Abstract
Haploinsufficiency of progranulin (PGRN) gene (GRN) causes familial frontotemporal lobar degeneration (FTLD), and modulates an innate immune response in humans and mouse models. GRN polymorphism may be linked to late-onset Alzheimer’s disease (AD). However, PRGN’s role in AD pathogenesis is unknown. Here, we show PGRN inhibits amyloid β (Aβ) deposition. Selectively reducing microglial PGRN in AD mice impaired phagocytosis and increased plaque load threefold. Lentivirus-mediated PGRN overexpression lowered plaque load in AD mice with aggressive amyloid plaque pathology. Aβ plaque load correlated negatively with levels of hippocampal PGRN, showing PGRN’s dose-dependent inhibitory effects on plaque deposition. PGRN also protected against Aβ toxicity. Reducing microglial PGRN exacerbated cognitive deficits in AD mice. Lentivirus-mediated PGRN overexpression prevented spatial memory deficits and hippocampal neuronal loss in AD mice. PGRN’s protective effects against Aβ deposition and toxicity have important therapeutic implications. We propose enhancing PGRN as a potential treatment for PGRN-deficient FTD and AD.
Progranulin (PGRN) is a secreted pleiotropic growth factor expressed in peripheral organs and the central nervous system. In brain, PGRN is expressed in neurons and microglia1–3. Loss-of-function mutations in the GRN gene are an important cause of familial frontotemporal lobar degeneration with TAR DNA-binding protein 43 (TDP-43)-positive inclusions (FTLD-TDP)4–6. Mouse models of PGRN deficiency exhibit abnormal neuronal and behavioral phenotypes7–10 and increased susceptibility to neuronal loss after toxin-induced injury11. How GRN mutations lead to neurodegeneration via haploinsufficiency is unknown. Recent studies focused on PGRN’s role in modulating neuroinflammation7–9. PGRN-deficient mice exhibit increased expression of proinflammatory cytokines and exacerbated microglial activation and astrogliosis7–10.
An elevated proinflammatory state contributes to pathogenesis of neurodegenerative diseases, including AD12–17. Genome-wide association studies linked CR1, CD33, and TREM2 to elevated risk for late-onset AD, implicating innate immunity in AD etiology18–19. GRN polymorphisms associated with reduced PGRN levels also modify risk for AD20–22. Individuals with a single base-pair deletion that causes a frame-shift mutation had clinical presentations that resembled AD or amnestic mild cognitive impairment23. While PGRN deficiency is predominantly associated with TDP-43 pathology, some GRN mutation carriers, especially those with apolipoprotein E4, exhibit AD pathology, including amyloid aggregation, neurofibrillary tangles, and TDP-43 mislocalization24.
Although PGRN deficiency is associated with higher risk for AD, PGRN levels are upregulated in microglia surrounding plaques in AD patients25–26. Whether this increase is protective or detrimental in AD pathogenesis is unknown.
Here, we report that brain PGRN levels were reduced in mice expressing human amyloid precursor protein (APP) with familial AD-linked mutations (FAD) before plaque deposition. To investigate how reduced PGRN levels contribute to AD pathogenesis, we ablated or overexpressed Grn in APP mouse models and examined how neuronal, behavioral, and inflammatory processes were affected. We investigated the role of microglia-derived PGRN by selectively ablating PGRN in myeloid cells of APP mice, and found that PGRN enhances microglia-mediated phagocytosis and protects against Aβ-mediated neurotoxicity and cognitive deficits, suggesting increased PGRN as a novel therapeutic strategy for AD.
RESULTS
Regulation of PGRN expression in APP transgenic mice
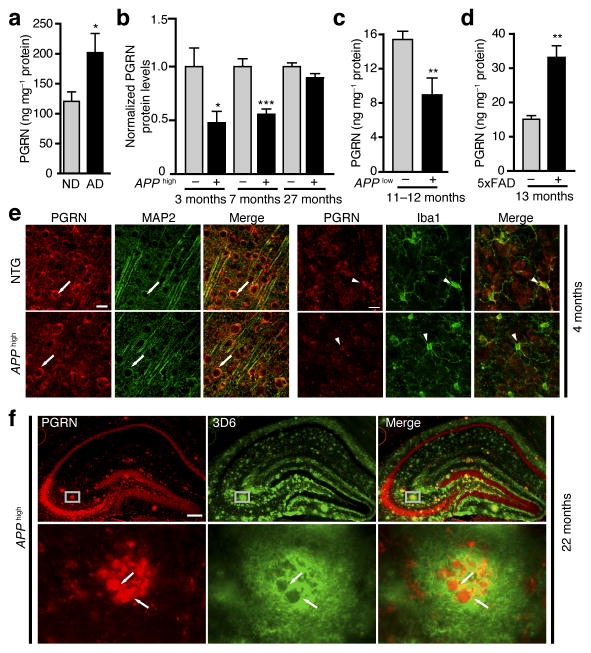
PGRN deficiency is associated with increased risk for AD20–22. We first examined if PGRN levels are altered in AD (Supplementary Table-1) using a human PGRN ELISA (Supplementary Fig. 1a–c). PGRN levels were significantly higher in AD brains than non-demented controls, consistent with elevated PGRN expression around plaques (Fig. 1a). We then established a mouse PGRN ELISA assay (Supplementary Fig. 1d,e) and measured PGRN in PDAPPSw,Ind J20 (APPhigh) mice at different stages of plaque deposition27. Interestingly, PGRN levels were significantly lower in the cortices of 3- and 7-month-old APPhigh mice with minimal plaque deposition, but not in those of 27-month-olds with extensive plaque pathology (Fig. 1b).
Figure 1.
Differential expression of PGRN in AD brains and in young and aged APP transgenic mice. (a) ELISA measurement of human PGRN protein levels in brains of AD patients and non-demented controls (n = 12, n = 11). *, P < 0.05, unpaired student’s t test. ND, non-demented. See Supplementary Table-1 for sample information. (b) ELISA measurement of mouse PGRN levels in APPhigh mice and littermate controls (n = 6, n = 7, n = 10, n = 8, n = 9, n = 10, from left to right). *, P < 0.05 by unpaired student’s t test; ***, P < 0.001 by Mann-Whitney non-parametric test. (c) ELISA measurement of PGRN levels in 11–12-month-old APPlow mice (n = 10, n = 9, respectively). **, P < 0.01 by Mann-Whitney non-parametric test. (d) ELISA measurement of PGRN levels in 13-month-old 5xFAD mice (n = 3, n = 6 respectively). **, P < 0.01 by unpaired student’s t test. (e) PGRN immunoreactivity is detected in both neurons (MAP2, arrow) and microglia (Iba1, arrow head) in non-transgenic (NTG) and APPhigh mice. Scale bar 25 μm. (f) PGRN immunoreactivity in amyloid plaques in aged APPhigh mice. Representative images of colocalization of PGRN (red) with amyloid plaques (anti-3D6, green) in the hippocampus of 22-month-old APPhigh mice. Bottom panels are higher magnification images of top panels. Scale bar 200 μm. Values are mean ± SEM (a–d).
To further investigate differential regulation of PGRN in AD mice with or without plaques, we measured PGRN in PDGF-APPSw,Ind J9 (APPlow) mice, a mild amyloid model developing plaques at 10 months of age27, and in 5xFAD mice, an aggressive amyloid model developing plaques at 2 months of age28. Levels of PGRN protein were reduced in 11–12-month-old APPlow mice with limited plaque pathology27, but elevated in 13-month-old 5xFAD mice with extensive dense-core plaques25 (Fig. 1c,d). By immunohistochemical analysis, PGRN was expressed in neurons and microglia, regardless of human APP (hAPP) expression (Fig. 1e). PGRN immunoreactivity exhibited remarkable colocalization with plaques at elevated levels (Fig. 1f). Moreover, levels of PGRN mRNA were upregulated in 13-month-old 5xFAD mice, consistent with the same observation in aged APP/PS1 and BriAb42 mice with aggressive plaque deposition25 (Supplementary Fig. 2). In contrast, reducing PGRN protein levels in APPhigh or APPlow mice lacking extensive plaques was not accompanied by reduced PGRN mRNA (Supplementary Fig. 2).
PGRN deficiency exacerbates Aβ-dependent cognitive deficits
To test our hypothesis that PGRN deficiency contributes to AD pathogenesis, we ablated PGRN in APPlow mice, which express low hAPP levels and exhibit no overt behavioral deficits29. PGRN deficiency did not affect APP processing in APPlow mice, as total APP and α- or β-CTFs in cortex and hippocampus were similar (Supplementary Fig. 3). Levels of Aβ1-x and Aβ1–42, which correlated with age, were not affected by PGRN ablation either (Supplementary Fig. 3).
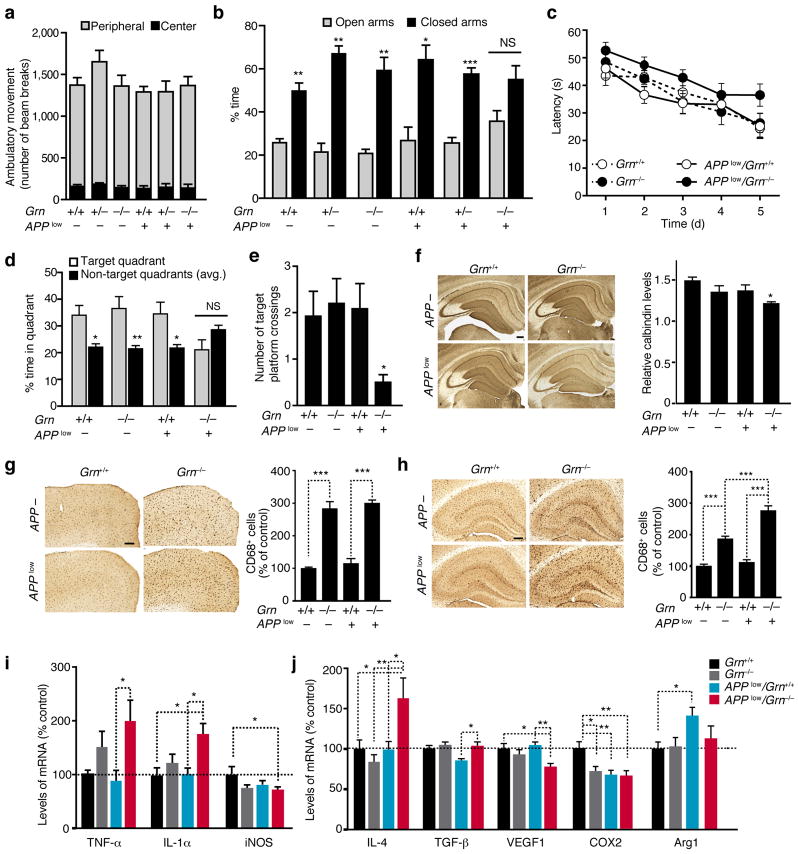
We next investigated how PGRN deficiency affects AD-related behaviors. In the open field, APPlow mice showed normal levels of activity regardless of Grn genotype (Fig. 2a). In the elevated plus maze, all genotypes showed preference to closed arms except for APPlow/Grn−/− mice, which exhibited no preference. Thus, PGRN deficiency induced a disinhibition phenotype in APPlow mice that is reminiscent of that observed in APPhigh mice29 (Fig. 2b). The Morris water maze test was used to examine spatial learning and memory. Swim speed and latency in the visible platform trials were similar for all genotypes (Supplementary Fig. 4). APPlow and Grn−/− mice learned similarly as Grn+/+ mice; the modest spatial learning deficits of APPlow/Grn−/− mice were not significant (Fig. 2c). Importantly, while mice lacking PGRN or expressing hAPP/Aβ alone exhibited normal spatial memory, APPlow/Grn−/− mice were significantly impaired in spatial memory retention: they exhibited no preference for the target quadrant and crossed the target platform fewer times than Grn−/− mice in the probe trial (Fig. 2d,e). These findings suggest that PGRN deficiency exacerbates cognitive deficits in mouse models of mild AD.
Figure 2.
PGRN deficiency exacerbates Aβ-mediated behavioral and neuronal deficits and modulates the innate immunity in 9–13-month-old APPlow mice. (a) Locomotor activity in the open field (n = 10 mice/genotype). (b) Time spent in the open and closed arms of the elevated plus maze (n = 10 mice/genotype, *, P < 0.05, **, P < 0.01, ***, P < 0.001, paired student’s t-test). NS, not significant. (c–e) Spatial learning and memory in MWM. (c) No significant differences were detected in spatial learning by longitudinal mixed effects model with linear time trend. (d,e) Probe trial performance after 24 h. (d) Time spent in target quadrant. *, P < 0.05, **, P < 0.01, paired student’s t-test. There was a trend towards interaction of the APP and Grn genotype variables for target quadrant % time, P = 0.06 (n = 13, n = 12, n = 12, n = 11). (e) Total number of target platform crossings. *P < 0.05, two-way ANOVA, Bonferroni posthoc analyses (n = 12, n = 12, n = 11, and n = 10). (f) (left) Calbindin immunostaining in the hippocampus. Scale bar, 200 μm. (right) Quantification of calbindin levels (n = 13, n = 12, n = 12, n = 11). *, P < 0.05, two-way ANOVA, Tukey-Kramer posthoc analyses. Variances were significantly different between groups. (g, h) CD68 immunoreactivity in cortex (g) and hippocampus (h). (left) CD68+ immunostaining of cortical (g) or hippocampal (h) sections. Scale bar, 200 μm. (right) Quantification of CD68+ immunoreactivity (n = 13, n = 12, n = 12, n = 11), ***, P < 0.001, two-way ANOVA, Tukey-Kramer posthoc analyses. Variances were significantly different between groups. There was a significant interaction between the APP and Grn genotype variables in hippocampal CD68+ immunoreactivity,*** P < 0.001. (i, j) qRT-PCR measurements of levels of M1 (i) or M2 (j) inflammatory markers from the cortex (n = 7, n = 7, n = 5, n = 5), *, P < 0.05, **, P < 0.01 by two-way ANOVA, Tukey-Kramer posthoc analyses. Variances were significantly different between groups for TNF-α and iNOS. There was a significant interaction between the APP and Grn genotype variables in IL-4, VEGF1, and COX2, * P < 0.05.
Levels of calbindin in the molecular layer of dentate gyrus (DG) are depleted in APPhigh mice, and the extent of depletion correlates with spatial memory deficits30. In APPlow mice, calbindin levels were unaffected, consistent with their relatively normal cognitive function (Fig. 2f). Deleting PGRN in APPlow mice lowered calbindin immunoreactivity in the DG, consistent with impaired spatial memory in these mice.
PGRN modulates innate immunity in APP transgenic mice
PGRN deficiency exacerbates microgliosis in the brain7,11. CD68 immunoreactivity, a marker of activated microglia, was elevated by PGRN deficiency in the cortex and hippocampus, regardless of APP expression (Fig. 2g,h). In the hippocampus, APPlow/Grn−/− mice exhibited stronger CD68 immunoreactivity than APPlow or Grn−/− mice (Fig. 2h). PGRN deficiency elevated levels of the proinflammatory markers TNF-α and IL-1α only with hAPP expression (Fig. 2i). Levels of iNOS, an inflammatory marker, were significantly reduced in APPlow/Grn−/− mice. PGRN deficiency with hAPP expression also altered expression of anti-inflammatory M2 markers, increasing levels of IL-4 and TGF-β while decreasing those of VEGF1 and COX2 (Fig. 2j).
Microglial PGRN deficiency increases plaque deposition and impairs phagocytosis
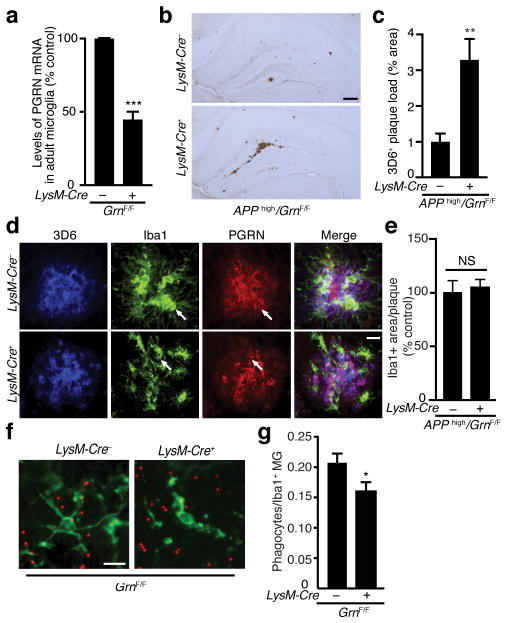
PGRN is expressed in neurons and microglia. The potent immunomodulation induced by PGRN deficiency suggests an important role of microglia-derived PGRN. To examine how microglial PGRN modulates Aβ pathology, we selectively reduced PGRN in microglia of APPhigh mice. Unlike APPlow mice, which develop sparse plaque pathology even when they are 12 months of age, APPhigh mice express higher hAPP/Aβ levels and develop plaques at around 6–7 months27. LysM-Cre mice were crossed with GrnF/F mice to drive PGRN deletion in cells of myeloid origin31. CD11b+ microglia were isolated from adult mice by myelin depletion and separation with CD11b-conjugated microbeads32. By FACS analysis, 84–90% of isolated cells were CD11b+ microglia (Supplementary Fig. 5). LysM-Cre expression led to 50% reduction of PGRN mRNA in CD11b+ microglia from adult mouse brain, as measured with qRT-PCR (Fig. 3a).
Figure 3.
Microglial PGRN deficiency increases plaque deposition and impairs phagocytosis. (a) Quantification of PGRN mRNA in microglia isolated from adult GrnF/F mice with real-time RT-PCR. n = 4 (pooled from 10 mice), n = 3 (pooled from 8 mice), ***, P < 0.001, unpaired student t-test. Variances were significantly different between groups. (b–c) Effects of microglial PGRN deficiency on plaque load in APPhigh mice. (b) Representative photomicrograph of amyloid plaques detected with 3D6 antibody. Scale bar, 200 μm. (c) Quantification of 3D6-positive plaque load in APPhigh mice with normal or deficient microglial PGRN, n = 8, n = 9, **, P < 0.01, unpaired student t-test. Variances were significantly different between groups. (d) Colocalization of PGRN (red) with microglia (anti-IbaI, green) and amyloid deposition (3D6, blue). Scale bar, 10 μm. (e) Quantification of the number of Iba1+ microglia per 3D6+ plaque in APPhigh mice with normal or deficient microglial PGRN (n = 9, n = 10). (f–g) Phagocytosis of fluorescent beads by LysM-Cre+/GRNF/F microglia. (f) Representative photomicrograph of acute slice sections from 8-month-old mice incubated with fluorescent beads. (g) Quantification of the number of Iba1-positive phagocytes normalized to total number of Iba1-positive cells, n = 34 fields of view from 3 mice, n = 35 fields of view from 4 mice, *, P < 0.05, mixed effects model. Scale bar, 10 μm.
Surprisingly, reducing microglial PGRN by 50% led to a striking increase in hippocampal plaque load in 7-month-old APPhigh mice (Fig. 3b,c). These effects on plaque deposition are not due to Aβ metabolism or hAPP processing since levels of soluble Aβ1-x or Aβ1–42, α-CTF, β-CTF, and full-length APP were unaffected by reducing microglial PRGN in 4-month-old APPhigh mice before plaque deposition (Supplementary Fig. 6).
PGRN overexpression enhances microglial recruitment surrounding the injection site33. To determine if microglial PGRN deficiency impairs microglial recruitment to amyloid plaques, we quantified Iba1 immunoreactivity surrounding the plaques and found no difference (Fig. 3d,e). In primary microglia, endocytosis of Aβ was enhanced by recombinant PGRN33. We next performed a phagocytosis assay with acute brain slices from adult GrnF/F mice with or without expression of LysM-Cre (Fig. 3f,g). Fewer PGRN-deficient microglia (LysM-Cre/GrnF/F) phagocytosed fluorescent beads than wildtype microglia (GrnF/F), suggesting that PGRN deficiency in microglia impairs phagocytosis in the adult brain (Fig. 3f,g).
Microglial PGRN protects against Aβ toxicity
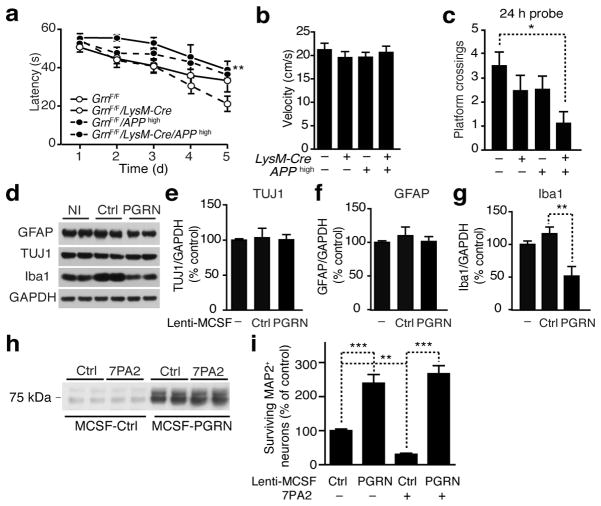
APPhigh mice exhibit robust spatial learning and memory deficits at 6 months of age. To investigate if microglia-derived PGRN affects Aβ-mediated cognitive deficits, we performed MWM at 4 months of age at the onset of behavioral deficits in APPhigh mice. At this age before plaque deposition, Aβ levels did not differ with or without microglial PGRN (Supplementary Fig. 6), precluding differences in Aβ levels as a confounding factor. APPhigh and microglial PGRN-deficient mice exhibited strong trends in learning deficits, but only APPhigh mice lacking microglial PGRN exhibited a significant deficit in spatial learning (Fig. 4a). The increased latency was not due to differences in swim speed (Fig. 4b). Also, APPhigh mice lacking microglial PGRN crossed the target platform fewer times than wildtype mice in the 24-h probe trial, consistent with a protective effect of microglial PGRN against Aβ-mediated cognitive deficits (Fig. 4c).
Figure 4.
Microglial PGRN protects against Aβ toxicity. (a–c) Spatial learning and memory in MWM in APPhigh mice. (a) Learning curve to locate the hidden platform. **, P < 0.01, by longitudinal mixed effects model with linear time trend. (b) Average swim speed across all training days. (c) Number of target platform crossings after 24 h (n = 11, n = 10, n = 9, n = 7, *, P < 0.05 by two-way ANOVA, Tukey-Kramer posthoc analyses). (d–g) Effects of viral PGRN overexpression in mixed cortical cultures. (d) Representative western blots of cell lysates from primary cultures infected with Lenti-MCSF-Ctrl or Lenti-MCSF-PGRN. Non-infected (NI). (e–g) Quantification of TUJ1 (e), GFAP (f), and Iba1 (g). n = 6 from three independent experiments performed in duplicates, **, P < 0.01 by one-way ANOVA, Tukey-Kramer posthoc analyses. (h–i) Effects of microglial PGRN on Aβ toxicity in vitro. (h) Representative western blot of supernatants from mixed cortical cultures infected with Lenti-MCSF-Ctrl or Lenti-MCSF-PGRN treated with Aβ oligomers (7PA2). (i) Aβ toxicity measured by survival of MAP2+ neurons. n = 84, n = 83, n = 82, n = 82 fields of view from two independent experiments, **, P < 0.01, ***, P < 0.001, by mixed effects model and multiple comparison correction using the method of Holm.
To directly examine the protective effects of microglial PGRN on Aβ toxicity, we infected mixed cortical cultures, which contained neurons, microglia, and astrocytes34, with a lentiviral vector encoding Grn under the macrophage colony stimulating factor (MCSF) promoter34. Compared with control virus, infection of Lenti-MCSF-PGRN did not affect the number of neurons and astroglia, as indicated by similar levels of TUJ1 and GFAP (Fig. 4d–f). Interestingly, overexpression of PGRN in microglia reduced the levels of Iba-1 significantly (Fig. 4g), consistent with suppression of microgliosis by endogenous PGRN in vivo35. To test the effects of microglial PGRN on Aβ toxicity, mixed primary cultures were treated with oligomeric Aβ from a cell line that constitutively expresses mutant APP (7PA2)36, and surviving neurons were identified by MAP2 staining. PGRN levels were higher in the supernatants of cells infected with Lenti-MCSF-PGRN, regardless of 7PA2 treatment (Fig. 4h). Aβ treatment significantly decreased surviving MAP2+ neurons; microglial PGRN overexpression markedly enhanced neuronal survival (Fig. 4i).
PGRN overexpression decreases plaque load in 5xFAD mice
To determine if PGRN protects against Aβ pathology and Aβ toxicity in vivo, we injected lentiviral vectors expressing mouse PGRN (Lenti-PGRN) into 5xFAD mice, which express FAD-linked mutant APP and presenilin28. Compared with APPlow and APPhigh mice, 5xFAD mice develop more aggressive plaque pathology earlier and exhibit significant neurodegeneration, particularly in the neocortex and subregions of the hippocampus28,37–38.
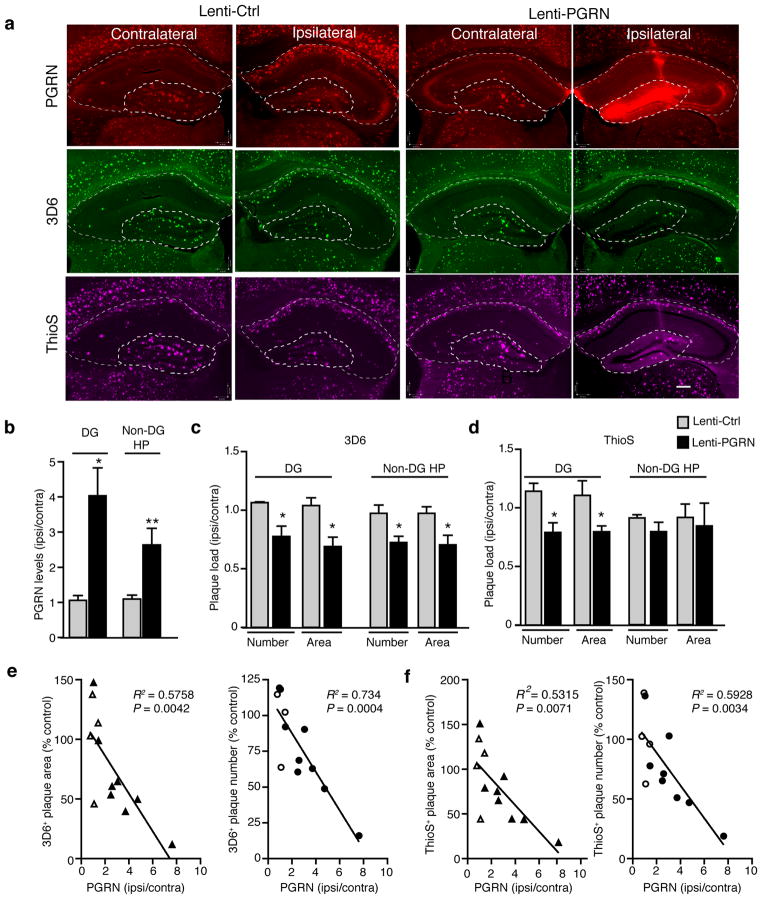
5xFAD mice were unilaterally injected with similar titers of Lenti-Control or Lenti-PGRN at 2 months of age and analyzed 3 months later. PGRN immunoreactivity was much stronger in the ipsilateral hippocampus than the contralateral side, especially in the DG where the injection was targeted (Fig. 5a,b). Injection of Lenti-PGRN significantly decreased 3D6- and ThioS-positive plaque load in the ipsilateral DG, quantified by the number of plaques and total plaque area; control virus had no effect (Fig. 5c,d). In non-DG hippocampus with weaker PGRN immunoreactivity, 3D6-, but not ThioS-, positive plaque load was lower in the ipsilateral side. Moreover, PGRN levels, which differed mostly from variability in virus-mediated expression, correlated negatively with 3D6- and ThioS-positive plaque load, as quantified by the number or total area of plaques in the DG (Fig. 5e,f).
Figure 5.
PGRN overexpression in hippocampus decreases amyloid plaque load in 5xFAD mice. (a) Representative images of the contralateral and ipsilateral hemispheres of Lenti-Ctrl- and Lenti-PGRN-injected 5xFAD mice. Dashed lines indicate boundaries used for quantification of dentate gyrus (DG) and non-dentate gyrus hippocampal (non-DG HP) areas. PGRN (red), 3D6+ plaques (green), and ThioS+ plaques (magenta). Scale bar, 200 μm. (b) Quantification of PGRN immunostaining normalized to the uninjected contralateral hemisphere for Lenti-Ctrl- or Lenti-PGRN-injected mice. Two Lenti-PGRN-injected mice lacking sufficient overexpression (i.e. < 2 fold) were excluded. n = 4 (Lenti-Ctrl), n = 6 (Lenti-PGRN), *, P < 0.05, **, P < 0.01, unpaired student’s t-test. (c–d) Quantification of 3D6+ (c) and ThioS+ (d) plaque load by number and area. Two Lenti-PGRN injected mice lacking sufficient overexpression (i.e. < 2 fold) were excluded. n = 4 (Lenti-Ctrl), n = 6 (Lenti-PGRN)*, P < 0.05, **, P < 0.01, unpaired student’s t-test. Welch’s correction was applied for DG 3D6+ plaque number to account for significantly different variance between groups. (e–f) Pearson correlation analyses of PGRN levels (normalized to the contralateral side) with the number or area of 3D6+ (e) or ThioS+ (f) amyloid plaques in the dentate gyrus. Open triangle or circle (Lenti-Ctrl), closed triangle or circle (Lenti-PGRN). n = 12.
PGRN overexpression prevents neuron loss and memory deficits
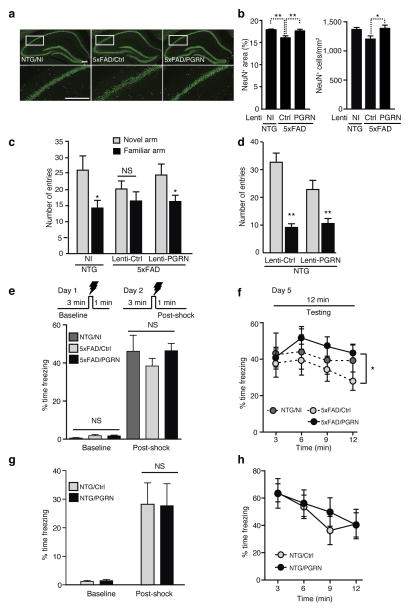
To further determine if overexpressing PGRN protected against neurodegeneration and cognitive deficits in 5xFAD mice, we injected equivalent amounts of Lenti-PGRN or Lenti-Ctrl virus bilaterally into the DG of 2-month-old 5xFAD mice, and examined them three months later. 5xFAD mice injected with Lenti-Ctrl contained fewer NeuN-positive neurons than NTG mice in the hippocampus (Fig. 6a,b). Overexpressing PGRN with Lenti-PGRN injection abolished the loss of hippocampal NeuN-positive neurons (Fig. 6a,b), and led to a trend toward increase in calbindin levels in the CA1, but not DG, region of 5xFAD mice (Supplementary Fig. 7). These findings support the protective role of PGRN against Aβ-mediated neuronal deficits in vivo.
Figure 6.
Lentiviral PGRN overexpression prevents neuronal loss and hippocampus-dependent memory deficits in 5xFAD mice. (a,b) Effects of PGRN overexpression on neuronal loss. (a) Representative images of NeuN staining in hippocampus. Scale bar, 200 μm. Bottom panels are higher magnification images of top panels. (b) Quantification of number and area of NeuN+ cells in the hippocampus. n = 12, n = 14, n = 14 from 6 (NI), 7 (Lenti-Ctrl), or 7 (Lenti-PGRN) mice, *, P < 0.05, **, P < 0.01, mixed effects model and multiple comparison correction using the method of Holm. (c,d) Cued Y-maze spatial memory performance in PGRN-overexpressing 5xFAD (c) or non-transgenic (d) mice. n = 9, n = 12, n = 13, *, P < 0.05; NS, not significant, paired student’s t-test (c). Lenti-Ctrl-injected 5xFAD mice were analyzed by Wilcoxon signed rank test of pairs. n = 6, n = 7, **, P < 0.01, paired student’s t-test (d). (e–h) Contextual fear conditioning in 5xFAD (e,f) or non-transgenic (g,h) mice. (e,g) Quantification of freezing time on Day 1 for 3 min pre-shock (baseline) and on Day 2 for 1 min post-shock. n = 7, n = 12, n = 13 (e); n = 6, n = 7 (g). (f,h) Quantification of freezing time in 3-min intervals on Day 5 testing. n = 7, n = 12, n = 13, *, P < 0.05, by analysis of min 6–12 by mixed effects model and Tukey-Kramer post-hoc analysis (f). Two mice from the NTG/NI group were statistical outliers (> 2 S.D.) due to low freezing during testing, thus excluded from analysis. n = 6, n = 7, not significant (NS) by analysis of min 6–12 by mixed effects model and Tukey-Kramer post-hoc analysis (h).
We next examined PGRN’s effects on hippocampal function in the cued Y-maze, where mice were allowed to freely explore two of three arms, identifiable with spatial cues. Mice were introduced into the same start arm and allowed to explore all three arms 2 hours later. Control non-injected mice with normal spatial memory preferred the novel arm (Fig. 6c). 5xFAD mice injected with Lenti-Ctrl showed no preference. Those injected with Lenti-PGRN preferred the novel arm, exhibiting enhanced spatial memory (Fig. 6c). PGRN overexpression had no effect on NTG controls (Fig. 6d). In a contextual fear-conditioning test, 5xFAD and NTG mice injected with Lenti-PGRN or Lenti-Ctrl showed similar baseline and post-shock freezing during the training period (Fig. 6e,g). During testing 3 days later, PGRN overexpression induced higher levels of freezing than control-injected 5xFAD mice, but had no effect on NTG mice (Fig. 6f,h).
Control experiments were performed to determine if Lenti-PGRN affected non-hippocampus-dependent behaviors. 5xFAD mice exhibited largely normal working memory, as measured by the percentage of alterations in the small Y-maze. Overexpressing PGRN did not affect working memory in 5xFAD or NTG mice (Supplementary Fig. 8a,b). In the elevated plus maze, overexpressing PGRN did not affect 5xFAD mice or NTG controls; 5xFAD mice exhibited a disinhibition phenotype, regardless of PGRN overexpression (Supplementary Fig. 8c,d). Thus, hippocampal PGRN expression protected against Aβ-mediated spatial and contextual fear deficits with little effect on non-hippocampus-dependent functions.
DISCUSSION
Here, we show that PGRN protects against Aβ toxicity and Aβ deposition in AD mouse models. Ablating Grn exacerbated cognitive and neuronal/synaptic deficits and induced aberrant immune responses. PGRN deficiency in microglia impaired phagocytosis and increased plaque load. Moreover, PGRN overexpression reduced plaque load and ameliorated neuronal loss and cognitive deficits. These findings established a causal role of PGRN in AD pathogenesis, consistent with the genetic association of PGRN deficiency with increased risk of late-onset AD.
Unlike previous studies on plaque-dependent PGRN regulation25–26, we examined plaque-independent PGRN regulation. In three AD mouse models, PGRN protein was downregulated when plaque deposition was minimal, but elevated in mice with extensive dense-core plaques. Upregulation of PGRN by dense-core plaques likely occurs at the transcriptional level whereas plaque-independent downregulation involves post-transcriptional or –translational mechanisms. The differential mechanisms could result from soluble Aβ oligomers versus fibrils, which could regulate PGRN expression via different mechanisms. Microglial PGRN could be upregulated by Aβ fibrils associated with plaques; Aβ oligomers could downregulate PGRN expression in neurons and microglia via alteration of signaling pathways. For example, PGRN expression is downregulated by toll-like receptor ligands (e.g., LPS and poly-IC) and proinflammatory cytokines (e.g., IL-1/IFNγ)39. Related pathways may be involved in downregulating PGRN expression in AD mouse models at early disease stages. PGRN expression could also be regulated by non-Aβ factors, including C-terminal hAPP fragments and associated downstream signaling. Moreover, PGRN could be cleaved by proteases (e.g., matrix metalloproteinases) into granulins that exert divergent biological functions39–40. Further studies will be needed to establish whether enhanced aberrant cleavage of PGRN contributes to PGRN deficiency in AD.
Our study demonstrated a causative role of PGRN deficiency in AD pathogenesis in vivo. Deleting Grn in APPlow mice, which do not exhibit AD-related behavioral and neuronal changes due to low levels of hAPP expression, led to deficits similar to those of APPhigh mice. These findings suggest that downregulating PGRN early on renders the brain vulnerable to Aβ toxicity. One striking phenotype of PGRN deficiency is accelerated lipofuscinosis in human41 and mouse42–43. However, accelerated lipofuscinosis is unlikely to underlie the exacerbated Aβ-mediated deficits and toxicity in APPlow/Grn−/− mice. Selectively reducing microglial PGRN in APP mice, which does not accelerate lipofuscinosis, led to exacerbated plaque deposition and cognitive deficits. PGRN overexpression also did not affect lipofuscinosis directly. Age-dependent lipofuscinosis can be measured by autofluorescence44–45. We injected Lenti-PGRN or Lenti-Ctrl into the hippocampus of 16–20-month-old non-transgenic mice and showed that PGRN overexpression had no effect on aging-dependent lipofuscinosis (Supplementary Fig. 9).
The protective effects of PGRN are not likely to be Aβ-specific; PGRN exerts neuroprotection in a number of acute and chronic neuronal injury models. In zebrafish, overexpressing PGRN rescued axonopathy induced by PGRN knockdown or mutant TDP-43 overexpression46. PGRN overexpression also promoted cell survival against apoptotic challenge in cultured motor neuron cell lines47. PGRN could activate survival pathways in neurons directly or in a non-cell-autonomous manner. Targeted expression of PGRN in microglia in mixed cortical cultures strongly protected against Aβ toxicity. Phospho-tau remained unchanged by PGRN overexpression in wildtype and 5xFAD mice, suggesting the protective effects are not tau-dependent (Supplementary Fig. 10). Extracellular PGRN can stimulate the activation of the MEK/ERK and PI3K/Akt pathways to protect against glutamate toxicity and neurotoxins in cultured neurons48. Further studies will be needed to establish whether these pathways are involved in protecting against Aβ toxicity in vivo.
PGRN deficiency in microglia reduced the phagocytosis of beads in adult brain, suggesting phagocytosis of Aβ aggregates is impaired. Indeed, microglial PGRN deficiency drastically increased plaque load in APPhigh mice without affecting soluble Aβ levels. In complementary experiments, PGRN overexpression lowered plaque load, and the amount of PGRN overexpression correlated negatively with plaque load in a model of aggressive plaque deposition (5xFAD). These findings indicate that microglia-derived PGRN exerts an anti-amyloidogenic effect in vivo, likely via enhanced phagocytosis. Our findings are consistent with the finding that recombinant PGRN increased Aβ internalization by primary microglia33. In contrast to microglia, PGRN-deficient peritoneal macrophages showed increased phagocytosis of apoptotic cells49 while PGRN-deficient bone-marrow-derived macrophages exhibited normal phagocytosis7. Given the many differences between microglia and macrophages50–52, it is not surprising that they are regulated differentially by PGRN deficiency. Moreover, since different receptors mediate phagocytosis in microglia, the uptake of beads, apoptotic cells, or Aβ might require different mechanisms. Various pathways could underlie the regulation of microglial phagocytosis by PGRN. Previous studies showed that inhibiting NF-κB restored the ability of pro-inflammatory cytokine-induced microglia to phagocytose Aβ in culture53–54. Ongoing studies will determine if inhibition of NF-κB signaling would restore normal function in PGRN-deficient microglia.
Our finding that PGRN plays a significant protective role in AD pathogenesis suggests that PGRN-enhancing therapies developed for the treatment of FTLD could also be beneficial for AD. However, the differential roles of neuronal and microglial PGRN are poorly understood, and further study is needed to determine how each exerts its neuroprotective and plaque-reducing effects.
ONLINE METHODS
Mice
Mice were housed in a pathogen-free barrier facility with a 12 h light/dark cycle and ad libitum access to food and water. All animal procedures were carried out under University of California, San Francisco, Institutional Animal Care and Use Committee-approved guidelines.
Grn was genetically ablated from APPlow mice by crossing PDAPPSw,Ind J9 mice (C57BL/6)27 with Grn−/− mice11 to generate APPlow/Grn+/− mice, which were further crossed with Grn+/− mice to generate Grn+/+, Grn+/−, Grn−/−, APPlow/Grn+/+, APPlow/Grn+/− and APPlow/Grn−/− mice. To remove Grn from myeloid cells, GrnF/F mice11 were crossed with mice expressing Cre recombinase under the control of the lysozyme M (LysM) promoter31 (Jackson Laboratory, Bar Harbor, Maine). LysM-Cre+/GrnF/F mice were crossed with APPhigh (PDAPPSw,Ind J20) mice27 to generate LysM-Cre−/GrnF/F, LysM-Cre+/GrnF/F, APPhigh/LysM-Cre−/GrnF/F, and APPhigh/LysM-Cre+/GrnF/F mice. We obtained 5xFAD mice (034840-JAX), which overexpress human APP with the Swedish (K670N, M671L), Florida (I716V), and London (V717I) mutations and human PS1 with M146L and L286V mutations28, and non-transgenic (NTG) controls from the Mutant Mouse Regional Resource Centers. Gender-balanced groups were used for all experiments except for NTG control group of 5xFADmice; only male mice were used in this group. Sample size for each experiment was determined based on previous experiences with these models.
Brain harvest and immunohistochemistry
Brains were perfused with 0.9% saline, followed by fixation with 4% PFA for 24 h and incubation in 30% sucrose for 48 h. Brains were sectioned into 30 μm slices on a Leica SM2010R freezing microtome, permeabilized in Tris-buffered saline with 0.5% Triton X-100 and blocked with 10% normal donkey serum. For 3,3′-diaminobenzidine (DAB) immunohistochemical staining, sections were quenched to suppress endogenous peroxidase activity, followed by incubation with anti-calbindin (CB38, 1:15,000, Swant, Switzerland), anti-CD68 (MCA1957GA, 1:500, Serotec, Raleigh, NC), or biotinylated 3D6 (5 μg/ml, Elan, South San Francisco, CA). Immunoreactivity was detected with 0.25 mg/ml DAB with 0.01% H2O2. Images were acquired on a brightfield Leica DM5000B microscope with a Leica DFC310 FX camera. For thioflavin S (ThioS) staining, sections were incubated in 0.25% filtered potassium permanganate, 2% K2O5S2 with 1% oxalic acid, and 0.015% filtered ThioS in 50% ethanol. For immunofluorescence staining, sections were incubated with anti-Iba1 (#019–19741, 1:500, Wako, Osaka, Japan), 3D6 (5 μg/ml, Elan), anti-NeuN (MAB377, 1:500, Millipore), or anti-MAP2 (MAB3418, 1:300, Millipore), Immunoreactive structures were detected with Alexa Fluor donkey anti-mouse or rabbit secondary antibodies in the 405-, 488-, 555-, or 647-nm range (Invitrogen, Carlsbad, CA). PGRN expression was detected after antigen retrieval with 10 mM citrate (pH 6.0) for 30 min at 90°C. Sections were blocked in TBS containing 10% calf serum (Sigma, St. Louis, MO), 3% bovine serum albumin (BSA), 1% glycine, and 0.4% Triton X-100, incubated with anti-PGRN (AF2557, 1:200, R&D Systems), and detected with Alexa Fluor donkey anti-sheep (405 or 555 nm). Immunofluorescent images were acquired on a Nikon ECLIPSE Ti 2000 spinning disk confocal microscope.
Volocity software (Perkin Elmer, Waltham, MA) was used for immunofluorescent quantification, and ImageJ software was used for DAB immunohistochemical quantification, as described with minor modifications55–56. Regions of interest were drawn around the dentate gyrus and non-dentate gyrus hippocampal regions, and 3D6- and ThioS-positive immunoreactivity was quantified in each sub-region. For plaque analysis of Lenti-PGRN-injected mice, two mice were excluded due to a lower than two-fold increase in PGRN; however, these mice were included for correlational analyses. For quantification of NeuN staining, images were acquired on a Keyence BZ-9000 fluorescence microscope (Keyence Corporation of America, Itasca, IL) and stitched images were analyzed with BZ Analyzer software for deconvoluted fluorescence quantification. Experimenters quantifying immunoreactive structures were blind to the genotype and treatment of the mice.
Quantification of lipofuscin
Hippocampi of 16–20-month-old non-transgenic 5xFAD mice were unilaterally injected with Lenti-Ctrl or Lenti-PGRN, and mice were sacrificed seven weeks later. Brains were fixed and sectioned as described above, followed by PGRN staining with sheep anti-PGRN/anti-sheep Alexa-488 secondary. Autofluorescence imaging of lipofuscin was identified as overlapping signal in both Cy3 and Cy5 filter sets on a Keyence BZ-9000 digital fluorescence microscope. Percent area of PGRN immunoreactivity and autofluorescent material was quantified using Volocity software.
Western blot and ELISA
For western blot analysis of cortical full-length APP/APP fragments and tau, cortices were lysed with a Dounce homogenizer in a 10x volume of radio-immunoprecipitation assay (RIPA) buffer containing 50 mM Tris (pH 7.4), 150mM NaCl, 0.25% sodium deoxycholate, 1% Nonidet P-40, 1mM EDTA, and protease and phosphatase inhibitors. Lysates were sonicated briefly and centrifuged at 18,000 x g at 4°C for 15 min, and the supernatant was collected. Protein concentrations from brain lysates were determined by the Bradford method, and 60 μg of proteins were resolved on a 4–12% SDS-PAGE gel, transferred to nitrocellulose membrane, blocked with 5% milk, and probed for full-length APP and APP fragments with 8E5 (Elan) or anti-CT15 (a kind gift from E.H. Koo, University of California at San Diego, CA), total tau with Tau 5 (Life Technologies), or phospho-tau with PHF1 (a kind gift from Peter Davies, Albert Einstein Medical College, NY). Anti-GAPDH (MAB374, 1:5000, Millipore, Billerica, MA) was used as a loading control. For western blot analysis of hippocampal APP and APP fragments, guanidine lysates were precipitated with 90% ethanol at −20° C for 24 h and centrifuged at 18,000 x g for 15 min. The pellets were subsequently washed with 90% ethanol and air-dried. After addition of sample buffer and boiling at 95°C for 10 min, samples were run on 4–20% Tris-Glycine gels (EC6025BOX, Life Technologies), transferred to PVDF membrane (#162–0177, Bio-Rad), and blotted with CT15 or 8E5.
For mouse PGRN ELISA, high-binding 96-well EIA/RIA plates (Corning, Corning, NY) were coated overnight with a polyclonal antibody raised against mouse PGRN (2 μg/ml) and blocked with 1% BSA for 1 h at 37°C. Plates were incubated with brain lysates (1:50) or mouse recombinant PGRN (0–16 ng/ml, AF2557, R&D Systems) for 1 h at 37°C, followed by sequential incubation with an anti-PGRN antibody (0.2 μg/ml, R&D Systems), anti-sheep biotinylated IgG (1:5000, R&D Systems) and streptavidin-horseradish peroxidase (HRP) conjugate (1:10,000, Thermo Fisher Scientific). Similarly, for full-length human PGRN ELISA, 96-well plates were coated overnight with a monoclonal antibody raised against C-terminal PGRN (0.77 μg/ml) and blocked with 1% BSA for 1 h at 37°C. Plates were incubated with RIPA-homogenized brain lysates (1:50) or human recombinant PGRN (0–32 ng/ml, 2420, R&D Systems) for 1 h at 37°C, followed by sequential incubation with an N-terminal monoclonal PGRN antibody (1.5 μg/ml), anti-mouse biotinylated IgG (1:5000, Vector Laboratories, Burlingame, CA) and streptavidin-HRP conjugate (1:10,000, Thermo Fisher Scientific). For validation of the human PGRN ELISA, human lymphoblast cell lines from de-identified human subjects were obtained from the UCSF Memory and Aging Center. Lines from five separate heterozygous GRN-mutation carrying subjects and five non-mutation carrying siblings were grown in DMEM with 10% FCS at equal cell densities for 72 h. Conditioned medium was collected and cells were lysed in RIPA buffer. For both mouse and human PGRN ELISA, the reactions were developed at room temperature with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific), quenched with 1 N HCl, and read at 450 nm on a SpectraMax M5 spectrophotometer (Molecular Devices, Sunnyvale, CA). For Aβ ELISA, hippocampal and cortical tissues were lysed with guanidine buffer and measured by ELISA as described57. Antibody 266 (for Aβ1-x) and 21F12 (for Aβ1–42) were used as capture antibodies, and biotin-conjugated 3D6 was used as detection antibody (Elan).
Adult microglia isolation
Adult microglia were isolated as described with minor modifications32. Briefly, brains were perfused with ice-cold PBS, chopped and minced with a razor blade in 5 ml of RPMI with L-glu medium containing 6% collagenase and 3 U/ml dipase. After 45 min incubation with DNase I (0.2 U) at 37°C, enzymes were deactivated with an equal volume of PBS containing 2.5 mM EDTA and 1% fetal bovine serum (FBS). Cells were triturated thoroughly and passed through a 70 μm filter, centrifuged at 300 x g for 5 min at 18°C, and resuspended in fluorescence-activated cell sorting (FACS) buffer (500 mM EDTA, 0.5% BSA, 0.09% sodium azide in PBS). Myelin-removal beads (Miltenyi Biotec, Auburn, CA) were added for 15 min at 4°C. After centrifugation, cells were resuspended in FACS buffer and magnetically separated with the autoMACS pro-separator per the manufacturer’s protocol (Miltenyi Biotec). After myelin removal, cells from two to three brains per genotype were combined, selected for CD11b+ with CD11b microbeads (Miltenyi Biotec), and separated with autoMACS. Microglial purity was determined by incubating CD11b+ cells with an anti-CD11b-FITC antibody (1:200, Miltenyi Biotec) for 30 min on ice, followed by flow cytometry on FACS AriaII (BD Biosciences).
Quantitative Real-Time PCR
To measure PGRN mRNA from adult microglia, CD11b+ cells were purified as described and resuspended in RLT buffer with 1% β-mercaptoethanol (β-ME). To measure mRNA of cytokines, cortical brain tissue was homogenized with a 21G needle in RLT buffer with 1% β-ME. RNA was isolated per the RNeasy isolation kit instructions (Qiagen, Valencia, CA), and the remaining DNA in the sample was removed by incubation with RNase-free DNase (Ambion, Austin, TX). RNA was converted to cDNA by the TaqMan reverse transcription (RT) kit (Applied Biosystems, Foster City, CA). Real-time RT-PCR was performed on the ABI7900HT sequence detector (Applied Biosystems) with SYBR Green (Applied Biosystems). The relative gene expression was quantified as 2−ΔCt. The mean value of replicates of each sample was expressed as the threshold cycle (Ct) at which the level of fluorescence begins to increase rapidly. The amount of gene expression was calculated as the difference (ΔCT) between the CT value of the sample for target gene and the CT value of that sample for the endogenous control GAPDH (ΔCt = Ct(target gene) – Ct(GAPDH)). The relative amount of gene expression for each target gene was determined by 2−ΔCt and expressed as the fold-change compared with control. The primers used for real-time RT-PCR are listed in Supplementary Table 2.
Primary culture
Preparation of primary cultures was performed as described58. Briefly, cortices from P0 or P1 rat pups were isolated and plated at 160,000 cells ml−1 on glass coverslips in Dulbecco’s modified Eagle’s medium, 10% FBS, GlutaMAX, 100 units/ml penicillin, and 100 μg ml−1 streptomycin. Cultures were infected with equal amount (~1 x 107 PFU) of Lenti-MCSF-Ctrl or Lenti-MCSF-PGRN at DIV 5, followed by treatment with 7PA2 conditioned medium59–60 at DIV 7–9 for 48–72 h. Cells were fixed with 4% PFA and immunostained with anti-MAP2 antibody (1:300; Millipore). Surviving MAP2+ neurons were counted on a Leica DM5000B microscope. The experimenter who scored MAP2+ neurons was blinded to treatment of the cultures. For western blot analysis of cell-type specific markers, cultures were infected with equal amount of lentivirus at DIV5 and harvested at DIV7. Cells were immunoblotted with anti-GFAP (MAB3402, 1:5000, Millipore), TUJ1 (MRB-435P, 1:1000, Covance, Princeton, NJ), Iba1 (1:1000, Wako), or GAPDH (MAB374, 1:5000, Millipore).
Phagocytosis assay
The phagocytic activity of adult microglia in the hippocampus of acute brain slices was analyzed as described61. Brains from LysM-Cre+/GRNF/F or LysM-Cre−/GRNF/F mice were washed in carbogen-saturated (95% O2, 5% CO2) artificial cerebrospinal fluid (ACSF) containing (in mM): NaCl 126; KCl 2.5; MgSO4 1.3; CaCl2 2.5; NaH2PO4 1.25; NaHCO3 26; D-glucose 10; pH 7.4 (all from Sigma). Coronal slices (130 μm) were prepared using a vibratome (Microm, Walldorf, Germany) at 4°C, and allowed to rest in ACSF buffer at room temperature for 1 h before incubation with red fluorescent carboxylated microspheres (1 μm diameter, FluoSpheres®, Invitrogen, 1:100) in PBS (Cellgro) containing 4.5 g l−1 D-glucose (Sigma) for 60 min at 37°C. The slices were washed and fixed with 4% PFA. To visualize microglia, slices were permeabilized (2% Triton-X, 2% BSA, 10% donkey serum in PBS) and incubated with anti-Iba-1 (1:750, Wako), followed by donkey anti-rabbit Alexa 488 (1:250, Invitrogen) and Hoechst 33258 (Sigma-Aldrich; 1:10000). Imaging was performed with a Nikon ECLIPSE Ti 2000 spinning disk confocal microscope with a 20x objective. Slice-spanning Z-stack images (1 μm step-size) were acquired and analyzed using ImageJ MacBiophotonics cell counter plugin.
Stereotaxic injections
Lentiviruses were prepared by cotransfection of lentiviral shuttle vector encoding mouse PGRN and packaging vectors into HEK293T cells, followed by ultracentrifugation of viral supernatant as described62. Mice were anesthetized with Avertin (250 mg/kg) by intraperitoneal injection, and secured on a stereotaxic frame (Kopf Instruments, Tujunga, CA). 3 μl of Lenti-Ctrl or Lenti-PGRN lentivirus was delivered to the DG at a rate of 0.5 μl min−1 (AP -2.1, ML ±1.7, DV -2.1). Mice were assigned into gender- and age-matched treatment groups using a randomized block design. For mice subject to plaque analyses, mice were unilaterally injected with Lenti-Ctrl or Lenti-PGRN at 2 months of age and harvested 3 months later. The pre-set criterion for exclusion from analysis was a less than twofold increase in ipsilateral PGRN immunoreactivity compared to the contralateral side. For mice subject to behavioral tests, Lenti-Ctrl or Lenti-PGRN was bilaterally injected at 2 months of age and tested behaviorally at 4 months of age, before harvest at 5 months of age.
Morris water maze
Behavior experiments were conducted during daylight hours. The water maze consisted of a pool (122 cm in diameter) containing opaque water (20 ± 1°C) and a platform (10 cm in diameter) submerged 1.5 cm. Hidden platform training (days 1–5) consisted of 10 sessions (two per day, 2 h apart), each with two trials. The mouse was placed into the pool at alternating drop locations for each trial. A trial ended when the mouse located the platform and remained motionless on the platform for 5 s, for a maximum of 60 s per trial. Performance was measured with EthoVision video-tracking (Noldus Information Technology). Probe trials were conducted 1 and 5 days after training. Visible platform training, where the platform was cued with a mounted black-and-white striped mast, was conducted for four sessions after the conclusion of probe trials. Pre-set criteria for exclusion from analysis included floating and thigmotaxic behaviors, neither of which was observed in current studies.
Elevated plus maze
The elevated plus maze consisted of two open and two enclosed arms elevated 63 cm above the ground (Hamilton-Kinder, Poway, CA). Mice were first allowed to habituate in the testing room under dim light for 30 min. During testing, mice were placed at the junction between the open and closed arms of the maze and allowed to explore for 10 min. The distance traveled and time spent in the open and closed arms were determined. The maze was thoroughly cleaned with 70% ethanol between testing sessions.
Cued Y-maze
The cued version of the Y-maze assesses short-term memory of the familiarity of a specific context63. For the initial training trial, the mouse was placed at the end of one arm of a large Y-maze designated “start arm” (arm length: 50 cm). The third arm of the maze was blocked off, allowing the mouse to explore two of the three arms freely (“start arm” and “familiar arm”) for 5 min. Each arm contained spatial cues. Two h later, the mouse was placed back into the maze in the “start arm,” and allowed to explore all three arms with the third arm unblocked (“novel arm”). Movements in and out of each arm were tracked using automated tracking software (Noldus). Testing was performed under dim lighting, and the apparatus was cleaned with 70% ethanol between trials.
Small Y-maze
Mice were placed in the center of a small Y-maze (arm length: 15 cm) and spontaneous alternation was recorded in a single continuous 6-min trial by a live observer. Each of the three arms was designated a letter A–C, and entries into the arms were recorded (i.e., A, B, C, B, A). The percent of spontaneous alternation was calculated over the total number of entries possible. Testing was performed under dim lighting, and the apparatus was cleaned with 70% ethanol between trials.
Contextual fear conditioning
Experiments were conducted in Quick Change Test chambers (Med Associates), which were cleaned with 1% acetic acid to provide a strong contextual odor. Mice underwent two days of training where the mouse was placed in the chamber and allowed to acclimate for 3 min. After 3 min, a 0.4-mA foot shock was administered for 2 s. The mouse remained in the chamber for an additional 1 min. Baseline freezing was recorded for the entire 3-min pre-shock interval on Day 1. The percent time freezing was also recorded for the 1 min after the shock on Day 2. For testing on Day 5, mice were placed in the chamber for 12 min, and percent time freezing was measured in 3-min intervals. Mice that did not show freezing behavior after shock on Day 1 were excluded from analysis.
Statistical analysis
Data were analyzed with GraphPad Prism v.5 (GraphPad, San Diego, CA) or R (F Foundation for Statistical Computing, Vienna, Austria). Differences between means were assessed with paired or unpaired Student’s t test, one-way or two-way analysis of variance, followed by post hoc testing of all pairwise comparisons among genotypes (with Tukey-Kramer correction for multiple comparisons), or by mixed effects model, as indicated. Pearson’s correlation coefficients were used to quantify the linear relationship between two variables. The Shapiro-Wilk test of normality was applied to all data sets, and in cases where the data did not demonstrate a normal distribution, non-parametric tests were used to analyze statistical differences. The Mann-Whitney test was used for unpaired t-tests, the Wilcoxon matched pairs test was used for paired comparisons, and the Kruskal-Wallis test was used for ANOVAs. An F test or Bartlett’s test was performed to determine significant differences in variances between groups for t-tests and ANOVAs, respectively. Multivariate log-linear regression analyses performed in R were used to assess age-adjusted Aβ levels. Linear mixed effects models fit using the R package lme464 were used to compare learning curves in MWM, random intercepts and linear time slopes for each mouse accounted for the correlation among repeated observations; P-values for differences between groups were calculated by inverting the simulated confidence intervals around the differences65. For cluster analyses of multiple measurements from individual mice, we fit linear mixed effects models, and random intercepts for each mouse accounted for the correlation among repeated observations; P-values were corrected for multiple comparisons using the method of Holm. For all experiments, values that were not within the pre-determined criterion of two standard deviations from the mean were considered statistical outliers and excluded from analyses. All available samples or animals were included for statistical analysis unless otherwise noted.
Supplementary Material
Acknowledgments
We thank M. Finucane for statistical analyses, J. Lau and W. Weiss for technical advice, B. Miller and L. Mitic for discussions, G. Howard and A. Lucido for editorial review, J. Carroll and T. Roberts for graphics assistance, V. Shen and R. Chen for technical assistance, and L. Goss for administrative assistance. This work was supported in part by the Consortium for Frontotemporal Dementia (L.G and R.V.F), US National Institutes of Health (1R01AG036884 and R01AG030207 to L.G.), and the Stephen D. Bechtel Jr. Foundation. S.S.M. is supported by US National Institutes of Health fellowship F32NS076239, and L.H.M. is supported by US National Institutes of Health fellowship F31AG034793.
Footnotes
Author Contributions
L.G, S.S.M., and S.-W.M. conceived the project and designed the experiments. S.S.M, S.-W.M., G.K., Y.Z., C.W., Y.L., and R.A. conducted experiments. L.H.M., L.P.E., M.E.W., L.M. and R.V.F. developed experimental tools or mouse models. S.S.M. and L.G. wrote the manuscript.
References
- 1.Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48:999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- 2.Daniel R, Daniels E, He Z, Bateman A. Progranulin (acrogranin/PC cell-derived growth factor/granulin-epithelin precursor) is expressed in the placenta, epidermis, microvasculature, and brain during murine development. Dev Dyn. 2003;227:593–599. doi: 10.1002/dvdy.10341. [DOI] [PubMed] [Google Scholar]
- 3.Petkau TL, et al. Progranulin expression in the developing and adult murine brain. J Comp Neurol. 2010;518:3931–3947. doi: 10.1002/cne.22430. [DOI] [PubMed] [Google Scholar]
- 4.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 5.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 6.Gass J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 7.Yin F, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2010;207:117–128. doi: 10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petkau TL, et al. Synaptic dysfunction in progranulin-deficient mice. Neurobiol Dis. 2012;45:711–722. doi: 10.1016/j.nbd.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Yin F, et al. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: a mouse model of frontotemporal dementia. FASEB J. 2010 doi: 10.1096/fj.10-161471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filiano AJ, et al. Dissociation of Frontotemporal Dementia-Related Deficits and Neuroinflammation in Progranulin Haploinsufficient Mice. J Neurosci. 2013;33:5352–5361. doi: 10.1523/JNEUROSCI.6103-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martens LH, et al. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest. 2012;122:3955–3959. doi: 10.1172/JCI63113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyama H, et al. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeer PL, McGeer EG. The inflammatory response system of brain: implications for therapy of Alzheimer and other neurodegenerative diseases. Brain Res Brain Res Rev. 1995;21:195–218. doi: 10.1016/0165-0173(95)00011-9. [DOI] [PubMed] [Google Scholar]
- 14.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 16.McGeer PL, Rogers J, McGeer EG. Inflammation, anti-inflammatory agents and Alzheimer disease: the last 12 years. J Alzheimers Dis. 2006;9:271–276. doi: 10.3233/jad-2006-9s330. [DOI] [PubMed] [Google Scholar]
- 17.Frank-Cannon TC, Alto LT, McAlpine FE, Tansey MG. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seshadri S, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouwers N, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology. 2008;71:656–664. doi: 10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- 21.Cortini F, et al. Novel exon 1 progranulin gene variant in Alzheimer’s disease. Eur J Neurol. 2008;15:1111–1117. doi: 10.1111/j.1468-1331.2008.02266.x. [DOI] [PubMed] [Google Scholar]
- 22.Viswanathan J, et al. An association study between granulin gene polymorphisms and Alzheimer’s disease in Finnish population. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:747–750. doi: 10.1002/ajmg.b.30889. [DOI] [PubMed] [Google Scholar]
- 23.Kelley BJ, et al. Alzheimer disease-like phenotype associated with the c.154delA mutation in progranulin. Arch Neurol. 2010;67:171–177. doi: 10.1001/archneurol.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry DC, et al. Progranulin Mutations as Risk Factors for Alzheimer Disease. JAMA Neurol. 2013:1–5. doi: 10.1001/2013.jamaneurol.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereson S, et al. Progranulin expression correlates with dense-core amyloid plaque burden in Alzheimer disease mouse models. J Pathol. 2009;219:173–181. doi: 10.1002/path.2580. [DOI] [PubMed] [Google Scholar]
- 26.Gliebus G, Rosso A, Lippa CF. Progranulin and beta-amyloid distribution: a case report of the brain from preclinical PS-1 mutation carrier. Am J Alzheimers Dis Other Demen. 2009;24:456–460. doi: 10.1177/1533317509346209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mucke L, et al. High-level neuronal expression of abeta 1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oakley H, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin J, et al. Fyn kinase induces synaptic and cognitive impairments in a transgenic mouse model of Alzheimer’s disease. J Neurosci. 2005;25:9694–9703. doi: 10.1523/JNEUROSCI.2980-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palop JJ, et al. Neuronal depletion of calcium-dependent proteins in the dentate gyrus is tightly linked to Alzheimer’s disease-related cognitive deficits. Proc Natl Acad Sci USA. 2003;100:9572–9577. doi: 10.1073/pnas.1133381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 32.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickford F, et al. Progranulin is a chemoattractant for microglia and stimulates their endocytic activity. Am J Pathol. 2011;178:284–295. doi: 10.1016/j.ajpath.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, et al. SIRT1 Protects against Microglia-dependent Amyloid-{beta} Toxicity through Inhibiting NF-{kappa}B Signaling. J Biol Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 35.Yin F, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2009 doi: 10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh DM, et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 37.Eimer WA, Vassar R. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Abeta42 accumulation and Caspase-3 activation. Mol Neurodegener. 2013;8:2. doi: 10.1186/1750-1326-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jawhar S, Trawicka A, Jenneckens C, Bayer TA, Wirths O. Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Abeta aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol Aging. 2012;33:196 e129–140. doi: 10.1016/j.neurobiolaging.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 39.Suh HS, Choi N, Tarassishin L, Lee SC. Regulation of progranulin expression in human microglia and proteolysis of progranulin by matrix metalloproteinase-12 (MMP-12) PLoS One. 2012;7:e35115. doi: 10.1371/journal.pone.0035115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cenik B, Sephton CF, Kutluk Cenik B, Herz J, Yu G. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem. 2012;287:32298–32306. doi: 10.1074/jbc.R112.399170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith KR, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed Z, et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177:311–324. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filiano AJ, et al. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J Neurosci. 2013;33:5352–5361. doi: 10.1523/JNEUROSCI.6103-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottis P, et al. Human and rat brain lipofuscin proteome. Proteomics. 2012;12:2445–2454. doi: 10.1002/pmic.201100668. [DOI] [PubMed] [Google Scholar]
- 45.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 46.Laird AS, et al. Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS One. 2010;5:e13368. doi: 10.1371/journal.pone.0013368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan CL, et al. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci. 2009;10:130. doi: 10.1186/1471-2202-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, et al. Extracellular progranulin protects cortical neurons from toxic insults by activating survival signaling. Neurobiol Aging. 2011;32:2326 e2325–2316. doi: 10.1016/j.neurobiolaging.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kao AW, et al. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc Natl Acad Sci U S A. 2011;108:4441–4446. doi: 10.1073/pnas.1100650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan XD, et al. Microglial phagocytosis induced by fibrillar beta-amyloid is attenuated by oligomeric beta-amyloid: implications for Alzheimer’s disease. Mol Neurodegener. 2011;6:45. doi: 10.1186/1750-1326-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koenigsknecht J, Landreth G. Microglial phagocytosis of fibrillar beta-amyloid through a beta1 integrin-dependent mechanism. J Neurosci. 2004;24:9838–9846. doi: 10.1523/JNEUROSCI.2557-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller-Steiner S, et al. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 56.Sun B, et al. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron. 2008;60:247–257. doi: 10.1016/j.neuron.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson-Wood K, et al. Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc Natl Acad Sci U S A. 1997;94:1550–1555. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, et al. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J Biol Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 59.Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 60.Walsh DM, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 61.Krabbe G, et al. Functional impairment of microglia coincides with Beta-amyloid deposition in mice with Alzheimer-like pathology. PLoS One. 2013;8:e60921. doi: 10.1371/journal.pone.0060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun B, Gan L. Manipulation of gene expression in the central nervous system with lentiviral vectors. Methods Mol Biol. 2011;670:155–168. doi: 10.1007/978-1-60761-744-0_12. [DOI] [PubMed] [Google Scholar]
- 63.Hughes RN. The value of spontaneous alternation behavior (SAB) as a test of retention in pharmacological investigations of memory. Neurosci Biobehav Rev. 2004;28:497–505. doi: 10.1016/j.neubiorev.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 65.Altman DG, Bland JM. How to obtain the P value from a confidence interval. BMJ. 2011;343:d2304. doi: 10.1136/bmj.d2304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.