Induction of tetraploidy through cleavage failure induces G1 arrest and senescence in primary mammalian cells but not in immortal cells. Induction of senescence occurs without DNA damage, and the capacity to become senescent appears to be a prerequisite of tetraploid arrest.
Abstract
Tetraploidy can arise from various mitotic or cleavage defects in mammalian cells, and inheritance of multiple centrosomes induces aneuploidy when tetraploid cells continue to cycle. Arrest of the tetraploid cell cycle is therefore potentially a critical cellular control. We report here that primary rat embryo fibroblasts (REF52) and human foreskin fibroblasts become senescent in tetraploid G1 after drug- or small interfering RNA (siRNA)-induced failure of cell cleavage. In contrast, T-antigen–transformed REF52 and p53+/+ HCT116 tumor cells rapidly become aneuploid by continuing to cycle after cleavage failure. Tetraploid primary cells quickly become quiescent, as determined by loss of the Ki-67 proliferation marker and of the fluorescent ubiquitination-based cell cycle indicator/late cell cycle marker geminin. Arrest is not due to DNA damage, as the γ-H2AX DNA damage marker remains at control levels after tetraploidy induction. Arrested tetraploid cells finally become senescent, as determined by SA-β-galactosidase activity. Tetraploid arrest is dependent on p16INK4a expression, as siRNA suppression of p16INK4a bypasses tetraploid arrest, permitting primary cells to become aneuploid. We conclude that tetraploid primary cells can become senescent without DNA damage and that induction of senescence is critical to tetraploidy arrest.
INTRODUCTION
During cell proliferation, maintenance of the integrity of the genome is of paramount importance. For this reason, multiple cell cycle checkpoints assure the proper completion of preceding stages of the cell cycle before the next stage ensues. These regulatory mechanisms protect cells from the consequences of DNA damage, premature termination of DNA replication, and progression into anaphase before chromosomes are properly aligned and under tension at the metaphase plate.
Of equal importance to preservation of euploidy, cells must properly complete cytokinesis to ensure correct distribution of chromatin to daughter cells. Despite these controls, aneuploidy and chromosomal instability are characteristic of the great majority of human cancers (Cahill et al., 1999) and are linked to the progressive development of high-grade, invasive tumors (Sandberg, 1977; Rabinovitch et al., 1989; Giaretti, 1994).
Tetraploidy—the inheritance of twice the normal number of chromosomes—can arise as a result of pathological processes such as chromosome nondisjunction (Shi and King, 2005), telomere dysfunction (Davoli et al., 2010; Davoli and de Lange, 2012), adenomatous polyposis coli (APC) mutation (Caldwell et al., 2007), or abnormal cell fusion (Duelli et al., 2005). Because tetraploid cells inherit twice the normal complement of centrosomes (Borel et al., 2002; Margolis et al., 2003; Quintyne et al., 2005), they can rapidly proceed to aneuploidy by production of multipolar spindles at the next mitosis, with one centrosome at each spindle pole driving random chromosome segregation into aneuploid daughter cells (Andreassen et al., 1996, 2001; Margolis et al., 2003). Alternatively, if multiple centrosomes cluster to form a proper bipolar division (Borel et al., 2002; Meraldi et al., 2002; Kwon et al., 2008; Leber et al., 2010), tetraploid cells may generate aneuploidy by exiting mitosis with lagging chromosomes (Ganem and Pellman, 2007).
A critical question is to what extent cells can control or suppress the cell cycle after cleavage failure. Although preservation of an intact genome is important to the organism, the extent to which tetraploid cells have the capacity to arrest remains unclear.
Several laboratories have found that nontransformed mammalian cells cease proliferating immediately after becoming tetraploid (Wright and Hayflick, 1972; Andreassen et al., 2001; Yang et al., 2004; Duelli et al., 2005; Fujiwara et al., 2005), whereas transformed cells continue cycling and proceed to aneuploidy (Andreassen et al., 2001; Duelli et al., 2005). Our results showed that arrest induced by cleavage failure occurs immediately in G1 of the next cycle (Andreassen et al., 2001). Others found that nontransformed cells inefficiently arrest after cleavage failure (Uetake and Sluder, 2004; Shi and King, 2005; Krzywicka-Racka and Sluder, 2011). This disparity requires an explanation, which may lie in the molecular details of the G1 checkpoint machinery.
Although arrest in tetraploid G1 requires p53 (Andreassen et al., 2001; Fujiwara et al., 2005), it is not strictly p53 dependent, as primary mouse embryo fibroblasts (MEFs) with intact p53 but with triple knockout of the Rb pocket protein family (pRb, p107, p130) escape tetraploid arrest (Borel et al., 2002; Lohez et al., 2003). In accord with lack of strict dependence on p53 response, it appears that tetraploid arrest in G1 does not induce a DNA damage response (Fujiwara et al., 2005). The capacity of cells to arrest in G1 when disruptions of mitosis or cell cleavage induce tetraploidy is potentially important to the control of tumor growth, as it represents the last opportunity for tetraploid cells to avoid aneuploidy.
Here we examine the induction of cell cycle arrest by cleavage failure in both rodent and human cell lines. Both rat and human primary cells arrest indefinitely in G1 immediately after induction of tetraploidy, and arrest occurs through the induction of senescence. The induction of senescence requires both p53 and pRb pathways and is particularly dependent on p16INK4a function (Beausejour et al., 2003), as suppression of p16INK4a permits bypass of tetraploidy arrest.
Assuming failure of cell cleavage induces senescence only in primary cells with intact p16INK4a or pRb function, our results predict that immortalized and transformed cells, which routinely suppress p16INK4a or pRb function (Okamoto et al., 1994; Dickson et al., 2000; Beausejour et al., 2003) and are unable to enter senescence (Serrano et al., 1996), are also unable to arrest when tetraploid.
Our work suggests that competence to become senescent is an absolute requirement for the prolonged arrest of primary tetraploid cells and that senescence invariantly follows induction of tetraploidy in primary cells but not in immortalized cells.
RESULTS
Effect of cell cleavage failure on the cell cycle of primary and transformed cells
Unsynchronized rat embryo fibroblast primary cells (REF52) and large SV-40 T-antigen transformed–REF52 variants (TAGs) were exposed to dihydroxycytochalasin B (DCB; Figure 1), an inhibitor of actin assembly (Aubin et al., 1981; Martineau et al., 1995) that blocks mammalian cell cleavage at concentrations >4 μM (Lohez et al., 2003), or to blebbistatin, a myosin II inhibitor (Straight et al., 2003; Figure 2). Both inhibitors effectively suppress cytokinesis, generating tetraploid cells. In our previous studies on tetraploidy, we used synchronous cell populations. The use of unsynchronized cells in the present work was designed to avoid any possible contribution of DNA damage (Wong and Stearns, 2005; Uetake and Sluder, 2010; Ganem and Pellman, 2012) induced by synchronization before induction of tetraploid cell arrest.
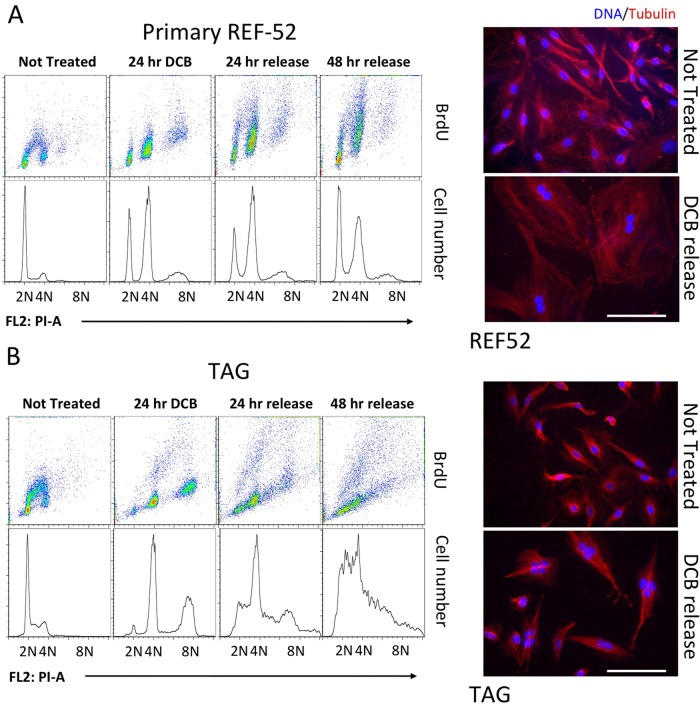
FIGURE 1:
Response of REF52 and TAG cells to DCB-induced tetraploidy. Both REF52 (A) and TAG (B) cells were exposed to 10 μM DCB for 24 h and then released from DCB for the indicated times while remaining subconfluent. Cells were then harvested at the times shown and subjected to flow cytometry to follow DNA content (lower line plots) and BrdU incorporation (upper dot plots). DNA content marks indicate 2N unreplicated cells, 4N replicated cells, and 8N cells that have proceeded through another replication cycle after becoming tetraploid. Cells that do not align with the marks are aneuploid. BrdU arcs indicate DNA replication during 0.5-h exposure to BrdU. Microscopy images show microtubules (red) and DNA (DAPI, blue) in both nontreated (NT) cells and cells released from DCB for 48 h. Note binucleate REF52 and multinucleate TAG cells after DCB release. Scale bars, 40 μm.
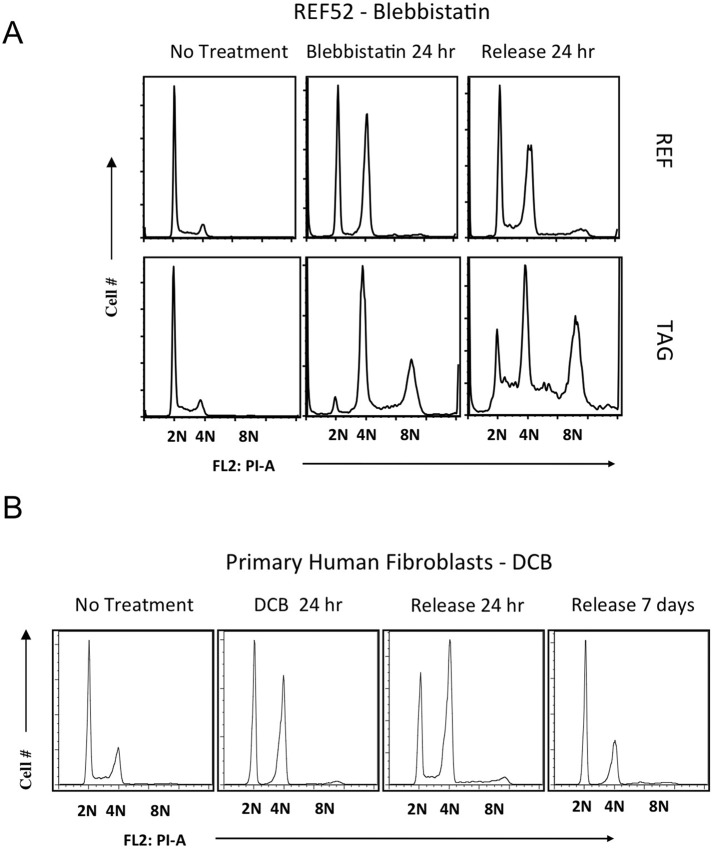
FIGURE 2:
Response of REF52 and TAG cells to blebbistatin-induced tetraploidy and of HFF to DCB-induced tetraploidy. (A) REF52 and TAG cells were exposed to the myosin II inhibitor blebbistatin (100 μM) for 24 h, as for DCB in Figure 1, and then released from drug for the indicated times while remaining subconfluent. Flow cytometry shows DNA content. (B) HFF cells at low passage were exposed to 10 μM DCB for 48 h and then released for the indicated times. Flow cytometry plots show population distribution relative to DNA content, indicated as 2N, 4N, and 8N, where 8N represents tetraploid cells that have continued to cycle.
Approximately half the initially asynchronous population had 4N DNA content after 24-h exposure to either DCB or blebbistatin, as analyzed by flow cytometry, whereas half had 2N DNA content (Figures 1 and 2) as previously demonstrated (Lohez et al., 2003). During drug treatment, REF52 did not incorporate 5-bromo-2’-deoxyuridine (BrdU; Figure 1A), indicating lack of DNA synthesis. The persistent 2N peak and lack of DNA replication exist during DCB exposure because, as previously demonstrated, even minimal suppression of actin assembly induces a transient and reversible G1 (2N) arrest in primary fibroblasts (Lohez et al., 2003). In contrast, untreated controls had a predominantly 2N profile and exhibited a robust BrdU arc between 2N and 4N, indicating active DNA replication.
On release from DCB, a BrdU arc reappeared between 2N and 4N, indicating restoration of the euploid cell cycle. In contrast, 4N cells were largely unable to proceed to 8N and showed little BrdU incorporation. The 4N population thus remained arrested after DCB release, whereas the transiently arrested 2N population reestablished the proliferating population. A small 8N peak appeared during the first 24 h of drug exposure, suggesting that an initial 4N-to-8N bypass created a small 8N subpopulation that did not go on to divide (Figure 3 and Supplemental Video S1). After DCB release, the population exhibited many binucleate cells not present before treatment (Figure 1A, right).
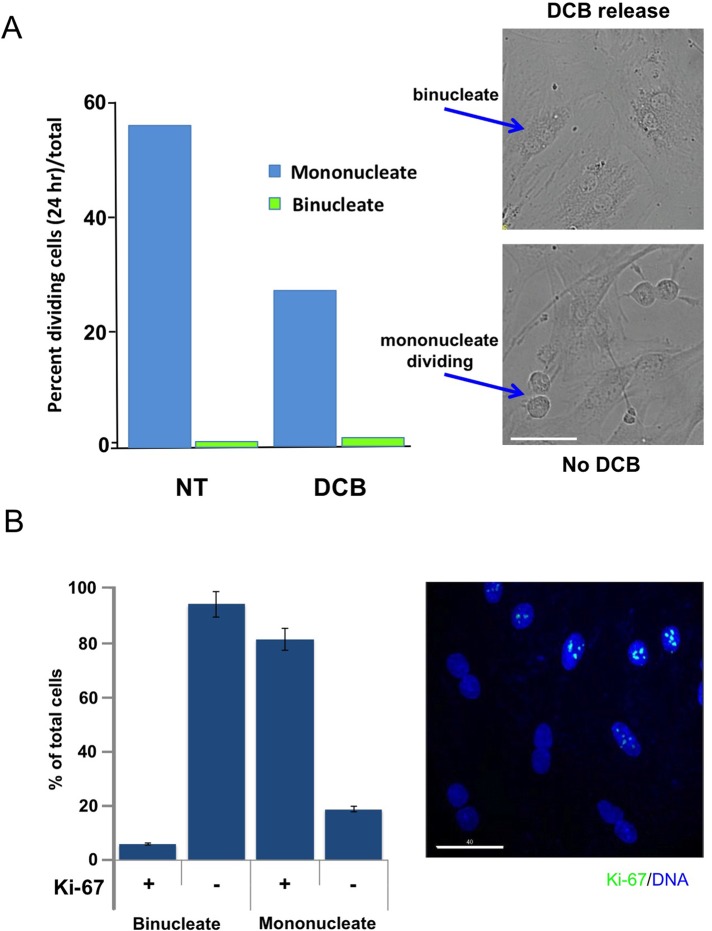
FIGURE 3:
Quantitation of mitosis in mononucleate and binucleate cells. (A) REF52 cells were either untreated or exposed to 10 μM DCB for 24 h and then released from drug. Cells were then recorded by DeltaVision deconvolution video microscopy at 400×, and the number of cells undergoing mitosis relative to the total cells was counted from random field video recordings over a 24 h period. Mononucleate cells and binucleate cells were separately scored relative to the total cells of their respective classes. N ≥ 300 cells per lane. Values indicate the percentage of mononucleate or binucleate cells that divided in 24 h. Right, image captures from videos of DCB-treated and then released binucleate cells (top) and euploid controls (bottom). The euploid image includes two anaphases. (B) Ki-67, a cell proliferation marker, is specifically absent from binucleate cell nuclei. HFF were exposed to 10 μM DCB for 24 h and then released from drug. Mononucleate and binucleate cells were quantitated as Ki-67 positive or negative in three independent experiments. More than 300 mononucleate or binucleate cells were counted in each experiment. Results are expressed as mean ± SD. Representative IF image of anti–Ki-67 stain (green, enhanced green fluorescent protein secondary antibody) shows binucleate cells without Ki-67. DNA counterstain is DAPI. Scale bar, 40 μm.
The outcome with blebbistatin (Figure 2A) was comparable in detail to results with DCB. During drug treatment, many 2N cells did not proceed in the cell cycle, whereas the rest failed in cleavage and accumulated as a 4N population. The transient 2N arrest with either DCB or blebbistatin suggests that suppressed lamellipodial motility, rather than suppression of actin assembly per se, induced euploid G1 cell cycle arrest in nontransformed cells (Dang and Gautreau, 2012). These results contrast with the claim that blebbistatin does not induce transient G1 arrest in euploid primary cells (Krzywicka-Racka and Sluder, 2011). The increasing prominence of the 2N peak during recovery indicates that the transiently arrested 2N cells recover and reestablish a euploid population.
Primary human foreskin fibroblasts (HFFs) at low passage responded to DCB (Figure 2B) in a manner that paralleled the response of low-passage REF52 cells (Lohez et al., 2003). In HFF cells plated on fibronectin and treated with DCB, a predominantly 2N euploid population was restored 7 d after release from DCB. The 4N population remained arrested, as ungated flow cytometry indicated that few cells had >4N DNA content at this time, and the absence of a <2N population in ungated flow cytometry indicated no appreciable cell death.
Video recordings of primary cells released from DCB after 24-h exposure and recorded in the first 24 h of recovery indicate that binucleate cells are abundant. Although the cells are healthy and motile, they do not undergo mitosis (Supplemental Video S1). Of importance, video recordings were done in the absence of blue light, known to interfere with cell cycle progression (Uetake and Sluder, 2004). In striking contrast, untreated controls exhibit many mitotic events in the same time course (Supplemental Video S2), confirming that recording conditions do not inhibit mitosis. The cells used in all our experiments were grown on a lawn of fibronectin to determine whether it modifies the induction of tetraploid arrest (Uetake and Sluder, 2004). We found no notable effect of fibronectin on the outcome compared with growth on poly-d-lysine–coated surfaces. Quantitation of the percentage of cells undergoing mitosis, from multiple videos, confirmed that virtually no binucleate cells underwent division during 24 h of recovery from DCB, in contrast to mononucleate cells in the same culture dishes (Figure 3A).
To confirm that binucleate cells were not cycling, we exposed HFF to DCB for 24 h, released them from DCB for 24 h, and then stained them for Ki-67 nuclear antigen, a proliferation marker (Scholzen and Gerdes, 2000). In the mixed population of mononucleate and binucleate cells on the same slide, Ki-67 was specifically absent from the nuclei of binucleate cells, whereas it gave a strong positive signal in mononucleate cell nuclei (Figure 3B).
TAG cells responded to DCB in a notably different manner than primary REF52 or HFF cells. After 24 h exposure to drug, there was no 2N G1 subpopulation, and the cells predominantly exhibited 4N-to-8N DNA content, with a prominent 8N peak (Figure 1B). At 24 and 48 h of release from DCB, the transformed cells were actively proliferating and increasingly aneuploid. Microscopic images of treated cells confirm the flow cytometry data, indicating that TAG cells, unlike REF52, became multinucleate and highly aneuploid after release from DCB (Figure 1B). Similarly, TAG cells became highly aneuploid within 24 h of release from blebbistatin (Figure 2A).
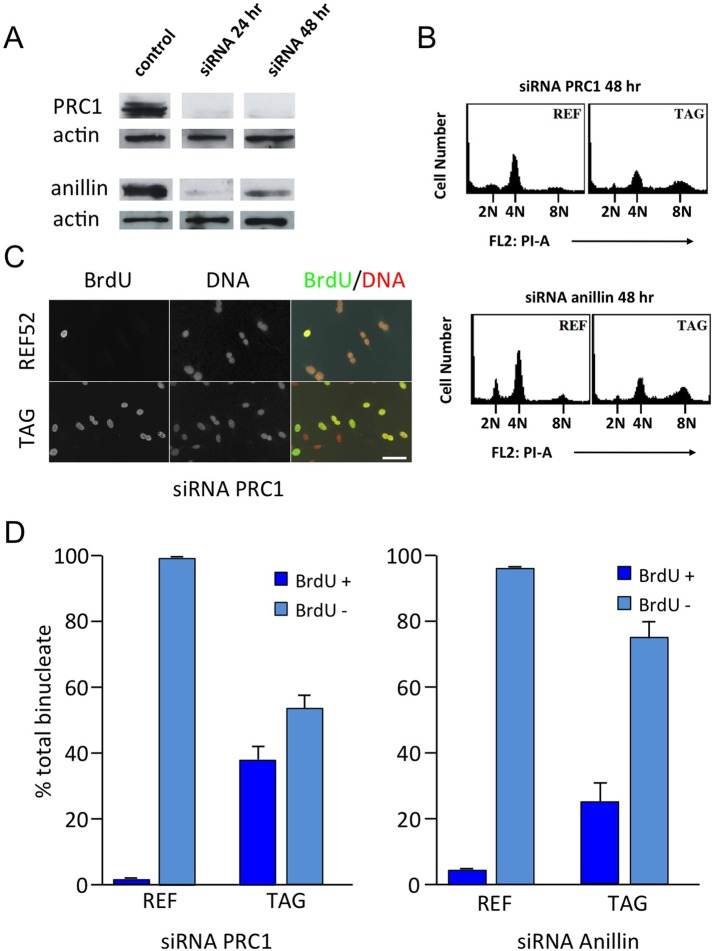
To confirm that cell cycle arrest of primary tetraploid cells was not a nonspecific consequence of drug exposure, we suppressed cell cleavage in REF52 and TAG cells by small interfering RNA (siRNA) targeting of two proteins required for cytokinesis, protein required for cytokinesis 1 (PRC1; Mollinari et al., 2005) and anillin (Oegema et al., 2000). Western blots confirmed that knockdown was effective at 24 and 48 h posttransfection (Figure 4A). By 48 h, REF52 that were transfected with siRNA to PRC1 or anillin had largely accumulated at 4N, whereas identically treated TAG cells had largely proceeded to 8N (Figure 4B). Immunofluorescence assays showed a substantial accumulation of binucleate cells at 48 h (Figure 4C), as was also evident in wide-angle anti-tubulin images of control and PRC1 siRNA–treated cells (Supplemental Figure S2). Quantitation of results from several microscopic assays confirmed that the REF52 population accumulated little BrdU, whereas TAG cells were substantially positive (Figure 4D), confirming that primary cells made tetraploid by siRNA suppression of cell cleavage did not undergo DNA replication, whereas TAG cells did.
FIGURE 4:
REF52 cells arrest in tetraploid G1 after PRC1 or anillin ablation. (A) Western blots confirm that either PRC1 or anillin had been effectively depleted at 24 and 48 h after siRNA transfection. Actin is a loading control. Extracts from each experimental condition were run on the same gel, but gel lanes were not contiguous, as indicated by separations. (B) At 48 h after siRNA ablation of PRC1 (top) or anillin (bottom), REF52 largely accumulated at 4N, whereas identically treated TAG cells substantially proceeded to 8N. (C) Immunofluorescence of BrdU incorporation into REF52 and TAG cells transfected with siRNA to PRC1. In both cases, binucleate cells are abundant. Both REF52 and TAG were recorded at identical microscope settings. Only TAG cells incorporated BrdU after becoming binucleate. DNA was stained with propidium iodide. Scale bar, 50 μm. (D) Microscopic quantitation of BrdU incorporation into binucleate cells after PRC1 or anillin knockdown. Cells were transfected with siRNA to PRC1 or anillin, and the binucleate cells were quantitated at 48 h for BrdU incorporation. Binucleate REF52 show little BrdU incorporation, whereas binucleate TAG cells are highly positive for BrdU. Graphs are averages from three different assays for each condition. Results are expressed as mean ± SD.
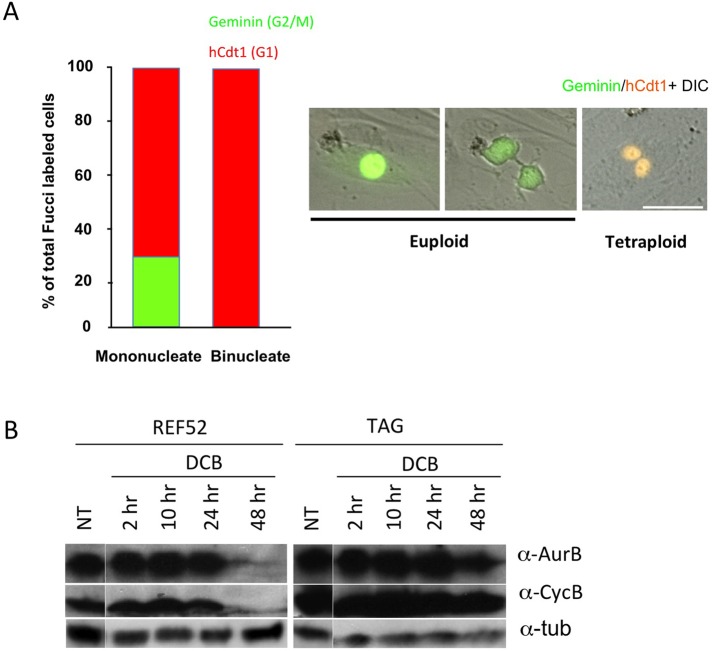
As a further confirmation that tetraploid primary cells are unable to cycle, we used a fluorescent ubiquitination-based cell cycle indicator (FUCCI) expression assay to assess the cell cycle distribution of HFF cells 48 h after release from DCB-induced tetraploidy. The FUCCI assay (Sakaue-Sawano et al., 2008) uses lentivirus vector coexpression of mAG-hGem, a green fluorescent marker for geminin expression, which is specific to the G2/M phase of the cell cycle, and of mKO2-hCdt1, an orange-red fluorescent marker for Cdt1 expression, which is specific to the G0/G1 phase of the cell cycle. Results confirm that binucleate tetraploid HFFs were uniformly in G0/G1, whereas mononucleate euploid cells in the same dishes were in both early and late phases of the cell cycle (Figure 5, A and B).
FIGURE 5:
Binucleate primary cells lose G2/M markers. (A) HFF cells were transfected with lentivirus expressing fluorescent geminin and hCdt1 (chromatin licensing and DNA replication factor 1), markers for G2/M and G1, respectively. After ascertaining that the cells were fluorescent, cells were exposed to 10 μM DCB for 24 h and then were recorded for 24 h after drug release. Video images from DeltaVision microscope recordings were quantitated for cells positive for geminin or hCdt1 markers. In the same culture chamber, mononucleate cells were positive for the G2/M geminin marker, while binucleate cells were negative. Both populations were positive for hCdt1. Data are composite from three independent experiments. Right, microscopy images show geminin (green) mononucleate cells and an hCdt1 (orange) positive binucleate cell. Scale bar, 40 μm. (B) Tetraploid REF52 cells lose late cell cycle markers. REF52 and TAG cells were exposed to 10 μM DCB for 24 h, released for 6 h, and then exposed again to DCB for 24 h to maximize REF52 tetraploidy. Western blots of cell extracts taken at the indicated times after final drug release show that REF52 cells have lost Aurora B and cyclin B by 48 h of release, whereas TAG cells continue to express these late-cell-cycle proteins. α-Tubulin serves as a loading control. NT gel lanes were from the same gel as treated samples, but they were not contiguous, as indicated by separations.
Consistent with FUCCI and Ki-67 results, Western blots of two late-cell-cycle markers, Aurora B and cyclin B1, indicated that late-cell-cycle markers were greatly diminished by 48 h of release from DCB-induced tetraploidy in REF52 cells but remained present in paired large T-antigen–transformed TAG cells (Figure 5C).
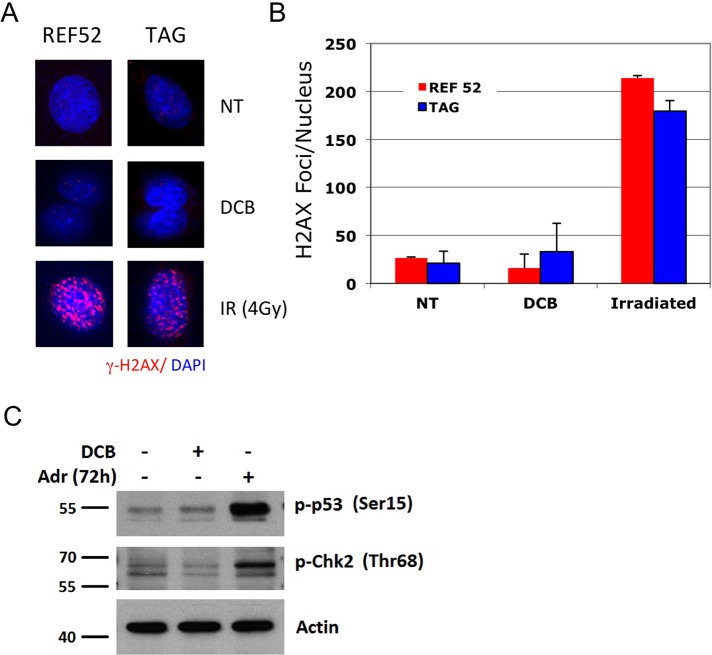
Failure of cell cleavage should not affect the integrity of the genome, which segregates without error during mitosis before cleavage failure. There was, indeed, no evidence for DNA damage in tetraploid G1-arrested cells, as indicated by analysis of the DNA damage marker phosphorylated histone 2 variant H2AX (Paull et al., 2000) in tetraploid nuclei (Figure 6). The signal strength of γ-H2AX foci in nuclei closely matched that of controls (Figure 6A), as did the average number of foci per nucleus (Figure 6B). For these experiments, the microscope gain was set high in order to capture any positive foci in controls or DCB-treated cells, yielding a background signal in both conditions. By contrast, a modest level of γ-irradiation yielded abundant H2AX phosphorylation (Figure 6, A and B). In accord with these results, the Ser-15 residue, phosphorylated on p53 in response to DNA damage (Giaccia and Kastan, 1998), was not phosphorylated in DCB-treated HFF cells (Figure 6C). For comparison we show the phosphorylation of Ser-15 on p53 in response to DNA damage induced by exposure to Adriamycin. Similarly, tetraploidy does not provoke another response to DNA damage—phosphorylation of checkpoint kinase 2 (Chk2) on Thr-68 (Ahn et al., 2000; Figure 6C).
FIGURE 6:
Tetraploidy arrest does not involve DNA damage. (A) Both REF52 and TAG cells were exposed to 10 μM DCB for 24 h and then released for 24 h and stained for γ-H2AX, a marker for cell response to DNA damage (Paull et al., 2000). The counterstain for DNA is DAPI. Binucleate cells of either cell type show no intense DNA damage response. γ-Irradiation (4Gy) of REF52 and TAG cells serves as a positive control. All images were taken from the same experiment, using identical image capture settings. (B) Quantitation of the number of γ-H2AX foci present in nuclei, visualized with microscopy, using procedures detailed in Materials and Methods. All foci that were above a set intensity threshold were automatically counted. The intensity of foci in irradiated nuclei was substantially greater than in other conditions, as seen in A. Results are expressed as mean ± SD. Foci within at least 100 nuclei were counted in each condition. (C) Western blot of DNA-damage markers in HFF cells, phospho-Ser-15 p53, and phospho-Thr-68 Chk2, demonstrating that these markers are not elevated after tetraploidy induction by two rounds of 10 μM DCB treatment compared with control random cycling cells. Adriamycin induction of DNA damage 72 h after drug treatment is the positive control for DNA damage response (Adr). Actin is a loading control.
Because we avoided synchronization procedures earlier in the cell cycle that might initiate a DNA damage response (Wong and Stearns, 2005; Uetake and Sluder, 2010; Ganem and Pellman, 2012), these results show that failure in the completion of cytokinesis does not, in itself, provoke DNA damage.
Primary tetraploid cells become senescent
Our results demonstrate that REF52 and HFF primary cells arrest in G0/G1 when tetraploid and that the failure to reenter the cell cycle is stable. This status suggests that the cells have permanently lost the capacity to proliferate and therefore become senescent. To confirm continuing quiescence, we assayed tetraploid cells for the persistence of primary cilia, a marker of cell quiescence (Tucker et al., 1979; Pugacheva et al., 2007; Gerdes et al., 2009). To assess the induction of senescence, we assayed for expression of a well-established cell senescence marker, senescence-associated β-galactosidase (SA-β-gal), an enzyme activity associated with the senescent phenotype (Dimri et al., 1995).
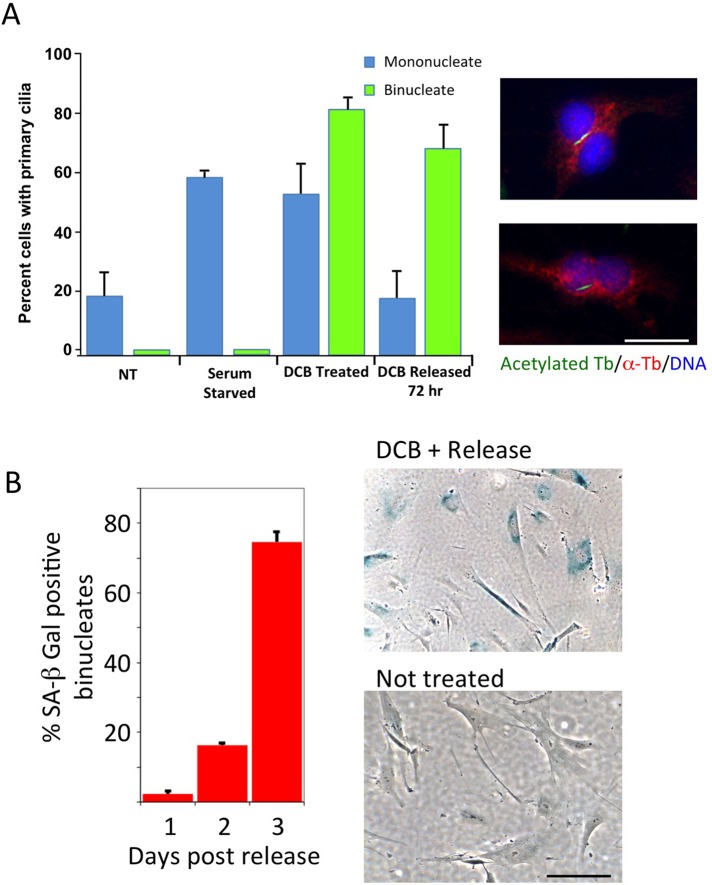
Nontransformed cells in culture assemble primary cilia during cell cycle exit, and disassembly occurs during cell cycle reentry (Seeley and Nachury, 2010). Tubulin acetylation is required for stability of primary cilia and serves as a marker (Pugacheva et al., 2007). Acetylated tubulin–stained primary cilia were evident in mononucleate serum-starved HFF cells and in mononucleate cells in the presence of DCB (Figure 7A), indicating G1 quiescence in the presence of the drug. After release from DCB, the mononucleate HFF cells reverted to control levels. Binucleate cells, also positive during exposure to DCB, remained ciliated 72 h after DCB release, indicating a sustained G0 block (Figure 7A). Images show representative ciliated binucleate HFF cells (Figure 7A).
FIGURE 7:
Tetraploid HFFs express quiescence and senescence markers. (A) Primary cilia, markers of cell quiescence, become abundant in HFF cells that have been exposed to 10 μM DCB for 24 h (DCB treated), indicating quiescence of both mononucleate and binucleate cells. By 72 h of release from DCB, the transiently arrested mononucleate population returns to nontreated (NT) levels, whereas the binucleate population remains ciliated. Serum-starved G0 HFFs serve as a positive control. Right, immunofluorescence of primary cilia in binucleate HFF cells, visualized with antibody to acetylated α-tubulin (green; Pugacheva et al., 2007). Counterstains were total α-tubulin (red) and DNA (DAPI, blue). Scale bar, 20 μm. (B) Tetraploid HFFs are positive for SA-β-galactosidase by 3 d after release from DCB. HFFs were treated with 10 μM DCB for 24 h and released. Cells were harvested and assayed for SA-β-gal activity at time points indicated. The percentage SA-β-gal–positive binucleate cells was quantitated in three independent experiments. Results are expressed as mean ± SD. Right, images of binucleate cells stained for SA-β-gal activity at 3 d of release compared with untreated controls. Scale bar, 40 μm.
When assayed for the level of expression of SA-β-gal, HFF cells exposed to DCB and released for 3 d were increasingly SA-β-gal positive (Figure 7B), indicating induction of senescence. An image enlargement shows that positive cells are binucleate (Supplemental Figure S1).
After tetraploid arrest, primary cells exhibit senescence markers characteristic of Raf-induced senescence
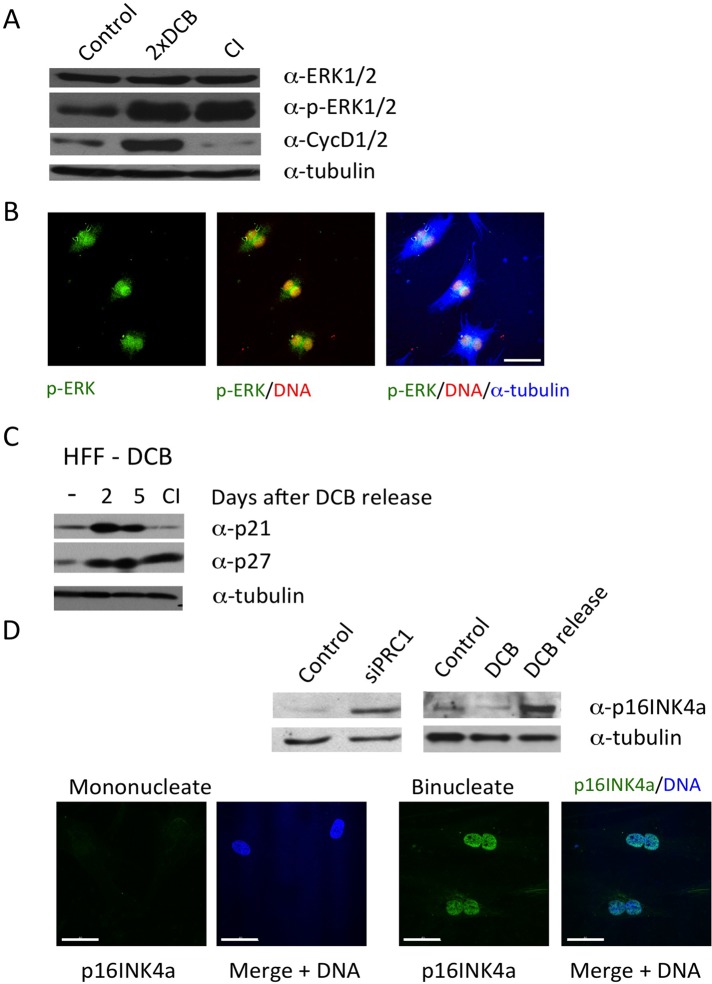
Oncogenic Ras and Raf transform immortalized cells but cause primary cells to instead enter premature senescence (Lin et al., 1998; Zhu et al., 1998; Meloche and Pouyssegur, 2007). The induction of premature senescence by oncogenic Ras or Raf is accompanied by characteristic protein changes, including high levels of extracellular signal-regulated kinase (ERK) 1/2 phosphorylation and elevated expression of G1 cyclin D1 (Zhu et al., 1998). Induction of tetraploidy in primary cells has a similar effect, causing ERK1/2 to become highly phosphorylated at MEK substrate residues (Figure 8A). Senescence induced by oncogenic Raf in primary cells also drives expression of cyclin D1 (Zhu et al., 1998), and cyclin D1 is elevated in primary cells that have been made tetraploid (Figure 8A), in contrast to contact-inhibited cells, which show little cyclin D1 expression. For these experiments, cells were doubly exposed to DCB to enrich for a 4N quiescent population, as described in Materials and Methods.
FIGURE 8:
Changes in protein expression in tetraploid arrest. (A) HFF cells were exposed to 10 μM DCB for 24 h, released for 6 h, and then exposed again to DCB for 24 h to maximize tetraploidy. Extracts were harvested for assay 2 d after DCB release. ERK1/2 is highly phosphorylated (p-ERK1/2), and cells have a high level of cyclin D expression. There is no increase in ERK1/2 expression. Results were compared with contact-inhibited (CI) and random cycling controls. Samples are from the same extracts, and tubulin is a loading control. (B) Immunofluorescence of cells treated as in A shows that phospho-ERK (p-ERK) is perinuclear and intranuclear in binucleate cells. Counterstains are propidium iodide for DNA and anti–α-tubulin antibody. Scale bar, 40 μm. (C) HFF cells, doubly exposed to DCB as in A, were assayed at the times indicated after release and compared with CI and random cycling controls (–). DCB-treated cells were positive for p21waf1 and p27kip1. Tubulin is a loading control. (D) Western blot showing that tetraploidy induces expression of p16INK4a at 3 d after transfection of HFF cells with PRC1 siRNA or 2 d after release from 10 μM DCB (DCB release). Controls are cells transfected with scrambled siRNA or cells not treated with DCB. The DCB lane indicates a sample taken after 24 h in 10 μM DCB. Tubulin is a loading control. Bottom, microscope images show HFF cells released from 10 μM DCB for 24 h and stained for p16INK4a (green) and DNA (blue, DAPI). Binucleate cells are positive for p16INK4a, and mononucleate cells are negative. Microscope settings were constant for all images. Scale bars, 40 μm.
Phosphorylated ERK2 localizes both within the nucleus and in the perinuclear space (Figure 8B). As reported for Raf-induced senescence (Zhu et al., 1998), inhibition of ERK phosphorylation by exposure to the MEK inhibitor UO126 does not reverse tetraploidy-induced senescence (unpublished data). The molecular response to tetraploidy-induced senescence is therefore similar to that induced by oncogenic Raf.
Further, we found that the cell cycle arrest markers p21waf1 and p27kip1 become elevated in primary cells after release from DCB and remain persistent (Figure 8C). Elevated p21waf1 is also evident after induction of senescence in primary cells by oncogenic Raf (Zhu et al., 1998), and p27kip1 expression has been reported to suppress polyploidy after cleavage failure (Serres et al., 2012).
Dual control of tetraploidy arrest through p53 and p16INK4a
The activation of premature senescence in primary cells by Raf expression is regulated by both p53 and p16INK4a (Zhu et al., 1998), and p16INK4a is generally important to the induction of replicative senescence (Campisi, 2011). Although sustained arrest of primary tetraploid cells requires p53 (Andreassen et al., 2001; Fujiwara et al., 2005; Senovilla et al., 2009; Vitale et al., 2010), Rb controls are also evidently involved in regulating tetraploidy arrest. Consistent with a role for p16INK4a, which controls the Rb pathway (Sherr, 1996), we previously showed that triple knockout of pRb, p107, and p130 abrogates cell cycle arrest induced by tetraploidy in primary MEFs (Borel et al., 2002). p16INK4a, which is required for onset of senescence and is considered a senescence marker (Rayess et al., 2012; Salama et al., 2014), is induced in primary cells made tetraploid by either transfection of siRNA to PRC1 or exposure to DCB (Figure 8D). Induction of p16INK4a arises with a short time lag after exposure to DCB, as found previously for p16INK4a induction after DNA damage (Robles and Adami, 1998; Johmura et al., 2014). Immunofluorescence shows strong p16INK4a labeling in DCB-treated binucleate cells, whereas mononucleate cells have only background stain (Figure 8D).
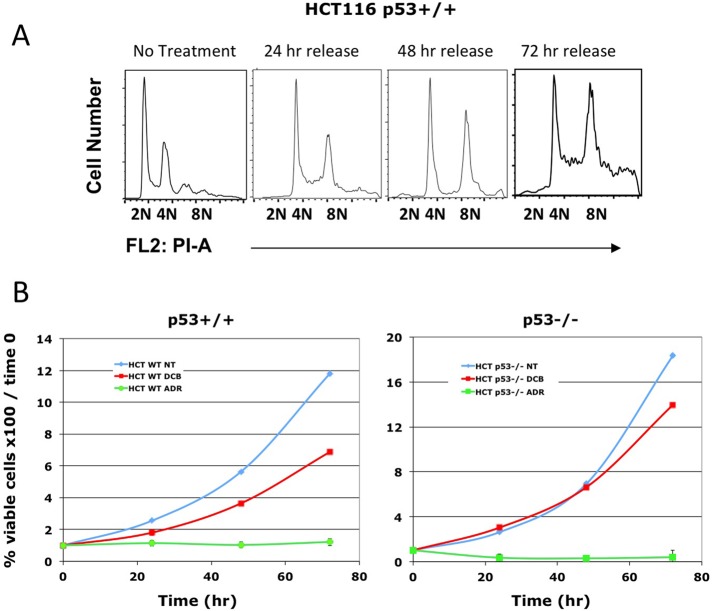
To demonstrate a dual requirement for p53 and p16INK4a in the induction of senescence by tetraploidy, we assayed p53-competent HCT116 colon carcinoma cells. We found that HCT116 did not arrest when made tetraploid but continued first to 8N and then to aneuploidy (Figure 9A). In fact, p53+/+ HCT116 colon carcinoma cells continued to proliferate at the same pace as p53−/− HCT116 (Figure 9B). In contrast, exposure of the same cells to the DNA damage agent Adriamycin effectively blocked proliferation. Of importance, although HCT116 cells express wild-type p53, they do not express p16INK4a (Myohanen et al., 1998). These results indicate that expression of intact p53 is not sufficient to induce arrest in tetraploid cells.
FIGURE 9:
Treatment with DCB does not arrest HCT116 p53+/+ cell proliferation. (A) HCT116 p53+/+ cells were treated with 10 μM DCB for 24 h and then released for the indicated times. The HCT116 cells proceed from tetraploidy to aneuploidy during 72 h of release. (B) HCT116 p53+/+ and HCT116 p53−/− were exposed to 10 μM DCB or 2 μg/ml Adriamycin (Skoufias et al., 2004) for 24 h and then released. Cell counts were then performed at the indicated times. Cells continue to proliferate when tetraploid and aneuploid, regardless of p53 status, but do not recover from Adriamycin.
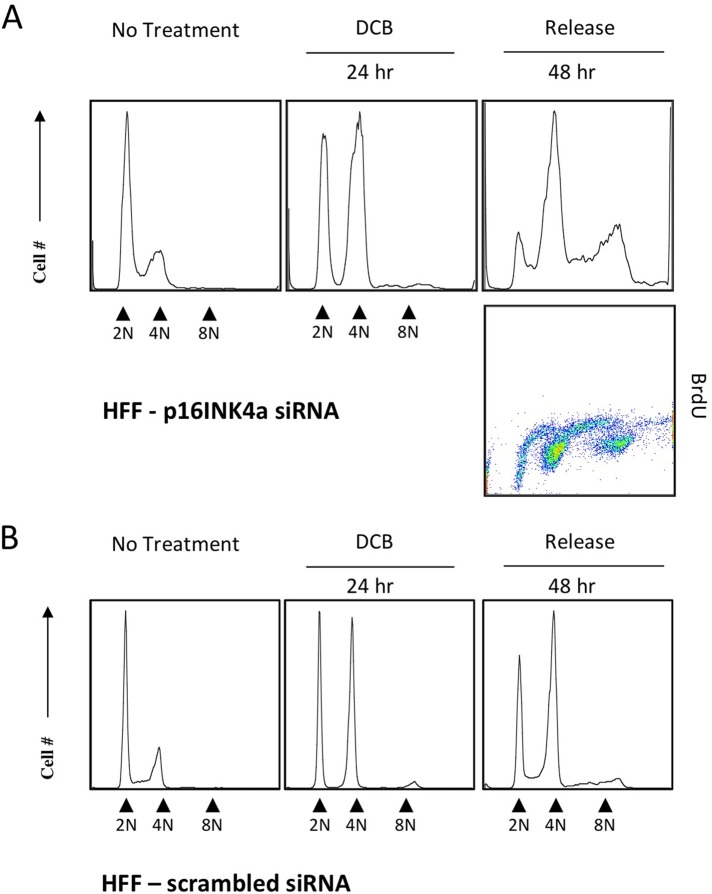
To test directly the importance of p16INK4a in tetraploidy arrest, we transfected HFFs with siRNA to p16INK4a and 48 h later exposed the cells to DCB for 24 h. After siRNA suppression of p16INK4a, tetraploid HFF cells did not remain arrested in G1 but continued to cycle, creating a prominent 8N population (Figure 10) and evident induction of aneuploidy. Both euploid and tetraploid cells exhibited extensive incorporation of BrdU. Control treatment with scrambled siRNA yielded a cell distribution like that shown for HFF cells treated with DCB (Figure 2). The result demonstrates that the capacity to arrest in G1 when tetraploid depends on an intact p16INK4a response and that its activation is thus one of the components required for the induction of senescence in response to the onset of tetraploidy in primary cells.
FIGURE 10:
Suppression of p16INK4a abrogates tetraploid arrest of HFF cells. (A) HFFs were transfected with siRNA to p16INK4a, exposed to 10 μM DCB for 48 h, and then assayed with flow cytometry. p16INK4a siRNA–transfected cells accumulated at 2N and 4N during DCB treatment. By 48 h of release from DCB, p16INK4a siRNA suppression substantially abrogated tetraploid arrest, and both euploid and tetraploid siRNA-transfected HFF cells were positive for BrdU incorporation (dot plot). (B) HFF cells transfected with scrambled siRNA as a control, treated as described with DCB, and assayed with flow cytometry.
DISCUSSION
The importance of tetraploidy arrest
Tetraploidy—the presence of twice the normal number of chromosomes—is an ominous state in mammalian tissues. In many human carcinomas, cells with tetraploid DNA content arise as an early step in tumorigenesis that precedes the formation of aneuploid cells (Margolis et al., 2003; Scrittori et al., 2005; Storchova and Kuffer, 2008). Aneuploidy and chromosomal instability in turn are characteristic of the great majority of human cancers (Cahill et al., 1999) and are linked to the progressive development of high-grade, invasive tumors. Aneuploidy can arise from tetraploid cells, regardless of whether subsequent cell divisions are bipolar or multipolar. We previously showed that cells competent to continue cycling when tetraploid either proceed to a multipolar mitosis with separated spindle poles (Borel et al., 2002) or, with nearly equal likelihood, cluster their centrosomes to create a bipolar spindle (Borel et al., 2002). Gross aneuploidy results from multipolar spindle mitosis, but tetraploid cells proceeding through bipolar mitosis with clustered centrosomes are prone to induction of aneuploidy through improper merotelic chromosome attachments and chromosome loss in anaphase (Ganem et al., 2009). Flow cytometry confirms that continued cycling of tetraploid TAG and HCT116 p53+/+ cells creates a mixture of tetraploid and highly aneuploid daughter cells (Figures 1, 2, and 10).
Tetraploidy can arise through any of several mitotic errors, including chromosome nondisjunction (Shi and King, 2005), mitotic slippage (Minn et al., 1996; Lanni and Jacks, 1998; Brito and Rieder, 2006), as a result of critically short telomeres (Davoli and de Lange, 2012), or through cleavage failure consequent to aberrant expression of APC (Caldwell et al., 2007), LATS1/2 (Iida et al., 2004; Aylon et al., 2006), or BRCA1 (Schlegel et al., 2003). Although many tetraploid-transformed cells that become aneuploid may die, the survivors can acquire either favorable mutations or chromosome profiles, with consequences for tumor development.
The presence of prolonged tetraploidy before aneuploidy is of central importance to cancer progression in multiple tumor types (Davoli and de Lange, 2011). It has therefore been an important but unresolved issue whether euploid nontransformed cells have mechanisms to prevent tetraploid proliferation and induction of aneuploidy, and it is important to understand how tumor cells evade these controls. The existence of controls that can arrest nontransformed tetraploid cells has been considered controversial due to reports that nontransformed tetraploid cells only partially arrest (Uetake and Sluder, 2004; Shi and King, 2005; Krzywicka-Racka and Sluder, 2011) or that tetraploid arrest is attributable to DNA damage induced by experimental manipulation (Wong and Stearns, 2005). In contrast, several laboratories reported that nontransformed mammalian cells cease proliferating immediately after becoming tetraploid (Wright and Hayflick, 1972; Andreassen et al., 2001; Yang et al., 2004; Duelli et al., 2005; Fujiwara et al., 2005). We believe that the present study, demonstrating induction of senescence in tetraploid primary cells, offers a resolution of these disparate results.
The question arises of why our results are at variance from those of the Sluder laboratory (Uetake and Sluder, 2004; Krzywicka-Racka and Sluder, 2011). Given the repeated demonstration by several laboratories that nontransformed cells rapidly arrest when tetraploid (Wright and Hayflick, 1972; Andreassen et al., 2001; Yang et al., 2004; Duelli et al., 2005; Fujiwara et al., 2005) and the observation that hTERT immortalized cells only partially arrest (Shi and King, 2005), we propose that the critical difference may be that cells must be competent to senesce in order to arrest in tetraploid state and that hTERT immortalized cells have suppressed the p16INK4a response and do not senesce (Dickson et al., 2000; Noble et al., 2004). We note that the work from the Sluder laboratory showing tetraploid bypass in nontransformed cells used hTERT-RPE1 cells for a substantial part of one study (Uetake and Sluder, 2004) and hTERT-RPE1 for all the nontransformed cells in another (Krzywicka-Racka and Sluder, 2011).
The requirement for primary cell status in tetraploidy arrest
Senescence is defined as the persistent arrest of cell proliferation in the presence of nutrients (Lin et al., 1998; Zhu et al., 1998), and induction of senescence requires the intact function of both the p53 and pRb pathways (Lin et al., 1998). Tetraploid primary fibroblasts subjected to fluorescence-activated cell sorting cannot proliferate, whereas sorted diploid cells from the same population can (Fujiwara et al., 2005). In contrast, hTERT-1 human fibroblasts, immortalized by hTERT expression, continue to cycle when tetraploid (Shi and King, 2005). hTERT-1 cells lack functional p16INK4a, a key protein in the activation of the pRb response (Dickson et al., 2000). In general, suppression of p16INK4a is critical to immortalization of hTERT-expressing cells (Kiyono et al., 1998; Dickson et al., 2000).
Primary REF52 cells arrest in tetraploid G1 after DCB exposure, whereas p53 mutant and large T-antigen–transformed REF52 continue cycling (Andreassen et al., 2001). Further, wild-type mouse epithelial cells do not proliferate when tetraploid (Senovilla et al., 2009), whereas p53-deficient cells continue to cycle (Vitale et al., 2010). Suppression of survivin triggers cell cleavage failure, causing primary IMR-90 and RPE cells to arrest in tetraploid G1, but p53 depletion abrogates the arrest and drives endoreduplication (Yang et al., 2004). However, it is also clear that suppression of the pRb pocket proteins is sufficient to abrogate tetraploid arrest in p53-competent cells, as mouse embryo fibroblasts lacking pRb, p107, and p130 but wild type for p53 fail to arrest when they spontaneously become tetraploid (Borel et al., 2002) or become tetraploid after drug induction (Lohez et al., 2003).
Tetraploidy controls in the organism
Tetraploidy is normally incompatible with mammalian embryonic development. In a routine technique, mutant mice are produced by mixture of diploid and tetraploid cells to form a chimeric blastocyst. In resulting embryos, the tetraploid cells are restricted to extraembryonic tissue, whereas the epiblast becomes entirely euploid (Nagy et al., 1990, 1993; Eakin and Behringer, 2003). Thus tolerance of tetraploidy is limited to specific extraembryonic cells. Exceptions exist in both mice and humans (Ganem and Pellman, 2007), but the vast majority of embryos eliminate polyploid cells during early development (Nagy et al., 1993). Given our results with p16INK4a, it is noteworthy that tetraploid cells persist in the epiblast until gastrulation in mouse (Mackay and West, 2005), coincident with the time when pRb-dependent G1 controls initiate (Egashira et al., 2011). Although certain adult mammalian cell types become tetraploid or polyploid during terminal differentiation (Ganem and Pellman, 2007), the absence of tetraploidy is the rule in continuously replicating cells, likely due to specific constraints, the molecular nature of which remains to be determined.
Senescence induction without DNA damage
A distinction of the present study from our previous work on tetraploidy lies in the avoidance of cell synchronization steps before induction of tetraploidy, by either drug or siRNA suppression of critical cell cleavage proteins. In this way, we avoided cell presynchronization, which has been critiqued as a possible cause of DNA damage, a possible alternative trigger for the arrest of tetraploid cells (Wong and Stearns, 2005). In this study, we find no evidence for a DNA damage response in tetraploid cells, as confirmed by the absence of phosphorylated histone H2AX, phospho-Ser-15 p53, or phospho-Thr-68 Chk2.
Indeed, with respect to the absence of DNA damage, the senescence induced by tetraploidy appears to be distinct from the majority of other premature senescence mechanisms. In contrast to tetraploidy-induced senescence, premature senescence in response to oncogene activation or critically short telomeres is preceded by a period of hyperproliferation and accumulation of DNA damage, accompanied by a robust DNA damage checkpoint response (Takai et al., 2003; Bartkova et al., 2006; Di Micco et al., 2006; Nardella et al., 2011), which is followed by late onset of senescence. In fact, premature senescence has been characterized as a DNA damage response (von Zglinicki et al., 2005; Di Micco et al., 2006).
In contrast, senescence induction in response to Raf or Ras overexpression in nontransformed cells (Serrano et al., 1996, 1997; Zhu et al., 1998) resembles the tetraploidy response. Both involve ERK1/2 phosphorylation and cyclin D1 overexpression in the absence of proliferation, and both occur only in primary cells. Further parallels include the rapid onset of senescence (within 3 d after induction), a requirement for the parallel function of p16INK4a and p21waf1, and the absence of a DNA damage response during activation (Jeanblanc et al., 2012). In the broader picture, induction of senescence in the absence of DNA damage occurs as an essential part of organism patterning during embryogenesis (Storer et al., 2013). It will be of great interest to address whether there are common mechanisms that underlie induction of senescence without DNA damage in nontransformed cells, either in the course of normal development or as a result of becoming tetraploid.
MATERIALS AND METHODS
Cell culture
HFFs (Hs-27) were obtained from the stem cell core facility at the Sanford-Burnham Institute (La Jolla, CA) and were used at passages 9–11. HCT116 p53+/+ and p53−/−cells were the kind gift of Bert Vogelstein (Johns Hopkins Medicine, Baltimore, MD). Rat primary REF52 and TAG cell lines were as reported previously (Andreassen et al., 2001). REF52 were used at passages 18–20.
HFF, REF52, and TAG cell lines were grown as a monolayer in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and maintained in a humid incubator at 37°C in a 5% CO2 environment. HCT116 was grown in McCoy's 5A medium (Life Technologies, Carlsbad, CA) with 10% fetal bovine serum and maintained as described.
Drugs used to suppress cytokinesis were the myosin II inhibitor blebbistatin (100 μM; Straight et al., 2003) and DCB (10 μM), which were added to cell culture from 100× stocks in dimethyl sulfoxide, filtered for sterility, and kept frozen until use. The concentration of DCB used (10 μM) was the lowest concentration that completely suppresses cytokinesis, in our experience (Lohez et al., 2003). Cytochalasin D, also used for tetraploidy analysis (Uetake and Sluder, 2004), suppresses cytokinesis at an eightfold lower concentration than DCB (Atlas and Lin, 1978). We routinely used DCB to avoid secondary effects on glucose metabolism (Atlas and Lin, 1978).
Double DCB synchronization of HFF cells
To obtain highly synchronous 4N HFF cells, randomly cycling cells were exposed to 10 μM DCB for 24 h, released for 6 h, exposed to DCB again for 24 h, and then released and harvested at indicated times for assays.
Flow cytometry
For flow cytometry, cells were harvested using trypsin-EDTA and centrifuged at 500 × g for 5 min at room temperature. The supernatant was aspirated and cells were trypsinized, washed, and fixed in −20ºC methanol for a minimum of 20 min and stored at −20ºC until time of staining. For analysis, cells were washed with 1× phosphate-buffered saline (PBS), stained with a propidium iodide (PI) solution (0.1% Triton X-100, 200 μg/ml DNase-free RNase A, and 3.0 μM PI), and analyzed on a Becton-Dickinson Excalibur flow cytometer using FloJo software (Ashland, OR). For BrdU incorporation, medium was removed from cells and replaced with fresh, warm medium containing 10 μM BrdU (B5002; Sigma-Aldrich, St. Louis, MO), and incubated for 1 h. Cells were then fixed and prepared for flow cytometry. For BrdU staining, fixed cells were prepared for PI stain and additionally incubated with fluorescein isothiocyanate (FITC)–conjugated anti-BrdU antibody (Becton Dickinson, San Jose, CA) as previously described (Andreassen et al., 2001).
Immunofluorescence microscopy
For immunofluorescence (IF), cells were grown on fibronectin-coated (20 μg/ml human foreskin fibroblast fibronectin [F2518; Sigma-Aldrich]) coverslips. For fixation, coverslips were washed and exposed to 4% paraformaldehyde/PBS at room temperature. After fixation, coverslips were washed three times and blocked for 1 h in blocking buffer (0.3% Triton X-100, 5% normal goat serum diluted in PBS). Primary antibodies in 0.3% Triton X-100 and 1% bovine serum albumin in PBS were incubated on coverslips at 4ºC overnight in a humid chamber. Coverslips were then washed three times in PBS, and secondary antibodies were added and incubated for 1 h at room temperature. Coverslips were then washed once and incubated in antibody dilution buffer containing 4′,6-diamidino-2-phenylindole (DAPI) or PI for 10 min at room temperature. Coverslips were then washed three times in PBS, dipped briefly in double-distilled H2O, air-dried, and mounted with Clarion mounting medium. Images were taken using a DeltaVision deconvolution microscope equipped with an automated stage. A minimum of 15 0.2-μm z-sections were taken per field. Images were then deconvolved using softWoRx software (Applied Precision, Issaquah, WA). Where indicated, to stain DNA, DAPI (Life Technologies) was used at 300 nM and PI (Invitrogen) at 1.0 μM after DNase-free RNase pretreatment.
We used the softWoRx two-dimensional polygon tool for microscopic quantitation of γ-H2AX foci within each nucleus. The threshold was manually set so that overlaid polygons reflected single spots as they appeared to the eye. The threshold was then held constant for data capture of untreated, DCB-treated, and irradiated samples.
Primary cilia were quantitated by counting the percentage of total cells in random fields for the presence of cilia, as determined by anti–acetylated tubulin antibody stain, counting as positive all stained linear elements adjacent to nuclei.
Antibodies used for IF included anti–α-tubulin (B512; Sigma-Aldrich), anti–α-tubulin rat monoclonal antibody (mAb; YL1/2; ab6160; Abcam, Cambridge, UK), or anti–acetylated α-tubulin mouse mAb (clone 6-11B-1; Sigma-Aldrich), anti–phospho-ERK1/2 (anti–phospho-Thr-202/Tyr-204; 4377; Cell Signaling, Beverly, MA), anti–Ki-67 (rabbit mAb; 9129; Cell Signaling), anti-p16INK4a (mouse mAb 11104; Immuno-Biological Laboratories, Gunma, Japan), and anti–γ-H2AX (4411-PC; Trevigen, Gaithersburg, MD). Secondary fluorescent antibodies were from Cell Signaling.
Time-lapse microscopy
For video microscopy, HFF, REF52, and TAG cell lines were seeded into four- or eight-chamber glass slides coated with 20 μg/ml fibronectin. For these experiments, we used L-15 medium for optimal buffering over prolonged periods. As appropriate, samples were exposed to 10 μM DCB for 24 h and then released from drug before video recording. At the time of release, slides were mounted on a DeltaVision deconvolution microscope equipped with an automatic stage and CO2 and temperature control. Images were taken in the differential interference contrast (DIC) channel every 20 min, using the point revisit function, for up to 3 d, as described (Panopoulos et al., 2011). A minimum of 10 fields were selected for each condition and experiment.
HFF cells infected with lentiviruses to express FUCCI markers were plated in four- or eight-chamber glass slides, exposed to 10 μM DCB, and then released into L-15 medium for video microscopy, and images were taken in the DIC, tetramethylrhodamine, and FITC channels every 20 min during a period of 24 h.
SA-β-galactosidase assay
HFF cells were seeded on fibronectin-treated glass coverslips as described for IF. At the time of harvest, cells were washed once with PBS at room temperature and then fixed in 0.2% glutaraldehyde/PBS for 5 min. After fixation, coverslips were washed three times with PBS for 5 min each at room temperature. Coverslips were then stained for activity for 24 h at 37ºC in 0.1 mg/ml X-gal, 150 mM NaCl, 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 40 mM sodium phosphate, pH 6.0, and then imaged by phase contrast microscopy.
Treatment with siRNA targeting PRC1, anillin, and p16INK4a
To target rat PRC1 and anillin, we used rat PRC1 and anillin siRNA from Dharmacon-GE (Lafayette, CO). For knockdown of PRC1 or anillin, cells were transfected 1 d after plating in 60-mm dishes when they displayed an estimated 40–45% confluency, and cells were collected for analysis 24 or 48 h after transfection. For BrdU analysis, cells were harvested 48 h after transfection and stained for BrdU incorporation as described.
To target p16INK4a, preverified siRNA and scrambled siRNA were obtained from Qiagen (Valencia, CA). Twenty-four hours after transfection, cells were exposed to 10 μM DCB for 24 h and then released for 48 h and assayed by flow cytometry. All siRNA transfections used 30 nM siRNA (final), introduced with Lipofectamine RNAiMax.
Immunoblotting
Cell lysates (25 μg/well) were resolved by PAGE, transferred to polyvinylidene fluoride membranes, and detected using chemiluminescence (Thermo Scientific Pierce, Rockford, IL). All primary antibodies were used at 1:1000. Appropriate secondary horseradish peroxidase–conjugated antibodies were used at 1:10,000. Antibodies used were p21waf1 (C-19; Santa Cruz Biotechnology, Dallas, TX), p27kip1 (610241; BD Transduction Lab, San Jose, CA), α-tubulin (B511; Sigma-Aldrich), anti-ERK1/2 (9102; Cell Signaling), anti–phospho-ERK1/2 (4377; Cell Signaling), anti–γ-H2AX (4411-PC; Trevigen), anti–cyclin D1/2 (05-362; Upstate Biotechnology, Lake Placid, NY), anti-anillin (C. Field, Harvard Medical School, Boston, MA; Oegema et al., 2000), anti–phospho-Ser-15 p53 (12571; Cell Signaling), anti–phospho-Thr-68 Chk2 (2197; Cell Signaling), anti-p16INK4a (mouse mAb 11104; Immuno-Biological Laboratories, Minneapolis, MN), and PRC1 antibody described previously (Mollinari et al., 2002).
Cell proliferation assay
After treatment with DCB for 24 h and release, HCT116 cells were suspended at the times indicated in a precise volume and an aliquot placed on a calibrated slide. Viable cells, identified by trypan blue exclusion, were counted in phase contrast.
γ-Irradiation
Randomly cycling REF52 and TAG cells received 4 Gy, using a 137Cs γ-irradiator at 2 Gy/min, and then were processed for IF using anti-γ-H2AX antibody, DAPI stain, and IF procedures described earlier.
Lentivirus production and infection of HFF
Lentiviral particles were obtained by transfecting human embryonic kidney HEK293 cells (seeded in 15-cm dishes) using FUCCI lentiviral constructs (L-CDT and L-GMN), as described earlier (Sakaue-Sawano et al., 2008). To infect HFF cells, 5 × 103 cells were seeded on four- or eight-well chambered glass slides, coated with fibronectin, and incubated with 1 ml of each viral supernatant in the presence of Polybrene, 10 μg/ml. After 24 h of incubation with lentivirus, fresh medium with or without 10 μM DCB was added, and cells were seeded onto four- or eight-well chambered glass slides, where they were subjected to live-cell imaging with a DeltaVision deconvolution microscopy unit.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01GM068107 and R01GM088716 (R.L.M.) and R01CA108947 (R.F.).
Abbreviations used:
- BrdU
5-bromo-2′-deoxyuridine
- DAPI
4′,6-diamidino-2-phenylindole
- DCB
dihydroxycytochalasin B
- DIC
differential interference contrast
- ERK
extracellular signal-regulated kinase
- FITC
fluorescein isothiocyanate
- FUCCI
fluorescent ubiquitination-based cell cycle indicator
- HFF
human foreskin fibroblast
- IF
immunofluorescence
- mAb
monoclonal antibody
- PRC1
protein required for cytokinesis 1
- REF52
primary rat embryo fibroblasts
- SA-β-gal
senescence-associated β-galactosidase
- siRNA
small interfering RNA
- TAG
SV-40 large T-antigen–transformed primary rat embryo fibroblast
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0844) on August 20, 2014.
REFERENCES
- Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Martineau SN, Margolis RL. Chemical induction of mitotic checkpoint override in mammalian cells results in aneuploidy following a transient tetraploid state. Mutat Res. 1996;372:181–194. doi: 10.1016/s0027-5107(96)00138-8. [DOI] [PubMed] [Google Scholar]
- Atlas SJ, Lin S. Dihydrocytochalasin B. Biological effects and binding to 3T3 cells. J Cell Biol. 1978;76:360–370. doi: 10.1083/jcb.76.2.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin JE, Osborn M, Weber K. Inhibition of cytokinesis and altered contractile ring morphology induced by cytochalasins in synchronized PtK2 cells. Exp Cell Res. 1981;136:63–79. doi: 10.1016/0014-4827(81)90038-0. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel F, Lohez OD, Lacroix FB, Margolis RL. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci USA. 2002;99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- Caldwell CM, Green RA, Kaplan KB. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol. 2007;178:1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence: putting the paradoxes in perspective. Curr Opin Genet Dev. 2011;21:107–112. doi: 10.1016/j.gde.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang I, Gautreau A. Evidence for a cell cycle checkpoint that senses branched actin in the lamellipodium. BioEssays. 2012;34:1021–1024. doi: 10.1002/bies.201200119. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli DM, Hearn S, Myers MP, Lazebnik Y. A primate virus generates transformed human cells by fusion. J Cell Biol. 2005;171:493–503. doi: 10.1083/jcb.200507069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin GS, Behringer RR. Tetraploid development in the mouse. Dev Dynam. 2003;228:751–766. doi: 10.1002/dvdy.10363. [DOI] [PubMed] [Google Scholar]
- Egashira A, Kano K, Naito K. Preimplantation-embryo-specific cell-cycle regulation is attributable to a low expression of retinoblastoma protein rather than its phosphorylation. J Reprod Dev. 2011;57:492–499. doi: 10.1262/jrd.10-170o. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131:437–440. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–881. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Giaretti W. A model of DNA aneuploidization and evolution in colorectal cancer. Lab Invest. 1994;71:904–910. [PubMed] [Google Scholar]
- Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, Honda S, Kosai K, Kawasuji M, Pallas DC, et al. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 2004;23:5266–5274. doi: 10.1038/sj.onc.1207623. [DOI] [PubMed] [Google Scholar]
- Jeanblanc M, Ragu S, Gey C, Contrepois K, Courbeyrette R, Thuret JY, Mann C. Parallel pathways in RAF-induced senescence and conditions for its reversion. Oncogene. 2012;31:3072–3085. doi: 10.1038/onc.2011.481. [DOI] [PubMed] [Google Scholar]
- Johmura Y, Shimada M, Misaki T, Naiki-Ito A, Miyoshi H, Motoyama N, Ohtani N, Hara E, Nakamura M, Morita A, et al. Necessary and sufficient role for a mitosis skip in senescence induction. Mol Cell. 2014;55:73–84. doi: 10.1016/j.molcel.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- Krzywicka-Racka A, Sluder G. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J Cell Biol. 2011;194:199–207. doi: 10.1083/jcb.201101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, Wagner L, Ho AD, Salisbury JL, Boutros M, et al. Proteins required for centrosome clustering in cancer cells. Sci Transl Med. 2010;2:33ra38. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohez OD, Reynaud C, Borel F, Andreassen PR, Margolis RL. Arrest of mammalian fibroblasts in G1 in response to actin inhibition is dependent on retinoblastoma pocket proteins but not on p53. J Cell Biol. 2003;161:67–77. doi: 10.1083/jcb.200208140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay GE, West JD. Fate of tetraploid cells in 4N <–>2N chimeric mouse blastocysts. Mech Dev. 2005;122:1266–1281. doi: 10.1016/j.mod.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- Martineau SN, Andreassen PR, Margolis RL. Delay of HeLa cell cleavage into interphase using dihydrocytochalasin B: retention of a postmitotic spindle and telophase disc correlates with synchronous cleavage recovery. J Cell Biol. 1995;131:191–205. doi: 10.1083/jcb.131.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53–/– cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Boise LH, Thompson CB. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Saoudi Y, Jablonski SA, Perard J, Yen TJ, Margolis RL. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol Biol Cell. 2005;16:1043–1055. doi: 10.1091/mbc.E04-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myohanen SK, Baylin SB, Herman JG. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nat Rev. Cancer. 2011;11:503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- Noble JR, Zhong ZH, Neumann AA, Melki JR, Clark SJ, Reddel RR. Alterations in the p16(INK4a) and p53 tumor suppressor genes of hTERT-immortalized human fibroblasts. Oncogene. 2004;23:3116–3121. doi: 10.1038/sj.onc.1207440. [DOI] [PubMed] [Google Scholar]
- Oegema K, Savoian MS, Mitchison TJ, Field CM. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto A, Demetrick DJ, Spillare EA, Hagiwara K, Hussain SP, Bennett WP, Forrester K, Gerwin B, Serrano M, Beach DH, et al. Mutations and altered expression of p16INK4 in human cancer. Proc Natl Acad Sci USA. 1994;91:11045–11049. doi: 10.1073/pnas.91.23.11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos A, Howell M, Fotedar R, Margolis RL. Glioblastoma motility occurs in the absence of actin polymer. Mol Biol Cell. 2011;22:2212–2220. doi: 10.1091/mbc.E10-10-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Rabinovitch PS, Reid BJ, Haggitt RC, Norwood TH, Rubin CE. Progression to cancer in Barrett's esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. International journal of cancer. J Int Cancer. 2012;130:1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles SJ, Adami GR. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene. 1998;16:1113–1123. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg AA. Chromosome markers and progression in bladder cancer. Cancer Res. 1977;37:2950–2956. [PubMed] [Google Scholar]
- Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–991. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Scrittori L, Skoufias DA, Hans F, Gerson V, Sassone-Corsi P, Dimitrov S, Margolis RL. A small C-terminal sequence of Aurora B is responsible for localization and function. Mol Biol Cell. 2005;16:292–305. doi: 10.1091/mbc.E04-06-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Galluzzi L, Vivet S, Joza N, Younes AB, Rello-Varona S, Castedo M, Kroemer G. p53 represses the polyploidization of primary mammary epithelial cells by activating apoptosis. Cell Cycle. 2009;8:1380–1385. doi: 10.4161/cc.8.9.8305. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Serres MP, Kossatz U, Chi Y, Roberts JM, Malek NP, Besson A. p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J Clin Invest. 2012;122:844–858. doi: 10.1172/JCI60376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, Yosef R, Pilpel N, Krizhanovsky V, Sharpe J, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155:1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint.”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T, Saretzki G, Ladhoff J, d'Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126:111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Hayflick L. Formation of anucleate and multinucleate cells in normal and SV 40 transformed WI-38 by cytochalasin B. Exp Cell Res. 1972;74:187–194. doi: 10.1016/0014-4827(72)90496-x. [DOI] [PubMed] [Google Scholar]
- Yang D, Welm A, Bishop JM. Cell division and cell survival in the absence of survivin. Proc Natl Acad Sci USA. 2004;101:15100–15105. doi: 10.1073/pnas.0406665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.