Binding to microtubules of Xenopus microtubule-associated protein 4 (XMAP4) negatively regulates dynein-dependent movement of membrane organelles and positively regulates kinesin-2–based movement. Phosphorylation reduces binding of XMAP4 to microtubules and therefore regulates the direction of microtubule-based transport.
Abstract
Microtubule (MT)-based transport of organelles driven by the opposing MT motors kinesins and dynein is tightly regulated in cells, but the underlying molecular mechanisms remain largely unknown. Here we tested the regulation of MT transport by the ubiquitous protein MAP4 using Xenopus melanophores as an experimental system. In these cells, pigment granules (melanosomes) move along MTs to the cell center (aggregation) or to the periphery (dispersion) by means of cytoplasmic dynein and kinesin-2, respectively. We found that aggregation signals induced phosphorylation of threonine residues in the MT-binding domain of the Xenopus MAP4 (XMAP4), thus decreasing binding of this protein to MTs. Overexpression of XMAP4 inhibited pigment aggregation by shortening dynein-dependent MT runs of melanosomes, whereas removal of XMAP4 from MTs reduced the length of kinesin-2–dependent runs and suppressed pigment dispersion. We hypothesize that binding of XMAP4 to MTs negatively regulates dynein-dependent movement of melanosomes and positively regulates kinesin-2–based movement. Phosphorylation during pigment aggregation reduces binding of XMAP4 to MTs, thus increasing dynein-dependent and decreasing kinesin-2–dependent motility of melanosomes, which stimulates their accumulation in the cell center, whereas dephosphorylation of XMAP4 during dispersion has an opposite effect.
INTRODUCTION
Intracellular transport is essential for the delivery of membrane-bound organelles, RNA granules, and chromosomes to specific cellular destinations and is critical for diverse biological processes such as mitosis, membrane trafficking, cell locomotion, and spatial organization of the cytoplasm (Lane and Allan, 1998; Caviston and Holzbaur, 2006; Akhmanova and Hammer, 2010; Walczak et al., 2010; Kapitein and Hoogenraad, 2011; Stehbens and Wittmann, 2012; Foley and Kapoor, 2013). Tracks for long-range intracellular transport are provided by cytoplasmic microtubules (MTs) organized into a polarized radial array with minus ends clustered in the cell center and plus ends pointing to the cell surface. The inherent polarity of MTs is recognized by MT motors bound to the surface of cargo organelles and cytoplasmic particles. These motors—kinesins and dynein—use the energy of ATP hydrolysis to move cargoes specifically to the plus or minus ends of MTs (Vale, 2003). Kinesins generally support transport to the MT plus ends (Goldstein, 2001; Hirokawa et al., 2009; Verhey et al., 2011), whereas dynein is exclusively minus-end directed (Hook and Vallee, 2006).
Opposing MT motors are usually bound to the surface of the same cargo organelles, which results in the discontinuous (“saltatory”) pattern of their movement, with stochastic transitions among three states: uninterrupted displacements to the minus or plus ends, and pauses (Gross, 2004; Welte, 2004; Jolly and Gelfand, 2011). The balance between the plus and minus end–directed runs, which determines the direction of MT transport, is tightly regulated in cells. This regulation can involve changes in the numbers or the activation state of kinesin and dynein motors associated with cargoes (Kamal and Goldstein, 2002; Karcher et al., 2002; Verhey and Hammond, 2009; Akhmanova and Hammer, 2010; Barlan et al., 2013b) or modification of MT transport tracks (Verhey and Hammond, 2009). The state of MT tracks is influenced by specific posttranslational modifications of tubulin subunits (Liao and Gundersen, 1998; Reed et al., 2006; Dunn et al., 2008; Cai et al., 2009; Konishi and Setou, 2009) or nonmotor microtubule-associated proteins (MAPs).
MAPs are a group of proteins that bind along the length of MTs, promote their assembly, and stabilize MTs from disassembly (Olmsted, 1986; Matus, 1988). Tau, microtubule-associated protein 2 (MAP2), and MAP4 are structurally related MAPs that have similar MT-binding domains but different projection domains that extend from the MT surface. These MAPs act as obstacles for MT motors in general and kinesin motors in particular and negatively influence their motility by decreasing the frequency of attachment to MTs and the average length of MT runs (Atherton et al., 2013). In contrast to tau/MAP2/MAP4, doublecortin and MAP7/ensconsin stimulate interaction of kinesin motors with MTs. Doublecortin and its homologue, doublecortin-like kinase 1, selectively enhance the affinity of the kinesin-3 Kif1A to MTs (Liu et al., 2012), and MAP7/ensconsin increases the frequency of productive encounters of the kinesin-1 motors with MTs (Sung et al., 2008; Metzger et al., 2012; Barlan et al., 2013a). The ability of MAPs to enhance or inhibit motility of specific kinesin motors makes them likely targets for signaling cascades that regulate the direction of MT transport. The recruitment of MAPs to MTs is controlled by phosphorylation, which negatively affects their ability to bind MTs (Lee, 1993; Avila et al., 1994; Mandell and Banker, 1996; Drewes et al., 1998; Cassimeris and Spittle, 2001). Therefore phosphorylation-dependent changes in affinity of MAPs to MTs would be expected to affect the amount of plus end–directed motility of kinesin cargoes and thus the overall direction of MT-based transport. However, whether MAPs regulate the direction of MT transport in cells has not been directly tested.
In this study, we examined the regulation of MT transport by MAPs using Xenopus melanophores as an experimental system. In these cells, thousands of membrane-bound pigment granules move along radial MTs to the cell center (pigment aggregation) or the periphery (pigment dispersion) by means of cytoplasmic dynein and kineisn-2, respectively (Nascimento et al., 2003; Kashina and Rodionov, 2005; Aspengren et al., 2009). Uniform distribution of pigment granules at later stages of dispersion involves movement along randomly arranged actin filaments (Rodionov et al., 1998; Rogers and Gelfand, 1998). The direction of transport is regulated by changes in the levels of a single second messenger cAMP, which alters the length of plus- and minus-end MT runs of pigment granules. An increase in cAMP levels induces an increase in the length of plus-end runs generated by kinesin-2 and a concurrent decrease in the length of minus-end runs produced by dynein and leads to pigment dispersion (Gross et al., 2002; Rodionov et al., 2003; Zaliapin et al., 2005). A reduction in cAMP levels triggers opposite changes in the length of granule runs and results in pigment aggregation (Gross et al., 2002; Rodionov et al., 2003; Zaliapin et al., 2005). cAMP levels affect the activity of protein kinase A (PKA) and other protein kinases that act downstream of PKA in the signaling cascades that regulate the motility of pigment granules (Rodionov et al., 2003; Deacon et al., 2005; Ikeda et al., 2011). We hypothesized that if MAPs regulate pigment granule transport, their phosphorylation levels should change in response to signals that trigger aggregation or dispersion of pigment granules. To test this hypothesis and identify MAPs involved in the regulation of pigment granule transport, we compared phosphoproteomic profiles of melanophores in the two signaling states.
We identified Xenopus MAP4 (XMAP4) as a protein whose phosphorylation levels significantly increased during pigment aggregation. We found that overexpression of XMAP4 did not affect dispersion of pigment granules but markedly reduced the rate of their aggregation, and this effect was explained by the shortening of MT minus-end runs. In a marked contrast to overexpression, removal of XMAP4 from MTs by microinjection of cells with a blocking antibody inhibited dispersion of pigment granules by shortening plus-end granule runs but did not affect their aggregation. Phosphomimetic mutant of XMAP4 had reduced abilities to bind MTs and inhibit aggregation of pigment granules. On the basis of these results, we propose a model for the regulation of MT-based transport of pigment granules in melanophores in which reversible binding of XMAP4 to MTs determines the direction of MT-based pigment granule movement.
RESULTS
XMAP4 is phosphorylated during pigment aggregation
To gain insight into the regulation of pigment transport in melanophores and understand the role of MAPs in this regulation, we compared the phosphoproteomic profiles of cells stimulated to aggregate or disperse pigment granules. Phosphopeptides in unfractionated lysates of melanophores were enriched on iron immobilized metal ion affinity chromatography or with TiO2 resin. We identified >5000 unique phosphopeptides whose abundance increased in response to aggregation or dispersion signals. These peptides were derived from 2045 different proteins. Quantitative analysis of the phosphoproteomic data revealed 62 proteins whose phosphorylation levels changed in response to aggregation or dispersion stimuli more than fourfold. Among them were seven cytoskeleton-related proteins and only one structural MAP, XMAP4, whose phosphorylation increased during pigment aggregation. We cloned XMAP4 by PCR using cDNA synthesized from total RNA isolated from Xenopus melanophores as a template and a pair of primers specific to the published nucleotide sequence of XMAP4 from Xenopus oocytes. The amino acid sequence of the melanophore-specific XMAP4 was identical to the sequence of XMAP4 from oocytes, except for a deletion of 57 amino acid residues at the C-terminus and insertion of 10 amino acid residues in the middle of the molecule. We identified the amino acid residues phosphorylated during pigment aggregations as Thr-758 and Thr-762 located in the proline-rich region of the MT-binding domain (Figure 1). Phosphorylation of XMAP4 at Thr-758 and Thr-762 in melanophores stimulated to aggregate pigment increased more than fivefold compared with cells with dispersed pigment granules. Previous work showed that these threonines were targets of p34cdc2 and MAP kinases known to reduce the ability of mammalian MAP4 to bind MTs in HeLa cells (Ookata et al., 1995, 1997; Shiina and Tsukita, 1999; Kitazawa et al., 2000; Chang et al., 2001). Taken together, these results indicate that XMAP4 is phosphorylated in melanophores in response to pigment aggregation signals and suggest that this phosphorylation reduces the affinity of XMAP4 to MTs.
FIGURE 1:
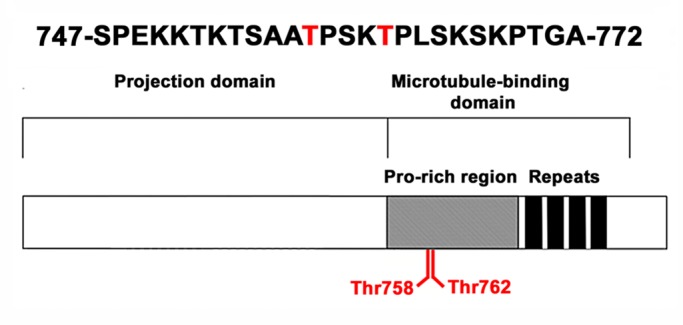
XMAP4 is phosphorylated at threonine residues located in the proline-rich region of the MT-binding domain. Top, amino acid sequence of the XMAP4 phosphopeptide identified in the phosphoproteomic screen; Thr-758 and Thr-762 are shown in red; bottom, domain organization of the XMAP4 molecule; the molecule of XMAP4 is composed of an N-terminal projection domain, which protrudes from the MT surface, and a C-terminal MT-binding domain that contains a proline-rich region and four imperfect amino acid repeats (Aizawa et al., 1991; Chapin and Bulinski, 1991); Thr-758 and Thr-762 map to the proline-rich region of the MT-binding domain.
Overexpression of XMAP4 slows aggregation of pigment granules
Phosphorylation during pigment aggregation suggested that XMAP4 was directly or indirectly involved in the regulation of pigment granule transport. To understand the role of XMAP4 in transport of pigment granules, we examined the effect of XMAP4 overexpression on pigment aggregation and dispersion. XMAP4 was expressed in melanophores as an enhanced green fluorescent protein (EGFP) fusion to identify the overexpressing cells. Fluorescence microscopy indicated that, as expected, EGFP-XMAP4 bound cytoplasmic MTs (Supplemental Figure S1). To measure the effects of EGFP-XMAP4 overexpression on pigment granule transport, we stimulated pigment aggregation or dispersion and quantified the fractions of cells with aggregated, partially responded, or dispersed pigment granules 15 min after stimulation. Nontransfected melanophores or cells expressing EGFP were used as controls. We found that overexpression of EGFP-XMAP4 did not affect significantly pigment granule dispersion (Figure 2A) but dramatically inhibited their aggregation, as evidenced from the approximately eightfold and approximately twofold increases in the fractions of cells whose pigment granules remained completely and partially dispersed, respectively, as compared with control nontransfected or EGFP-expressing cells (Figure 2B). However, 1 h after application of pigment aggregation stimuli, the fractions of cells with aggregated or partially responded pigment granules approached control levels (Figure 2C), indicating that overexpression of EGFP-XMAP4 did not block pigment aggregation completely but instead slowed accumulation of pigment granules in the cell center. We conclude that overexpression of EGFP-XMAP4 significantly reduces the rate of pigment granule aggregation.
FIGURE 2:
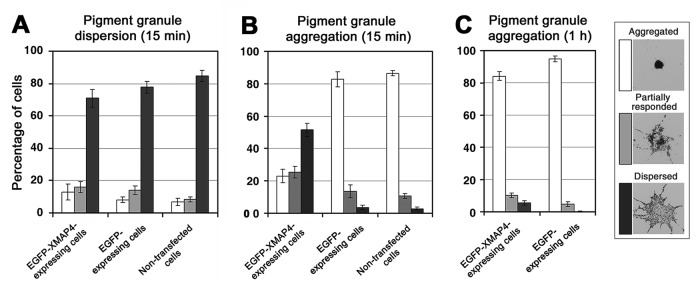
Expression of XMAP4 reduces the rate of pigment aggregation. Quantification of response to dispersion (A) or aggregation (B, C) stimuli applied for 15 min (A, B) or 1 h (C) to melanophores expressing EGFP-XMAP4 (left set of bars), EGFP (middle sets of bars in A and B, right set in C), or nontransfected melanophores (right sets of bars in A and B); data are expressed as the percentages of cells with aggregated (white bars), partially responded (gray bars), or dispersed (black bars) pigment granules. Expression of EGFP-XMAP4 does not affect dispersion but significantly slows aggregation of pigment granules, as evidenced by the increase in the fractions of cells with dispersed or partially responded pigment granules 15 min after the stimulation of pigment aggregation.
Inhibition of pigment granule aggregation in cells overexpressing EGFP-XMAP4 is not explained by reduced rate of transfer of pigment granules from actin filaments onto MTs
Our data showed that overexpression of EGFP-XMAP4 dramatically delayed aggregation of pigment granules. Pigment aggregation includes two independent steps—the transfer of pigment granules from actin filaments onto MTs, and subsequent transport along MTs to the cell center. Either step could be inhibited by the overexpression of EGFP-XMAP4. Our previous work showed that the transfer step involves capture of pigment granules by growing MT tips and that the +TIP protein CLIP170 is essential for this process (Lomakin et al., 2009, 2011). It was therefore possible that EGFP-XMAP4 inhibited pigment granule capture by suppressing MT dynamics or preventing accumulation of CLIP-170 at the MT plus ends. In support of this possibility, overexpression of MAP4 in mammalian cells stabilized cytoplasmic MTs (Nguyen et al., 1997; Holmfeldt et al., 2002), and MAP1b, a MAP structurally related to MAP4, reduced the levels of MT-associated EB1, an adaptor protein that facilitates binding of CLIP-170 to MT tips (Tortosa et al., 2013). We therefore sought to determine whether overexpression of EGFP-XMAP4 inhibited pigment granule aggregation by suppressing MT dynamics or reducing binding of CLIP-170 to the plus ends of MTs.
To examine the effect of EGFP-XMAP4 overexpression on MT dynamics, we compared parameters of MT dynamic instability in melanophores induced to aggregate pigment granules overexpressing EGFP-XMAP4 and EGFP by injecting Cy3-tubulin into EGFP-positive cells, tracking tips of growing and shortening MTs, and decomposing tip trajectories into periods of growth, shortening, and pauses. We detected small but significant differences in the parameters of MT dynamic instability between the EGFP-XMAP4- and EGFP-expressing cells (Table 1). In particular, the average lengths of growth and shortening episodes were slightly reduced and the average duration of pauses increased in the melanophores overexpressing EGFP-XMAP4 (Table 1). To determine whether these changes affected the rate of pigment granule aggregation, we used a stochastic computational model for pigment aggregation that we previously developed (Lomakin et al., 2009, 2011). The model computes kinetics of pigment granule aggregation based on the statistics of bidirectional movement of pigment granules, the probability of pigment granule capture by growing MT ends, and parameters of MT dynamic instability and allows one to estimate independent effects of changes in MT dynamics on the pigment aggregation rate (Zaliapin et al., 2005; Lomakin et al., 2009, 2011). The output of the model is the kinetics of gray level decrease, which reflects how fast the cytoplasm becomes increasingly transparent as pigment granules accumulate in the cell center. We first tested the model by performing experimental measurements of gray level decrease in cells expressing EGFP and comparing experimental data with the results of simulations that used the parameters of MT dynamic instability and pigment granule movement measured in these cells (Tables 1 and 2). This comparison showed a close match between the experimental and computed kinetic curves (Supplemental Figure S2). These results, in agreement with our previous analyses (Zaliapin et al., 2005; Lomakin et al., 2009, 2011), demonstrated that the computational model closely reproduced the kinetics of pigment aggregation. We next simulated aggregation of pigment granules using parameters of MT dynamic instability measured in melanophores overexpressing EGFP-XMAP4 (Table 1) without changing the granule movement statistics. These computer simulations generated kinetics of gray level decrease (Figure 3A, open squares) that was close to the kinetics measured for the EGFP-expressing cells (Figure 3A, open circles), with a half-time 4.36 ± 0.19 min. The experimentally measured half-time of gray level decrease estimated for melanophores overexpressing EGFP-XMAP4 (Figure 3A, black squares) was 27.78 ± 4.77 min, much longer than the half-time computed on the basis of dynamic instability parameters. Therefore changes in MT dynamics could not explain the dramatic inhibition of pigment aggregation observed in the EGFP-XMAP4–overexpressing cells.
TABLE 1:
Parameters of MT dynamic instability in melanophores with aggregated pigment expressing EGFP-XMAP4 or EGFP.
| Dynamic parameter | EGFP-XMAP4–expressing cells | EGFP-expressing cells |
|---|---|---|
| Growth length (μm) | 1.06 ± 0.09* | 1.41 ± 0.10* |
| Growth rate (μm/s) | 0.09 ± 0.01 | 0.11 ± 0.01 |
| Shortening length (μm) | 0.73 ± 0.09* | 1.27 ± 0.11* |
| Shortening rate (μm/s) | 0.09 ± 0.01* | 0.13 ± 0.01* |
| Catastrophe frequency (s−1) | 0.048 ± 0.001* | 0.041 ± 0.001* |
| Rescue frequency (s−1) | 0.053 ± 0.001* | 0.047 ± 0.001* |
| Pause duration (s) | 5.19 ± 0.31* | 3.96 ± 0.22* |
| Number of analyzed MTs | 40 | 41 |
| Number of analyzed cells | 20 | 14 |
Values are marked with an asterisk if the difference of the mean parameter value between EGFP-XMAP4– and EGFP-expressing cells is significant at the 0.05 level, according to a one-way analysis of variance test.
TABLE 2:
Parameters of MT-based movement of pigment granules during pigment aggregation in melanophores expressing EGFP-XMAP4 or EGFP.
| Movement parameter | EGFP-XMAP4–expressing cells | EGFP-expressing cells |
|---|---|---|
| Velocity of minus-end runs (nm/s) | 401.5 ± 7.5 | 386.6 ± 8.5 |
| Length of minus-end runs (nm) | 117.1 ± 4.5* | 161.9 ± 8.6* |
| Velocity of plus-end runs (nm/s) | 355.5 ± 9.2 | 337.9 ± 10.1 |
| Length of plus-end runs (nm) | 36.8 ± 0.7* | 39.5 ± 1.2* |
| Duration of pauses (s) | 0.69 ± 0.03 | 0.66 ± 0.03 |
| Number of analyzed pigment granules | 121 | 80 |
Values are marked with an asterisk if the difference of the mean parameter value between EGFP-XMAP4– and EGFP-expressing cells is significant at the 0.05 level, according to a one-way analysis of variance test.
FIGURE 3:
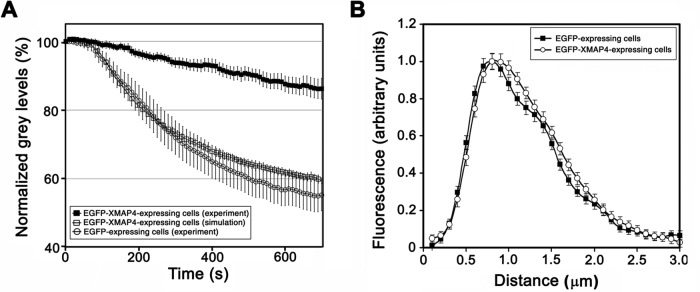
Inhibition of pigment aggregation in melanophores overexpressing XMAP4 cannot be explained by changes in MT dynamics or loss of CLIP-170 from the MT tips. (A) Comparison of kinetics of pigment aggregation measured in cells expressing EGFP-XMAP4 (black squares) or computed based on parameters of MT dynamic instability measured in EGFP-expressing (white circles) or EGFP-XMAP4–expressing cells (white squares); changes in parameters of MT dynamic instability caused by the overexpression of EGFP-XMAP4 cannot account for inhibition of pigment aggregation in the EGFP-XMAP4–expressing cells. (B) Profiles of CLIP-170 fluorescence at MT plus ends normalized by maximum fluorescence and averaged for melanophores expressing EGFP (black squares) or EGFP-XMAP4 (white circles); expression of EGFP-XMAP4 does not significantly change the distribution of the CLIP-170 fluorescence at the MT plus ends.
To determine whether EGFP-XMAP4 displaced CLIP-170 from MT plus ends, we immunostained melanophores overexpressing EGFP or EGFP-XMAP4 with a CLIP-170 antibody and used images of the immunostained cells to generate normalized profiles of the CLIP-170 fluorescence at MT plus ends (Figure 3B). Quantitative analysis of the fluorescence profiles showed that the average length of MT segments decorated with CLIP-170 was similar between the EGFP- and EGFP-XMAP4–overexpressing melanophores (0.67 ± 0.05 and 0.68 ± 0.06 μm, respectively). Absolute values of CLIP-170 fluorescence per MT end were also similar between melanophores expressing EGFP and EGFP-XMAP4 (331 ± 13 and 428 ± 15 arbitrary units, respectively). Thus overexpression of EGFP-XMAP4 did not remove CLIP-170 from the MT plus ends. Taken together, the results of these experiments indicate that inhibition of pigment aggregation in melanophores overexpressing EGFP-XMAP4 is not explained by the reduced transfer of pigment granules from actin filaments onto MTs during pigment aggregation.
Overexpression of EGFP-XMAP4 reduces the length of minus-end MT runs of pigment granules during pigment aggregation
Our data indicated that overexpression of EGFP-XMAP4 did not affect capture of pigment granules by growing MT tips during pigment aggregation. During pigment aggregation, granules captured by MTs run to the MT minus ends focused in the cell center. We therefore tested whether overexpression of EGFP-XMAP4 affected minus end–directed granule motility by tracking individual pigment granules and decomposing motion trajectories into plus- and minus-end runs and pauses. For pigment granule tracking, we used melanophores expressing EGFP-XMAP4 at low levels, which aggregated pigment granules at a slightly reduced rate. This partial inhibition of pigment aggregation allowed us to estimate parameters of bidirectional granule movement. Control experiments involved tracking of pigment granules in melanophores expressing EGFP. Comparison of pigment granule movement parameters between the EGFP- and EGFP-XMAP4–expressing cells showed that EGFP-XMAP4 expression did not change the average velocity or length of plus-end granule runs (Table 2). However, expression of EGFP-XMAP4 reduced the average length of minus-end runs to 72% of the control levels, without affecting their velocity (Table 2). For comparison with pigment aggregation, we also performed tracking of pigment granules in melanophores expressing EGFP-XMAP4 or GFP shortly after the stimulation of pigment granule dispersion. Our previous work indicates that at early stages of pigment dispersion, granules move exclusively along the radial MT tracks (Zaliapin et al., 2005; Slepchenko et al., 2007). We did not find any significant differences in minus-end run length or other granule movement statistics between populations of melanophores expressing EGFP-XMAP4 and EGFP (Supplemental Table S1). We conclude that expression of EGFP-XMAP4 does not significantly affect parameters of pigment granule movement during their dispersion and inhibits pigment aggregation by reducing the length of minus-end runs of pigment granules.
Removal of XMAP4 from MTs selectively inhibits dispersion of pigment granules
To further understand the role of XMAP4 in pigment transport, we next tested whether removal of XMAP4 from MTs affected aggregation or dispersion of pigment granules. To remove XMAP4 from MTs, we produced antibodies against the XMAP4 microtubule-binding domain (MBD). Such antibodies have been shown to block binding of mammalian MAP4 to MTs in vivo and in vitro (Wang et al., 1996). We performed several control experiments to confirm that our antibodies against XMAP4 MBD were specific and had the predicted blocking effect.
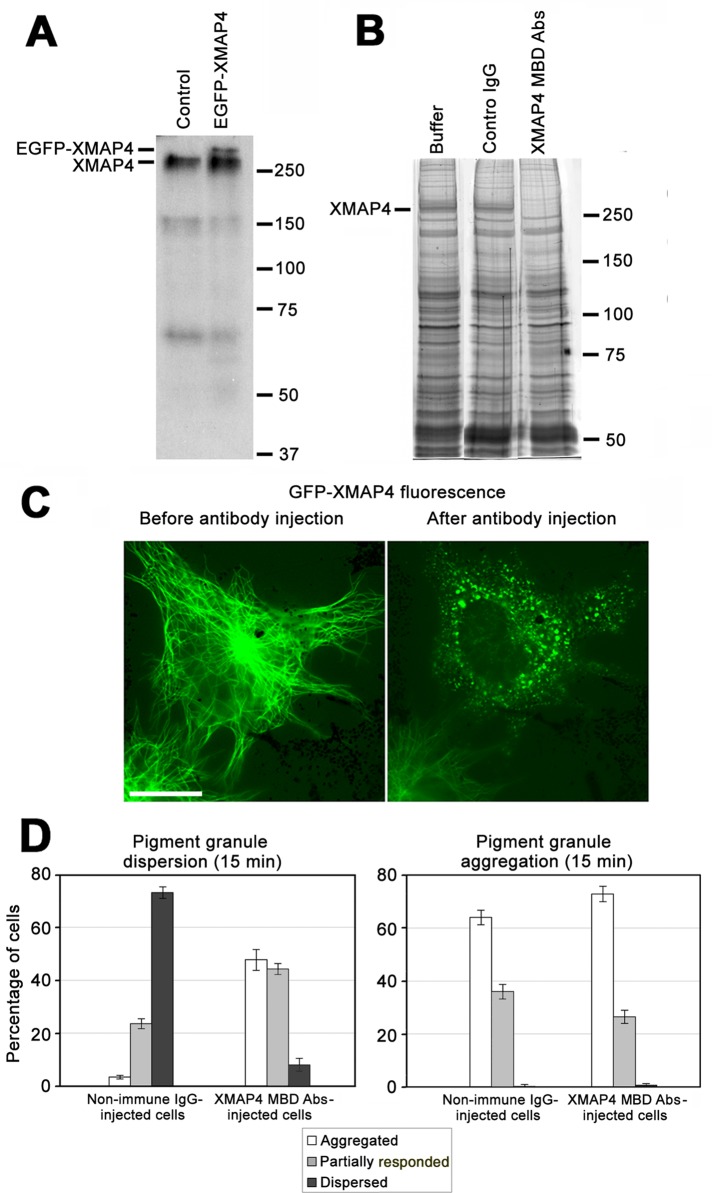
To examine whether MBD antibodies were specific for XMAP4, we performed immunoblotting with whole-cell extracts. In extracts of control, nontransfected melanophores the antibodies recognized a major band with apparent molecular weight ∼250 kDa (Figure 4A, left). The additional lower–molecular weight bands bound to MBD antibodies (Figure 4A) likely represented degradation products of XMAP4, since the corresponding proteins cosedimented with MTs assembled in cell extracts (unpublished data). In extracts of melanophores overexpressing EGFP-XMAP4, MBD antibodies recognized an additional band with electrophoretic mobility characteristic of the EGFP-XMAP4 (Figure 4A, right). The apparent molecular weights of the immunoreactive proteins were higher than the values predicted for the XMAP4 or GFP-XMAP4 based on the amino acid sequence. However, previous studies showed that the mobility of mammalian MAP4 on SDS gels was also unusually slow (Aizawa et al., 1990; West et al., 1991; Chapin et al., 1995). Therefore immunoblotting experiments showed that MBD antibodies were specific for the XMAP4.
FIGURE 4:
Displacement of XMAP4 from MTs by injection of MBD antibodies inhibits dispersion but not aggregation of pigment granules. (A) Immunoblotting with MBD antibodies of whole-cell extracts of control nontransfected cells (left) or melanophores overexpressing EGFP-XMAP4 (right); MBD antibodies recognize the XMAP4 band in whole-cell extracts of control cells and an additional EGFP-XMAP4 band in whole-cell extracts of EGFP-XMAP4–overexpressing cells. (B) Coomassie-stained SDS gels of pelleted MTs assembled in whole-cell extracts preincubated without added IgG (left), in the presence of control nonimmune IgG (middle), or antibodies against XMAP4 MBD (right); preincubation of whole-cell extracts with MBD antibodies prevents cosedimentation of XMAP4 but not other MAPs with MTs. (C) Live images of a melanophore expressing EGFP-XMAP4 before (left) and 30 min after (right) injection of antibodies against XMAP4 MBD; scale bar, 20 μm; the antibody injection completely removes XMAP4 from the MTs. (D) Quantification of response to dispersion (left) or aggregation (right) stimuli of melanophores microinjected with nonimmune IgG or antibodies against XMAP4 MBD. Microinjection of MBD antibodies does not significantly affect pigment aggregation but markedly inhibits pigment dispersion, as evidenced by increases in the fractions of cells with aggregated or partially responded pigment granules compared with melanophores microinjected with nonimmune IgG.
To test whether antibodies against MBD blocked binding of XMAP4 to MTs in vitro, we assembled MTs in cell extracts in the presence or absence of control nonimmune immunoglobulin G (IgG) or MBD antibodies, pelleted MTs through a glycerol cushion to separate soluble and MT-bound MAPs, and examined the composition of MAPs in the MT pellets. MTs assembled in cell extracts bound numerous MAPs (Figure 4B, left), and preincubation of extracts with nonimmune IgG did not change composition of MAPs in the MT pellets. However, preincubation with MBD antibodies removed from MT pellets a single band with electrophoretic mobility characteristic of XMAP4 (Figure 4B, right). Western blots confirmed that XMAP4 was absent from the pellets of MTs assembled in the presence MBD antibodies but not control nonimmune IgG (Supplemental Figure S3). Therefore MBD antibodies completely and selectively blocked the binding of XMAP4 to MTs in vitro.
To determine whether MBD antibodies removed XMAP4 from MTs in cells, we transfected melanophores with EGFP-XMAP4 and microinjected EGFP-positive cells with MBD antibodies or control nonimmune IgG. We found that, as expected, EGFP-XMAP4 was bound along the length MTs (Figure 4C, left) and that microinjection of antibodies against MBD caused rapid (20 min) and complete loss of MT fluorescence and redistribution of the EGFP signal into bright spots randomly distributed throughout the cytoplasm (Figure 4C, right). These spots likely represented EGFP-XMAP4–MBD antibody complexes. Microinjection of control nonimmune IgG did not affect MT localization of EGFP-XMAP4 (unpublished data), which indicated that the blocking effect of the MBD antibodies was highly specific. Immunofluorescence staining with anti-tubulin antibody showed that the MT network appeared normal in the MBD antibody–injected cells (Supplemental Figure S4). Furthermore, antibody microinjection did not significantly affect parameters of MT dynamic instability. The lengths of growth and shortening episodes were slightly reduced in antibody-injected cells compared with melanophores injected with nonimmune IgG (Supplemental Table S2). However, growth and shortening of MTs were affected to about the same extent, and therefore these changes in MT dynamics could not cause significant reduction in MT density. These results are in line with published data that showed that depleting MAP4 from MTs had no detectable effect on the distribution or dynamics of MTs in human fibroblasts and monkey kidney epithelial cells (Wang et al., 1996). Therefore microinjection of antibodies against MBD into melanophores abolished binding of XMAP4 to MTs without changing MT organization or dynamics.
We next tested whether removal of XMAP4 from MTs affected pigment granule transport. We injected melanophores with MBD antibodies or nonimmune IgG, stimulated pigment aggregation or dispersion, and estimated the fractions of cells with aggregated, partially responded, or dispersed pigment granules. Microinjection of antibodies against MBD significantly inhibited pigment dispersion, as evidenced by the approximately ninefold increase in the fraction of cells whose response to dispersion signal was completely inhibited and a twofold increase in the fraction of cells that showed partial inhibition compared with melanophores microinjected with nonimmune IgG (Figure 4D, left). Aggregation of pigment granules was not significantly affected, given that the fractions of cells with completely or partially aggregated pigment granules were similar between populations of melanophores microinjected with MBD antibodies or nonimmune IgG (Figure 4D, right). Furthermore, microinjection of MBD antibodies did not significantly change parameters of bidirectional granule movement during pigment aggregation (Supplemental Table S3). Thus removal of XMAP4 from MTs selectively inhibited dispersion of pigment granules.
Dispersion of pigment granules in XMAP4 antibody–injected cells could be inhibited through decrease in the length or velocity of plus-end granule runs, increase in the length or velocity of minus-end runs, or both. To determine how depletion of XMAP4 from MTs affected parameters of bidirectional granule movement, we microinjected cells with MBD antibodies or control nonimmune IgG and recorded and tracked individual pigment granules shortly after the application of a pigment dispersion stimulus. To allow for the tracking of pigment granules, we decreased the amounts of MBD antibodies and nonimmune IgG delivered into the cytoplasm during microinjection by reducing microinjection time by ∼50%. Reduced cytoplasmic levels of MBD antibodies slowed pigment dispersion but did not block it completely, as evidenced by the increased fraction of cells with partially dispersed pigment granules compared with melanophores injected with control nonimmune IgG (Supplemental Figure S5). Partial inhibition of pigment granule dispersion made possible measurements of the velocity and length of bidirectional granule runs. Comparison of granule movement statistics between cells microinjected with MBD antibodies and control IgG showed that MBD antibodies reduced pigment granule runs in both directions and that the average length of runs was affected more significantly than run velocity, reaching ∼70% of the control values (Table 3). Further, the average length of plus- and minus-end runs was reduced to about the same extent, but the net effect was more prominent in the case of longer plus-end runs, which shortened by ∼30 nm (Table 3). We conclude that XMAP4 positively regulates motility of pigment granules along MTs in both directions and stimulates pigment dispersion by increasing the length of plus-end granule runs.
TABLE 3:
Parameters of MT-based movement of pigment granules during pigment dispersion in melanophores microinjected with antibody against XMAP4 MT-binding domain or control IgG.
| Movement parameter | XMAP4 antibody–injected cells | Control IgG-injected cells |
|---|---|---|
| Velocity of minus-end runs (nm/s) | 250.3 ± 9.9 | 272.0 ± 10.1 |
| Length of minus-end runs (nm) | 53.8 ± 1.5* | 74.4 ± 4.1* |
| Velocity of plus-end runs (nm) | 250.7 ± 7.5 | 271.2 ± 7.6 |
| Length of plus-end runs (nm) | 111.3 ± 4.1* | 141.5 ± 5.5* |
| Duration of pauses (s) | 1.1 ± 0.1 | 1.1 ± 0.1 |
| Number of analyzed pigment granules | 98 | 116 |
Values are marked with an asterisk if the difference of the mean parameter value between XMAP4 antibody and IgG-injected cells is significant at the 0.05 level, according to a one-way analysis of variance test.
XMAP4 interacts with the p150Glued subunit of the dynactin complex but does not bind subunits of MT motor proteins
Our data showed that overexpression of EGFP-XMAP4 dramatically reduced the rate of pigment aggregation by shortening the length of minus-end runs of pigment granules, whereas removal of EGFP-XMAP4 from MTs suppressed pigment dispersion mainly by reducing the length of plus-end granule runs. Given that minus-end runs of pigment granules are generated by dynein (Nilsson and Wallin, 1997) and plus-end runs by kinesin-2 (Tuma et al., 1998), these results indicated that XMAP4 bound to MTs negatively regulated dynein motility and positively regulated motility of kinesin-2. MAPs are known to block travel of motor proteins, including dynein (Lopez and Sheetz, 1993; Vershinin et al., 2007, 2008; Dixit et al., 2008; Markus et al., 2012), along MTs, and therefore the negative effect of XMAP4 overexpression on dynein motility is likely explained by steric interference with the movement of dynein motors by EGFP-XMAP4 molecules bound to the MT surface. However, tethering of dynein to MTs mediated by XMAP4 could enhance an inhibitory effect. On the contrary, positive regulation of kinesin-2 suggests direct or indirect interaction of this motor protein with XMAP4. In support of a role for MAP–motor interaction in stimulation of kinesin motility, docking the motor domain of kinesin-3 Kif1A to MTs by doublecortin was shown to enhance transport of Kif1A cargoes in neurons (Liu et al., 2012). Therefore, to further understand the mechanisms of regulation of MT motor proteins by XMAP4, we sought to determine whether XMAP4 interacts with kinesin-2 or dynein or the dynactin complex that mediates their binding to pigment granules.
To investigate whether XMAP4 interacts with MT motor proteins or the dynactin complex, we immunoprecipitated XMAP4 from whole-cell extracts and probed immunoprecipitates with antibodies specific for kinesin-2, dynein, or dynactin subunits. To avoid competition of XMAP4 binding partners with antibodies during immunoprecipitation, we expressed EGFP-XMAP4 in melanophores and used rabbit antibodies specific for the EGFP tag for immunoprecipitation. To confirm the presence of EGFP-XMAP4 in the immunoprecipitates, we probed them with mouse monoclonal EGFP antibodies. We found that, as expected, immunoprecipitates contained a protein band with electrophoretic mobility consistent with the mobility of EGFP-XMAP4 (Figure 5A). We also found, however, that antibodies against Kif3A subunit of kinesin-2 or cytoplasmic dynein IC, which recognized relevant bands in whole-cell extracts, did not react with immunoprecipitates (Figure 5A), suggesting that neither kinesin-2 nor dynein bound XMAP4 with high affinity in whole-cell extracts. In contrast to antibodies specific for MT motor proteins, antibody raised against the 150Glued subunit of the dynactin complex revealed in immunoprecipitates a protein with appropriate electrophoretic mobility (Figure 5A). To examine whether endogenous MAP4 also bound p150Glued and to exclude the possibility that the presence of p150Glued in EGFP antibody immunoprecipitates could be explained by nonspecific binding of p150Glued to EGFP, we pulled down XMAP4 from extracts of nontransfected cells using the XMAP4 MBD antibodies. Once again, we detected the p150Glued band in the immunoprecipitate obtained with MBD antibodies (Figure 5B). We conclude that XMAP4 binds the p150Glued subunit of the dynactin complex but not subunits of MT motor proteins.
FIGURE 5:
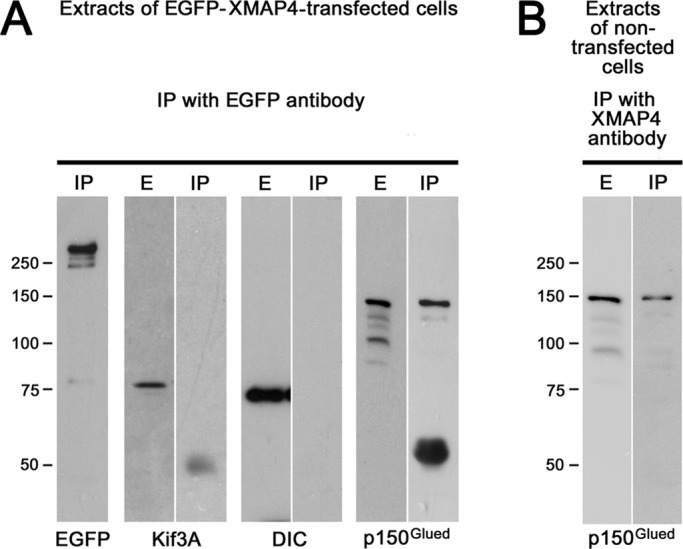
XMAP4 binds dynactin but not MT motor proteins. (A) Whole-cell extracts or immunoprecipitates of EGFP-XMAP4 with rabbit polyclonal antibodies against EGFP probed with mouse monoclonal antibodies specific for EGFP (EGFP), Kif3A subunit of kinesin-2 (Kif3A), dynein IC (DIC), or the p150Glued subunit of the dynactin complex (p150Glued); p150Glued but not kinesin-2 or dynein coimmunoprecipitates with EGFP-XMAP4. (B) Immunoprecipitate of the endogenous XMAP4 with antibodies against XMAP4 MBD probed with an antibody against p150Glued; p150Glued coimmunoprecipitates with endogenous XMAP4. E, whole-cell extract; IP, immunoprecipitate.
Phosphorylation decreases affinity of XMAP4 for MTs and reduces inhibition of pigment granule aggregation
Our phosphoproteomic profiling indicated that aggregation signals significantly increased phosphorylation of Thr-758 and Thr-763 located in the MT-binding domain of the XMAP4 molecule (Figure 1). It is known that phosphorylation of the MT-binding domain inhibits binding of mammalian MAP4 to MTs (Illenberger et al., 1996; Kitazawa et al., 2000; Chang et al., 2001). Furthermore, threonine residues in the mammalian MAP4 that correspond to the XMAP4 Thr-758 and Thr-762 are phosphorylated by p34cdc2 and MAP protein kinases during the transition from interphase to mitosis, and this phosphorylation correlates with a decrease in the ability of MAP4 to bind cytoplasmic MTs (Ookata et al., 1995, 1997; Shiina and Tsukita, 1999). We hypothesized that phosphorylation of XMAP4 at Thr-758 and Thr-762 reduces the binding of XMAP4 to MTs, removing physical barriers in the path of dynein motors moving pigment granules to the cell center, thus accelerating pigment aggregation.
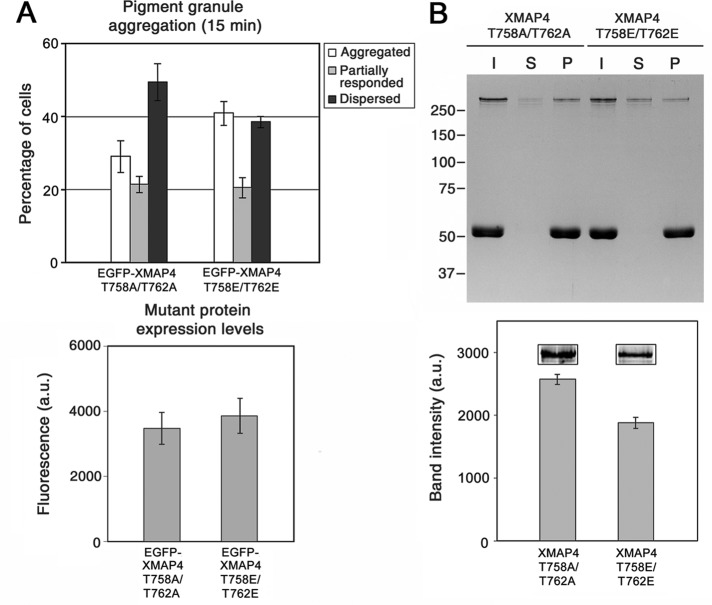
To determine whether phosphorylation of XMAP4 at Thr-758 and Thr-762 affects aggregation of pigment granules, we compared the response to aggregation signal of melanophores overexpressing the EGFP-tagged Thr-758E/Thr-756E phosphomimetic or Thr-758A/Thr-762A nonphosphorylatable XMAP4 mutants. Measurement of pigment aggregation responses showed that the phosphomimetic XMAP4 mutant had a reduced inhibitory effect on pigment aggregation compared with the nonphosphorylatable mutant, as evidenced by the statistically significant (p = 0.03) increase in the fraction of cells with aggregated pigment granules from 29 to 41%, concomitant with a decrease in the fraction of melanophores with dispersed pigment (Figure 6A). This effect could not be explained by a difference in the expression levels of mutant proteins (Figure 6B). As expected, overexpression of phosphomimetic or nonphosphorylatable XMAP4 mutants did not significantly affect pigment dispersion (Supplemental Figure S6). Therefore phosphorylation at Thr-758 and Thr-756 reduced the inhibitory effect of XMAP4 on pigment aggregation.
FIGURE 6:
Phosphorylation of XMAP4 reduces inhibition of pigment aggregation and decreases binding to MTs. (A) Top, quantification of the response to a pigment aggregation stimulus of melanophores overexpressing nonphosphorylatable (left set of bars) or phosphomimetic (right set of bars) mutants or XMAP4; data are expressed as the percentages of cells with aggregated (white bars), partially responded (gray bars), or dispersed (black bars) pigment granules. Bottom, comparison of the levels of expression of XMAP4 mutants based on the EGFP fluorescence. Overexpression of the phosphomimetic XMAP4 mutant has a weaker inhibitory effect compared with the nonphosphorylatable mutant, as evidenced by a smaller fraction of cells with dispersed and larger fraction of cells with aggregated pigment granules compared with melanophores overexpressing the XMAP4 nonphosphorylatable mutant; this difference cannot be explained by the reduced expression levels of the phosphomimetic mutant, given that the mutants are expressed at approximately the same levels. (B) Cosedimentation of recombinant nonphosphorylatable and phosphomimetic XMAP4 mutant proteins with MTs in vitro. Top, Coomassie-stained gel, which shows protein composition of input (XMAP4 mutant protein–MT mixtures before centrifugation; I), supernatants (S), and MT pellets (P). Bottom, measurement of relative amounts of nonphosphorylatable (left) and phosphomimetic (right) mutant proteins in MT pellets using quantitative immunoblotting; the data represent averages for the values measured in three independent experiments; inset, representative images of the XMAP4 mutant bands. The amount of the phosphomimetic mutant protein in the MT pellets is significantly smaller, and in the supernatants larger, than the nonphosphorylatable mutant, which indicates reduced binding to MTs.
To test whether phosphorylation at Thr-758 and Thr-756 affected binding of XMAP4 to MTs, we expressed hexahistidine (6xHis)-tagged phosphomimetic and nonphosphorylatable mutants in baculovirus, purified recombinant proteins by chromatography on Ni-nitrilotriacetic acid (NTA) agarose (Supplemental Figure S7), and compared their ability to cosediment with MTs in vitro.
For the MT cosedimentation, we combined purified XMAP4 phosphomimetic or nonphosphorylatable mutant proteins with paclitaxel-stabilized MTs to attain a final concentration equimolar to tubulin and pelleted MTs by high-speed centrifugation. We found that the Thr-758E/Thr-756E band was reduced in the MT pellets and increased in the supernatants compared with the Thr-758A/Thr-756A band (Figure 6B, top). Measurement the amounts of XMAP4 mutant proteins in the MT pellets using quantitative immunoblotting confirmed that intensity of the Thr-758E/Thr-756E band was on average ∼1.4-fold lower than that of the Thr-758A/Thr-756A band (Figure 6B). Therefore binding to MTs of the phosphomimetic mutant protein compared with the nonphosphorylatable mutant protein was significantly decreased. We conclude that phosphorylation at Thr-758 and Thr-756 reduces binding of XMAP4 to MTs and partially relieves inhibition of pigment aggregation in the overexpressing cells.
Our phosphoproteomic profiling indicated that phosphorylation of XMAP4 is increased during pigment granule aggregation, and the results of MT cosedimentation experiments showed that this phosphorylation reduced binding of XMAP4 to MTs. Therefore the levels of endogenous MT-bound XMAP4 should decrease during pigment aggregation. We therefore compared the levels of MT-bound XMAP4 in melanophores stimulated to aggregate or disperse pigment granules by measuring XMAP4 fluorescence after immunostaining with antibodies against XMAP4 MBD. To reduce background fluorescence, we extracted cells with 0.5% Triton X-100 before fixation. We found that the endogenous XMAP4 was associated with MTs in both signaling states. However, XMAP4 MT fluorescence in cells stimulated to disperse pigment granules appeared brighter than in melanophores induced to disperse pigment granules (Supplemental Figure S8). Averaging measurements of XMAP4 MT fluorescence in 20 cells in each signaling state confirmed that the brightness of MTs was higher in cells treated to disperse than aggregate pigment granules (60.9 ± 1.39 vs. 45.6 ± 1.44 arbitrary units, respectively; mean ± SEM; n = 100). This result was highly statistically significant (p = 1.4 × 10−12). We conclude that pigment aggregation signals that induce XMAP4 phosphorylation partially remove XMAP4 from MTs.
DISCUSSION
In this study, we identified XMAP4 as an important regulator of MT-based transport of pigment granules in Xenopus melanophores. This conclusion is based on several lines of experimental evidence. First, overexpression of XMAP4 significantly slowed pigment aggregation, without affecting pigment dispersion. This effect could be explained by a reduced length of minus-end MT runs of pigment granules in overexpressing cells. Second, removal of XMAP4 from MTs did not influence pigment aggregation but suppressed pigment dispersion by reducing the length of plus-end granule runs. Finally, binding of XMAP4 to MTs and the inhibition of pigment granule aggregation by overexpression of XMAP4 were reduced by using phosphomimetic mutations. Therefore our work for the first time demonstrates that phosphorylation-dependent changes in the binding of a structural MAP to MTs regulate the direction of MT transport of membrane organelles.
Our data show that overexpression of XMAP4 reduces the average length of minus-end MT runs of pigment granules generated by dynein. This result is in line with the results of other studies that indicate that MAP4 inhibits dynein motility. Overexpression of MAP4 in interphase mammalian cells prevents clustering of the Golgi cisternae in the cell center, which requires dynein activity (Bulinski et al., 1997). Similarly, MAP4 suppresses dynein-dependent sliding of astral MTs at the cell cortex essential for positioning of the mitotic spindle and inhibits gliding of MTs powered by dynein in vitro (Samora et al., 2011). On the other hand, motility of single dynein molecules along MTs in vitro was relatively insensitive to decoration of MTs with fragments of tau, a MAP structurally similar to the MAP4 (Dixit et al., 2008; Vershinin et al., 2008). However, in living cells, dynein motors moving pigment granules have to overcome significant viscous drag and therefore might be more susceptible to obstacles on the MT surface. Further, XMAP4 might be a more efficient inhibitor of dynein motility than MT-binding fragments of tau used in the in vitro motility assays. Therefore it is likely that XMAP4 inhibits minus end–directed MT movement of pigment granules during pigment aggregation by blocking the path of dynein motors.
In a marked contrast to pigment aggregation, overexpression of XMAP4 does not inhibit dispersion of pigment granules, which requires the activity of kinesin-2 (Tuma et al., 1998). Thus, unlike dynein, kinesin-2 is capable of moving along MTs bearing XMAP4. The lack of effect on kinesin-2–driven transport might be explained by the particular isoform of XMAP4 expressing in Xenopus melanophores. Only the five-repeat isoform, and not the three- or four-repeat isoforms, of mammalian MAP4 suppresses gliding MTs driven by kinesin-1 in in vitro motility assays (Tokuraku et al., 2007), and it is possible that kinesin-2 shares with kinesin-1 insensitivity to the MAP4 isoforms with reduced numbers of MT-binding repeats. Therefore specific isoforms of XMAP4 might selectively affect motility of MT motors similar to yeast protein She1, which inhibits movement of dynein but not kinesins along MT tracks (Markus et al., 2012). However, it is also possible that movement along MTs with bound XMAP4 or other MAPs is an inherent property of kinesin-2, as the unique neck-linker region allows kinesin-2 to navigate obstacles on the MT surface (Hoeprich et al., 2014). Unlike kinesin-1 or cytoplasmic dynein, which take straight paths, kinesin-2 family members are capable of switching protofilaments (Brunnbauer et al., 2012). The sideways steps may let kinesin-2 bypass molecules of XMAP4 and other MAPs extending from the MT wall (Hoeprich et al., 2014).
We unexpectedly discovered that XMAP4 positively regulates plus end–directed runs of pigment granules during pigment dispersion. This conclusion is based on the results of experiments that involved microinjection of blocking antibodies that removed XMAP4 from MTs. In Xenopus melanophores, plus-end runs of pigment granules have been shown to be generated by kinesin-2 (Tuma et al., 1998). Therefore our data suggest that, similar to MAP7/ensconsin, which activates kinesin-1 (Sung et al., 2008; Metzger et al., 2012; Barlan et al., 2013a), and doublecortin/doublecortin-like kinase, which facilitates transport of membrane vesicles driven by Kif1A (Liu et al., 2012), XMAP4 stimulates kinesin-2 motility. Doublecortin increases the length of MT runs of Kif1A cargoes by enhancing binding of the Kif1A motor domain to MTs (Liu et al., 2012). We hypothesize that XMAP4 might stimulate kinesin-2 motility by a similar mechanism. Our pull-down experiments indicate that, unlike doublecortin, which forms a complex with Kif1A, XMAP4 does not binds kinesin-2 directly. However, we instead observe an interaction between XMAP4 and the p150Glued subunit of the dynactin complex, an adaptor to MT motor proteins on the granule surface (Deacon et al., 2003). Mammalian MAP4 also immunoprecipitates p150Glued from cell extracts, which indicates that the interaction between the two proteins is highly conserved (Samora et al., 2011). We hypothesize that transient attachment of XMAP4 to p150Glued during granule movement may keep kinesin-2 in proximity to the MT wall, reducing the detachment rate and therefore increasing the length of uninterrupted runs of relatively nonprocessive kinesin-2 motors (Muthukrishnan et al., 2009).
Surprisingly, our data indicate that effects of XMAP4 on granule runs in each direction are different in the two signaling states. Overexpression of XMAP4 does not increase the average length of plus-end runs during pigment granule aggregation as would be expected in light of a positive effect of XMAP4 on plus-end run length during dispersion of pigment granules. Similarly, removal of XMAP4 from MTs during granule dispersion reduced the length of minus-end granule runs, which is apparently inconsistent with the minus-end run length decrease during granule aggregation in XMAP4-overexpressing cells. We hypothesize that the observed differences in effects of XMAP4 on granule runs during pigment aggregation and dispersion might be explained by variations in sensitivity to XMAP4 by the active and inactive states of MT motors. In the fully active state typical for pigment aggregation, dynein might be highly sensitive to XMAP4 obstacles on the MT surface. On the contrary, in an inactive state characteristic of pigment dispersion, dynein processivity is enhanced by XMAP4, presumably through interaction of XMAP4 with p150Glued on the granule surface. Similarly, XMAP4 enhances plus end–directed granule runs generated by active kinesin-2 motors during pigment dispersion, but binding of XMAP4 to MTs might be insufficient to activate kinesin-2 motility during aggregation of pigment granules. It is therefore likely that reversible binding of XMAP4 to MTs cooperates with independent regulation of the activities of MT motors to control the direction of MT-based transport of pigment granules.
Our data show that during pigment aggregation, XMAP4 is phosphorylated on Thr-758 and Thr-762 located in the proline-rich region of the MBD. This phosphorylation reduces the affinity of XMAP4 binding to MTs, as evidenced by the decreased cosedimentation of the phosphomimetic mutant protein with MTs in vitro. These data are consistent with the result of studies that show that phosphorylation of MBD causes detachment of MAP4 from MTs in vivo and in vitro (Ookata et al., 1995; Illenberger et al., 1996; Drewes et al., 1997; Shiina and Tsukita, 1999; Kitazawa et al., 2000; Chang et al., 2001; Chinnakkannu et al., 2010). In mammalian cells, Thr-758 and Thr-762 are phosphorylated during transition to mitosis by p34cdc2 and mitogen-activated protein kinases, which induce dissociation of MAP4 from MTs (Ookata et al., 1995; Shiina and Tsukita, 1999). The identity of protein kinases that phosphorylate XMAP4 in Xenopus melanophores is unknown, but it is clear that their activities must be stimulated by the decrease in cytoplasmic levels of cAMP and activity of PKA, which trigger aggregation of pigment granules (Daniolos et al., 1990; Rodionov et al., 2003). Our previous work indicates that PKA and other components of the signaling cascades that regulate MT motors are bound to the surface of pigment granules (Kashina et al., 2004). We suggest that protein kinases that phosphorylate XMAP4 are bound together with PKA to the MT surface. Attachment of signaling enzymes to MTs might be mediated by the XMAP4 itself or other MAPs distributed along the MT length. In support of this idea, MAP4 and MAP2 are known to mediate binding of p34cdc2 kinase (Ookata et al., 1995) and PKA (Obar et al., 1989) to MTs. Scaffolding of signaling enzymes by MAPs would bring protein kinases in proximity to their substrates and allow for the rapid release of MAP4 and other MAPs from MTs, clearing a path for dynein motors during pigment aggregation.
On the basis of our results, we propose a hypothesis for the regulation of MT-based transport of pigment granules by XMAP4 (Figure 7). We suggest that binding of XMAP4 to MTs creates obstacles in the path of dynein motors and therefore negatively regulates dynein-dependent motility of pigment granules. Conversely, attachment of XMAP4 to MTs positively regulates kinesin-2–based motility due to the interaction of XMAP4 with p150Glued bound to pigment granules, which increases the length of plus-end runs by keeping the kinesin-2 motor domain close to the MT surface. Phosphorylation of XMAP4 on Thr-758 and Thr-762 during aggregation reduces the binding of XMAP4 to MTs, thus increasing minus end–directed and decreasing plus end–directed motility of pigment granules, causing their accumulation in the cell center (Figure 7, left), whereas dephosphorylation of XMAP4 during pigment dispersion has the opposite effect (Figure 7, right).
FIGURE 7:
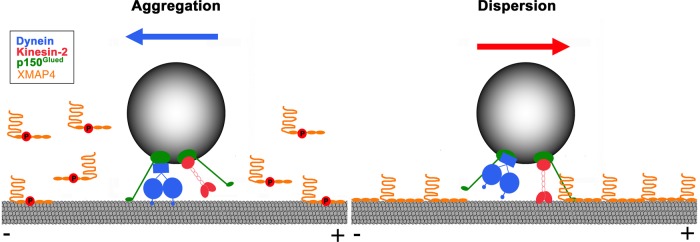
Hypothesis for the regulation of MT-based transport of pigment granules by XMAP4. XMAP4 negatively regulates minus end–directed MT transport of pigment granules by blocking the movement of dynein motors along MTs and positively regulates plus end–directed, kinesin-2–based transport through interaction with the granule-bound p150Glued. This interaction increases processivity of kinesin-2 motors by keeping them in proximity to the MT surface. Phosphorylation during pigment aggregation reduces binding of XMAP4 to MTs, thus increasing minus end–directed and decreasing plus end–directed motility of pigment granules (left), which stimulates their accumulation at the cell center, whereas dephosphorylation of XMAP4 during pigment dispersion has an opposite effect (right).
MATERIALS AND METHODS
Cell culture
Xenopus melanophores (Kashina et al., 2004) were cultured in Xenopus tissue culture medium (70% L15 medium supplemented with 0.2 mg/ml penicillin, 0.2 mg/ml streptomycin, 5 μg/ml insulin, and 10% heat-inactivated fetal bovine serum) at 27°C. Before stimulation of aggregation or dispersion, melanophores were transferred into serum-free 70% L15 medium and incubated for at least 1 h at 27°C. Aggregation of pigment granules was induced by treatment with 10−8 M melatonin. For dispersion of pigment granules, cells were treated with 10−8 M melanocyte-stimulating hormone (MSH).
Quanatification of aggregation and dispersion responses of melanophores
To determine the fractions of cells with aggregated, partially responded, or dispersed pigment, melanophores were treated with melatonin or MSH for 15 min or 1 h and fixed with formaldehyde. The numbers of cells with aggregated, partially responded, or dispersed pigment granules were determined by counting cells in each category by phase-contrast microscopy as described previously (Kashina et al., 2004).
To determine kinetics of pigment aggregation, time series of bright-field images of melanophores treated with melatonin were acquired with 10-s time intervals. Integrated pixel values within cell outlines were determined for each of the acquired images using the region measurement tool of MetaMorph image acquisition and analysis software (Molecular Devices, Downingtown, PA). The percentage of gray levels was calculated for each image as described previously (Lomakin et al., 2011). Data were averaged across the recorded cells and plotted as a function of time.
Mass spectrometry
Melanophores grown in dense monolayers in two 75-cm2 flasks were transferred into 70% serum-free L15 medium for 60 min, stimulated with either melatonin or MSH for 60 min, washed twice with 10 ml of ice-cold 0.7× phosphate-buffered saline (PBS), and lysed on ice by adding 1 ml of lysis buffer containing 8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.2, one tablet of protease inhibitor cocktail (Roche Applied Sciences, Indianapolis, IN) per 10 ml of lysis buffer, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell lysates were sonicated and clarified by centrifugation. Proteins were reduced and alkylated and digested with trypsin as described previously (Villen and Gygi, 2008). After desalting and strong cation exchange (SCX) chromatography, phosphopeptides were selectively enriched by a combination of SCX chromatography and immobilized metal affinity chromatography (Villen and Gygi, 2008). Liquid chromatography–tandem mass spectrometry (MS/MS) was performed on an LTQ Orbitrap hybrid mass spectrometer (Thermo Fisher, San Jose, CA). MS/MS spectra were searched via the SEQEST algorithm with the target-decoy strategy (Elias and Gygi, 2007) against a Xenopus laevis open reading frame database. Ascore algorithm (Beausoleil et al., 2006) was used to assign phosphorylation site localization.
XMAP4 cloning and mutagenesis
DNA for the XMAP4 from Xenopus melanophores was amplified by PCR using as a template cDNA synthesized with SuperScriptII reverse transcriptase (Life Technologies, Carlsbad, CA) from the total melanophore RNA. The following set of primers designed against the X. laevis XMAP4 (National Center for Biotechnology Information accession number NP_001083770.1) was used for the DNA amplification: CCGGCTCGAGCATGGCGGACCTTGGAC (forward primer) and CGCCCCGCGGTTAGATGCTTGTCTCTGG (reverse primer). The PCR product was cloned into pEGFP-C2 vector (Clontech, Mountain View, CA) using XhoI and SacII restriction sites and verified by sequencing. Amino acid sequence of the cloned protein was identical to the sequence published for XMAP4 except that it had a 10–amino acid insert (AEVLSAPIPE) between the amino acid residues 63 and 73 and a 57–amino acid deletion between the amino acid residues 1056–1112 in the C-terminal region of the molecule. Several independent amplification experiments produced PCR products with the same nucleotide sequence, which indicated that the amplified DNA encoded the principal isoform of XMAP4 expressed in melanophores.
Phosphomimetic (T758E/T762E) and nonphosphorylatable (T758A/T762A) mutants of XMAP4 were generated using QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA).
Expression of XMAP4 mutant proteins in baculovirus
Recombinant phosphomimetic and nonphosphorylatable XMAP4 mutant proteins were expressed in baculovirus using the Bac-to-Bac Baculovirus expression system (Life Technologies). To facilitate purification of recombinant proteins and enable detection of protein expression, 6xHis and mCherry tags were added to the N-termini of the XMAP4 sequences. 6xHis/mCherry tag DNA was amplified by PCR using mCherry N1 mammalian expression vector (Clontech) as a template and cloned into pFastBac1 vector (Life Technologies) using BamH1 and EcoRI restriction sites. PCR-amplified XMAP4 mutant DNAs were cloned into pFastBac1 vector downstream of the 6xHis/mCherry tag using SpeI and XhoI restriction sites. Recombinant bacmid and baculovirus stocks were produced according to instructions provided by the kit manufacturer. For protein expression, Sf9 cells were infected with baculovirus and incubated for 48 h in Sf-900 II medium (Life Technologies) containing 50 U/ml penicillin and 50 μg/ml streptomycin. Cells were pelleted at 1000 × g for 10 min at 4°C and washed with PBS. Cell pellets were resuspended in the lysis buffer (20 mM Tris, 500 mM NaCl, 10 mM imidazole, 1 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.5 mM MgATP, 1 mM PMSF, pH 7.5) and incubated on ice for 45 min for the cell lysis. Cell lysates were clarified by centrifugation at 100,000 × g for 30 min at 4°C and used for purification of recombinant proteins by chromatography on Ni-NTA Agarose (Qiagen, Valencia, CA) according to instructions provided by the manufacturer.
Cell transfection
Melanophores were transfected using GeneCellin DNA transfection reagent (Bulldog Bio, Portsmouth, NH) according to instructions provided by the manufacturer. After transfection, cells were incubated for 2–4 d at 27°C for protein expression.
Production of antibodies against XMAP4
For production of antibodies against the MBD of XMAP4, a fragment of DNA that encoded amino acids 722–1228 of XMAP4 was amplified by PCR using GGAATTCCATATGGCA GAACCT GCTGCTGCAGC as forward primer and CCGCTCGAGTTA GATGCTTGT CTCTGG TATTAG as reverse primer. The PCR product was cloned into the bacterial expression vector pCold I DNA (Clontech) using the Nde and XhoI restriction sites. Recombinant protein was expressed in Escherichia coli according to instructions provided by the vector manufacturer, purified by affinity chromatography on Ni-NTA agarose (Qiagen), dialyzed against PBS, and used for rabbit immunization. High-titer antisera were produced by Bio-Synthesis (Lewisville, TX). Polyclonal antibodies were purified from antisera by affinity chromatography on a CNBr-activated Sepharose 4B (Sigma-Aldrich, St. Louis, MO) with covalently attached antigen. Purified antibodies were dialyzed against microinjection buffer and concentrated by ultrafiltration.
Polyclonal antibodies against XMAP4 projection domain were generated by immunization of rabbits with the synthetic peptide GCDDDDVKEPKNKSERSAAPHD manufactured by Bio-Synthesis, which corresponded to the amino acid residues 56–75 of XMAP4. Antibodies were purified by affinity chromatography of antisera on SulfoLink resin (Thermo Scientific, Rockford, IL) with covalently attached peptide antigen.
Cosedimentation of recombinant XMAP4 mutant proteins with MTs
MTs were assembled by polymerization of purified porcine brain tubulin (Cytoskeleton, Denver, CO) in the presence of paclitaxel (20 μM; Sigma-Aldrich), and recombinant XMAP4 mutant proteins were added at 1:1 molar ratio to tubulin. After incubation for 30 min at room temperature, MTs were pelleted by centrifugation through a glycerol cushion at 140,000 × g for 40 min at 25°C, and the MT pellets were resuspended in SDS–PAGE sample buffer. The amounts of XMAP4 mutant proteins in the MT pellets were measured by quantitative immunoblotting.
Immunoprecipitation and immunoblotting
For immunoprecipitation, whole-cell extracts were incubated for 2 h at room temperature with rabbit affinity-purified anti-GFP antibodies (ab290; Abcam, Cambridge, MA) or antibodies against XMAP4 MBD bound to protein A agarose (Thermo Scientific). Agarose beads were pelleted and washed, and adsorbed proteins were eluted by the treatment of beads with SDS–PAGE sample buffer for 5 min at 100°C.
Immunoblotting was performed by the method described in Towbin et al. (1979). Blots were stained with mouse monoclonal GFP antibodies (ab1218; Abcam), p150Glued subunit of the dynactin complex (610473; BD Biosciences, San Jose, CA), cytoplasmic dynein IC (74.1; Covance, Princeton, NJ), or kinesin-2 motor subunit (K2.4; Covance). Immunoreactive bands were detected with SuperSignal West Femto maximum-sensitivity substrate (Thermo Scientific, Waltham, MA). To measure the amounts of recombinant XMAP4, nonphosphorylatable and phosphomimetic mutant proteins cosedimented with MTs, blots of MT pellets were successively incubated with antibodies against XMAP4 MBD and IRDye800-conjugated affinity-purified anti-mouse IgG (Rockland Immunochemicals, Gilbertsville, PA), and the intensity of the infrared signal was quantified with the Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE).
Microinjection
Pressure microinjection of antibodies against XMAP4 MBD (8–10 mg/ml), nonimmune rabbit IgG (12 mg/ml), and Cy3-labeled porcine brain tubulin (6–7 mg/ml) was performed using the PLI-100 Pico-Liter microinjection system (Harvard Apparatus, Hollistson, MA) as described previously (Semenova and Rodionov, 2007; Ikeda et al., 2010).
Immunofluorescence staining
For immunofluorescence staining of MTs, melanophores microinjected with antibody against XMAP4 MBD were briefly rinsed with PBS, fixed with glutaraldehyde, permeabilized with PBS containing 1% Triton X-100 (Ted Pella, Redding, CA), and sequentially incubated with mouse monoclonal antibodies against α-tubulin (1:200, DM1A; Cedarlane Laboratories, Burlington, NC) and goat anti-mouse antibodies conjugated with Alexa Fluor 488 (1:200; Invitrogen). Immunofluorescence staining of MTs was followed by the incubation cells with goat anti-rabbit antibodies conjugated with rhodamine (1:200; KPL, Gaithersburg, MD), which allowed us to identify the microinjected cells. For immunofluorescence staining of CLIP170 comets, melanophores were fixed in cold methanol, postfixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, and immunostained with antibodies specific for CLIP-170 N-terminus (no. 2221; 1:200; Hoogenraad et al., 2000) and goat anti-rabbit antibodies conjugated with Alexa 488 (1:200; Invitrogen).
For double immunofluorescence staining with XMAP4 MBD and tubulin antibodies, melanophores treated for 5 min with melatonin or 3 min with MSH were briefly rinsed with PBS and placed for 2 min in permeabilization buffer containing 60 mM 1,4-piperazinediethanesulfonic acid, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM ethylene glycol tetraacetic acid, 2 mM MgCl2, 20 mM paclitaxel, and 0.5% Triton X-100. After permeabilization, cells were washed with PBS, fixed with 1% glutaraldehyde for 30 min, treated with NaBH4 to quench the unreacted glutaraldehyde, and blocked with 2% solution of bovine serum albumin. Cells were then sequentially incubated with affinity-purified antibodies against XMAP4 MBD (0.7 μg/ml), goat anti-rabbit antibodies conjugated with rhodamine (1:200; KPL), mouse monoclonal antibodies against α-tubulin (1:200, DM1A; Cedarlane Laboratories), and goat-anti-mouse antibodies conjugated with Alexa Fluor 488 (1:200, Invitrogen). Immunostained cells were mounted in Aqua-Poly/Mount medium (Polysciences, Warrington, PA).
Image acquisition and analysis
Fluorescence images of melanophores were acquired using a Nikon Eclipse Ti inverted microscope equipped with a Plan Apochromat 100×/1.4 numerical aperture objective lens and Andor iXon EM-CCD sensor (Andor Technology, Windsor, CT) driven by MetaMorph image acquisition and analysis software (Molecular Devices, Downingtown, PA). MT dynamics was measured by acquiring time series of images of Cy3-labeled MTs, as described previously (Semenova and Rodionov, 2007). To reduce photobleaching and photodamage, cells were treated with the oxygen-depleting agent Oxyrase (Oxyrase Company, Mansfield, OH) before image acquisition. Tips of individual MTs were manually tracked using MetaMorph software. Parameters of MT dynamic instability were determined by decomposing the trajectories of MT ends into phases of growth, shortening, and pauses using multiscale trend analysis (Lomakin et al., 2009). The length of CLIP-170 comets and the amount of CLIP-170 bound to MT ends were measured as described previously (Lomakin et al., 2011). For quantification of expression levels of XMAP4 mutants, images of 100 cells expressing GFP-tagged phosphomimetic or nonphosphorylatable mutant were acquire using a fluorescein filter set, and the values for average gray levels within the cell outlines were determined using the Region Measurements Tool of MetaMorph software. For quantification of levels of XMAP4 bound to MTs, images of 20 cells stimulated with either melatonin or MSH immunostained with XMAP4 MBD antibodies and rhodamine-conjugated secondary antibodies were acquired using a rhodamine filter set, and the MetaMorph Region Measurements Tool was used to quantify average fluorescence over five ∼1-μm MT segments in each cell. After background subtraction, the results of fluorescence measurements were averaged separately for melanophores aggregating or dispersing pigment granules. The movement of individual pigment granules was recorded and analyzed as described previously (Zaliapin et al., 2005). Time-lapse sequences of aggregating pigment granules were acquired 10 min after the application of melatonin. Dispersing pigment granules were recorded within 7.5 min after the treatment with MSH. At this early stage of dispersion, transport of pigment granules was shown to be exclusively MT dependent (Zaliapin et al., 2005; Slepchenko et al., 2007).
Stochastic computational model for pigment aggregation
Computational modeling of pigment aggregation was performed as described previously (Lomakin et al., 2009, 2011).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM62290 (to V.I.R.) and P41RR013186 (to V.I.R. and A.C.).
Abbreviations used:
- MAP
microtubule-associated protein
- MBD
microtubule-binding domain
- MT
microtubule
- XMAP
Xenopus microtubule-associated protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0022) on August 20, 2014.
REFERENCES
- Aizawa H, Emori Y, Mori A, Murofushi H, Sakai H, Suzuki K. Functional analyses of the domain structure of microtubule-associated protein-4 (MAP-U) J Biol Chem. 1991;266:9841–9846. [PubMed] [Google Scholar]
- Aizawa H, Emori Y, Murofushi H, Kawasaki H, Sakai H, Suzuki K. Molecular cloning of a ubiquitously distributed microtubule-associated protein with Mr 190,000. J Biol Chem. 1990;265:13849–13855. [PubMed] [Google Scholar]
- Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspengren S, Hedberg D, Skold HN, Wallin M. New insights into melanosome transport in vertebrate pigment cells. Int Rev Cell Mol Biol. 2009;272:245–302. doi: 10.1016/S1937-6448(08)01606-7. [DOI] [PubMed] [Google Scholar]
- Atherton J, Houdusse A, Moores C. MAPping out distribution routes for kinesin couriers. Biol Cell. 2013;105:465–487. doi: 10.1111/boc.201300012. [DOI] [PubMed] [Google Scholar]
- Avila J, Dominguez J, Diaz-Nido J. Regulation of microtubule dynamics by microtubule-associated protein expression and phosphorylation during neuronal development. Int J Dev Biol. 1994;38:13–25. [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol. 2013a;23:317–322. doi: 10.1016/j.cub.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K, Rossow MJ, Gelfand VI. The journey of the organelle: teamwork and regulation in intracellular transport. Curr Opin Cell Biol. 2013b;25:483–488. doi: 10.1016/j.ceb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Brunnbauer M, Dombi R, Ho TH, Schliwa M, Rief M, Okten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Bulinski JC, McGraw TE, Gruber D, Nguyen HL, Sheetz MP. Overexpression of MAP4 inhibits organelle motility and trafficking in vivo. J Cell Sci. 1997;110:3055–3064. doi: 10.1242/jcs.110.24.3055. [DOI] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Spittle C. Regulation of microtubule-associated proteins. Int Rev Cytol. 2001;210:163–226. doi: 10.1016/s0074-7696(01)10006-9. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Holzbaur EL. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Chang W, Gruber D, Chari S, Kitazawa H, Hamazumi Y, Hisanaga S, Bulinski JC. Phosphorylation of MAP4 affects microtubule properties and cell cycle progression. J Cell Sci. 2001;114:2879–2887. doi: 10.1242/jcs.114.15.2879. [DOI] [PubMed] [Google Scholar]
- Chapin SJ, Bulinski JC. Non-neuronal 210 x 10(3) Mr microtubule-associated protein (MAP4) contains a domain homologous to the microtubule-binding domains of neuronal MAP2 and tau. J Cell Sci. 1991;98:27–36. doi: 10.1242/jcs.98.1.27. [DOI] [PubMed] [Google Scholar]
- Chapin SJ, Lue CM, Yu MT, Bulinski JC. Differential expression of alternatively spliced forms of MAP4: a repertoire of structurally different microtubule-binding domains. Biochemistry. 1995;34:2289–2301. doi: 10.1021/bi00007a025. [DOI] [PubMed] [Google Scholar]
- Chinnakkannu P, Samanna V, Cheng G, Ablonczy Z, Baicu CF, Bethard JR, Menick DR, Kuppuswamy D, Cooper GT. Site-specific microtubule-associated protein 4 dephosphorylation causes microtubule network densification in pressure overload cardiac hypertrophy. J Biol Chem. 2010;285:21837–21848. doi: 10.1074/jbc.M110.120709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniolos A, Lerner AB, Lerner MR. Action of light on frog pigment cells in culture. Pigment Cell Res. 1990;3:38–43. doi: 10.1111/j.1600-0749.1990.tb00260.x. [DOI] [PubMed] [Google Scholar]
- Deacon SW, Nascimento A, Serpinskaya AS, Gelfand VI. Regulation of bidirectional melanosome transport by organelle bound MAP kinase. Curr Biol. 2005;15:459–463. doi: 10.1016/j.cub.2004.12.074. [DOI] [PubMed] [Google Scholar]
- Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, Vaughan KT, Gelfand VI. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Mandelkow EM. MAPs, MARKs and microtubule dynamics. Trends Biochem Sci. 1998;23:307–311. doi: 10.1016/s0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Dunn S, Morrison EE, Liverpool TB, Molina-Paris C, Cross RA, Alonso MC, Peckham M. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121:1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev. Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS. Kinesin molecular motors: transport pathways, receptors, and human disease. Proc Natl Acad Sci USA. 2001;98:6999–7003. doi: 10.1073/pnas.111145298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SP. Hither and yon: a review of bi-directional microtubule-based transport. Phys Biol. 2004;1:R1–R11. doi: 10.1088/1478-3967/1/2/R01. [DOI] [PubMed] [Google Scholar]
- Gross SP, Tuma MC, Deacon SW, Serpinskaya AS, Reilein AR, Gelfand VI. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev. Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Hoeprich GJ, Thompson AR, McVicker DP, Hancock WO, Berger CL. Kinesin's neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys J. 2014;106:1691–1700. doi: 10.1016/j.bpj.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P, Brattsand G, Gullberg M. MAP4 counteracts microtubule catastrophe promotion but not tubulin-sequestering activity in intact cells. Curr Biol. 2002;12:1034–1039. doi: 10.1016/s0960-9822(02)00897-7. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A, Grosveld F, De Zeeuw CI, Galjart N. Functional analysis of CLIP-115 and its binding to microtubules. J Cell Sci. 2000;113:2285–2297. doi: 10.1242/jcs.113.12.2285. [DOI] [PubMed] [Google Scholar]
- Hook P, Vallee RB. The dynein family at a glance. J Cell Sci. 2006;119:4369–4371. doi: 10.1242/jcs.03176. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Semenova I, Zhapparova O, Rodionov V. Melanophores for microtubule dynamics and motility assays. Methods Cell Biol. 2010;97:401–414. doi: 10.1016/S0091-679X(10)97021-0. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Zhapparova O, Brodsky I, Semenova I, Tirnauer JS, Zaliapin I, Rodionov V. CK1 activates minus-end-directed transport of membrane organelles along microtubules. Mol Biol Cell. 2011;22:1321–1329. doi: 10.1091/mbc.E10-09-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illenberger S, Drewes G, Trinczek B, Biernat J, Meyer HE, Olmsted JB, Mandelkow EM, Mandelkow E. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J Biol Chem. 1996;271:10834–10843. doi: 10.1074/jbc.271.18.10834. [DOI] [PubMed] [Google Scholar]
- Jolly AL, Gelfand VI. Bidirectional intracellular transport: utility and mechanism. Biochem Soc Trans. 2011;39:1126–1130. doi: 10.1042/BST0391126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Goldstein LS. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–68. doi: 10.1016/s0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Hoogenraad CC. Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol Cell Neurosci. 2011;46:9–20. doi: 10.1016/j.mcn.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Karcher RL, Deacon SW, Gelfand VI. Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 2002;12:21–27. doi: 10.1016/s0962-8924(01)02184-5. [DOI] [PubMed] [Google Scholar]
- Kashina A, Rodionov V. Intracellular organelle transport: few motors, many signals. Trends Cell Biol. 2005;15:396–398. doi: 10.1016/j.tcb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Semenova IV, Ivanov PA, Potekhina ES, Zaliapin I, Rodionov VI. Protein kinase A, which regulates intracellular transport, forms complexes with molecular motors on organelles. Curr Biol. 2004;14:1877–1881. doi: 10.1016/j.cub.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Kitazawa H, Iida J, Uchida A, Haino-Fukushima K, Itoh TJ, Hotani H, Ookata K, Murofushi H, Bulinski JC, Kishimoto T, et al. Ser787 in the proline-rich region of human MAP4 is a critical phosphorylation site that reduces its activity to promote tubulin polymerization. Cell Struct Funct. 2000;25:33–39. doi: 10.1247/csf.25.33. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Lane J, Allan V. Microtubule-based membrane movement. Biochim Biophys Acta. 1998;1376:27–55. doi: 10.1016/s0304-4157(97)00010-5. [DOI] [PubMed] [Google Scholar]
- Lee G. Non-motor microtubule-associated proteins. Curr Opin Cell Biol. 1993;5:88–94. doi: 10.1016/s0955-0674(05)80013-4. [DOI] [PubMed] [Google Scholar]
- Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J Biol Chem. 1998;273:9797–9803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- Liu JS, Schubert CR, Fu X, Fourniol FJ, Jaiswal JK, Houdusse A, Stultz CM, Moores CA, Walsh CA. Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family proteins. Mol Cell. 2012;47:707–721. doi: 10.1016/j.molcel.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin AJ, Kraikivski P, Semenova I, Ikeda K, Zaliapin I, Tirnauer JS, Akhmanova A, Rodionov V. Stimulation of the CLIP-170–dependent capture of membrane organelles by microtubules through fine tuning of microtubule assembly dynamics. Mol Biol Cell. 2011;22:4029–4037. doi: 10.1091/mbc.E11-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin AJ, Semenova I, Zaliapin I, Kraikivski P, Nadezhdina E, Slepchenko BM, Akhmanova A, Rodionov V. CLIP-170-dependent capture of membrane organelles by microtubules initiates minus-end directed transport. Dev Cell. 2009;17:323–333. doi: 10.1016/j.devcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez LA, Sheetz MP. Steric inhibition of cytoplasmic dynein and kinesin motility by MAP2. Cell Motil Cytoskeleton. 1993;24:1–16. doi: 10.1002/cm.970240102. [DOI] [PubMed] [Google Scholar]
- Mandell JW, Banker GA. Microtubule-associated proteins, phosphorylation gradients, and the establishment of neuronal polarity. Perspect Dev Neurobiol. 1996;4:125–135. [PubMed] [Google Scholar]
- Markus SM, Kalutkiewicz KA, Lee WL. She1-mediated inhibition of dynein motility along astral microtubules promotes polarized spindle movements. Curr Biol. 2012;22:2221–2230. doi: 10.1016/j.cub.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Microtubule-associated proteins: their potential role in determining neuronal morphology. Annu Rev Neurosci. 1988;11:29–44. doi: 10.1146/annurev.ne.11.030188.000333. [DOI] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan G, Zhang Y, Shastry S, Hancock WO. The processivity of kinesin-2 motors suggests diminished front-head gating. Curr Biol. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AA, Roland JT, Gelfand VI. Pigment cells: a model for the study of organelle transport. Annu Rev Cell Dev Biol. 2003;19:469–491. doi: 10.1146/annurev.cellbio.19.111401.092937. [DOI] [PubMed] [Google Scholar]
- Nguyen HL, Chari S, Gruber D, Lue CM, Chapin SJ, Bulinski JC. Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J Cell Sci. 1997;110:281–294. doi: 10.1242/jcs.110.2.281. [DOI] [PubMed] [Google Scholar]
- Nilsson H, Wallin M. Evidence for several roles of dynein in pigment transport in melanophores. Cell Motil Cytoskeleton. 1997;38:397–409. doi: 10.1002/(SICI)1097-0169(1997)38:4<397::AID-CM9>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Obar RA, Dingus J, Bayley H, Vallee RB. The RII subunit of cAMP-dependent protein kinase binds to a common amino-terminal domain in microtubule-associated proteins 2A, 2B, and 2C. Neuron. 1989;3:639–645. doi: 10.1016/0896-6273(89)90274-2. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Microtubule-associated proteins. Annu Rev Cell Biol. 1986;2:421–457. doi: 10.1146/annurev.cb.02.110186.002225. [DOI] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Sugita M, Okuyama A, Murofushi H, Kitazawa H, Chari S, Bulinski JC, Kishimoto T. MAP4 is the in vivo substrate for CDC2 kinase in HeLa cells: identification of an M-phase specific and a cell cycle-independent phosphorylation site in MAP4. Biochemistry. 1997;36:15873–15883. doi: 10.1021/bi971251w. [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Rodionov VI, Hope AJ, Svitkina TM, Borisy GG. Functional coordination of microtubule-based and actin-based motility in melanophores. Curr Biol. 1998;8:165–168. doi: 10.1016/s0960-9822(98)70064-8. [DOI] [PubMed] [Google Scholar]
- Rodionov V, Yi J, Kashina A, Oladipo A, Gross SP. Switching between microtubule- and actin-based transport systems in melanophores is controlled by cAMP levels. Curr Biol. 2003;13:1837–1847. doi: 10.1016/j.cub.2003.10.027. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Gelfand VI. Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr Biol. 1998;8:161–164. doi: 10.1016/s0960-9822(98)70063-6. [DOI] [PubMed] [Google Scholar]
- Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- Semenova I, Rodionov V. Fluorescence microscopy of microtubules in cultured cells. Methods Mol Med. 2007;137:93–102. doi: 10.1007/978-1-59745-442-1_6. [DOI] [PubMed] [Google Scholar]
- Shiina N, Tsukita S. Mutations at phosphorylation sites of Xenopus microtubule-associated protein 4 affect its microtubule-binding ability and chromosome movement during mitosis. Mol Biol Cell. 1999;10:597–608. doi: 10.1091/mbc.10.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepchenko BM, Semenova I, Zaliapin I, Rodionov V. Switching of membrane organelles between cytoskeletal transport systems is determined by regulation of the microtubule-based transport. J Cell Biol. 2007;179:635–641. doi: 10.1083/jcb.200705146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Wittmann T. Targeting and transport: how microtubules control focal adhesion dynamics. J Cell Biol. 2012;198:481–489. doi: 10.1083/jcb.201206050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HH, Telley IA, Papadaki P, Ephrussi A, Surrey T, Rorth P. Drosophila ensconsin promotes productive recruitment of kinesin-1 to microtubules. Dev Cell. 2008;15:866–876. doi: 10.1016/j.devcel.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Tokuraku K, Noguchi TQ, Nishie M, Matsushima K, Kotani S. An isoform of microtubule-associated protein 4 inhibits kinesin-driven microtubule gliding. J Biochem. 2007;141:585–591. doi: 10.1093/jb/mvm063. [DOI] [PubMed] [Google Scholar]
- Tortosa E, Galjart N, Avila J, Sayas CL. MAP1B regulates microtubule dynamics by sequestering EB1/3 in the cytosol of developing neuronal cells. EMBO J. 2013;32:1293–1306. doi: 10.1038/emboj.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma MC, Zill A, Le Bot N, Vernos I, Gelfand V. Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J Cell Biol. 1998;143:1547–1558. doi: 10.1083/jcb.143.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev. Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Kaul N, Soppina V. Kinesin assembly and movement in cells. Annu Rev Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP. Multiple-motor based transport and its regulation by Tau. Proc Natl Acad Sci USA. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinin M, Xu J, Razafsky DS, King SJ, Gross SP. Tuning microtubule-based transport through filamentous MAPs: the problem of dynein. Traffic. 2008;9:882–892. doi: 10.1111/j.1600-0854.2008.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protocols. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nat Rev. Mol Cell Biol. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Peloquin JG, Zhai Y, Bulinski JC, Borisy GG. Removal of MAP4 from microtubules in vivo produces no observable phenotype at the cellular level. J Cell Biol. 1996;132:345–357. doi: 10.1083/jcb.132.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- West RR, Tenbarge KM, Olmsted JB. A model for microtubule-associated protein 4 structure. Domains defined by comparisons of human, mouse, and bovine sequences. J Biol Chem. 1991;266:21886–21896. [PubMed] [Google Scholar]
- Zaliapin I, Semenova I, Kashina A, Rodionov V. Multiscale trend analysis of microtubule transport in melanophores. Biophys J. 2005;88:4008–4016. doi: 10.1529/biophysj.104.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.