Abstract
Rett syndrome (RTT) is a neurodevelopmental disorder caused by mutations in the gene encoding MeCP2. By binding to methylated CpG dinucleotide promoter regions, MeCP2 acts as a transcriptional repressor, predicting that its absence might result in widespread aberrant gene transcription, leading to the RTT phenotype. Considering this potentially broad action of MeCP2 on expression and the complexity of the brain, especially during development, we approached the consequences of MeCP2 deficiency in a mouse model by using a temporal and regional proteomic strategy. We used the olfactory system (olfactory epithelium and bulb) because its attributes make it an excellent developmental model system. We find evidence of temporal and regional proteomic pattern differences between WT and MeCP2-deficient mice. It was possible to segregate these changes in protein expression into five biological function groups: cytoskeleton arrangement, chromatin modeling, energy metabolism, cell signaling, and neuroprotection. By combining the proteomic results with the RNA levels of the identified proteins, we show that protein expression changes are the consequence of differences in mRNA level or posttranslational modifications. We conclude that brain regions and ages must be carefully considered when investigating MeCP2 deficiency, and that not only transcription should be taken into account as a source for these changes, but posttranslational protein modifications as well.
Rett syndrome (RTT) is a neurodevelopment disorder characterized by seizures, motor dysfunction, and features of autism (1–3). Prominent neuropathological features include reductions in cortical thickness in multiple cerebral cortical regions, decreased neuronal soma size, a reduced number of identifiable synapses, dramatically decreased dendritic arborization with increased neuronal cell packing density, and predominance of immature neurons in the olfactory epithelium (4–9). The clinical course of RTT is dynamic, with onset of symptoms beginning at 6–18 months, followed by stabilization; these features suggest a disease process active during a specific developmental period (3).
RTT is an X chromosome-linked genetic disease caused by mutations in the MECP2 gene (10–12). Generation of mice deficient or mutated for the ortholog Mecp2 recapitulate many features of RTT (13–15). The gene product of MECP2, previously known as methyl-CpG-binding protein 2 (16) belongs to the methyl-CpG-binding protein group and acts, in complex with histone deacetylase and Sin 3, as a transcriptional repressor to silence gene expression through chromatin remodeling through the deacetylation of histones. Therefore, loss of MeCP2 function might be predicted to result in a widespread aberrant gene transcription state, leading to neuronal dysfunction and creating the phenotypic features of RTT. This hypothesis regarding the consequences of MeCP2 dysfunction engaged several groups to perform global transcriptional profiling (17–19). Although divergent results were obtained, the source of tissue and age were different between studies, making comparisons difficult. The consequences of loss of a transcription factor like MeCP2 may be complex, especially in an organ such as the brain. This pattern becomes more daunting in RTT, because it is a neurodevelopment disorder manifesting age-dependent clinical features.
We recently used the olfactory system as a developmental model (20) in which to analyze MeCP2 expression and the consequences of its deficiency (9, 21). We narrowed the onset of neuronal expression of Mecp2 to a time window preceding terminal maturation (21), and we found in olfactory biopsies from RTT patients a defect in terminal differentiation of olfactory receptor neurons (9). We hypothesized that the biochemical consequences of MeCP2 loss could be age- and region-related, and moreover transient in nature, given the dynamic clinical course of RTT (3). We considered that a proteomic differential display strategy would be more appropriate to approach the consequences of MeCP2 loss because it focuses on proteins, the active gene products.
By analyzing the proteome profile in the olfactory epithelium (OE) and olfactory bulb (OB), we found temporally and regionally dependent differences in protein expression profiles between Mecp2-null and WT mice. By using real-time RT-PCR, we confirmed differences in RNA levels for some of the differentially expressed proteins, whereas others changes were likely to be the result of posttranscriptional modifications or alterations in protein stability. Our results suggest that Mecp2 functions are spatiotemporally dynamic and transient, and that some consequences of MeCP2 loss occur independent of changes in transcription level.
Materials and Methods
The Mecp2-null mouse model used in these experiments was generated by the Cre LoxP recombination system to delete exon 3 of Mecp2 (13). WT mice were obtained from the same litters. Mice were killed by cervical dislocation and directly dissected. All experimental protocols were approved by The Johns Hopkins University Institutional Animal Care and Use Committee.
2D Electrophoresis. Tissues were immediately frozen in liquid nitrogen after dissection. Total proteins were extracted from OE and OB tissues of male mice of each genotype and age. Total protein samples were prepared in a urea buffer: 9 M urea/4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/40 mM Tris base/2% DTT. Lysates were centrifuged at 10,000 × g for 5 min and protein content was measured by Bradford assay; samples were prepared for electrophoresis, loading 50 μg per gel. Samples from individual mice were run in duplicate to assess reproducibility. Protein samples from three other mice were pooled by condition (age, region, and genotype) and run in duplicate. 2D electrophoresis was performed according to the method of O'Farrell (22) by Kendricks Labs (Madison, WI). Isoelectric focusing (first dimension) was carried out in glass tubes by using 2% pH 3.5–10 ampholines (Amersham Pharmacia) for 9,600 V·h. Fifty nanograms of an isoelectric focusing (IEF) internal standard, tropomyosin, was added to each sample. Tropomyosin was visualized as two polypeptide spots of similar pI; the lower spot of molecular weight 33,000 and pI 5.2 is marked with an arrow on the 2D gels. The pH gradient was determined by surface pH electrode. After equilibrium in SDS sample buffer (10% glycerol/50 nM DTT/2.3% SDS/0.0625 M Tris at pH 6.8), each tube gel was sealed to a stacking gel that overlayed a 10% acrylamide slab gel for electrophoretic mass separation (the second dimension). Proteins (Sigma) were added as molecular weight standards to the agarose sealing the tube gel to the slab gel. Protein spots were visualized by silver staining.
Limited Computerized Comparisons. Duplicate gels of those used to visualize proteins by silver staining were scanned with a laser densitometer (model PDSI, Molecular Dynamics). The scanner was checked for linearity with a calibrated neutral density filter set (Melles Griot, Irvine, CA). The images were analyzed by using progenesis software (Version 2002.01, Nonlinear Technology, Newcastle upon Tyne, U.K.) such that all major spots and all changing spots were outlined, quantified, and matched on all gels. The general method of computerized analysis for these pairs included automatic spot finding and quantification, automatic background subtraction (mode of nonspot), and automatic spot matching in conjunction with detailed manual checking of the spot finding and matching functions. Averaged gels were generated for each sample by progenesis software. The measurement of the fold change between conditions was calculated from the spot percentage, which is equal to spot integrated density (volume) expressed as a percentage of total density of all spots measured. Difference was defined by what fold change the percentage that a spot from the null mice differed from the spot's counterpart in the gel generated from WT mice.
Mass Spectrometry and Protein Identification. Proteins spots were cut with a spot-picker from Coomassie blue-stained gels by gel matching with the silver-stained gels. Mass spectrometry, spectral data acquisition, and database queries were performed by the protein chemistry core facility at Columbia University (New York). Protein bands were subjected to in-gel digestion with trypsin. One microliter of the digestion mixture was separated on a LC Packings nano-Lc on which the detector outlet was connected directly to the nanospray source of a Q-Tof mass spectrometer (Micromass, Manchester, U.K.). Peptides were eluted at a flow rate of 150 nl/min by a 2–80% acetonitrile gradient and 0.1% formic acid. Detected peptides were subjected to a MS/MS fragmentation, and resulting ions along with peptide masses were submitted to a mascot search for protein identification via mascot query at www.matrixscience.com.
RT-PCR. Total RNA samples were prepared from three additional male mice per genotype. Tissues were immediately frozen in liquid nitrogen and homogenized on dry ice. Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's protocols. Genomic DNA was digested with 1 unit of DNase I (Invitrogen). cDNA was produced by using the ThermoScript RT-PCR system kit (Invitrogen). Briefly, 1 μg of RNA was used in reaction with 50 ng of random hexamer primers, 1 mM dNTP mix, 40 units of RNaseOUT, and 1 unit of ThermoScript retrotranscriptase. Two units of RNase H was added at the end of the reactions.
Real-time PCR was carried out on an iCycler (Bio-Rad) by using a reaction mixture with SYBR Green as the fluorescent dye (Bio-Rad), a 1/10 vol of the cDNA preparation as template, and 500 nM each primer (sequences available upon request). The cycle used for PCR was as follows: 95°C for 180 sec 1 time; 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec 50 times; and 95°C for 60 sec 1 time. After completion, samples were subjected to a melting-curve analysis to confirm the amplification specificity. The change in fluorescence of SYBR Green dye was monitored in every cycle and the threshold cycle (CT) was calculated above the background for each reaction. For each cDNA sample, a ratio between the relative amounts of target gene and GAPDH was calculated to compensate for variations in quantity or quality of starting mRNA, as well as for differences in reverse transcriptase efficiency. The fold change in the target gene relative to the GAPDH endogenous control gene was determined by: fold change = 2Δ(ΔCT) where ΔCT = CT,target - CT,GAPDH and Δ(ΔCT) = ΔCT,Mecp2 KO - ΔCT,WT. RT-PCRs were run separately for each mouse in triplicate, and data were analyzed for statistical differences by the Mann–Whitney U test by using prism 4.0 software (GraphPad, San Diego).
Results
The protein profiles of OE and OB from either WT or Mecp2-null mice were compared at two postnatal ages. Our proteomic strategy was based on two hypotheses. First, given the heterogeneity of brain, its different regional times of maturation, and the function of MeCP2 as a potentially broad repressor of transcription, we reasoned that differences would be difficult to detect in whole brain without considering the parameters of time and region. Second, we hypothesized the largest differences would be observed when Mecp2 begins to be highly expressed. This time starts around 2 weeks after birth, corresponding to the maturation of the olfactory system (21, 23). Thus, we postulated that proteomic differences in Mecp2-null mice would be observed between the two regions of the olfactory system (OE and OB) as well as between 2 weeks (maturation) and 4 weeks (fully mature) after birth.
Protein Expression Profile Differences. Silver-stained 2D maps of OE and OB proteins at different times during development were obtained in a reproducible manner and detected ≈500 spots per sample (Fig. 1). Normalization of gels (loaded with the same amount of total protein), done according to the total quantity in valid spots, the total density in the gel, or the addition of an internal control, tropomyosin, allowed differences due to experimental procedures (protein loading and staining) to be eliminated, so that only significant variations related to the type of sample (WT or KO) remained. Comparisons of spot intensities, measured as spot volumes, were performed by matching averaged gels from WT and KO.
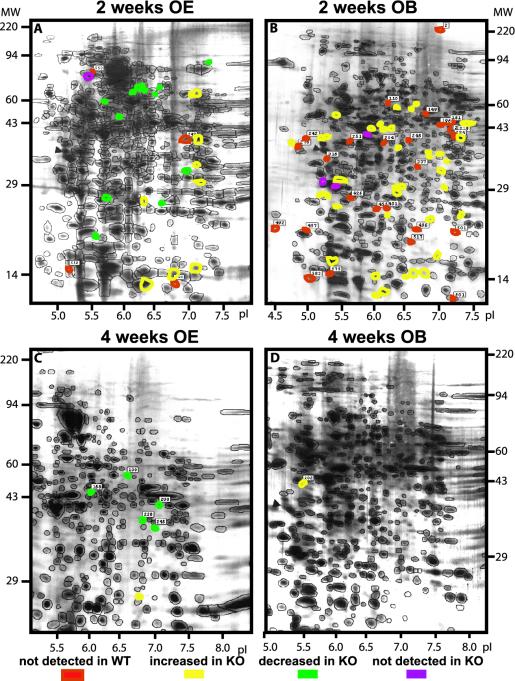
Fig. 1.
Difference images of averaged Mecp2-knockout (KO) vs. averaged WT 2D gels showing protein spot changes. The 2D electrophoresis and computerized comparisons were performed in OE (A and C) and OB (B and D) at the age of 2 (A and B)and4(C and D) weeks after birth. Most of the changes were observed at 2 weeks, in both OE and OB, whereas at 4 weeks after birth there were almost no differences between WT and KO tissues.
The majority of the spots (470–600 spots, depending on tissue and age) did not show significant intensity variations between WT and KO at any age or in any tissue. In the OE at the age of 2 weeks after birth, 27 spots (5% of detectable spots) were altered in intensity, 14 of which were decreased and 13 of which were increased. At the same age in the OB, 67 spots (12% of detectable spots) were altered in intensity, 3 of which were decreased and 64 of which were increased. Interestingly, at 4 weeks after birth, both the OE and OB displayed few changes. Five spots were decreased, whereas one spot was increased in intensity in the OE (1.2% of detectable spots), whereas one spot was decreased in the OB (0.2% of detectable spots). In all cases, differences were located throughout the 2D gels, covering small as well as large molecular weight domains and all of the pI ranges considered (Fig. 1).
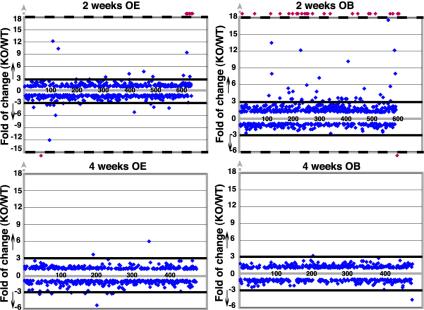
Fig. 2 shows the quantitative variations. Changes of a factor of 3 or greater were chosen as the basis for systematic comparisons. In the OE 2 weeks after birth, one spot was not detected whereas four were detected only in null mice compared with WT (Fig. 2, represented by red marks above or below the dashed lines). The range of differential spot intensities between WT and Mecp2-null varied from -12.1- to +12.2-fold. At 2 weeks in the OB, three spots were not detected whereas 25 were detected only in the null mice compared with WT mice. The range of differential spot intensities between WT and Mecp2 KO varied from -3.4- to +18.7-fold. At 4 weeks, in both OE and OB, no uniquely expressed proteins were detected in the null mice. Furthermore, the range of differential spot intensities between WT and Mecp2 KO was reduced compared with 2 weeks after birth, varying from -5.0- to +6.0-fold and -4.6-fold for OE and OB, respectively.
Fig. 2.
Scatter graph showing the fold difference (increase or decrease) of more than 500 polypeptides for Mecp2 KO and WT. Analysis was performed by using OE or OB tissue extracts at the ages of 2 and 4 weeks after birth. The differences were calculated from spot percentages (see Materials and Methods). Polypeptide spots detected only in the KO (above the top dashed line) or WT (below of the bottom dashed line) are indicated by red dots. Only polypeptide spots with a fold difference of >3 (increase) or <3 (decrease) were considered significant (above and below the solid lines).
Thus, when Mecp2 normally starts to be highly expressed in specific tissues, the absence of MeCP2 is reflected by proteomics changes in both tissues. These results also support our hypothesis that the variations in differentially expressed proteins are time-related.
Identification of Differentially Expressed Proteins. Identification was accomplished by Nano LC/MS/MS (Columbia University, New York). For some differential spots, identification on the Coomassie blue-stained preparative 2D gels was not successful because of the lack of signal for spots of low density.
Table 1 lists identified spots. The spot detected only in the WT was identified as GRP78 (78-kDa glucose-regulated protein). Although the same protein was also identified in the Mecp2-null mice, its molecular mass was increased to 85 kDa, suggesting that, in the null mice, GRP78 had a posttranslational modification. Other proteins expressed only in the null mouse include macrophage capping protein G (CAPG), histone 1H2aK, isovaleryl-CoA dehydrogenase, cytochrome b5, and M-chain creatine kinase. Spots with increased expression in the null mouse were identified as lamin C, histone H3, H2B, and GSTO1 (glutathione S-transferase omega 1). Surprisingly, the pI values of the histones, which are theoretically close to 11.0, were decreased to 7 in Mecp2-null mice. This discrepancy between theoretical and experimental pI values may be related to posttranslational modification affecting the positive charges of histones, probably acetylation; this hypothesis is supported by the fact that these histones were not detected where they would have migrated had they been deacetylated. CRMP2 (collapsin response mediator protein 2) had the highest decreased fold (-12.09) of expression in null compared with WT mice. Others spots specifically decreased included calretinin and the 14-3-3 proteins β and ζ.
Table 1.
Listing of the differentially expressed proteins identified by Nano LC/MS/MS
| Experimental
|
Data bank exploration
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Name | Fold | Mr/pI | Tissue | Function | Similarity | Tissue | Mr/pI | NCBI and SwissProt nos. |
| CRMP2 | -12.09 | 69.4/6.4 | OE | Cytoskeleton: promote microtubule assembly | Dehydropyrimidase | Brain | 62.2/6.0 | 6753676, O08553 |
| Lamin C | +12.17 | 64.9/7.2 | OE | Cytoskeleton: component of the nuclear lamina | Intermediate filament | Germ.cells | 65.5/6.4 | 1346414, P11516 |
| CAPG | +++ | 40.6/7.0 | OE | Cytoskeleton: blocks the barbed ends of actin filament | Villin/gelsolin | Large variety | 39.2/6.7 | 729023, P24452 |
| GRP78 | --- | 78.2/5.6 | OE | Neuroprotective: involved in stress response | Heat shock protein | Brain, liver | 70.4/5.0 | 109893, P20029 |
| +++ | 85/5.7 | |||||||
| GSTO1 | +4.73 | 30.8/7.3 | OE | Neuroprotective: Involved in stress response | GST superfamily | Large variety | 27.5/6.9 | 6754090, O09131 |
| Histone 1H2aK | +++ | 14.0/6.8 | OE | Chromatin: unknown | ? | ? | 14.1/11.0 | 20345513, Q8CGP7 |
| Histone H3 | +9.40 | 15.5/6.9 | OE | Chromatin: central role in nucleosome formation | Histone H3 | Large variety | 15.2/11.1 | 25025961, P16106 |
| Histone H2B | +9.40 | 15.5/6.9 | OE | Chromatin: regulates H3 methylation and gene silencing | Histone H2B | Large variety | 14.9/10.1 | 25025176 |
| Isovaleryl-CoA dehydrogenase | +++ | 40.6/7.0 | OE | Energy metabolism: catalyzes the conversion of isovaleryl-CoA to 3-methylcrotonyl-CoA | Acyl-CoA dehydro-genase | Large variety | 46.3/8.5 | 9789985, Q9JHI15 |
| Creatine kinase | +++ | 40.6/7.0 | OE | Energy metabolism: reversibly catalyzes the transfer of phosphate between ATP and various phosphogens | ATP guanido-phosphotransferase | Large variety | 43.0/6.6 | 90407, P07310 |
| +++ | 41.2/7.3 | OB | ||||||
| +3.08 | 40.9/7.4 | OB | ||||||
| Cytochrome b5 | +++ | 16.1/5.3 | OB | Energy metabolism: electron carrier for several membrane-bound oxygenases | Cytochrome b5 | ? | 15.1/4.9 | 13385268, P56395 |
| Calretinin | --- | 30.0/5.1 | OB | Cell signaling: calcium binding | Calbindin | Brain | 31.3/4.9 | 6970325, Q08331 |
| 14-3-3 protein ζ | --- | 30.0/5.1 | OB | Cell signaling: regulator of signaling mediated by PKC, tyrosine and trytophan hydroxylase | 14-4-3 | Brain | 27.7/4.7 | 1526539, P35215 |
| 14-3-3 protein β | --- | 30.0/5.1 | OB | Cell signaling: same as 14-3-3 protein ζ | 14-4-3 | Brain | 27.9/4.7 | 9055384, Q9CQV8 |
Names of protein, fold change, experimental molecular weights (Mr × 10—3)/pI and tissue source are indicated. Variations in spot intensities represented by fold are expressed as ratio of Mecp2 KO to WT. +++, Proteins specifically found in the KO Mecp2 mice; ---, proteins specifically absent from the Mecp2 KO mice. Data bank exploration of proteins includes: function, similarity, tissue detected, theoretical Mr × 10—3/pI, and data bank accession numbers [National Center for Biotechnology Information (NCBI) and SwissProt].
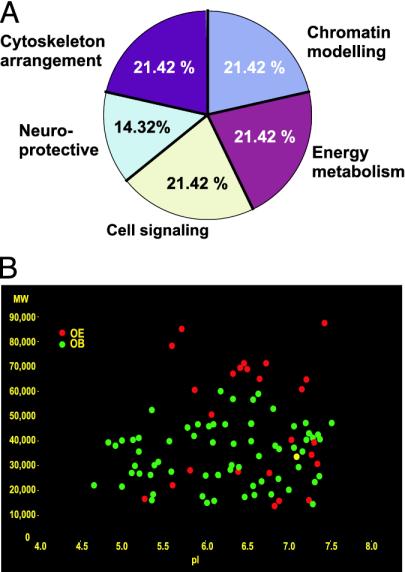
All of the proteins identified were not part of a single functional family of proteins, and their corresponding genes could not be assigned to one chromosomal location. However, it was possible to regroup them by their biological functions (Fig. 3A). Five main groups emerged, corresponding to cytoskeleton arrangement (CRMP2, CAPG, lamin C), chromatin modeling (histones H3, H2B, and 1H2AK), energy metabolism (cytochrome b5, isovaleryl-CoA dehydrogenase, creatine kinase), cell signaling (calretinin, 14-3-3 β and ζ), and neuroprotective (GRP78, GSTO1) functions.
Fig. 3.
(A) Percent distribution of the identified protein spots into functional groups. Cytoskeletal arrangement, chromatin modeling, and energy metabolism proteins represent the major changes observed. (B) Scatter graph showing matching coordinates (molecular weight and pI) of the protein changes detected at 2 weeks in the OE (red spots) and OB (green spots). Almost all of the spots detected to change have different coordinates with the exception of one strong match (molecular weight = 33,000, pI = 7.1) and a possible match (molecular weight = 41,000, pI = 7.4).
By comparing the coordinates (molecular weight and pI) of the detected spots significantly changed in both OE and OB tissues, we observed almost no matches in the spots detected in both OE and OB (Fig. 3B). Exceptions were found for two spots (molecular weight = 33,000, pI = 7.1, and molecular weight = 41,000 ± 0.3, pI = 7.4 ± 0.2, respectively), where the last one was identified as creatine kinase. This last observation supports our hypothesis on the presence of variations in the differentially expressed protein between regions of the nervous system, even when tissues are obtained at the same time.
Real-Time RT-PCR Analysis. The differences in protein expression levels between Mecp2-null mice and WT can occur as a result of a change in the level of translation, of modifications that occur posttranslationally, and/or of alterations in protein stability. The level of transcription was investigated for all of them (Fig. 4), except for GRP78, for which we found a change in gel coordinates.
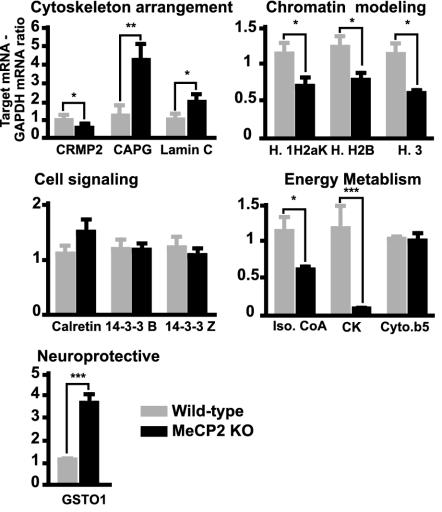
Fig. 4.
Bar chart representing the fold of RNA changes determined by real-time RT-PCR. Experiments were conducted on the cDNA (target gene) coding for the previously identified proteins by using the tissues in which the protein change was observed. Target genes are regrouped on the same x axis by their biological function. Gray bars represent WT mice and black bars Mecp2-null mice (KO). WT and KO results are represented as a ratio between the relative amounts of target gene and GAPDH. WT results were normalized to 1. Data were analyzed by unpaired Mann–Whitney test with 95% confidence intervals. Significant fold changes are represented by asterisks (*, P < 0.05; ***, P < 0.0001).
The RT-PCR analyses confirmed that there were statistically significant changes, and that the direction of the changes was consistent for the expression of CRMP2, CAPG, lamin C, and GSTO1 proteins (P = 0.014, 0.0037, 0.027, and 0.0004, respectively). The differences in RNA levels were smaller compared with the changes in protein levels. No significant changes were observed at the RNA levels for cytochrome b5, calretinin, or 14-3-3 β and ζ, suggesting for these proteins that posttranslational modifications and/or changes in protein stability occurred. Significant decreases in RNA levels were found for all histones (P = 0.036 for 1H2aK, 0.037 for H2B, and 0.020 for H3), for creatine kinase (P = 0.0007) and for isovaleryl-CoA dehydrogenase (P = 0.011). These decreases in RNA levels were discrepant compared with their respective protein levels. In these cases, we have to consider that the changes observed in the protein levels are the results of posttranslational modification, increased protein stability, or both.
Thus, the RT-PCR experiments revealed that mutation of Mecp2 led to discrete changes in RNA level, which can lead to changes at the level of protein expression, and second, that other proteome differences observed may be independent of a primary change in RNA levels.
Discussion
Proteomics allows for a large-scale analysis of proteins (24). It has been successfully used in differential display for comparison of changes in protein levels that happen in a wide range of diseases, including nervous system diseases (25, 26). In this study, we applied this technique to a mouse model of MeCP2 deficiency that may have implications for RTT.
Our results comparing the differences in protein profile patterns between WT and Mecp2-null mice in OE and OB at 2 and 4 weeks after birth suggest that the consequences (direct or indirect) of MeCP2 deficiency are transient, and differ over time and with respect to region. These differences would have been missed if the 2 weeks postnatal time had not been sampled, or if the two different regions of the olfactory system had been combined in our proteomics assay. Thus it revealed, at least for the olfactory system, a specific time and region for the participation of Mecp2 in the development. We suggest that these changes are relevant to the CNS. Maturation of the olfactory system occurs at 2 weeks after birth (27), the time when MeCP2 is highly expressed in this region (21). It is possible that the proteomic differences due to MeCP2 loss could be observed at other developmental times in the other regions of the nervous system in relation to the maturation of that specific region and the timing of expression of MeCP2 in that region. The fewer changes found at 4 weeks after birth compared with 2 weeks could be attributed to the capacity of the olfactory system to compensate after injury, or to changes in the status of any number of factors that regulate transcription, such as the transcription factors that are active, or DNA methylation and histone acetylation.
Some of the most prominent neuropathological features found in RTT and confirmed in this mouse model are decreased neuronal soma size and decreased or abnormal neuronal arborization (4–9, 13). These neuropathological features suppose cytoskeletal rear-rangement. Considering our proteomic and RT-PCR results, CRMP2 and CAPG could be candidates implicated in the molecular mechanisms responsible for the reduced and abnormal neuronal architecture. CRMP2 is known to promote tubulin assembly (28) and to be an intracellular messenger required for the growth cone collapse induced by semaphorin 3A (29). In the OE, CRMP2 is expressed in a population of neurons reaching their terminal maturity and connecting with mitral cells in the glomeruli of the OB (30). By consequence, down-regulation of the expression level of this protein could have a dramatic effect on the dendritic branching and axonal targeting in the maturing neurons. CapG, also known as GCap39, is an actin filament end capping protein, which influences cellular motility (31). Up-regulation of this protein could affect neuronal arborization. This deficiency in terminal neuronal differentiation is what we previously found in the olfactory system of RTT patients (9). Lamin C, which belongs to the intermediate filament family, is a component of the nuclear lamina (32). Lamin C is thought to provide a framework for the nuclear envelope and to interact with chromatin (33). We suppose that overexpression of lamin C might affect the nuclear structure and gene expression by its interaction with chromatin.
Histones play a central role in chromatin modeling and consequently in transcriptional silencing/activation. A striking feature of histones is their high content of positively charged side chains, which explains their high theoretical pI values. These positive charges are implicated in the binding of DNA, which blocks the access of transcription factors to promoter regions. Posttranslational modification of histones by acetylation is very common and important for neutralizing these positive charges and thus decreasing the binding of DNA/histones, which results in activation of transcription. It is known that MeCP2 deficiency produces hyperacetylation of histone H3 (15). The fact that we observed a neutral pI for histones supports the fact that, with mutation of Mecp2, not only histone 3, but also histones H2A and H2B are likely to be acetylated, as we observed shifts from a negative to a neutral pI for these protein as well. We also found that RNA levels of histones are decreased, which suggests that dysregulation of transcription or RNA stability of these histones also occurs.
Posttranslational modification of proteins is important in cell signaling. The functions of the 14-3-3 remain enigmatic (34), but they appear to play a critical role in cell signaling during neuronal differentiation (35) and olfactory learning (36). Posttranslationally modified forms of 14-3-3 β and ζ inhibit protein kinase C (37). Calretinin, another cell-signaling protein, used widely as a neuronal marker (38), is involved in regulating calcium pools critical for synaptic plasticity (39). Here, we found differences at their protein level, but not at their RNA level, suggesting some dysregulation of their posttranslational modification. Thus, all of these proteins whose expression levels are abnormal may contribute to the neuropathological features observed in RTT by affecting some signal transductions.
It is also widely recognized that metabolic disorders are associated with brain development disorders (40). In general, disturbances of the creatine kinase system have been observed in brain diseases (41, 42). In this regard, the dysregulation of protein expression that we found for M-creatine kinase, isovaleryl-CoA dehydrogenase, and cytochrome b5 can be placed in the context of changes that may contribute to the mental retardation and physical handicaps seen in RTT.
GSTO1 and GRP78 were modified in Mecp2-null mice. Both are implicated to have neuroprotective functions after stress response (43, 44). GSTO1 has been found to protect cells from oxidative stress by regulating the ryanodine receptor (45). Interestingly, GSTP1, another member of the GST family, is repressed by a methyl-CpG-binding domain (MBD) protein, MBD2 (46). GRP78, a 78-kDa member of the heat shock protein family (47), is expressed in the endoplasmic reticulum and is involved in the stress-induced unfolded protein response, which can occur in the setting of brain disorders (48). Although loss of MeCP2 does not lead to neuro-degeneration (13), these protein modifications suggest that neurons deficient in MeCP2 might experience increased stress.
We thus provide evidence of proteome profile differences incurred as a result of MeCP2 deficiency in a mouse model of RTT. It is now of importance to understand how these changes in the proteome develop. Future investigations into the molecular mechanisms that are responsible for such alterations that occur as a consequence of MeCP2 dysfunction or deficiency will need to consider temporal and regional aspects of MeCP2 function.
Acknowledgments
We thank Yajun Tu for assistance with real-time PCR and Amy Palmer, Alicia Degano, and Debby Cohen for comments. This work was supported by a grant from the International Rett Syndrome Association and National Institutes of Health grants from the National Institute of Neurological Disorders and Stroke and the National Institute on Deafness and Other Communication Disorders.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RTT, Rett syndrome; OE, olfactory epithelium; OB, olfactory bulb; KO, knockout.
References
- 1.Hagberg, B., Aicardi, J., Dias, K. & Ramos, O. (1983) Ann. Neurol. 14, 471-479. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, D. D. (1997) J. Neuropathol. Exp. Neurol. 56, 843-849. [DOI] [PubMed] [Google Scholar]
- 3.Naidu, S. (1997) Ann. Neurol. 42, 3-10. [DOI] [PubMed] [Google Scholar]
- 4.Jellinger, K., Armstrong, D., Zoghbi, H. Y. & Percy, A. K. (1988) Acta Neuropathol. 76, 142-158. [DOI] [PubMed] [Google Scholar]
- 5.Belichenko, P. V., Oldfors, A., Hagberg, B. & Dahlstrom, A. (1994) NeuroReport 5, 1509-1513. [PubMed] [Google Scholar]
- 6.Belichenko, P. V., Hagberg, B. & Dahlstrom, A. (1997) Acta Neuropathol. 93, 50-61. [DOI] [PubMed] [Google Scholar]
- 7.Bauman, M. L., Kemper, T. L. & Arin, D. M. (1995) Neuropediatrics 26, 105-108. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann, W. E., Taylor, C. V., Hohmann, C. F., Sanwal, I. B. & Naidu, S. (1997) Eur. Child Adolesc. Psychiatry 6, Suppl. 1, 75-77. [PubMed] [Google Scholar]
- 9.Ronnett, G., Leopold, D., Cai, X., Hoffbuhr, K., Moses, L., Hoffman, E. & Naidu, S. (2003) Ann. Neurol. 54, 206-218. [DOI] [PubMed] [Google Scholar]
- 10.Amir, R. E., Van den Veyver, I. B., Wan, M., Tran, C. Q., Francke, U. & Zoghbi, H. Y. (1999) Nat. Genet. 23, 185-188. [DOI] [PubMed] [Google Scholar]
- 11.Huppke, P., Laccone, F., Kramer, N., Engel, W. & Hanefeld, F. (2000) Hum. Mol. Genet. 9, 1369-1375. [DOI] [PubMed] [Google Scholar]
- 12.Bienvenu, T., Carrie, A., de Roux, N., Vinet, M. C., Jonveaux, P., Couvert, P., Villard, L., Arzimanoglou, A., Beldjord, C., Fontes, M., et al. (2000) Hum. Mol. Genet. 9, 1377-1384. [DOI] [PubMed] [Google Scholar]
- 13.Chen, R. Z., Akbarian, S., Tudor, M. & Jaenisch, R. (2001) Nat. Genet. 27, 327-331. [DOI] [PubMed] [Google Scholar]
- 14.Guy, J., Hendrich, B., Holmes, M., Martin, J. E. & Bird, A. (2001) Nat. Genet. 27, 322-326. [DOI] [PubMed] [Google Scholar]
- 15.Shahbazian, M., Young, J., Yuva-Paylor, L., Spencer, C., Antalffy, B., Noebels, J., Armstrong, D., Paylor, R. & Zoghbi, H. (2002) Neuron 35, 243-254. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, J. D., Meehan, R. R., Henzel, W. J., Maurer-Fogy, I., Jeppesen, P., Klein, F. & Bird, A. (1992) Cell 69, 905-914. [DOI] [PubMed] [Google Scholar]
- 17.Colantuoni, C., Jeon, O. H., Hyder, K., Chenchik, A., Khimani, A. H., Narayanan, V., Hoffman, E. P., Kaufmann, W. E., Naidu, S. & Pevsner, J. (2001) Neurobiol. Dis. 8, 847-865. [DOI] [PubMed] [Google Scholar]
- 18.Traynor, J., Agarwal, P., Lazzeroni, L. & Francke, U. (2002) BMC Med. Genet. 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tudor, M., Akbarian, S., Chen, R. Z. & Jaenisch, R. (2002) Proc. Natl. Acad. Sci. USA 99, 15536-15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon, C., Yoo, J. Y., Matarazzo, V., Sung, Y. K., Kim, E. J. & Ronnett, G. V. (2002) Proc. Natl. Acad. Sci. USA 99, 9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen, D. R., Matarazzo, V., Palmer, A. M., Tu, Y., Jeon, O. H., Pevsner, J. & Ronnett, G. V. (2003) Mol. Cell. Neurosci. 22, 417-429. [DOI] [PubMed] [Google Scholar]
- 22.O'Farrell, P. H. (1975) J. Biol. Chem. 250, 4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, H. & Greer, C. A. (2000) J. Comp. Neurol. 422, 297-311. [PubMed] [Google Scholar]
- 24.Pandey, A. & Mann, M. (2000) Nature 405, 837-846. [DOI] [PubMed] [Google Scholar]
- 25.Wulfkuhle, J. D., Liotta, L. A. & Petricoin, E. F. (2003) Nat. Rev. Cancer 3, 267-275. [DOI] [PubMed] [Google Scholar]
- 26.Morrison, R. S., Kinoshita, Y., Johnson, M. D., Uo, T., Ho, J. T., McBee, J. K., Conrads, T. P. & Veenstra, T. D. (2002) Mol. Cell Proteomics 1, 553-560. [DOI] [PubMed] [Google Scholar]
- 27.Hinds, J. W. & Hinds, P. L. (1976) J. Comp. Neurol. 169, 15-40. [DOI] [PubMed] [Google Scholar]
- 28.Fukata, Y., Itoh, T. J., Kimura, T., Menager, C., Nishimura, T., Shiromizu, T., Watanabe, H., Inagaki, N., Iwamatsu, A., Hotani, H. & Kaibuchi, K. (2002) Nat. Cell Biol. 4, 583-591. [DOI] [PubMed] [Google Scholar]
- 29.Pasterkamp, R. J. & Kolodkin, A. L. (2003) Curr. Opin. Neurobiol. 13, 79-89. [DOI] [PubMed] [Google Scholar]
- 30.Pasterkamp, R. J., De Winter, F., Holtmaat, A. J. & Verhaagen, J. (1998) J. Neurosci. 18, 9962-9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, T. D. & Borisy, G. G. (2003) Cell 112, 453-465. [DOI] [PubMed] [Google Scholar]
- 32.Hogner, D., Telling, A., Lepper, K. & Jost, E. (1984) Tissue Cell 16, 693-703. [DOI] [PubMed] [Google Scholar]
- 33.Worman, H. J. & Courvalin, J. C. (2002) Trends Cell Biol. 12, 591-598. [DOI] [PubMed] [Google Scholar]
- 34.Skoulakis, E. M. & Davis, R. L. (1998) Mol. Neurobiol. 16, 269-284. [DOI] [PubMed] [Google Scholar]
- 35.Broadie, K., Rushton, E., Skoulakis, E. M. & Davis, R. L. (1997) Neuron 19, 391-402. [DOI] [PubMed] [Google Scholar]
- 36.Skoulakis, E. M. & Davis, R. L. (1996) Neuron 17, 931-944. [DOI] [PubMed] [Google Scholar]
- 37.Aitken, A., Howell, S., Jones, D., Madrazo, J. & Patel, Y. (1995) J. Biol. Chem. 270, 5706-5709. [DOI] [PubMed] [Google Scholar]
- 38.Rogers, J. H. (1987) J. Cell Biol. 105, 1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwaller, B., Meyer, M. & Schiffmann, S. (2002) Cerebellum 1, 241-258. [DOI] [PubMed] [Google Scholar]
- 40.Kahler, S. G. & Fahey, M. C. (2003) Am. J. Med. Genet. 117C, 31-41. [DOI] [PubMed] [Google Scholar]
- 41.Wyss, M. & Kaddurah-Daouk, R. (2000) Physiol. Rev. 80, 1107-1213. [DOI] [PubMed] [Google Scholar]
- 42.Hemmer, W. & Wallimann, T. (1993) Dev. Neurosci. 15, 249-260. [DOI] [PubMed] [Google Scholar]
- 43.Kodym, R., Calkins, P. & Story, M. (1999) J. Biol. Chem. 274, 5131-5137. [DOI] [PubMed] [Google Scholar]
- 44.Reddy, R. K., Mao, C., Baumeister, P., Austin, R. C., Kaufman, R. J. & Lee, A. S. (2003) J. Biol. Chem. 278, 20915-20924. [DOI] [PubMed] [Google Scholar]
- 45.Dulhunty, A., Gage, P., Curtis, S., Chelvanayagam, G. & Board, P. (2001) J. Biol. Chem. 276, 3319-3323. [DOI] [PubMed] [Google Scholar]
- 46.Bakker, J., Lin, X. & Nelson, W. G. (2002) J. Biol. Chem. 277, 22573-22580. [DOI] [PubMed] [Google Scholar]
- 47.Munro, S. & Pelham, H. R. (1986) Cell 46, 291-300. [DOI] [PubMed] [Google Scholar]
- 48.Paschen, W. (2003) Cell Calcium 34, 365-383. [DOI] [PubMed] [Google Scholar]