Abstract
The synthesis of bioactive vitamin D requires hydroxylation at the 1α and 25 positions by cytochrome P450 enzymes in the kidney and liver, respectively. The mitochondrial enzyme CYP27B1 catalyzes 1α-hydroxylation in the kidney but the identity of the hepatic 25-hydroxylase has remained unclear for >30 years. We previously identified the microsomal CYP2R1 protein as a potential candidate for the liver vitamin D 25-hydroxylase based on the enzyme's biochemical properties, conservation, and expression pattern. Here, we report a molecular analysis of a patient with low circulating levels of 25-hydroxyvitamin D and classic symptoms of vitamin D deficiency. This individual was found to be homozygous for a transition mutation in exon 2 of the CYP2R1 gene on chromosome 11p15.2. The inherited mutation caused the substitution of a proline for an evolutionarily conserved leucine at amino acid 99 in the CYP2R1 protein and eliminated vitamin D 25-hydroxylase enzyme activity. These data identify CYP2R1 as a biologically relevant vitamin D 25-hydroxylase and reveal the molecular basis of a human genetic disease, selective 25-hydroxyvitamin D deficiency.
The metabolic pathway leading to the synthesis of active vitamin D involves three reactions that occur in different tissues (1). The pathway is initiated in the skin with the UV light-mediated cleavage of 5,7,-cholestadien-3β-ol to produce the secosteroid (3β,5Z,7E)-9,10-secocholesta-5,7,10(19)-trien-3-ol (vitamin D3). The second step occurs in the liver and is catalyzed by a cytochrome P450 (CYP) enzyme that hydroxylates carbon 25, producing the intermediate 25-hydroxyvitamin D3, which is the major circulatory form of the vitamin. The third and final step takes place in the kidney and involves 1α-hydroxylation by another CYP, producing 1α,25-dihydroxyvitamin D3. This product is a potent ligand of the vitamin D receptor (VDR) and mediates most of the physiological actions of the vitamin (1).
Although the chemical and enzymatic steps in the vitamin D3 biosynthetic pathway have been known for 30 years (1), the enzyme catalyzing the 25-hydroxylation step in the liver has never been identified. At least six CYPs can catalyze this reaction in vitro, including CYP2C11, CYP2D25, CYP3A4, CYP2J1, CYP27A1, and CYP2R1 (2–4). Of these CYPs, the two most viable candidates for the vitamin D 25-hydroxylase are CYP27A1 and CYP2R1 (2). Both enzymes are expressed in the liver and are conserved among species known to have an active vitamin D signaling pathway. However, mutations in the human and mouse genes encoding the mitochondrial CYP27A1 protein impair bile acid synthesis, but have no consequences for vitamin D metabolism (5–9). It is thus not clear whether the two vitamin D3 25-hydroxylases represent an example of biological redundancy in an important biosynthetic pathway or whether CYP2R1 alone or some unidentified enzyme fulfills this essential role.
In contrast to the uncertainty surrounding vitamin D3 25-hydroxylase, the renal enzyme responsible for 1α-hydroxylation of the vitamin is CYP27B1 (10). This mitochondrial protein is expressed in the proximal renal tubules where activation of vitamin D3 takes place (11–13). As predicted from the biosynthetic pathway, mutations in the gene encoding CYP27B1 cause selective 1α,25-dihydroxyvitamin D3 deficiency (also referred to as vitamin D-dependent rickets type I) in both humans (14) and mice (15), in which 25-hydroxyvitamin D3 accumulates in plasma and levels of 1α,25-dihydroxyvitamin D3 are low (16).
In the present study, we describe the molecular analysis of a patient with abnormally low plasma levels of 25-hydroxyvitamin D3 and classic symptoms of vitamin D deficiency, including skeletal abnormalities, hypocalcemia, and hypophosphatemia (17, 18). This individual is homozygous for a substitution mutation in exon 2 of the CYP2R1 gene on chromosome 11p15.2. The CYP2R1 mutation changes a leucine residue at position 99 to a proline and eliminates the vitamin D3 25-hydroxylase activity of CYP2R1.
Materials and Methods
Patient Description. The patient is a young man from Nigeria and was evaluated for bone abnormalities of the lower extremities appearing between 2 and 7 years of age (17). Before treatment, he had low normal serum calcium levels (2.00–2.32 mM; normal range 2.12–2.62 mM), low serum phosphate levels (0.84–0.87 mM; normal range 0.97–1.45 mM), elevated serum alkaline phosphatase levels (2,360–3,000 units/liter; normal range 100–320 units/liter), serum 1α,25-dihydroxyvitamin D levels (137–142 pM) that were in the normal range (48–182 pM), and low 25-hydroxyvitamin D levels (10–12 nM; normal 25–137 nM). Lymphocytes from this individual were immortalized and stored for future study when he was first seen in the clinic. He and his family have since been lost to follow-up.
DNA Sequencing. Genomic DNA was extracted from Epstein–Barr virus-transformed lymphocytes using standard techniques (19). Exons 2, 3, and 5 of the human CYP2R1 gene were amplified separately by PCR with Platinum Taq DNA polymerase (Invitrogen). Exon 4 was amplified by PCR using PfuTurbo Hotstart polymerase (Stratagene). The thermocycler program for exons 2–5 consisted of one cycle at 94°C for 60 s, 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 150 s, followed by one cycle at 72°C for 600 s. Exon 1 of the CYP2R1 gene was amplified by using the Advantage GC genomic PCR kit (BD Biosciences). The thermocycler program consisted of one cycle at 95°C for 60 s, 35 cycles at 94°C for 30 s, and 68°C for 180 s, followed by one cycle at 68°C for 180 s. The nine exons of the CYP27A1 gene were amplified by the PCR as described (20). PCR products from all amplification reactions were purified by low-speed centrifugation through a Centricon YM-100 filter device (Millipore) before DNA sequence analysis using fluorescently labeled dye terminators and a thermostable DNA polymerase on an A BI Prism 3100 Genetic Analyzer (Perkin–Elmer).
Genomic DNA also was extracted from white blood cells of 50 unaffected African-American subjects (21) and 50 Nigerian subjects (22) using standard techniques. Exon 2 of the CYP2R1 gene was amplified from each of these control DNAs by the PCR and was subjected to sequence analysis as described above.
Site-Directed Mutagenesis. A plasmid containing a full-length human CYP2R1 cDNA (GenBank/European Bioinformatics Institute accession no. AY323817) in the pCMV6 expression vector (GenBank/European Bioinformatics Institute accession no. AF239250) was the starting template DNA for mutagenesis, which was performed with the QuikChange kit (Stratagene). Nucleotide 296 of the CYP2R1 cDNA was mutated from thymine to cytosine using the sense oligonucleotide, 5′-TATGATGTAGTAAAGGAATGCCCTGTTCATCAAAGCGAAATTTTTG-3′, and its complement. The cDNA insert of the engineered expression vector was sequenced to confirm the presence of the desired base pair change and to ensure the absence of spurious mutations. The insert was then excised by digestion with the restriction enzymes SalI and NotI, and subcloned into an untreated pCMV6 expression vector.
TLC. Transfection of human embryonic kidney (HEK) 293 cells (American Type Culture Collection catalog no. 1573), incubation with 4.6 × 10-7 M [4-14C]vitamin D3, extraction of lipids from cells and medium, and resolution of vitamin D metabolites by TLC were performed as described (2). A solvent system of cyclohexane:ethyl acetate (3:2, vol/vol) was used for thin layer chromatography. Radiolabeled vitamin D metabolites were detected by phosphorimaging of TLC plates using Fuji BAS-TR2040 imaging plates (Fuji Medical Systems, Tokyo) and a Storm 820 imaging system (Amersham Pharmacia Biosciences, Piscataway, NJ).
VDR Activation Assay. Receptor activation assays were performed as described (2). Briefly, HEK 293 cells were grown in 96-well plates and were transfected with the indicated plasmid DNAs for a period of 8–10 h. Vitamin D ligands were then added to the medium in ethanol, and the cells cultured for an additional 16–20 h. Experiments were terminated by cell lysis and measurements of β-galactosidase and luciferase (LUC) enzyme activities with appropriate substrates were performed (2).
Three different VDR-LUC reporter gene systems were used (2). The galactose 4 (GAL4)-VDR/GAL4-LUC reporter gene system consisted of the pCMX/GAL4-VDR and pTK-MH100 × 4-LUC plasmids. pCMX/GAL4-VDR encodes a fusion protein composed of the GAL4 DNA-binding domain linked to the human VDR ligand-binding domain. pTK-MH100 × 4-LUC is a reporter plasmid with four GAL4 DNA response elements linked to a viral thymidine kinase (TK) promoter segment and the firefly LUC gene. The VDR/vitamin D response element (VDRE)-LUC reporter gene system consisted of an expression plasmid encoding a full-length human VDR, and the pSPPX3-TK-LUC plasmid, which is a reporter plasmid with three copies of the VDRE sequence from the mouse osteopontin gene linked to a viral TK promoter element and the LUC gene. The VDR/osteopontin-LUC reporter gene system consisted of the above human VDR expression plasmid coupled with the osteopontin-LUC plasmid, a reporter DNA containing 0.862 kb of the mouse osteopontin enhancer and promoter region linked to the LUC gene.
Results
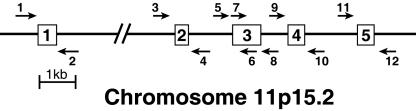
We first defined the exon–intron structure of the human CYP2R1 gene. As shown in Fig. 1, computer-assisted comparison of the cDNA and genomic DNA revealed that the CYP2R1 locus contains five exons separated by four introns that together span ≈15 kb of DNA on human chromosome 11p15.2. A series of six oligonucleotide primer pairs were designed and synthesized to allow amplification and sequencing through the PCR of individual exons and their immediate flanking regions (Fig. 1). PCRs with these oligonucleotide primers and genomic DNA isolated from a normal individual confirmed the predicted CYP2R1 gene structure. For genetic analysis of the CYP27A1 gene, oligonucleotides for amplification of the nine exons were synthesized as described (20), and optimal PCR conditions for their use were determined.
Fig. 1.
Structure of the human CYP2R1 gene. The predicted exon–intron structure of the human CYP2R1 gene is shown together with six pairs of oligonucleotides used to amplify and sequence fragments of the DNA. Exon 3 was amplified with primer pair 5, 8, and was sequenced with primers 5, 6, 7, and 8. The gene is drawn to scale with the exception of intron 1, which is estimated to be 9.1 kb in length. The autosomal location of the gene was deduced from the Human Genome database, which can be accessed at www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9606&chr=11.
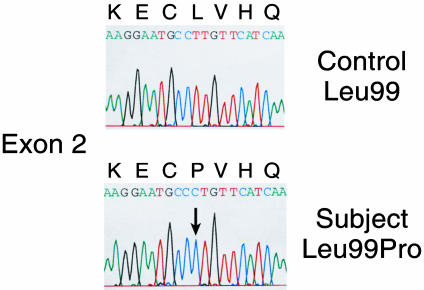
Genomic DNA from the patient with low 25-hydroxyvitamin D3 levels and control individuals was extracted and amplified by PCR using the CYP2R1 and CYP27A1 oligonucleotides, and individual products were subjected to DNA sequence analysis. These experiments revealed this individual is homozygous for a transition mutation (T → C) in exon 2 of the CYP2R1 gene, which is predicted to cause the substitution of a proline for leucine at position 99 (L99P) of the encoded protein (Fig. 2). Leucine 99 is conserved in the CYP2R1 enzymes of the puffer fish, mouse, and rat. The T → C change in exon 2 was not detected in the genomic DNAs of 50 control African-American subjects, suggesting that the nucleotide change represents a mutation rather than a polymorphism in the CYP2R1 gene. No mutations were detected in the CYP27A1 gene of this subject.
Fig. 2.
DNA sequence analysis of mutation in CYP2R1 vitamin D 25-hydroxylase. The partial DNA sequence of exon 2 from the CYP2R1 gene is shown from a normal individual (Upper) and from an individual with selective 25-hydroxyvitamin D deficiency (Lower). The codon specifying amino acid 99 is CTT in the normal gene and CCT (arrow) in the affected individual. The T → C transition mutation in the second nucleotide causes a change from leucine to proline at residue 99. All other nucleotides in the CYP2R1 gene of the proband were identical to those in normal individuals.
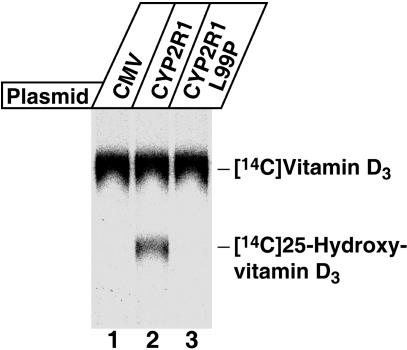
The biochemical consequences of the CYP2R1 L99P mutation for 25-hydroxylation were determined in assays by using [14C]vitamin D3 as a substrate (Fig. 3). When cells transfected with a control plasmid DNA were incubated with this radiolabeled substrate, no conversion to the 25-hydroxylated form was detected (lane 1). In contrast, expression of the normal CYP2R1 enzyme in the HEK 293 cells led to the formation of 25-hydroxyvitamin D3 (lane 2). The CYP2R1 enzyme with the L99P mutation was inactive (lane 3), and even after extended PhosphorImager analysis, showed no ability to 25-hydroxylate the substrate or to produce other products (data not shown).
Fig. 3.
Biochemical assay of CYP2R1 vitamin D 25-hydroxylase enzyme activity. HEK 293 cells were transfected with the indicated expression plasmids for a period of 18–20 h. Thereafter, the medium was made 4.6 × 10-7 M in [4-14C]vitamin D3 and the incubation continued for an additional 96 h. Lipids were extracted from cells and medium into chloroform:methanol (2:1, vol/vol), and vitamin D metabolites and standards were separated by TLC on 150-Å silica gel plates (Whatman, catalog no. 4855–821) in a solvent system containing cyclohexane:ethyl acetate (3:2, vol/vol). After development, radioactivity was detected by PhosphorImager analysis, and the positions to which authentic vitamin D3 and 25-hydroxyvitamin D3 migrated on the plate were determined by staining with iodine.
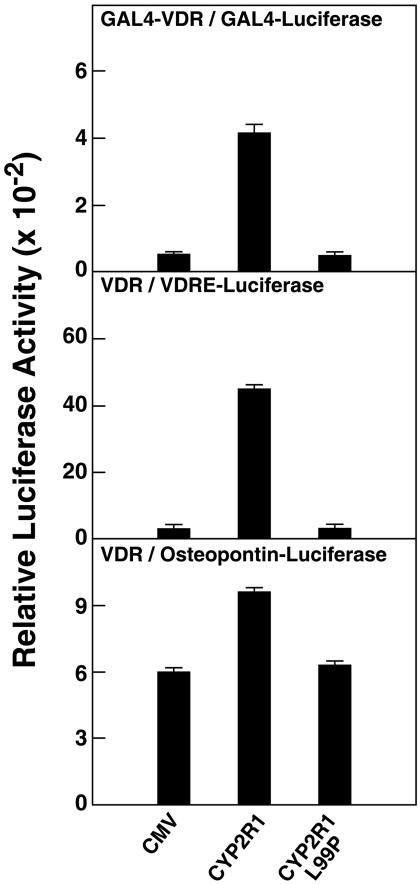
Because the amount of [14C]vitamin D3 substrate available for further biochemical assays was limiting, we next assessed the consequences of the L99P mutation for CYP2R1 enzyme activity by using functional assays. To this end, the abilities of the normal and mutant enzymes to activate the VDR were evaluated in three reporter gene systems in which VDREs were linked to transcription of a LUC gene. These experiments took advantage of the fact that 25-hydroxyvitamin D3 is a more potent ligand for the VDR than is nonhydroxylated vitamin D3 (2). As shown by the data in Fig. 4, when expression vectors specifying either normal or mutant CYP2R1 enzymes were transfected into cultured HEK 293 cells together with DNAs expressing different VDRs and their responsive genes, only cells expressing the normal CYP2R1 enzyme showed increased transcription from the LUC gene. The degree of stimulation varied with the system used, with the strongest activation measured for the intact VDR and a reporter gene containing three copies of a VDRE (Fig. 4 Middle). Here, the stimulus mediated by CYP2R1-catalyzed 25-hydroxylation of vitamin D3 was >40-fold over background, and no augmentation over baseline was observed with the L99P mutant protein.
Fig. 4.
L99P mutation in CYP2R1 causes loss of vitamin D3 activation. Expression vectors encoding no protein, the normal CYP2R1 enzyme (CYP2R1), or a version containing a proline substituted for leucine at amino acid 99 (CYP2R1L99P), were transfected in triplicate into HEK 293 cells together with the indicated VDR-reporter gene systems. The concentrations of vitamin D3 added to the medium ranged from 0.25 to 1.0 μM. After 16–20 h, cells were lysed and assayed for enzyme activities. Relative LUC activity was calculated by dividing units of LUC enzyme activity measured on a Dynex MLX luminometer by units of β-galactosidase enzyme activity measured on a Dynex Opsys MR spectrophotometer. Means ± SE of measurement were calculated and plotted in histogram form. The data are representative of two separate experiments carried out on different days.
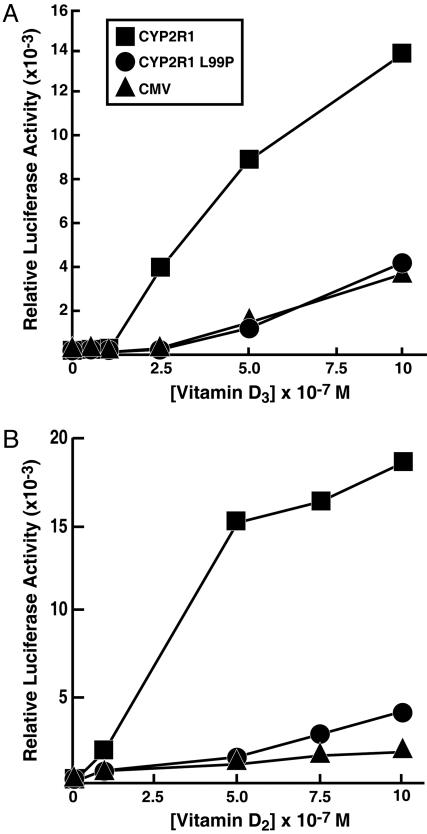
The reporter gene experiments shown in Fig. 4 were carried out at single concentrations of vitamin D3. To determine whether the CYP2R1 enzyme with the L99P mutation was active at higher concentrations of substrate, a dose–response curve was generated for the secosteroid. As shown in Fig. 5A, increasing the concentration of vitamin D3 in the medium from zero to 1 × 10-6 M led to an increase in LUC gene expression of ≈14-fold. When the L99P CYP2R1 enzyme was present, no stimulation over that in mock-transfected cells was observed. The stimulus detected in the control cells at higher vitamin D3 concentrations is attributable to endogenous CYP2R1 and CYP27A1 activities in the HEK 293 cells (2).
Fig. 5.
Response of normal and L99P CYP2R1 enzymes to increasing concentrations of vitamin D3 (A) and vitamin D2 (B). The indicated expression plasmids were introduced into HEK 293 cells together with DNAs constituting the VDR/VDRE-LUC reporter gene system. After an expression period (8 h), different amounts of the two forms of vitamin D were added to the medium and the incubation was continued for an additional 16 h. Thereafter, cells were lysed and assayed for LUC and β-galactosidase enzyme activities. Points on the graphs represent means of triplicate values established at each concentration of secosteroid. These experiments were repeated at least two times each.
A hallmark feature that distinguishes the microsomal CYP2R1 from the mitochondrial CYP27A1 is that the CYP2R1 enzyme 25-hydroxylates both vitamin D3 and (3β,5Z,7E,22E)-9,10-secoergosta-5,7,10(19),22-tetraen-3-ol (vitamin D2), whereas CYP27A1 only 25-hydroxylates vitamin D3 (2, 23, 24). The capacity of the CYP2R1 enzyme with the L99P mutation to 25-hydroxylate vitamin D2 is shown in Fig. 5B. As expected, the normal enzyme used this plant-derived secosteroid as a substrate, producing a ligand that activated the VDR. The mutant enzyme was far less active, but at the higher concentrations of vitamin D2 added to the medium (> 7.5 × 10-7 M), did provide a greater stimulus than that observed in mock-transfected cells. Similar modest stimulation was observed in three separate experiments. Together, the data shown in Fig. 5 suggested that the L99P CYP2R1 enzyme was incapable of hydroxylating vitamin D3 but retained residual activity against vitamin D2.
Discussion
The current data on mutation of the CYP2R1 enzyme are of interest from two points of view. First, they identify the microsomal CYP2R1 as a biologically important human vitamin D 25-hydroxylase, and second, they reveal the molecular basis of a human genetic disease, selective 25-hydroxyvitamin D3 deficiency.
At the enzyme level, it has been difficult to identify the biologically relevant vitamin D 25-hydroxylase for several reasons, including the existence of multiple candidate enzymes and the possibility of redundancy and compensation. Early studies indicated that 25-hydroxylase activity is present in both mitochondrial and microsomal subcellular fractions (25–27), and that the two enzymes exhibit different biochemical properties with respect to substrate affinity and capacity (28, 29). The mitochondrial enzyme was purified (30, 31), and after the cloning of its cDNA (32), revealed to be the CYP27A1 CYP that is also responsible for the 27-hydroxylation of sterol intermediates in bile acid synthesis (33). Shortly thereafter, mutations in the encoding gene located on human chromosome 2 were reported in patients with the cholesterol storage disorder cerebrotendinous xanthomatosis (5, 6). The finding of normal vitamin D metabolism in these subjects raised the redundancy issue (7).
Several different P450s were considered as candidates for the microsomal enzyme based on in vitro vitamin D 25-hydroxylase activity. One was the CYP2D25 enzyme purified from pig liver (34); however, this protein is not well conserved among species, as would be expected for an important vitamin D biosynthetic enzyme, and the closest human homologue (CYP2D6) does not have 25-hydroxylase activity (34). Furthermore, a molecular analysis by another group of the human CYP2D6 in the patient studied here failed to find mutations in the gene (35). Another candidate gene encoding the CYP3A4 enzyme also is normal in this individual (N.H.B., unpublished data). The rat CYP2C11 is a vitamin D 25-hydroxylase present in males only (36), and the fact that there is no known sexual dimorphism in vitamin D signaling means that this enzyme is unlikely to play a major biosynthetic role.
A fourth candidate for the microsomal enzyme is CYP2R1, which was identified in 2003 (2). CYP2R1 is preferentially expressed in the liver, is not sexually dimorphic, is highly conserved among species ranging from fish to human, and is equally active in the 25-hydroxylation of both vitamin D2 and vitamin D3 (2). These characteristics indicate that CYP2R1 is a biologically important vitamin D 25-hydroxylase, and the current data provide genetic evidence to support this assignment. CYP2R1 may be unique in fulfilling this role as the individual studied here has a normal CYP27A1 gene, and presumably normal enzyme activity, which were unable to compensate for the loss of CYP2R1. Whether the CYP27A1 enzyme plays any role as a vitamin D 25-hydroxylase remains moot; however, it is interesting to note that this subject responded to treatment with vitamin D and did have measurable levels of 1α,25-hydroxyvitamin D (17), suggesting that an alternate but nonessential pathway for the formation of 25-hydroxyvitamin D must exist.
With regard to human inborn errors, selective 25-hydroxyvitamin D3 deficiency is a rare autosomal recessive disorder characterized by impaired synthesis of an intermediate in the vitamin D biosynthetic pathway that results in endocrine defects in vitamin D signaling. In the patient studied here, the disease presented as vitamin D-dependent rickets associated with low-to-normal serum calcium and phosphate levels, elevated serum alkaline phosphatase activity, and most importantly, reduced levels of 25-hydroxyvitamin D. This individual was treated effectively with vitamin D2 therapy (3,000 units/day; ref. 17), confirming the absence of end-organ resistance and the presence of a functional VDR. The positive response to vitamin D2 may reflect the residual enzyme activity present with this substrate in the mutant CYP2R1 protein (Fig. 5) or conversion by an alternate vitamin D2 25-hydroxylase (3).
Multiple individuals have been described with selective 1α,25-dihydroxyvitamin D deficiency due to mutations in the gene encoding the CYP27B1 vitamin D 1α-hydroxylase (16, 37). Of the potential cases of selective vitamin D 25-hydroxylase deficiency that have been reported in the literature (17, 18, 35, 38–40), we were able to analyze two. The current studies indicate that the subject described by Casella et al. (17) has mutations in the CYP2R1 gene. The individual described by Zerwekh et al. (18) presented with a complex phenotype that was postulated to be due to mutations in both the vitamin D 25-hydroxylase and the VDR. We sequenced the exons of the CYP2R1 and CYP27A1 genes from this individual and found no mutations in either gene; however, biochemical and molecular analyses confirmed the presence of mutations in the VDR gene (41, 42).
It is not clear why mutations in the CYP27B1 vitamin D 1α-hydroxylase appear to be more common than those in the CYP2R1 vitamin D 25-hydroxylase. This discrepancy may represent an ascertainment bias as the subject studied here with CYP2R1 mutations had normal levels of circulating 1α,25-dihydroxyvitamin D. For reasons that are usually attributed to dietary supplementation or diminished clearance, many subjects with hereditary defects in calciferol metabolism have low-to-normal levels of 1α,25-dihydroxyvitamin D (16), which may mask mutations in other biosynthetic enzymes. The scarcity of 25-hydroxylase deficiency cases also may reflect genetic polymorphisms that allow alternate pathways to function in some individuals. Along these lines, it is of possible significance that the vitamin D 25-hydroxylase-deficient subject identified here was Nigerian. Individuals with dark skin require exposure to sunlight for longer periods of time to generate requisite levels of vitamin D3 as compared with those with less skin pigmentation and may be more sensitive to impaired synthesis of the vitamin (43–46). Furthermore, we sequenced exon 2 of the CYP2R1 gene in 50 control subjects from Nigeria (22), and identified one individual who was heterozygous for the L99P mutation. This result suggests that there may be a founder gene effect in Nigeria, a country in which the prevalence of rickets is high (47). Further studies in patients with selective 25-hydroxyvitamin D deficiency in Nigeria and elsewhere will be necessary to clarify these points.
Acknowledgments
We thank Daphne Head, Jeff Cormier, Erin Friedman, and Jill Fairless for technical assistance; Joe Zerwekh for [14C]vitamin D3 and access to patient DNA; and Helen Hobbs, Jay Horton, and Jean Wilson for critical reading of the manuscript. This work was supported by Robert A. Welch Foundation Grants I-0971 (to D.W.R.) and I-1275 (to D.J.M); National Institutes of Health Grants HL 20948 (to D.W.R.), GM 08014 (to J.B.C.), and DK 56603 (to N.H.B.); and the Howard Hughes Medical Institute (D.J.M.).
Abbreviations: vitamin D3, (3β,5Z,7E)-9,10-secocholesta-5,7,10(19)-trien-3-ol; vitamin D2, (3β,5Z,7E,22E)-9,10-secoergosta-5,7,10(19),22-tetraen-3-ol; CYP, cytochrome P450; GAL4, galactose 4; VDR, vitamin D receptor; VDRE, vitamin D response element; TK, thymidine kinase; HEK, human embryonic kidney; LUC, luciferase.
References
- 1.Jones, G., Strugnell, S. A. & DeLuca, H. F. (1998) Physiol. Rev. 78, 1193-1231. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, J. B., Motola, D. L., Mangelsdorf, D. J. & Russell, D. W. (2003) J. Biol. Chem. 278, 38084-38093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta, R. P., Hollis, B. W., Patel, S. B., Patrick, K. S. & Bell, N. H. (2004) J. Bone Miner. Res. 19, 680-688. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki, T., Izumi, S., Ide, H. & Ohyama, Y. (March 16, 2004) J. Biol. Chem., 10.1074/jbc.M312601200. [DOI] [PubMed]
- 5.Skrede, S., Bjorkhem, I., Kvittingen, E. A., Buchmann, M. S., Lie, S. O., East, C. & Grundy, S. (1986) J. Clin. Invest. 78, 729-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cali, J. J., Hsieh, C., Francke, U. & Russell, D. W. (1991) J. Biol. Chem. 266, 7779-7783. [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorkhem, I., Boberg, K. M. & Leitersdorf, E. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B., Kinzler, K. W. & Vogelstein, B. (McGraw–Hill, New York), Vol. 2, pp. 2961-2988. [Google Scholar]
- 8.Rosen, H., Reshef, A., Maeda, N., Lippoldt, A., Shpizen, S., Triger, L., Eggertsen, G., Bjorkhem, I. & Leitersdorf, E. (1998) J. Biol. Chem. 273, 14805-14812. [DOI] [PubMed] [Google Scholar]
- 9.Repa, J. J., Lund, E. G., Horton, J. D., Leitersdorf, E., Russell, D. W., Dietschy, J. M. & Turley, S. D. (2000) J. Biol. Chem. 275, 39685-39692. [DOI] [PubMed] [Google Scholar]
- 10.Takeyama, K., Kitanaka, S., Sato, T., Kobori, M., Yanagisawa, J. & Kato, S. (1997) Science 277, 1827-1830. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, M. Y., Wang, X., Wang, J. T., Compagnone, N. A., Mellon, S. H., Olson, J. L., Tenehouse, H. S., Miller, W. M. & Portale, A. A. (2002) Endocrinology 143, 587-595. [DOI] [PubMed] [Google Scholar]
- 12.Zehnder, D., Bland, R., Walker, E. A., Bradwell, A. R., Howie, A. J., Hewison, M. & Stewart, P. M. (1999) J. Am. Soc. Nephrol. 10, 2465-2473. [DOI] [PubMed] [Google Scholar]
- 13.Nykjaer, A., Dragnun, D., Walther, D., Vorum, H., Jacobsen, C., Herz, J., Melsen, F., Christensen, E. I. & Willnow, T. E. (1999) Cell 96, 507-515. [DOI] [PubMed] [Google Scholar]
- 14.Kitanaka, S., Takeyama, K., Murayama, A., Sato, T., Okamura, K., Nogami, M., Hasegawa, Y., Niimi, H., Yanagisawa, J., Tanaka, T. & Kato, S. (1998) N. Engl. J. Med. 338, 681-682. [DOI] [PubMed] [Google Scholar]
- 15.Kato, S. (2001) Endocrinology 142, 2734-2735. [DOI] [PubMed] [Google Scholar]
- 16.Liberman, U. A. & Marx, S. J. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B., Kinzler, K. W. & Vogelstein, B. (McGraw–Hill, New York), Vol. 3, pp. 4223-4240. [Google Scholar]
- 17.Casella, S. J., Reiner, B. J., Chen, T. C., Holick, M. F. & Harrison, H. E. (1994) J. Pediatr. 124, 929-932. [DOI] [PubMed] [Google Scholar]
- 18.Zerwekh, J. E., Glass, K., Jowsey, J. & Pak, C. Y. (1979) J. Clin. Endocrinol. Metab. 49, 171-175. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Leitersdorf, E., Reshef, A., Meiner, V., Levitzki, R., Schwartz, S. P., Dann, E. J., Berkman, N., Cali, J. J., Klapholz, L. & Berginer, V. M. (1993) J. Clin. Invest. 91, 2488-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mooser, V., Scheer, D., Marcovina, S. M., Wang, J., Guerra, R., Cohen, J. & Hobbs, H. H. (1997) Am. J. Hum. Genet. 61, 402-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper, R. S., Luke, A., Zhu, X., Kan, D., Adeyemo, A., Rotimi, C., Bouzekri, N., Ward, R. & Rorimi, C. (2002) Hypertension 40, 629-633. [DOI] [PubMed] [Google Scholar]
- 23.Holmberg, I., Berlin, T., Ewerth, S. & Bjorkhem, I. (1986) Scand. J. Clin. Lab. Invest. 46, 785-790. [DOI] [PubMed] [Google Scholar]
- 24.Guo, Y.-D., Strugnell, S. A., Back, D. W. & Jones, G. (1993) Proc. Natl. Acad. Sci. USA 90, 8668-8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya, M. H. & DeLuca, H. F. (1974) Arch. Biochem. Biophys. 160, 58-62. [DOI] [PubMed] [Google Scholar]
- 26.Bjorkhem, I. & Holmberg, I. (1978) J. Biol. Chem. 253, 842-849. [PubMed] [Google Scholar]
- 27.Madhok, T. C. & DeLuca, H. F. (1979) Biochem. J. 184, 491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukushima, M., Susuki, Y., Tohira, Y., Nishi, Y., Susuki, M., Sasaki, S. & Suda, T. (1976) FEBS Lett. 65, 211-214. [DOI] [PubMed] [Google Scholar]
- 29.Bjorkhem, I., Holmberg, I., Oftebro, H. & Pedersen, J. I. (1980) J. Biol. Chem. 255, 5244-5249. [PubMed] [Google Scholar]
- 30.Wikvall, K. (1984) J. Biol. Chem. 259, 3800-3804. [PubMed] [Google Scholar]
- 31.Dahlback, H. & Wikvall, K. (1988) Biochem. J. 252, 207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson, S., Davis, D. L., Dahlback, H., Jornvall, H. & Russell, D. W. (1989) J. Biol. Chem. 264, 8222-8229. [PubMed] [Google Scholar]
- 33.Russell, D. W. (2003) Annu. Rev. Biochem. 72, 137-174. [DOI] [PubMed] [Google Scholar]
- 34.Hosseinpour, F. & Wikvall, K. (2000) J. Biol. Chem. 275, 34650-34655. [DOI] [PubMed] [Google Scholar]
- 35.Lin, C. J., Wijesuriya, S. D., Abdullah, M. A., Casella, S. J. & Miller, W. L. (2003) Mol. Genet. Metab. 80, 469-472. [DOI] [PubMed] [Google Scholar]
- 36.Andersson, S., Holmberg, I. & Wikvall, K. (1983) J. Biol. Chem. 258, 6777-6781. [PubMed] [Google Scholar]
- 37.Kitanaka, S., Takeyama, K., Murayama, A. & Kato, S. (2001) Endocr. J. 48, 427-432. [DOI] [PubMed] [Google Scholar]
- 38.Asami, T., Kawasaki, T. & Uchiyama, M. (1995) Acta Paediatr. Jpn. 37, 182-188. [DOI] [PubMed] [Google Scholar]
- 39.Nutzenadel, W., Mehls, O. & Klaus, G. (1995) J. Pediatr. 126, 676-677. [DOI] [PubMed] [Google Scholar]
- 40.Abdullah, M. A., Salhi, H. S., Bakry, L. A., Okamoto, E., Abomelha, A. M., Stevens, B. & Mousa, F. M. (2002) J. Pediatr. Endocrinol. Metab. 15, 1017-1025. [DOI] [PubMed] [Google Scholar]
- 41.Griffin, J. E. & Zerwekh, J. E. (1983) J. Clin. Invest. 72, 1190-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitfield, G. K., Selznick, S. H., Haussler, C. A., Hsieh, J.-C., Galligan, M. A., Jurutka, P. W., Thompson, P. D., Lee, S. M., Zerwekh, J. E. & Haussler, M. R. (1996) Mol. Endocrinol. 10, 1617-1631. [DOI] [PubMed] [Google Scholar]
- 43.Bell, N. H., Greene, A., Epstein, S., Oexmann, M. J., Shaw, S. & Shary, J. (1985) J. Clin. Invest. 76, 470-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norman, A. W. (1998) Am. J. Clin. Nutr. 67, 1108-1110. [DOI] [PubMed] [Google Scholar]
- 45.Kreiter, S. R., Schwartz, R. P., Kirkman, H. N., Charlton, P. A., Calikoglu, A. S. & Davenport, M. L. (2000) J. Pediatr. 137, 153-157. [DOI] [PubMed] [Google Scholar]
- 46.Nesby-O'Dell, S., Scanlon, K. S., Cogswell, M. E., Gillespie, C., Hollis, B. W., Looker, A. C., Allen, C., Doughertly, C., Gunter, E. W. & Bowman, B. A. (2002) Am. J. Clin. Nutr. 76, 187-192. [DOI] [PubMed] [Google Scholar]
- 47.Thacher, T. D., Fischer, P. R., Pettifor, J. M., Lawson, J. O., Isichei, C. O. & Chan, G. M. (2000) J. Pediatr. 137, 367-373. [DOI] [PubMed] [Google Scholar]