003) Pediatr. Pulm. 25, Suppl., 104-105. [Google Scholar]
- 30.Dean, T. P., Dai, Y., Shute, J. K., Church, M. K. & Warner, J. O. (1993) Pediatr. Res. 34, 159-161. [DOI] [PubMed] [Google Scholar]
- 31.Richman-Eisenstat, J. B., Jorens, P. G., Hebert, C. A., Ueki, I. & Nadel, J. A. (1993) Am. J. Physiol. 264, L413-L418. [DOI] [PubMed] [Google Scholar]
- 32.Francoeur, C. & Denis, M. (1995) Inflammation 19, 587-598. [DOI] [PubMed] [Google Scholar]
- 33.Bedard, M., McClure, C. D., Schiller, N. L., Francoeur, C., Cantin, A. & Denis, M. (1993) Am. J. Respir. Cell Mol. Biol. 9, 455-462. [DOI] [PubMed] [Google Scholar]
- 34.Ruef, C., Jefferson, D. M., Schlegel-Haueter, S. E. & Suter, S. (1993) Eur. Respir. J. 6, 1429-1436. [PubMed] [Google Scholar]
- 35.Dimango, E. Zar, H. J., Bryan, R. & Prince, A. (1995) J. Clin. Invest. 96, 2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonfield, T. L., Konstan, M. W., Burfeind, P., Panuska, J. R., Hilliard, J. B. & Berger, M. (1995) Am. J. Respir. Cell Mol. Biol. 13, 257-261. [DOI] [PubMed] [Google Scholar]
- 37.Bonfield, T. L., Panuska, J. R., Konstan, M. W., Hilliard, K. A., Hilliard, J. B., Ghnaim, H. & Berger, M. (1995) Am. J. Respir. Crit. Care Med. 152, 2111-2118. [DOI] [PubMed] [Google Scholar]
- 38.DiMango, E., Ratner, A. J., Bryan, R., Tabibi, S. & Prince, A. (1998) J. Clin. Invest. 101, 2598-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabary, O., Zahm, J. M., Hinnrasky, J., Couetil, J. P., Cornillet, P., Guenounou, M., Gaillard, D., Puchelle, E. & Jacquot, J. (1998) Am. J. Pathol. 153, 921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briars, G. L., Dean, T. P., Murphy, J. L., Rolles, C. J. & Warner, J. O. (1995) Arch. Dis. Child 73, 74-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tirouvanziam, R., de Bentzmann, S., Hubeau, C., Hinnrasky, J., Jacquot, J., Peault, B. & Puchelle, E. (2000) Am. J. Respir. Cell Mol. Biol. 23, 121-127. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann, E., Dittrich-Breiholz, O., Holtmann, H. & Kracht, M. (2002) J. Leukocyte Biol. 72, 847-854. [PubMed] [Google Scholar]
- 43.Kelly, R. A. & Smith, T. W. (1996) in Goodman and Gilman's The Pharmacological Basis of Therapeutics, eds. Hardman, J. G., Limbird, L. E., Molinoff, P. B., Ruddon, R. W. & Gilman, A. G. (McGraw–Hill, New York), 9th Ed., pp. 809-838.
- 44.Zeitlin, P. L., Lu, L., Hwang, T-C, Rhim, J., Cutting, G. R., Keiffer, K. A., Craig, R. & Guggino, W. B. (1991) Am. J. Respir. Cell Mol. Biol. 4, 313-319. [DOI] [PubMed] [Google Scholar]
- 45.Flotte, T. R., Afione, S. A., Solow, R., Drumm, M. L., Markakis, D., Guggino, W. B., Zeitlin, P. L. & Carter, B. J. (1993) J. Biol. Chem. 268, 3781-3790. [PubMed] [Google Scholar]
- 46.Srivastava, M., Eidelman, O. & Pollard, H. B. (1999) Mol. Med. 5, 753-767. [PMC free article] [PubMed] [Google Scholar]
- 47.Paweletz, C. P., Charboneau, L., Bichsel, V. E., Simone, N. L., Chen, T., Gillespie, J. W., EmmertBuck, M. R., Roth, M. J., Petricoin, E. F. & Liotta, L. A. (2001) Oncogene 20, 1981-1989. [DOI] [PubMed] [Google Scholar]
- 48.Nishizuka, S., Charboneau, L., Young, L., Major, S., Reinhold, W. C., Wlatham, M., Kouros-Mehr, H., Bussey, K. J., Lee, J. K., Espina, V., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 14229-14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada, Y., Mukhopadhyay, A., Kundu, G. C., Mahabeleshwar, G. H. & Singh-Aggarwal, B. B. (2003) J. Biol. Chem. 278, 24233-24241. [DOI] [PubMed] [Google Scholar]