Loss of γ-cytoplasmic actin induces epithelial-to-myofibroblast transition (EmyT), which depends on activation of SRF and its cofactor, MRTF, formin-mediated actin polymerization, and activated Rho GTPase. This demonstrates a unique role of γ-cytoplasmic actin in regulating the epithelial phenotype and the suppression of EmyT.
Abstract
Transdifferentiation of epithelial cells into mesenchymal cells and myofibroblasts plays an important role in tumor progression and tissue fibrosis. Such epithelial plasticity is accompanied by dramatic reorganizations of the actin cytoskeleton, although mechanisms underlying cytoskeletal effects on epithelial transdifferentiation remain poorly understood. In the present study, we observed that selective siRNA-mediated knockdown of γ-cytoplasmic actin (γ-CYA), but not β-cytoplasmic actin, induced epithelial-to-myofibroblast transition (EMyT) of different epithelial cells. The EMyT manifested by increased expression of α-smooth muscle actin and other contractile proteins, along with inhibition of genes responsible for cell proliferation. Induction of EMyT in γ-CYA–depleted cells depended on activation of serum response factor and its cofactors, myocardial-related transcriptional factors A and B. Loss of γ-CYA stimulated formin-mediated actin polymerization and activation of Rho GTPase, which appear to be essential for EMyT induction. Our findings demonstrate a previously unanticipated, unique role of γ-CYA in regulating epithelial phenotype and suppression of EMyT that may be essential for cell differentiation and tissue fibrosis.
INTRODUCTION
The formation of epithelial layers represents a key step in the development of multicellular organisms. Epithelia protect from external pathogens and other noxious environmental injury and help to create the unique architecture and biochemical composition of different internal organs. The majority of epithelial cells become well differentiated by acquiring unique structural characteristics, including cell–cell junctions and apicobasal cell polarity, which allow for the formation of barriers and directional transport of fluid and solutes. However, differentiated epithelial cells retain significant phenotypic plasticity and can be dedifferentiated or transdifferentiated into other cell types (Nieto, 2013). Such phenotypic plasticity is essential for normal tissue morphogenesis but can also contribute to the progression of various diseases. For example, epithelial-to-mesenchymal transition (EMT) is considered a key mechanism of metastatic dissemination of tumor cells (De Craene and Berx, 2013; Tam and Weinberg, 2013), whereas epithelial-to-myofibroblast transition (EMyT) can be important for the development of fibrosis in chronically inflamed tissues (Quaggin and Kapus, 2011; Lee and Nelson, 2012). Hence elucidating mechanisms of epithelial transdifferentiation is essential for a thorough understanding of the pathogenesis of human diseases.
A common and characteristic feature of EMT and EMyT is rearrangement of the actin cytoskeleton (Yilmaz and Christofori, 2009; Sandbo and Dulin, 2011). This rearrangement involves dismantling the apical and perijunctional actin bundles that are characteristic of epithelial cells and assembling prominent basal F-actin fibers abundant in fibroblasts and myofibroblasts (Yilmaz and Christofori, 2009; Le Bras et al., 2012). On the biochemical level, epithelial transdifferentiation results in switching from epithelial-specific to fibroblast- or smooth muscle–specific types of different actin-binding proteins (Sandbo and Dulin, 2011). Such cytoskeletal rearrangements are not just phenotypic consequences of EMT or EMyT, but instead are important mechanisms that drive epithelial transdifferentiation. Indeed, actin filaments are essential for generation, acceleration, and transduction of signals and forces that affect cell fate and differentiation (Olson and Nordheim, 2010). Furthermore, actin itself is known to be a potent regulator of gene expression by interacting with a number of transcriptional factors and chromatin-remodeling proteins in the cytoplasm and the nucleus (Olson and Nordheim, 2010; Visa and Percipalle, 2010). However, the roles of actin in regulating epithelial differentiation remain poorly understood.
Epithelial cells express cytoplasmic β- and γ- actins (β-CYA and γ-CYA, respectively). These two actin isoforms have almost identical sequences that differ in only four amino acids in close proximity to their N-terminus (Khaitlina, 2001; Perrin and Ervasti, 2010). Despite the high degree of sequence similarity, β-CYA and γ-CYA have different biochemical properties in vitro (Bergeron et al., 2010) and different localization in vivo (Belyantseva et al., 2009; Dugina et al., 2009; Tondeleir et al., 2009; Baranwal et al., 2012). Furthermore, several recent studies revealed nonredundant roles of β-CYA and γ-CYA in regulating cell growth, motility, and epithelial junctions (Belyantseva et al., 2009; Bunnell and Ervasti, 2010; Bunnell et al., 2011; Perrin and Ervasti, 2010; Baranwal et al., 2012). Evidence suggests that β-CYA and γ-CYA can control gene expression in different cell types. For example, early studies of β-CYA and γ-CYA overexpression in myoblasts demonstrated decreased transcription of endogenous actins and matrix adhesion proteins (Lloyd et al., 1992; Schevzov et al., 1995). Furthermore, recent gene arrays and proteomic studies of embryonic fibroblasts isolated from either β-CYA– or γ-CYA–null mice revealed significant alterations to the transcription program after deletion of individual cytoplasmic actins (Bunnell et al., 2011; Tondeleir et al., 2012). Nevertheless, the involvement of cytoplasmic actin isoforms in regulating gene expression and transdifferentiation of epithelial cells has not been investigated. This study demonstrates a previously unanticipated and unique role of γ-CYA in regulating EMyT in human epithelial cells via mechanisms involving transcriptional activity of serum response factor, polymerization of actin filaments, and stimulation of Rho GTPase.
RESULTS
γ-CYA, but not β-CYA, depletion induces expression of myofibroblast markers in human lung epithelial cells
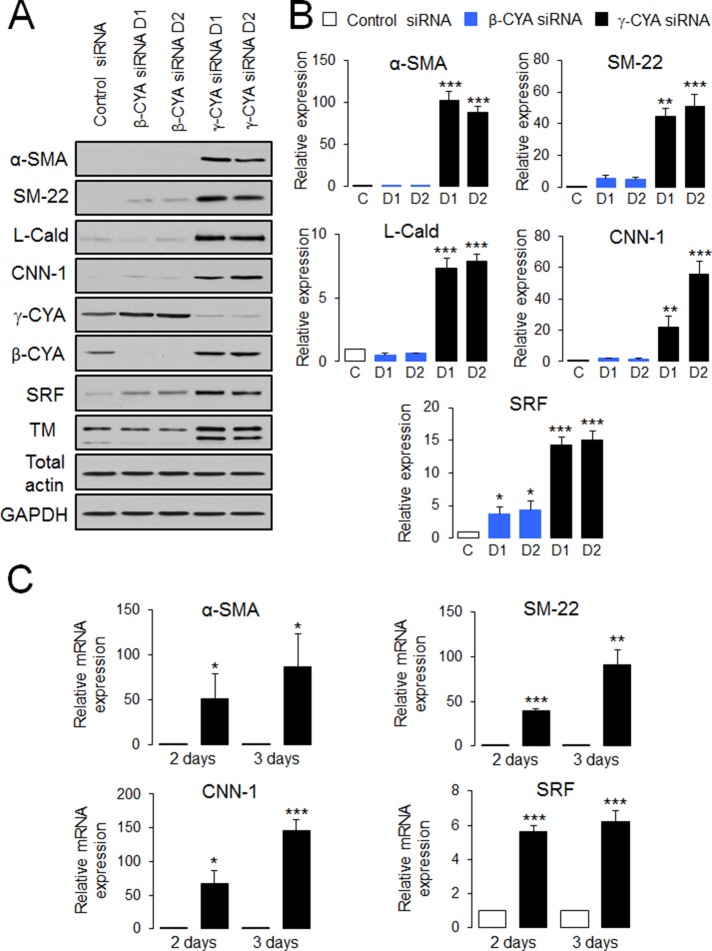
Because the actin cytoskeleton can regulate development of EMT by switching expression from epithelial-specific to mesenchyme-specific sets of cytoskeletal proteins (Zheng et al., 2008; Beach et al., 2011), we initially asked whether cytoplasmic actin isoforms can affect the development of EMT. To answer this question, we used A549 human lung epithelial cells treated with transforming growth factor-β (TGF-β), which represents a common in vitro model of EMT (Kasai et al., 2005). As expected, 48-h exposure to TGF-β (10 ng/ml) induced a prominent morphological change and biochemical signature characteristic of EMT, including elongated cell shape (Supplemental Figure S1A), the cadherin switch (down-regulation of E-cadherin and up-regulation of N-cadherin), and induction of mesenchymal protein markers such as vimentin and fibronectin (Supplemental Figure S1C). Knockdown of either β-CYA or γ-CYA using actin isoform–specific small interfering RNA (siRNA) SmartPools did not show consistent effects on TGF-β–dependent changes in the expression of epithelial and mesenchymal markers (Supplemental Figure S1C). Unexpectedly, loss of γ-CYA but not β-CYA induced marked expression of α-smooth muscle actin (α-SMA) in the absence of TGF-β (Supplemental Figure S1C). Because α-SMA induction was not accompanied by significant up-regulation of other mesenchymal markers such as N-cadherin and fibronectin, we hypothesized that γ-CYA depletion triggers a specific subset of EMT known as epithelial-to-myofibroblast transformation (EMyT). EMyT is characterized by induction of contractile cytoskeletal proteins abundant in myofibroblasts and smooth muscle cells (Sandbo and Dulin, 2011). To test this idea, we depleted either γ-CYA or β-CYA in A549 epithelial cells by using individual actin isoform–specific siRNA duplexes and subsequently examined expression of different EMyT markers on day 4 posttransfection. Transfection with different siRNA duplexes resulted in efficient and specific depletion of targeted actin isoforms, with >97 and 83% decrease in β-CYA and γ-CYA expression, respectively (Figure 1A and unpublished data). Of interest, neither knockdown significantly affected the total level of actin, due to compensatory up-regulation of the remaining CYA isoform (Figure 1A). Using densitometric analysis of control and β-CYA–depleted cell lysates (no α-SMA induction) probed with either actin isoform–specific or total-actin antibodies, we estimated that control A549 cells express ∼45% γ-CYA and 55% β-CYA.
FIGURE 1:
Downregulation of γ-CYA selectively stimulates expression of EMyT markers in lung epithelial cells. A549 epithelial cells were transfected with control or β-CYA– or γ-CYA–specific siRNA duplexes (D1 and D2), and expression of EMyT markers was analyzed by immunoblotting (A, B) and quantitative real-time RT-PCR (C) at different times after transfection. Immunoblots are quantified by densitometric analysis, and protein expression is calculated relative to the control siRNA–treated group. mRNA expression of all EMyT markers is normalized by the expression of a housekeeping gene. Data are presented as mean ± SE (n = 3); *p < 0.05, **p < 0.005, ***p < 0.0005.
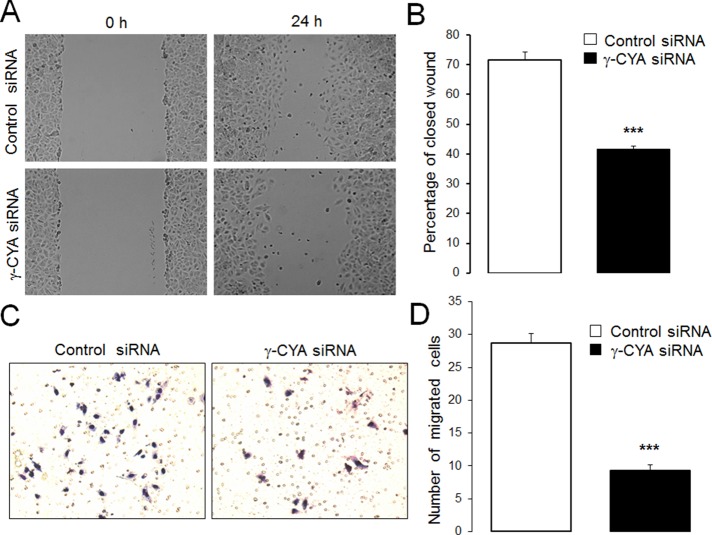
Specific depletion of γ-CYA caused dramatic induction of all studied EMyT markers, including α-SMA, SM-22, L-caldesmon (L-Cald), calponin-1 (CNN-1), and tropomyosin (TM), whereas β-CYA depletion did not stimulate expression of these contractile/cytoskeletal proteins (Figure 1, A and B). Induction of contractile proteins in γ-CYA–depleted A549 cells was due to up-regulation of their mRNA expression, evident as early as day 2 posttransfection (Figure 1C). Note that we obtained similar results after depleting γ-CYA with six different siRNA duplexes targeting the coding and untranslated parts of its mRNA (Figure 1 and unpublished data). Furthermore, induction of these contractile proteins after γ-CYA knockdown was detected in other types of epithelial cells, such as 293HEK (kidney), PANK1 (pancreatic), and SW13 (thyroid) cells, indicating that this is not a response unique to A549 cells (Supplemental Figure S2). To further distinguish between “classic” EMT, which alters cell shape and increases cell motility, and EMyT, which does not yield a promotile phenotype, we examined the effect of γ-CYA depletion on morphology and migration of A549 cells. Loss of γ-CYA resulted in the formation of rounded, well-spread cells that did not resemble the typical protrusive, spindle-shaped cells induced by TGF-β treatment (Supplemental Figure S1, A and B). Furthermore, γ-CYA depletion resulted in significant decrease in wound closure and Matrigel invasion compared with control siRNA–treated A549 cells (Figure 2). Our data are consistent with a previous study demonstrating attenuated migration of γ-CYA–depleted neuroblastoma cells (Shum et al., 2011). Taken together, these results reveal a previously unrecognized effect of γ-CYA depletion manifested by the EMyT-like transdifferentiation of human epithelial cells.
FIGURE 2:
Depletion of γ-CYA inhibits epithelial cell migration. (A, B) Control and γ-CYA–depleted A549 cell monolayers were mechanically wounded on day 3 posttransfection. To examine the rate of cell motility, the cell-free area was measured at 0 and 24 h after wounding; percentage of closed wound area was calculated as described in Materials and Methods. (C, D) Control and γ-CYA–depleted A549 cells were subjected to the Matrigel invasion assay. Data are presented as mean ± SE (n = 3); ***p ≤ 0.0005.
Induction of contractile proteins in γ-CYA–depleted cells depends on serum response factor
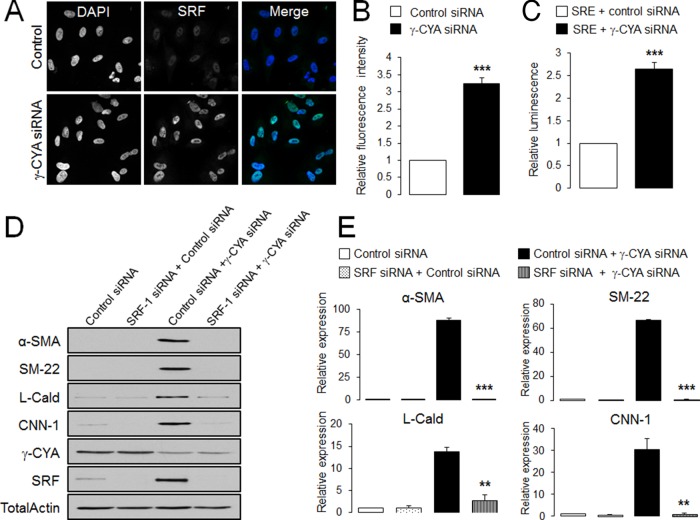
Next we sought to investigate potential molecular pathways leading to EMyT in γ-CYA–deficient epithelial cells. Given the fact that induction of contractile proteins during EMyT is mediated by a specific transcription factor called serum response factor (SRF), we asked whether SRF plays a role in γ-CYA–dependent EMyT. First we examined SRF expression by using immunoblotting, immunofluorescence analysis, and quantitative reverse transcription (RT) PCR. All these techniques demonstrated dramatic up-regulation of SRF expression in γ-CYA–depleted cells (Figures 1, A and C, and 3, A and B), where SRF was predominantly localized in the nucleus. Of note, down-regulation of β-CYA also increased levels of SRF protein, although in lower magnitude compared with γ-CYA–depleted cells (4- and 15-fold respectively; Figure 1, A and B). Loss of γ-CYA not only increased SRF level, but it also increased its activity as indicated by a luciferase-based assay of SRF-dependent promoters (Figure 3C). To establish a causal role of SRF in induction of EMyT, we performed siRNA-mediated knockdown of this transcriptional factor in control and γ-CYA–deficient A549 cells. Co-knockdown of SRF and γ-CYA dramatically attenuated expression of α-SMA, SM22, L-Cald, and CNN-1 caused by γ-CYA depletion (Figure 3, D and E). Together these data identified SRF as a key mediator of EMyT in γ-CYA–deficient epithelial cells.
FIGURE 3:
SRF mediates induction of EMyT markers in γ-CYA–depleted epithelial cells. (A, B) A549 cells transfected with either control or γ-CYA siRNAs were immunolabeled for SRF and counterstained with a nuclear dye, DAPI, on day 4 posttransfection. Intensity of nuclear SRF signal was quantified with image analysis. (C) A549 cells stably expressing SRE luciferase reporter were transfected with either control or γ-CYA siRNAs and cotransfected with a Renilla luciferase plasmid. Dual luciferase assay was performed 2 d posttransfection. The luminescence signal of SRF-dependent promoters was normalized by the Renilla luciferase signal. (D, E) A549 cells were subjected to sequential transfections with one of the following siRNA pairs: control–control, SRF–control, control–γ-CYA, and SRF–γ-CYA. Expression of targeted proteins and EMyT markers was determined by immunoblotting on day 3 after the second transfection. Data are presented as mean ± SE (n = 3); **p < 0.005, ***p < 0.0005.
Induction of contractile proteins of γ-CYA–depleted cells is regulated by MRTF
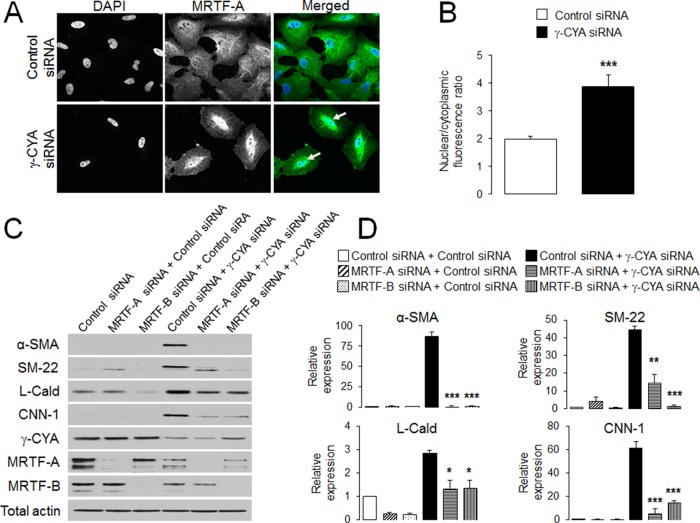
SRF is a multifunctional transcription factor that is known to regulate the expression not only of cytoskeletal proteins but also of so-called “early response” genes such as c-Fos, JunD, and Erg-1 (Soh et al., 1999; Lee et al., 2010b). Therefore we asked whether increased SRF expression after γ-CYA depletion resulted in ubiquitous activation of different SRF targets. Quantitative RT-PCR analysis showed that, in contrast to contractile proteins, mRNA levels of early-response genes were significantly decreased by γ-CYA knockdown (Supplemental Figure S3). This result indicates a specific shift of SRF activity toward transcriptional up-regulation of myogenic markers. Similar transcriptional reprogramming observed in previous studies was associated with a family of specific SRF coactivators that includes myocardin and myocardin-related transcriptional factor A (MRTF-A) and MRTF-B, expressed in muscle and nonmuscle cells, respectively (Posern et al., 2002; Miralles et al., 2003; Wang et al., 2004). Therefore we next investigated whether MRTF is involved in γ-CYA–dependent EMyT. MRTF is known to dual localize in the nucleus and the cytoplasm, and it has to be translocated into the nucleus in order to stimulate gene expression (Miralles et al., 2003; Pawlowski et al., 2010). According to our immunofluorescence labeling and confocal microscopy analysis, loss of γ-CYA triggered a considerable translocation of endogenous MRTF-A into the nucleus, as manifested by an approximately twofold increase in its nuclear/cytoplasmic ratio of the MRTF signal (Figure 4, A and B). Similar nuclear translocation was observed for exogenously expressed, Flag-tagged MRTF-A, as well as for endogenous or exogenous MRTF-B (Supplemental Figure S4). Furthermore, dual MRTF-A–γ-CYA or MRTF-B–γ-CYA knockdown significantly attenuated induction of all EMyT markers as compared with γ-CYA–only depleted cells (Figure 4, C and D). Of interest, depletion of an individual MRTF isoform (either MRTF-A or MRTF-B) almost completely suppressed induction of several contractile proteins despite the presence of the other MRTF isoform. This most likely indicates that both isoforms cooperate in expressional up-regulation of contractile proteins and that a certain threshold concentration of MRTF is required in order to stimulate the EMyT.
FIGURE 4:
MRTF is essential for the induction of EMyT markers in γ-CYA–depleted epithelial cells. (A, B) A549 cells transfected with either control or γ-CYA siRNAs were immunolabeled for MRTF-A and counterstained with a nuclear dye, DAPI. Intensity of nuclear and cytoplasmic MRTF-A, determined by image analysis, was used to calculate the nuclear/cytoplasmic ratio. Arrows indicate nuclear localization of MRTF-A in γ-CYA–depleted cells. (C, D) A549 cells were subjected to sequential transfections with one of the following siRNA pairs: control–control, MRTF-A–control, MRTF-B–control, control–γ-CYA, MRTF-A–γ-CYA, and MRTF-B–γ-CYA. Expression of targeted proteins and EMyT markers was determined by immunoblotting on day 3 after the second transfection. Data are presented as mean ± SE (n = 3); *p < 0.05, **p < 0.005, ***p < 0.0005.
EMyT in γ-CYA–depleted cells is associated with increased actin polymerization
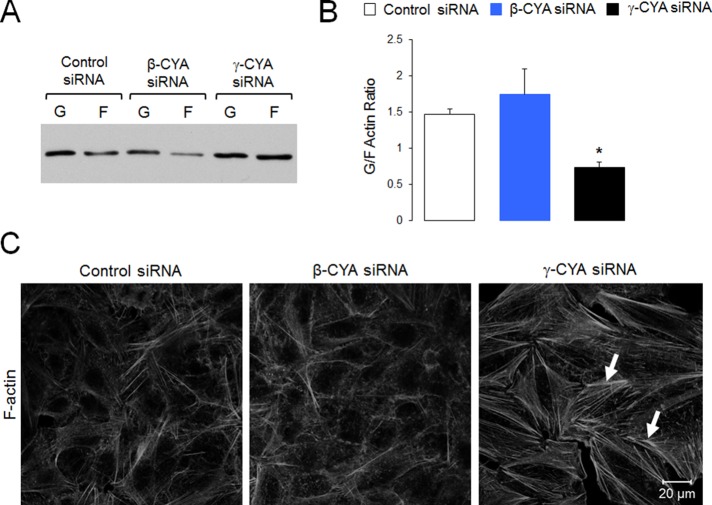
MRTF-dependent gene expression is regulated by the actin cytoskeleton (Posern and Treisman, 2006; Olson and Nordheim, 2010). Specifically, MRTF binds to G-actin, and such binding strongly inhibits MRTF activity (Sotiropoulos et al., 1999; Miralles et al., 2003; Vartiainen et al., 2007). As a result, decreasing G actin–MRTF interactions should result in MRTF activation. How does γ-CYA depletion selectively activate MRTF, whereas loss of β-CYA is inefficient? Three different scenarios can be envisioned. The first scenario implies decreased expression of total actin. The second scenario involves increased actin polymerization in γ-CYA–deficient cells, which depletes the G-actin pool available for MRTF binding. The final scenario involves differential inhibition of MRTF by different actin isoforms, with γ-CYA having a much stronger affinity for this transcription factor. Our results ruled out the first scenario, since γ-CYA depletion did not decrease the total actin level in A549 cells (Figure 1A). This is consistent with our previous findings in colonic epithelial cells and reflects compensatory up-regulation of β-CYA expression (Baranwal et al., 2012). We also examined actin polymerization status after γ-CYA and β-CYA depletion. Biochemical fractionation demonstrated a selective decrease in the relative amount of G-actin (decreased G-/F-actin ratio) in γ-CYA siRNA–treated A549 cells compared with the control group (Figure 5, A and B). In addition, fluorescence labeling revealed accelerated assembly of F-actin–based stress fibers after γ-CYA depletion, which is indicative of increased actin polymerization (Figure 5C, arrows). By contrast, β-CYA knockdown neither significantly decreased the G-/F-actin ratio nor induced stress fiber assembly (Figure 5).
FIGURE 5:
Depletion of γ-CYA but not β-CYA induces actin polymerization. A549 cells transfected with control or cytoplasmic actin isoform–specific siRNAs were subjected to detergent fractionation to determine the G-/F-actin ratio (A, B) or fluorescence labeling with Alexa 555–phalloidin to visualize actin filaments (C). Arrows highlight basal stress fibers in γ-CYA–depleted cells. Data are presented as mean ± SE (n = 3); *p < 0.05.
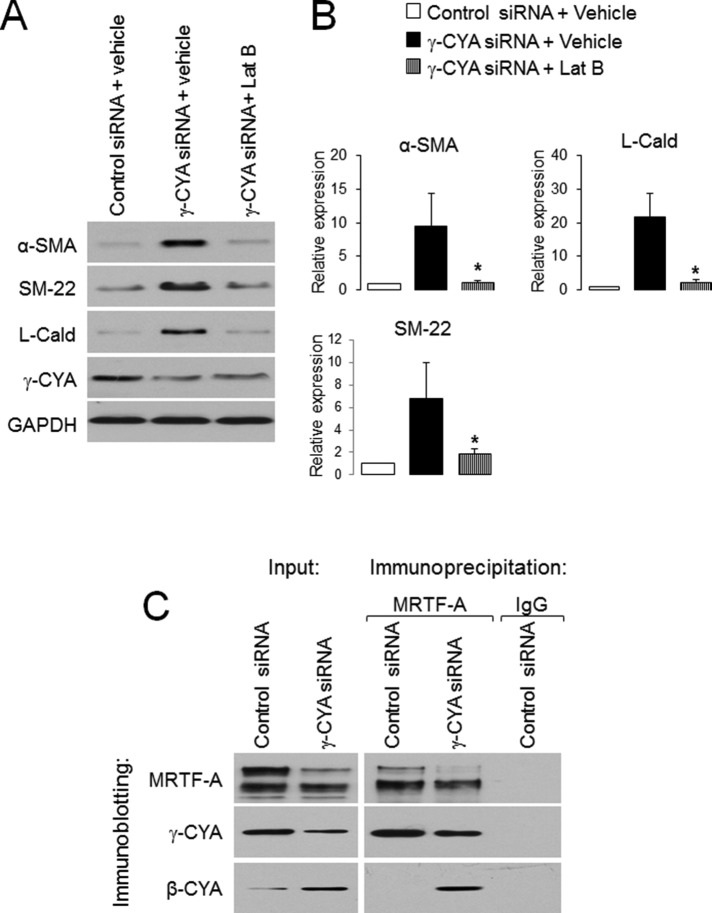
To gain functional insight into the role of monomeric actin in EMyT induction, we treated γ-CYA–depleted A549 cells with either vehicle or an actin-depolymerizing drug, latrunculin B (LatB; Morton et al., 2000). LatB treatment dramatically attenuated γ-CYA–dependent expressional up-regulation of contractile proteins, highlighting the significant role of G-actin sequestration in EMyT (Figure 6, A and B). Finally, we sought to compare interactions of two cytoplasmic actins with MRTF. MRTF-A was immunoprecipitated from total A549 epithelial cell lysates and subsequently probed with monoclonal antibodies selectively recognizing either γ-CYA or β-CYA. Remarkably, only γ-CYA coprecipitated with MRTF-A, whereas β-CYA was not precipitated with this transcriptional regulator (Figure 6C). Similarly, γ-CYA but not β-CYA was detected in immunoprecipitates obtained with SRF antibody (unpublished data). In contrast, MRTF-A antibody pulled down β-CYA from γ-CYA–depleted A549 cell lysates (Figure 6C). These data suggest that γ-CYA and β-CYA can compete for MRTF binding and that MRTF preferentially binds to γ-CYA in control A549 cells. Of interest, loss of γ-CYA appears to decrease total expression of MRTF-A protein (Figures 4C and 6C). This provides additional indirect arguments in support of preferential MRTF–γ-CYA interactions that not only inactivate but might also stabilize this transcriptional regulator in epithelial cells.
FIGURE 6:
EMyT induction in γ-CYA–depleted epithelial cells is controlled by actin polymerization, and γ-CYA preferentially interacts with MRTF. (A, B) γ-CYA–depleted A549 cells were treated for 24 h with either vehicle or the F-actin–depolymerizing drug Lat B (1 μM), and expression of EMyT markers was determined by immunoblotting. Data are presented as mean ± SE (n = 3); *p < 0.05. (C) Association of MRTF-A with γ-CYA and β-CYA in control and γ-CYA–depleted A549 cells was examined using immunoprecipitation with anti–MRTF-A antibody.
Inhibition of formins and RhoA signaling suppresses EMyT in γ-CYA–depleted epithelial cells
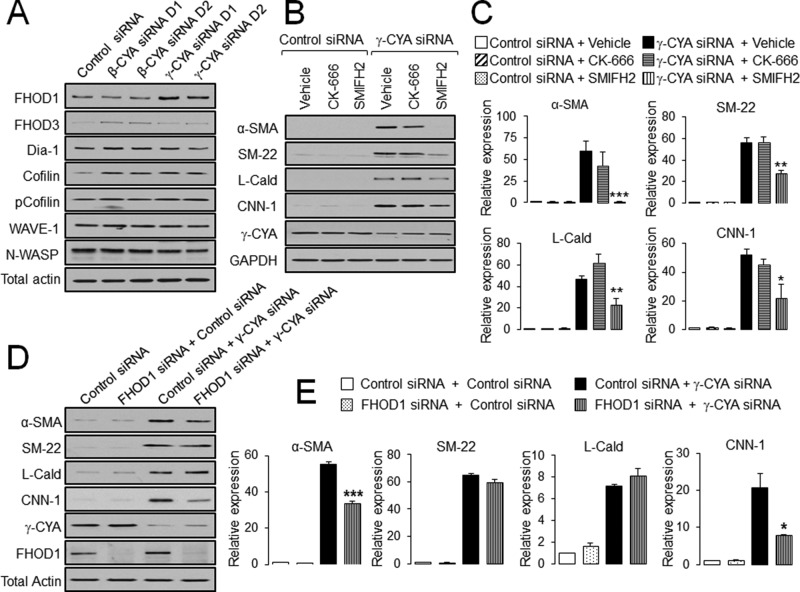
Because increased actin polymerization appears to be involved in the EMyT of γ-CYA–depleted epithelial cells, we investigated mechanisms underlying this process. A large number of actin-binding proteins can stimulate polymerization of either linear or branched actin filaments (Campellone and Welch, 2010). Immunoblotting of several well-characterized actin-polymerizing factors showed a selective increase in the expression of formin homology 2 domain containing 1 (FHOD1) in γ-CYA–depleted A549 cells (Figure 7A). According to densitometric analysis, FHOD1 protein level increased (3.0 ± 0.8)- and (2.5 ± 0.6)-fold in cells treated with γ-CYA siRNA duplex 1 and 2, respectively (n = 3, p < 0.05). To examine the involvement of different modes of actin polymerization in EMyT induction, we treated γ-CYA–depleted cells for 24 h with a pharmacological inhibitor of formins, SMIFH2 (50 μM; Rizvi et al., 2009), an inhibitor of Arp2/3 polymerization, CK-666 (50 μM; Nolen et al., 2009), or vehicle. The formin inhibitor significantly suppressed induction of all EMyT markers, whereas inhibition of the Arp2/3 complex was ineffective (Figure 7B). To investigate the specific role of FHOD1, we compared the effects of γ-CYA and FHOD1/γ-CYA knockdowns on EMyT induction. Depletion of FHOD1 only partially reversed the induction of α-SMA and CNN-1 without having significant effects on SM-22 and L-Cald expression (Figure 7C). These data indicate that up-regulation of FHOD1 alone does not explain the entire biochemical signature of EMyT of γ-CYA–depleted epithelial cells, which could be regulated by additional members of the formin family or other actin-binding proteins.
FIGURE 7:
Upregulation of EMyT markers in γ-CYA-depleted cells depends on formin-mediated actin polymerization. (A) A549 cells were transfected with control or β-CYA– or γ-CYA–specific siRNA duplexes, and expression of different actin-polymerizing proteins was analyzed by immunoblotting. (B, C) Control and γ-CYA–depleted A549 cells were treated for 24 h with either vehicle or pharmacological inhibitors of Arp2/3 complex (CK-666) and formin-dependent actin polymerization (SMIFH2). Effects of the inhibitors on induction of EMyT marker were determined by immunoblotting. (D, E) A549 cells were subjected to sequential transfections with one of the following siRNA pairs: control–control, FHOD1–control, control–γ-CYA, and FHOD1–γ-CYA. Expression of targeted proteins and EMyT markers was determined by immunoblotting on day 3 after the second transfection. Data are presented as mean ± SE (n = 3); *p < 0.05, **p < 0.005, ***p < 0.0005.
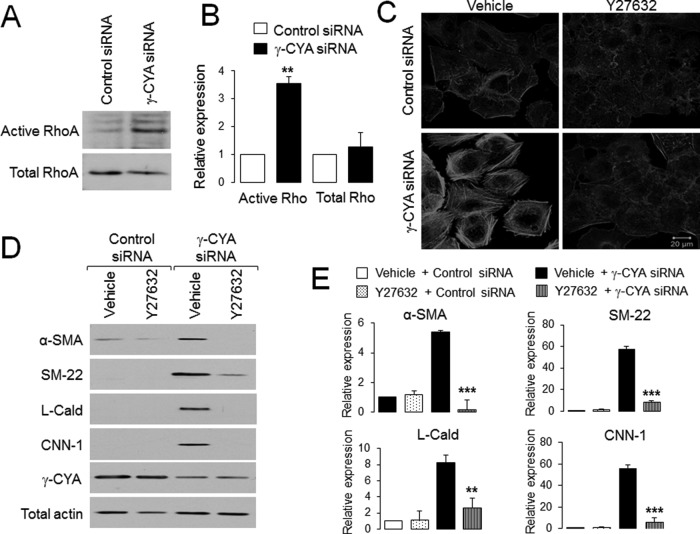
Formin-dependent actin polymerization is controlled by a small GTPase, RhoA (Goode and Eck, 2007). Furthermore, RhoA was shown to be essential for activation of an SRF/MRTF-dependent transcriptional program (Sotiropoulos et al., 1999; Mack et al., 2001; Miralles et al., 2003). Thus we next investigated the involvement of this GTPase in the observed EMyT response by using a Rho activation assay and inhibition of Rho signaling. Loss of γ-CYA resulted in a more than threefold increase in the amount of active RhoA without significant changes in total Rho expression (Figure 8, A and B). Furthermore Y27632, which is known to inhibit Rho-associated kinase (ROCK), suppressed induction of contractile proteins and assembly of F-actin bundles in γ-CYA–depleted A549 cells (Figure 8, C–E). Overall these results strongly suggest that Rho activation is essential for the EMyT induction caused by γ-CYA deficiency.
FIGURE 8:
Rho activation is essential for EMyT induction in γ-CYA–depleted cells. (A, B) Level of total and GTP-bound active RhoA in control and γ-CYA–depleted A549 cells was determined by a rhotekin pull-down assay and immunoblotting. (C–E) Control and γ-CYA–depleted A549 cells were treated for 24 h with either vehicle or ROCK inhibitor, Y27632, and assembly of stress fibers (C), as well as expression of different EMyT markers (D, E), was examined by fluorescence analysis and immunoblotting. Data are presented as mean ± SE (n = 3); **p < 0.005, ***p < 0.0005.
Weakening of cell–cell contacts is not responsible for induction of contractile proteins in γ-CYA–deficient epithelial cells
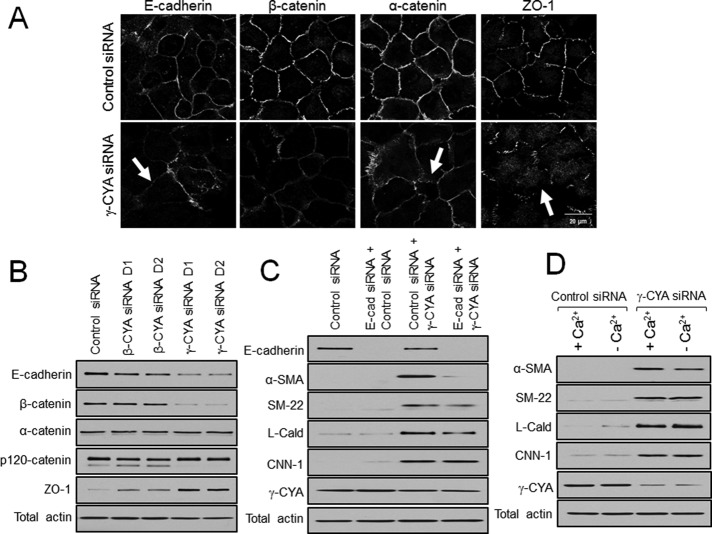
Next we asked whether up-regulation of contractile proteins can be causally linked to the loss of some epithelial features in γ-CYA–depleted A549 cells. Considering the compelling data that disruption of cell–cell junctions is capable of activating Rho and stimulating the EMyT of renal and mammary epithelia (Fan et al., 2007; Speight et al., 2013), we investigated the effects of junctional disassembly on transdifferentiation of γ-CYA depleted cells. First, we examined the integrity of cell–cell contacts along with the expression of major junctional proteins. Immunofluorescence analysis showed dramatic disruption of E-cadherin–based adherens junctions (AJs) and zonula occludens 1 (ZO-1)–based tight junctions (TJs) in γ-CYA–deficient A549 cells (Figure 9A, arrows). Furthermore, γ-CYA knockdown markedly decreased protein expression of E-cadherin and β-catenin and upregulated ZO-1 levels in A549 cells (Figure 9B). These data suggest that in poorly differentiated epithelial cells, such as A549, γ-CYA controls the integrity of both AJs and TJs, which contrasts with its functions in well-differentiated colonic epithelium, where γ-CYA was shown to selectively regulate TJ assembly (Baranwal et al., 2012). If down-regulation of cell–cell contacts contributes to transcriptional reprogramming of A549 cells, forceful junctional disassembly should accelerate expression of EMyT markers either in control or γ-CYA–depleted epithelial cells. This hypothesis was tested using two different approaches; one involved siRNA-mediated down-regulation of E-cadherin expression, and the other involved global disassembly of epithelial junctions by extracellular calcium depletion (Ivanov et al., 2004). Neither approach resulted in induction of EMyT in control A549 cells (Figure 9, C and D). Furthermore, in contrast to our prediction, loss of E-cadherin inhibited rather than accelerated expression of some contractile proteins (α-SMA and SM-22) in γ-CYA–depleted cells, whereas calcium depletion was entirely ineffective (Figure 9, C and D). Together these data strongly suggest that disruption of epithelial junctions plays no role in transcriptional reprogramming of γ-CYA–depleted cells.
FIGURE 9:
Disassembly of intercellular junctions does not contribute to induction of EMyT in γ-CYA–depleted epithelial cells. A549 cells were transfected with either control or cytoplasmic actin isoform–specific siRNAs. The integrity of cell–cell contacts (A) and expression of junctional proteins (B) were examined on day 4 posttransfection. Arrows highlight disassembly of intercellular junctions in γ-CYA–depleted cells. (C) A549 cells were subjected to sequential transfections with one of the following siRNA pairs: control–control, E-cadherin–control, control–γ-CYA, and E-cadherin–γ-CYA. Expression of targeted proteins and EMyT markers was determined by immunoblotting on day 3 after the second transfection. (D) Control and γ-CYA–depleted A549 cells were either incubated in normal cell culture medium (+Ca2+) or subjected to 24 h of extracellular calcium depletion (–Ca2+) and analyzed for expression of EMyT markers.
DISCUSSION
γ-CYA is essential for preserving the epithelial cell phenotype in vitro
Involved not only in the assembly of cytoskeletal structures but also in the control of gene expression in the nucleus, actin is a key regulator of cellular homeostasis (Posern and Treisman, 2006; Olson and Nordheim, 2010; Visa and Percipalle, 2010). Mammalian cells express six closely related actin isoforms. However, specific roles of these actin isoforms in regulating the transcriptional programs of different tissues remain poorly understood. The present study reveals a previously unanticipated and unique role of γ-CYA in maintaining the epithelial cell phenotype. This conclusion is based on our finding that depletion of γ-CYA in several types of epithelial cells induces an EMyT-like transition characterized by up-regulation of smooth muscle/myofibroblast-specific contractile cytoskeletal proteins (Figure 1 and Supplemental Figure S2). EMyT was accompanied by decreased transcription of genes involved in cell proliferation (Supplemental Figure S3) and alterations in cellular phenotype characterized by decreased cell motility and disassembly of cell–matrix adhesions (Figure 2 and unpublished data). Combined, the described phenomena suggest reprogramming of epithelial cells toward myogenic differentiation after the loss of γ-CYA expression (Figure 10). Such phenotypic reprogramming could be a common feature of different cell types, since up-regulation of α-SMA also accompanies γ-CYA depletion in epidermal and embryonic fibroblasts (Dugina et al., 2009; Bunnell and Ervasti, 2010). A surprising observation yielded by the present study is that induction of EMyT in epithelial cells appears to be a unique consequence of γ-CYA depletion and that down-regulation of β-CYA has only minor effects on contractile protein expression (Figure 1). Previous studies demonstrated noticeable induction of α-SMA and other smooth muscle proteins in embryonic fibroblasts isolated from β-CYA–null mice (Bunnell et al., 2011; Tondeleir et al., 2012) but showed lack of such induction after skeletal muscle–specific ablation of β-CYA (Prins et al., 2011). These contradictory results can be explained by different levels of γ-CYA and β-CYA in different cell types. For example, embryonic fibroblasts express large amounts of β-CYA and therefore could be more susceptible to EMyT induction after β-CYA depletion. In contrast, epithelial cells used in our study are characterized by high levels of γ-CYA (∼45% of the total actin), which acts as an efficient inhibitor of myogenic transdifferentiation.
FIGURE 10:
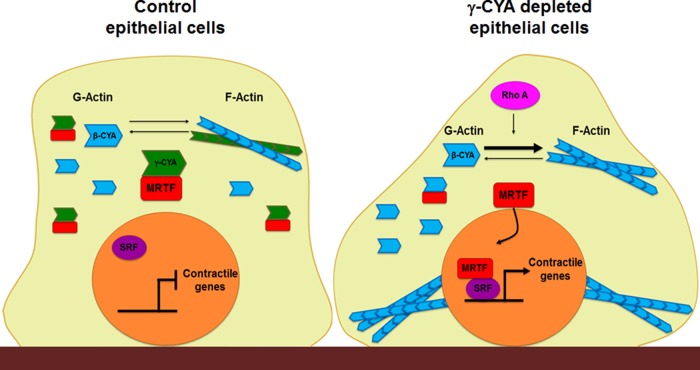
Schematic diagram of key molecular events that are likely to mediate myogenic transdifferentiation of γ-CYA–depleted epithelial cells. In control cells (left), MRTF preferentially interacts with monomeric γ-CYA in the cytoplasm, which prevents nuclear translocation and activation of this transcriptional regulator. As a result, nuclear SRF does not stimulate expression of contractile genes, and the myogenic transcriptional program remains inactive. Loss of γ-CYA triggers RhoA activation and Rho-mediated actin polymerization, which allow nuclear translocation of MRTF and subsequent formation of active MRTF/SRF complexes. This stimulates transcription of contractile/cytoskeletal genes, leading to EMyT.
Transcriptional reprogramming of γ-CYA–depleted epithelial cells is mediated by SRF and MRTF
Our data suggest that induction of EMyT in γ-CYA–depleted epithelial cells is regulated by SRF and its coactivator, MRTF (Figures 3 and 4). SRF is known to be a versatile transcriptional factor that controls the expression of hundreds of mammalian genes in a cell-specific manner (Miano et al., 2007). All SRF targets genes are characterized by the presence of consensus binding motifs known as GArG boxes (Posern and Treisman, 2006; Miano et al., 2007). Of interest, SRF can stimulate its own expression, and this positive feedback loop is likely to contribute to the considerable induction of SRF in γ-CYA–depleted epithelial cells (Figures 1 and 2). SRF is a crucial regulator of two distinct genetic programs, one involving genes responsible for cell growth and proliferation and the other comprising genes responsible for muscle differentiation (Miano et al., 2007; Olson and Nordheim, 2010; Mack, 2011). These transcriptional programs appear to be mutually exclusive and depend on specific SRF cofactors (Gineitis and Treisman, 2001). Thus cofactors of the Elk family shift SRF-dependent transcription toward activation of progrowth genes, whereas interactions with myocardin family cofactors, such as MRTF, stimulate a subset of genes responsible for muscle differentiation (Wang et al., 2002, 2004). Remarkably, γ-CYA knockdown stimulated expression of MRTF-responsible contractile genes (Figure 1) while inhibiting transcription of Elk1-dependent SRF targets (Supplemental Figure S3). This indicates a specific shift in the SRF-dependent transcription program toward MRTF-regulated genes. Furthermore, the role of MRTF in transcriptional reprogramming of γ-CYA–depleted epithelial cells is supported by our evidence of nuclear translocation of MRTF-A and MRTF-B under these conditions (Figure 4 and Supplemental Figure S4) and significantly diminished expression of contractile proteins after codepletion of γ-CYA with either MRTF isoform (Figure 4). A number of previous studies demonstrated modulation of MRTF-dependent gene expression by either overexpression of exogenous actin or drug-induced changes in actin polymerization (Sotiropoulos et al., 1999; Miralles et al., 2003; Vartiainen et al., 2007). Our study provides the first evidence that the MRTF/SRF-dependent transcriptional program can be regulated by more physiologically relevant changes in expression of endogenous actin.
EMyT induction in γ-CYA–depleted epithelial cells depends on actin polymerization
The observed expressional up-regulation of contractile genes in γ-CYA-depleted epithelial cells concurs well with a current paradigm that considers actin an inhibitor of the MRTF/SRF-dependent transcriptional program (Posern and Treisman, 2006; Miano et al., 2007; Olson and Nordheim, 2010). It has been shown that G-actin directly binds to the N-terminal RPEL motifs of MTRF (Mouilleron et al., 2011) and inhibits this transcriptional coactivator via two separate mechanisms. One mechanism involves blocking MRTF interactions with importins, which prevents its nuclear translocation (Pawlowski et al., 2010), whereas the other mechanism involves G-actin–dependent inhibition of MRTF activity in the nucleus (Vartiainen et al., 2007; Baarlink et al., 2013). Different signaling pathways or biochemical maneuvers that decrease G-actin levels result in stimulating MRTF/SRF-dependent gene expression (Sotiropoulos et al., 1999; Posern et al., 2002, 2004; Miralles et al., 2003). However, induction of EMyT in γ-CYA–depleted epithelial cells is unlikely to be a trivial consequence of diminished actin expression since it is not accompanied by a decrease in total actin level (Figure 1). Furthermore, depletion of highly homologous β-CYA fails to elicit similar transcriptional cell reprogramming. Taken together, these data highlight γ-CYA as a specific and potent suppressor of MRTF/SRF-dependent gene expression in human epithelia.
We believe that γ-CYA inhibits myogenic transdifferentiation of epithelial cells by two complementary mechanisms: controlling the G-/F-actin ratio and preferentially interacting with the MRTF/SRF complex. The first mechanism is supported by our data showing that depletion of γ-CYA, but not β-CYA, in A549 cells triggered actin polymerization, leading to a decrease in the G-actin level and assembly of prominent cytoplasmic stress fibers (Figure 5, A–C). LatB-induced depolymerization of actin filaments was sufficient to reverse the EMyT in γ-CYA–depleted cells, highlighting the crucial role of G-actin sequestration in such epithelial cell reprogramming (Figure 6). Of interest, even though the G/F ratio is lower after γ-CYA depletion, there appears to be a significant amount of G-actin, which does not prevent MRTF activation (Figure 5, A and B). It is likely that this G-actin is bound to other actin-binding proteins, which compete with its MRTF binding.
Increased actin polymerization after γ-CYA knockdown cannot be explained by the altered biochemical properties of actin per se because β-CYA forms less-stable filaments compared with γ-CYA (Bergeron et al., 2010; Baranwal et al., 2012). Hence γ-CYA depletion should initiate mechanisms promoting actin polymerization. One such mechanism involves the activation of formins, the proteins that initiate polymerization of linear actin cables (Goode and Eck, 2007). Several lines of evidence support the role of formins in the observed myogenic transdifferentiation. First, the EMyT caused by γ-CYA knockdown was accompanied by the appearance of prominent stress fibers, which is a typical consequence of formin activation (Figure 5C). Second, pharmacological inhibition of formin-dependent, but not Arp2/3-dependent, actin polymerization reversed the expression of EMyT markers in γ-CYA–depleted cells (Figure 7, C and D). Finally, expression of FHOD1 formin was selectively stimulated by loss of γ-CYA, and this event partially contributed to EMyT (Figure 7). Our findings agree with the current literature describing the roles of different formins (including FHOD1) in stimulating MRTF/SRF-dependent gene expression (Copeland and Treisman, 2002; Staus et al., 2011; Baarlink et al., 2013). Note that FHOD1 knockdown only partially inhibited EMyT in γ-CYA–depleted epithelial cells (Figure 7, D and E). This likely reflects either compensatory effects from other formins or the cooperative actions of different members of the large formin protein family in the observed epithelial cell transdifferentiation.
Another mechanism that can contribute to γ-CYA–dependent suppression of myogenic transdifferentiation of epithelial cells involves direct inhibition of MRTF by preferential γ-CYA binding. This mechanism is supported by our findings that endogenous γ-CYA, but not β-CYA, was immunoprecipitated with MRTF-A or SRF from control A549 epithelial cells and that the MRTF-A–β-CYA interaction was detectable only after γ-CYA depletion (Figure 6C and unpublished data). These results are unexpected, given several previous studies that demonstrated strong interactions between β-CYA and MRTF family proteins (Sotiropoulos et al., 1999; Posern et al., 2002, 2004; Miralles et al., 2003). Furthermore, recent crystallographic analysis revealed binding of MRTF-A RPEL motifs to the hydrophobic cleft between subdomains 1 and 3 of the actin monomer (Mouilleron et al., 2011), which has an identical amino acid sequence in both γ-CYA and β-CYA. It is worth mentioning that interactions between β-CYA and MRTF were previously examined in conditions of overexpression of one or both binding partners, but no previous studies compared the relative affinity of γ-CYA and β-CYA to MRTF. Because MRTF-A has several binding sites and can create a compact pentavalent complex with G-actin (Mouilleron et al., 2011), differences in γ-CYA and β-CYA self-oligomerization (Bergeron et al., 2010) may underline preferential binding of the former actin isoform to MRTF. Of interest, we found that β-CYA could interact with MRTF after γ-CYA depletion (Figure 6C), although such interaction did not inhibit nuclear translocation of MRTF and EMyT induction. It is possible that MRTF-γ-CYA and MRTF-β-CYA complexes include additional unique binding partners that modulate cellular localization and activity of MRTF. It has been reported that cytoplasmic actins may have unique binding partners despite their remarkable structural similarity. For example, γ-CYA is known to specifically associate with annexin V and BAP31 (Tzima et al., 2000; Ducret et al., 2003), whereas β-CYA selectively interacts with ezrin and βCap73 (Shuster and Herman, 1995; Shuster et al., 1996).
EMyT induction in γ-CYA–depleted cells is regulated by Rho independently of disrupted intercellular junctions
Our data strongly suggest that the observed myogenic transdifferentiation of epithelial cells is mediated by Rho signaling. Indeed, RhoA was significantly activated after γ-CYA knockdown (Figure 8, A and B), and pharmacological inhibition of Rho signaling blocked expression of all tested EMyT markers (Figure 8, C and D). Rho GTPases are well-known positive regulators of MRTF/SRF-mediated gene expression and may act in cytoskeletal-dependent and independent-fashions (Sotiropoulos et al., 1999; Mack et al., 2001; Miralles et al., 2003). Thus activated RhoA triggers a profound actin polymerization, thereby depleting the MRTF-inhibiting G-actin pool (Sotiropoulos et al., 1999; Mack et al., 2001). In addition, Rho is capable of directly interacting with and phosphorylating MRTF (Miralles et al., 2003). The observed inhibition of EMyT by Y27632 implicates Rho-ROCK-dependent stimulation of actin polymerization in the myogenic reprogramming of γ-CYA–depleted epithelial cells. Indeed, ROCK inhibition not only attenuated the expression of contractile proteins, but also blocked stress fiber formation (Figure 8), most likely by inhibiting formin-dependent nucleation and accelerating cofilin-dependent disassembly of actin filaments (Mack et al., 2001; Shum et al., 2011; Staus et al., 2011).
An important question that remains unanswered relates to the mechanisms of Rho activation in γ-CYA–depleted epithelial cells. One plausible mechanism may involve disruption of epithelial junctions. Although loss of cell–cell contacts per se does not induce myogenic reprogramming (Masszi et al., 2004, 2010), it does provide the necessary permissive signals for TGF-β–induced EMyT (Fan et al., 2007; Speight et al., 2013). Although loss of γ-CYA disrupted TJs and decreased expression of E-cadherin and β-catenin in A549 cells (Figure 9, A and B), junctional disassembly neither induced EMyT alone nor potentiated myogenic transdifferentiation of epithelial cells under our experimental conditions (Figure 9, C and D). Remarkably, down-regulation of E-cadherin even reversed some biochemical manifestations of EMyT in γ-CYA–depleted cells (Figure 9C), which agrees with a recent report implicating E-cadherin in the regulation of MRTF-SRF–dependent transcription in epithelial cells (Busche et al., 2010). Alternative mechanisms of Rho regulation in γ-CYA–depleted cells might involve increased expression of GTP exchange factors, as this occurs during TGF-β–induced EMyT (Busche et al., 2010).
What is the physiological relevance of the reported γ-CYA–dependent inhibition of myogenic transdifferentiation? Although little is known about expressional regulation of γ-CYA in different tissues, some evidence suggests that altered levels of this actin isoform may contribute to both normal development and disease states. First, γ-CYA is highly expressed in stem cells and proliferating myoblasts, but its expression becomes dramatically down-regulated during myoblast differentiation into myotubules (Lloyd and Gunning, 2002; Tondeleir et al., 2009; Drummond and Friderici, 2013). These data indicate that loss of γ-CYA expression can be essential for normal myogenesis. Second, down-regulation of γ-CYA may contribute to the development of tissue fibrosis in chronic inflammatory diseases and the drug resistance of certain types of cancer. Indeed, fibrosis is associated with induction of myofibroblasts that can originate from other cell types, including epithelial cells, endothelial cells, and pericytes (Willis et al., 2006; Ueha et al., 2012). Because γ-CYA is down-regulated in inflammatory conditions such as pancreatitis (Lee et al., 2010a), it is tempting to speculate that decreased expression of γ-CYA can contribute to transdifferentiation of epithelial or other cell types into myofibroblasts in chronically inflamed tissues. Decreased expression of γ-CYA is also characteristic of some types of aggressive tumors, such as HER-2/neu–expressing breast cancer cells (Oh et al., 1999), metastatic salivary gland carcinoma cells (Suzuki et al., 1998), and acute lymphoblastic leukemia (Verrills et al., 2006). Although the pathophysiological consequence of such expressional down-regulation is unknown, one study linked dysfunctions of γ-CYA with the development of drug resistance (Verrills et al., 2006). Clearly, more studies are needed to address possible pathophysiological implications of altered expression and activity of γ-CYA.
MATERIALS AND METHODS
Antibodies and other reagents
The following primary polyclonal (pAb) and monoclonal (mAb) antibodies were used to detect cytoskeletal, signaling, and junctional proteins: anti–calponin-1, L-caldesmon, p120-catenin, E-cadherin, and vimentin mAbs (BD Biosciences, San Jose, CA); anti–α-catenin mAb (Epitomic, Burlingame, CA); anti–total actin (MAB1501) and Rho mAbs (Millipore, Billerica, MA); anti–ZO-1 pAb (Invitrogen, Carlsbad, CA); anti–β-catenin, fibronectin pAbs, and anti-tropomyosin mAb (clone TM311; Sigma-Aldrich, St. Louis, MO); anti–SM-22 and N-cadherin pAbs (Abcam, Cambridge, MA); anti-MRTF-A/MLK1, MRTF-B/MLK2, and anti-mDia1 pAbs (Bethyl Laboratories, Montgomery, TX); and anti-SRF, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and lamin A/C pAbs (Santa Cruz Biotechnology, Santa Cruz, CA). Monoclonal antibodies selectively recognizing β-CYA, γ-CYA, and αSMA were described previously (Dugina et al., 2009). Alexa Fluor 488– and Alexa Fluor 555–conjugated donkey anti-mouse and goat anti-rabbit secondary antibodies and Alexa Fluor 555–phalloidin were obtained from Invitrogen. Horseradish peroxidase–conjugated goat anti-mouse and anti-rabbit antibodies were purchased from Bio-Rad Laboratories (Hercules, CA) and Thermo Scientific (Rockford, IL). p3XFLAG-MLK1 and p3XFLAG MKL2 plasmid were obtained from Addgene (Cambridge, MA). Latrunculin B, CK-666, SMIFH2, and Y27632 were obtained from EMD-Millipore, and TGF-β1 was purchased from R&B Systems (Minneapolis, MN). All other chemicals were obtained from Sigma-Aldrich.
Cell culture and calcium depletion
A549, 293HEK, PANK1, and SW13 cells were purchased from the American Type Culture Collection (Manassas, VA). A549 cells (passages 5–13) were cultured in DMEM/F12 supplemented with 10% fetal bovine serum (FBS), 15% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and penicillin-streptomycin. Other cell lines were propagated in a high-glucose DMEM supplemented with 10% FBS, HEPES, and antibiotics. Cells were plated on six-well plastic plates and collagen-coated coverslips for biochemical experiments and immunofluorescence labeling, respectively. To deplete extracellular calcium, A549 cells monolayers were washed twice with Eagle's MEM for suspension culture supplemented with 5 μM CaCl2, 10 mM HEPES, 14 mM NaHCO3, and 10% dialyzed FBS (designated here as S-MEM) and incubated in S-MEM for 24 h at 37°C.
Total cell lysate and immunoblotting
To obtain total cell lysates, cells were homogenized in RIPA buffer (20 mM Tris, 50 mM NaCl, 2 mM EDTA, 2 mM ethylene glycol tetraacetic acid [EGTA], 1% sodium deoxycholate, 1% Triton X-100 [TX-100], and 0.1% SDS, pH 7.4) containing protease inhibitor cocktail and phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich), and the obtained lysates were cleared by centrifugation (20 min at 14,000 × g). Protein concentration of total lysates and cellular fractions was determined using a bicinchoninic acid protein assay kit. Samples were diluted with 2× SDS sample buffer and boiled. SDS–PAGE electrophoresis and immunoblotting were performed using a standard protocol with equal amounts of total protein (10 μg) loaded per lane and transferred onto nitrocellulose membrane. Results shown are representative immunoblots of at least three independent experiments. Protein expression was quantified by densitometry, and signal intensities were calculated using ImageJ Software (National Institutes of Health, Bethesda, MD). Data are presented as normalized values, assuming the expression level of control siRNA–treated groups was at 1 arbitrary unit. Statistical analysis was performed with row densitometric data using Excel (Microsoft, Redmond, WA)
Immunoprecipitation
Cells were homogenized in immunoprecipitation (IP) buffer (50 mM 1,4-piperazinediethanesulfonic acid, 50 mM HEPES, 1 mM EDTA, 2 mM MgSO4, 1% TX-100, and 0.5% Igepal, pH 7.0) supplemented with proteinase inhibitor cocktail and phosphatase inhibitor cocktails 2 and 3. Cells were homogenized and centrifuged, and supernatants were precleared with protein A resin (GenScript, Piscataway, NJ) for 60 min at 4°C. Precleared lysates (1 mg) were then incubated overnight at 4°C with 5 μg of anti–MRTF-A, anti-SRF polyclonal antibodies or control rabbit immunoglobulin. Immunocomplexes were recovered by incubation with protein A–Sepharose beads for 2 h at 4°C with constant rotation. The extensively washed beads were boiled with 2× SDS sample buffer and pelleted by centrifugation. Equal volumes of supernatants (20 μl) were loaded into polyacrylamide gels and analyzed by electrophoresis and immunoblotting as described.
G-/F-actin fractionation
Quantification of G- and F-actin was performed by TX-100 fractionation of cellular actin, as previously described (Cramer et al., 2002). Briefly, epithelial monolayers were washed with HEPES-buffered Hanks’ balanced salt solution (HBSS+) buffer, and G-actin was extracted by gentle shaking for 5 min at room temperature with cytoskeleton stabilization buffer (10 mM 2-(N-morpholino)ethanesulfonic acid, 140 mM KCl, 3 mM MgCl2, 2 mM EGTA, 280 mM sucrose, pH 6.1) supplemented with 0.5% TX-100, proteinase inhibitor cocktail, and 1 μg/ml phalloidin to prevent filament disassembly. The TX-100–soluble G-actin fraction was mixed with an equal volume of 2× SDS sample buffer and boiled. Cells were then briefly washed with HBSS+ buffer, and the TX-100–insoluble F-actin fraction was collected by scraping cells in two volumes of SDS sample buffer, with subsequent homogenizing and boiling. The amount of actin in each fraction was determined by gel electrophoresis and immunoblotting using an antibody capable of recognizing all actin isoforms.
RNA interference
The siRNA-mediated knockdown of actin isoforms and other proteins of interest was carried out using Dharmacon siRNAs (Thermo Scientific) as previously described (Naydenov and Ivanov, 2010; Naydenov et al., 2014). Cytoplasmic actin isoforms were depleted by using individual siRNA duplexes targeting human β-CYA (duplex 1, GGGCAUGGGUCAGAAGGAU; duplex 2, AAACCUAACUUGCGCAGAA) and human γ-CYA (duplex 1, GAGAAGAUGACUCAGAUUA; duplex 2, GAGCCGUGUUUCCUUCCAU). SRF, MRTF-A, MRTF-B, E-cadherin, and FHOD1 were depleted using specific siRNA SmartPools. A noncoding siRNA duplex 2 was used as a control. Cells were transfected using DharmaFECT 1 reagent in Opti-MEM I medium (Invitrogen) according to the manufacturer's protocol, with a final siRNA concentration of 50 or 100 nM for single or dual knockdown, respectively. Cells were used in experiments on days 3 and 4 posttransfection. Cotransfection of γ-CYA siRNA (50 nM) and MRTF plasmids (100 ng) was performed using a DharmaFECT Duo transfection reagent (Dharmacon) according to the manufacturer's instructions. Cells were fixed and subjected to immunofluorescence analysis 48 h posttransfection.
Quantitative real-time RT-PCR
Total RNA was isolated from A549 cells using an RNeasy Mini kit (Qiagen, Valencia CA) including DNase treatment to remove traces of genomic DNA. Total RNA (1 μg) was reverse transcribed into cDNA using an iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using an iTaq Universal SYBR green Supermix (Bio-Rad) and the following primers: human α-SMA, 5′-AAAGACAGCTACGTGGGTGACGAA-3′ and 5′- TTCCCATGTCGTCCCAGTTGGTGAT-3′; human SM-22, 5′-TCCAGGTCTGGCTGAAGAATGG-3′ and 5′-CTGCTCCATCTGCTTGAAGACC-3′; human CNN-1, 5′-CCAACGACCTGTTTGAGAACACC-3′ and 5′-ATTTCCGCTCCTGCTTCTCTGC-3′; human c-Fos, 5′-GCCTCTCTTACTACCACTCACC-3′ and 5′-AGATGGCAGTGACCGTGGGAAT-3′; human Egr1, 5′-AGCAGCACCTTCAACCCTCAGG-3′ and 5′-GAGTGGTTTGGCTGGGGTAACT-3′; human JunD, 5′-ATCGACATGGACACGCAGGAGC-3′ and 5′-CTCCGTGTTCTGACTCTTGAGG-3′; and human SRF, 5′-TCACCTACCAGGTGTCGGAGTC-3′ and 5′-GTGCTGTTTGGATGGTGGAGGT-3′; as a control, we used specific primers for GAPDH: 5′-CATGTTCGTCATGGGTGTGAACCA-3′ and 5′-AGTGATGGCATGGACTGTGGTCAT-3′. Gene amplification was performed using a 7900HT Fast Real-Time PCR System (A&B Applied Biosystems, Carlsbad, CA) with the following reaction conditions: 1 cycle, 95°C for 120 s; and 40 cycles, 95°C for 1 s and 60°C for 20 s. Threshold cycle number for the gene of interest was calculated based on the amplification curve representing a plot of the fluorescence signal intensity versus the cycle number. Delta threshold cycle number was calculated as the difference between threshold cycle numbers of γ-CYA siRNA– and control siRNA–transfected cells, and each value was normalized using the difference in the threshold cycle number for the housekeeping gene amplification of the same samples.
Immunofluorescence labeling and confocal microscopy
A549 cells were fixed with 4% paraformaldehyde for 20 min at room temperature and permeabilized with 0.5% TX-100. Then the monolayers were washed with HBSS+, blocked with 1% bovine serum albumin in HBSS+ (blocking buffer) for 60 min at room temperature, and incubated for another 60 min with primary antibodies diluted in the blocking buffer. Cells were then washed, incubated for 60 min with Alexa Fluor–conjugated secondary antibodies, rinsed with blocking buffer, and mounted on slides with ProLong Gold Antifade Reagent (Invitrogen). F-actin and nuclei were labeled with Alexa 555–phalloidin and 4′,6-diamidino-2-phenylindole (DAPI), respectively. Fluorescently labeled cell monolayers were examined using a Zeiss LSM700 laser scanning confocal microscope (Zeiss Microimaging, Thornwood, NY). The Alexa Fluor 488 and 555 signals were imaged sequentially in frame-interlaced mode to eliminate cross-talk between channels. The images were processed using Zen 2011 software and Photoshop (Adobe, San Jose, CA). Signal intensity was measured by using Image J software after selecting a rectangular area of either nuclear or cytoplasmic signals. Images shown are representative of at least three experiments, with multiple images taken per slide.
RhoA activation assay
The amount of active GTP-bound Rho was determined using a Rho Activation Assay Kit (Millipore) according to the manufacturer's protocol. Briefly, cells were lysed in Mg2+ lysis/washing buffer, and GTP-bound RhoA was immunoprecipitated from cleared lysates with 25 μg of glutathione-agarose–bound, glutathione S-transferase (GST)–conjugated RhoA binding domain of rhotekin. Beads were washed, and the immunoprecipitates were analyzed using 15% SDS–PAGE. Membranes were probed with anti-Rho mAb. Initial cellular lysates were also loaded to probe total Rho expression.
SRF activity assay
Signal lentiviral SRE reporter assay, which expresses a luciferase gene driven by multiple SRE (GGATGTCCATATTAGGA) repeats, was purchased from SA Biosciences (Frederick, MD). Lentiviral transduction of A549 cells was performed as described previously (Kutner et al., 2009). Stable clones expressing SRE reporter vector (selected with 1.0 μg/ml puromycin) were transfected with control and γ-CYA siRNA, along with Renilla (pRL-TK) plasmid as an internal control. Dual luciferase reporter assay (Promega, Madison, WI) was performed 48 h posttransfection according to the manufacturer's protocol.
Wound-healing and Matrigel invasion assays
Cells, transfected with either control or γ-CYA–specific siRNAs, were grown to confluence and on day 3 posttransfection mechanically wounded by scratching the monolayers by dragging a 200-μl pipette tip across. The cells were washed twice with complete media, and images were acquired at 0 and 24 h after wounding using an inverted bright-field microscope equipped with a camera. The relative surface area traveled by the leading edge was calculated using TScratch software (Geback et al., 2009). Figure 2 shows representative images of three independent experiments performed in duplicate. The invasion assay was performed using commercially available BD Biocoat invasion chambers (BD Biosciences). Cells were trypsinized, resuspended in DMEM without serum, and added to the upper chamber at a concentration of 5 × 104 cells/chamber. In the lower chamber, complete growth medium containing 10% FBS as a chemoattractant was added, and cells were allowed to migrate for 24 h at 37°C. Cells were fixed and stained using a DIFF stain kit (IMEB, San Marcos, CA); nonmigrated cells were removed from the top of the membranes using cotton swabs. For both adhesion and invasion experiments, the number of cells was determined by manual count in 15 randomly selected microscopic images obtained from three independent experiments.
Statistics
Numerical values from individual experiments were pooled and expressed as a mean ± SEM throughout. Obtained numbers were compared by two-tailed Student's t test, with statistical significance assumed at p < 0.05.
Supplementary Material
Acknowledgments
We thank Alex Feygin for editing the manuscript. This work was supported by National Institutes of Health Grants RO1 DK083968 and R01 DK084953 to A.I.I. and Swiss National Science Foundation Grant 310030_125320 to C.C. N.G.N. is a recipient of Crohn's and Colitis Foundation of America Grant 254881.
Abbreviations used:
- AJ
adherens junction
- α-SMA
α-smooth muscle actin
- β-CYA
β-cytoplasmic actin
- CNN-1
calponin-1
- EMT
epithelial-to-mesenchymal transition
- EMyT
epithelial-to-myofibroblast transition
- FHOD1
formin homology 2 domain containing 1
- γ-CYA
γ-cytoplasmic actin
- L-Cald
L-caldesmon
- MRTF
myocardin-related transcriptional factor
- ROCK
Rho-associated kinase
- SRF
serum response factor
- TGF-β
transforming growth factor β
- TJ
tight junction
- TM
tropomyosin
- ZO-1
zonula occludens 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0815) on August 20, 2014.
*These authors contributed equally.
REFERENCES
- Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- Baranwal S, Naydenov NG, Harris G, Dugina V, Morgan KG, Chaponnier C, Ivanov AI. Nonredundant roles of cytoplasmic beta- and gamma-actin isoforms in regulation of epithelial apical junctions. Mol Biol Cell. 2012;23:3542–3553. doi: 10.1091/mbc.E12-02-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, Zheng Q, Keri RA, Reizes O, Bresnick AR, et al. Myosin II isoform switching mediates invasiveness after TGF-beta-induced epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2011;108:17991–17996. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva IA, Perrin BJ, Sonnemann KJ, Zhu M, Stepanyan R, McGee J, Frolenkov GI, Walsh EJ, Friderici KH, Friedman TB, et al. Gamma-actin is required for cytoskeletal maintenance but not development. Proc Natl Acad Sci USA. 2009;106:9703–9708. doi: 10.1073/pnas.0900221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron SE, Zhu M, Thiem SM, Friderici KH, Rubenstein PA. Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J Biol Chem. 2010;285:16087–16095. doi: 10.1074/jbc.M110.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell TM, Burbach BJ, Shimizu Y, Ervasti JM. beta-Actin specifically controls cell growth, migration, and the G-actin pool. Mol Biol Cell. 2011;22:4047–4058. doi: 10.1091/mbc.E11-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell TM, Ervasti JM. Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton. 2010;67:564–572. doi: 10.1002/cm.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busche S, Kremmer E, Posern G. E-cadherin regulates MAL-SRF-mediated transcription in epithelial cells. J Cell Sci. 2010;123:2803–2809. doi: 10.1242/jcs.061887. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nature reviews. Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JW, Treisman R. The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol Biol Cell. 2002;13:4088–4099. doi: 10.1091/mbc.02-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer LP, Briggs LJ, Dawe HR. Use of fluorescently labelled deoxyribonuclease I to spatially measure G-actin levels in migrating and non-migrating cells. Cell Motil Cytoskeleton. 2002;51:27–38. doi: 10.1002/cm.10013. [DOI] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- Drummond MC, Friderici KH. A novel actin mRNA splice variant regulates ACTG1 expression. PLoS Genet. 2013;9:e1003743. doi: 10.1371/journal.pgen.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Nguyen M, Breckenridge DG, Shore GC. The resident endoplasmic reticulum protein, BAP31, associates with gamma-actin and myosin B heavy chain. Eur J Biochem. 2003;270:342–349. doi: 10.1046/j.1432-1033.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- Dugina V, Zwaenepoel I, Gabbiani G, Clement S, Chaponnier C. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J Cell Sci. 2009;122:2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- Fan L, Sebe A, Peterfi Z, Masszi A, Thirone AC, Rotstein OD, Nakano H, McCulloch CA, Szaszi K, Mucsi I, et al. Cell contact-dependent regulation of epithelial-myofibroblast transition via the rho-rho kinase-phospho-myosin pathway. Mol Biol Cell. 2007;18:1083–1097. doi: 10.1091/mbc.E06-07-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geback T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. BioTechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J Biol Chem. 2001;276:24531–24539. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Ivanov AI, McCall IC, Parkos CA, Nusrat A. Role for actin filament turnover and a myosin II motor in cytoskeleton-driven disassembly of the epithelial apical junctional complex. Mol Biol Cell. 2004;15:2639–2651. doi: 10.1091/mbc.E04-02-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Respir Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitlina SY. Functional specificity of actin isoforms. Int Rev Cytol. 2001;202:35–98. doi: 10.1016/s0074-7696(01)02003-4. [DOI] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Le Bras GF, Taubenslag KJ, Andl CD. The regulation of cell-cell adhesion during epithelial-mesenchymal transition, motility and tumor progression. Cell Adh Migr. 2012;6:365–373. doi: 10.4161/cam.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Nelson CM. New insights into the regulation of epithelial-mesenchymal transition and tissue fibrosis. Int Rev Cell Mol Biol. 2012;294:171–221. doi: 10.1016/B978-0-12-394305-7.00004-5. [DOI] [PubMed] [Google Scholar]
- Lee J, Seo JH, Lim JW, Kim H. Membrane proteome analysis of cerulein-stimulated pancreatic acinar cells: implication for early event of acute pancreatitis. Gut Liver. 2010a;4:84–93. doi: 10.5009/gnl.2010.4.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Vasishtha M, Prywes R. Activation and repression of cellular immediate early genes by serum response factor cofactors. J Biol Chem. 2010b;285:22036–22049. doi: 10.1074/jbc.M110.108878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, Gunning P. Beta- and gamma-actin genes differ in their mechanisms of down-regulation during myogenesis. J Cell Biochem. 2002;84:335–342. doi: 10.1002/jcb.10014. [DOI] [PubMed] [Google Scholar]
- Lloyd C, Schevzov G, Gunning P. Transfection of nonmuscle beta- and gamma-actin genes into myoblasts elicits different feedback regulatory responses from endogenous actin genes. J Cell Biol. 1992;117:787–797. doi: 10.1083/jcb.117.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack CP. Signaling mechanisms that regulate smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol. 2011;31:1495–1505. doi: 10.1161/ATVBAHA.110.221135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack CP, Somlyo AV, Hautmann M, Somlyo AP, Owens GK. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem. 2001;276:341–347. doi: 10.1074/jbc.M005505200. [DOI] [PubMed] [Google Scholar]
- Masszi A, Fan L, Rosivall L, McCulloch CA, Rotstein OD, Mucsi I, Kapus A. Integrity of cell-cell contacts is a critical regulator of TGF-beta 1-induced epithelial-to-myofibroblast transition: role for beta-catenin. Am J Pathol. 2004;165:1955–1967. doi: 10.1016/s0002-9440(10)63247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masszi A, Speight P, Charbonney E, Lodyga M, Nakano H, Szaszi K, Kapus A. Fate-determining mechanisms in epithelial-myofibroblast transition: major inhibitory role for Smad3. J Cell Biol. 2010;188:383–399. doi: 10.1083/jcb.200906155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol. Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal. 2011;4:ra40. doi: 10.1126/scisignal.2001750. [DOI] [PubMed] [Google Scholar]
- Naydenov NG, Feygin A, Wang L, Ivanov AI. N-Ethylmaleimide-sensitive factor attachment protein alpha (alphaSNAP) regulates matrix adhesion and integrin processing in human epithelial cells. J Biol Chem. 2014;289:2424–2439. doi: 10.1074/jbc.M113.498691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov NG, Ivanov AI. Adducins regulate remodeling of apical junctions in human epithelial cells. Mol Biol Cell. 2010;21:3506–3517. doi: 10.1091/mbc.E10-03-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JJ, Grosshans DR, Wong SG, Slamon DJ. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucleic Acids Res. 1999;27:4008–4017. doi: 10.1093/nar/27.20.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev. Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski R, Rajakyla EK, Vartiainen MK, Treisman R. An actin-regulated importin alpha/beta-dependent extended bipartite NLS directs nuclear import of MRTF-A. EMBO J. 2010;29:3448–3458. doi: 10.1038/emboj.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, Ervasti JM. The actin gene family: function follows isoform. Cytoskeleton. 2010;67:630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Miralles F, Guettler S, Treisman R. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 2004;23:3973–3983. doi: 10.1038/sj.emboj.7600404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin′ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Prins KW, Call JA, Lowe DA, Ervasti JM. Quadriceps myopathy caused by skeletal muscle-specific ablation of beta(cyto)-actin. J Cell Sci. 2011;124:951–957. doi: 10.1242/jcs.079848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin SE, Kapus A. Scar wars: mapping the fate of epithelial-mesenchymal-myofibroblast transition. Kidney Int. 2011;80:41–50. doi: 10.1038/ki.2011.77. [DOI] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbo N, Dulin N. Actin cytoskeleton in myofibroblast differentiation: ultrastructure defining form and driving function. Transl Res. 2011;158:181–196. doi: 10.1016/j.trsl.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Lloyd C, Gunning P. Impact of altered actin gene expression on vinculin, talin, cell spreading, and motility. DNA Cell Biol. 1995;14:689–700. doi: 10.1089/dna.1995.14.689. [DOI] [PubMed] [Google Scholar]
- Shum MS, Pasquier E, Po'uha ST, O'Neill GM, Chaponnier C, Gunning PW, Kavallaris M. gamma-Actin regulates cell migration and modulates the ROCK signaling pathway. FASEB J. 2011;25:4423–4433. doi: 10.1096/fj.11-185447. [DOI] [PubMed] [Google Scholar]
- Shuster CB, Herman IM. Indirect association of ezrin with F-actin: isoform specificity and calcium sensitivity. J Cell Biol. 1995;128:837–848. doi: 10.1083/jcb.128.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster CB, Lin AY, Nayak R, Herman IM. Beta cap73: a novel beta actin-specific binding protein. Cell Motil Cytoskeleton. 1996;35:175–187. doi: 10.1002/(SICI)1097-0169(1996)35:3<175::AID-CM1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Soh JW, Lee EH, Prywes R, Weinstein IB. Novel roles of specific isoforms of protein kinase C in activation of the c-fos serum response element. Mol Cell Biol. 1999;19:1313–1324. doi: 10.1128/mcb.19.2.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Speight P, Nakano H, Kelley TJ, Hinz B, Kapus A. Differential topical susceptibility to TGFbeta in intact and injured regions of the epithelium: key role in myofibroblast transition. Mol Biol Cell. 2013;24:3326–3336. doi: 10.1091/mbc.E13-04-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staus DP, Blaker AL, Medlin MD, Taylor JM, Mack CP. Formin homology domain-containing protein 1 regulates smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2011;31:360–367. doi: 10.1161/ATVBAHA.110.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Nagata H, Shimada Y, Konno A. Decrease in gamma-actin expression, disruption of actin microfilaments and alterations in cell adhesion systems associated with acquisition of metastatic capacity in human salivary gland adenocarcinoma cell clones. Int J Oncol. 1998;12:1079–1084. doi: 10.3892/ijo.12.5.1079. [DOI] [PubMed] [Google Scholar]
- Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondeleir D, Lambrechts A, Muller M, Jonckheere V, Doll T, Vandamme D, Bakkali K, Waterschoot D, Lemaistre M, Debeir O, et al. Cells lacking beta-actin are genetically reprogrammed and maintain conditional migratory capacity. Mol Cell Proteomics. 2012;11:255–271. doi: 10.1074/mcp.M111.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondeleir D, Vandamme D, Vandekerckhove J, Ampe C, Lambrechts A. Actin isoform expression patterns during mammalian development and in pathology: insights from mouse models. Cell Motil Cytoskeleton. 2009;66:798–815. doi: 10.1002/cm.20350. [DOI] [PubMed] [Google Scholar]
- Tzima E, Trotter PJ, Orchard MA, Walker JH. Annexin V relocates to the platelet cytoskeleton upon activation and binds to a specific isoform of actin. Eur J Biochem. 2000;267:4720–4730. doi: 10.1046/j.1432-1327.2000.01525.x. [DOI] [PubMed] [Google Scholar]
- Ueha S, Shand FH, Matsushima K. Cellular and molecular mechanisms of chronic inflammation-associated organ fibrosis. Front Immunol. 2012;3:71. doi: 10.3389/fimmu.2012.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Verrills NM, Po′uha ST, Liu ML, Liaw TY, Larsen MR, Ivery MT, Marshall GM, Gunning PW, Kavallaris M. Alterations in gamma-actin and tubulin-targeted drug resistance in childhood leukemia. J Natl Cancer Inst. 2006;98:1363–1374. doi: 10.1093/jnci/djj372. [DOI] [PubMed] [Google Scholar]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harbor Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Willis BC, duBois RM, Borok Z. Epithelial origin of myofibroblasts during fibrosis in the lung. Proc Am Thoracic Soc. 2006;3:377–382. doi: 10.1513/pats.200601-004TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Safina A, Bakin AV. Role of high-molecular weight tropomyosins in TGF-beta-mediated control of cell motility. International journal of cancer. J Int Cancer. 2008;122:78–90. doi: 10.1002/ijc.23025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.