Actin-based protrusions are important for signaling and migration during development and homeostasis. Gain- and loss-of-function and quantitative approaches are used to define differential roles for the actin elongation factors Diaphanous and Enabled in regulating distinct protrusive behaviors in different tissues during Drosophila morphogenesis.
Abstract
Actin-based protrusions are important for signaling and migration during development and homeostasis. Defining how different tissues in vivo craft diverse protrusive behaviors using the same genomic toolkit of actin regulators is a current challenge. The actin elongation factors Diaphanous and Enabled both promote barbed-end actin polymerization and can stimulate filopodia in cultured cells. However, redundancy in mammals and Diaphanous’ role in cytokinesis limited analysis of whether and how they regulate protrusions during development. We used two tissues driving Drosophila dorsal closure—migratory leading-edge (LE) and nonmigratory amnioserosal (AS) cells—as models to define how cells shape distinct protrusions during morphogenesis. We found that nonmigratory AS cells produce filopodia that are morphologically and dynamically distinct from those of LE cells. We hypothesized that differing Enabled and/or Diaphanous activity drives these differences. Combining gain- and loss-of-function with quantitative approaches revealed that Diaphanous and Enabled each regulate filopodial behavior in vivo and defined a quantitative “fingerprint”—the protrusive profile—which our data suggest is characteristic of each actin regulator. Our data suggest that LE protrusiveness is primarily Enabled driven, whereas Diaphanous plays the primary role in the AS, and reveal each has roles in dorsal closure, but its robustness ensures timely completion in their absence.
INTRODUCTION
From bundled myofibrils in muscle to dynamic filopodia and lamellipodia in migratory axons, proper development requires cells to build distinct actin-based structures. A host of actin regulatory proteins govern the underlying geometries of actin structures, mediating elongation, nucleation, branching, capping, and severing (Pollard and Borisy, 2003). Elegant studies characterized biochemical properties and interactions of these regulators in vitro or in simple, single-cell systems, but we still lack a clear understanding of how they work together or separately to produce protrusions in vivo during development. Filopodia, first described on neuronal growth cones (Harrison 1910), were historically viewed as sensory structures of migratory cells, helping drive guided migration (Wood and Martin, 2002; Gupton and Gertler, 2007). A century later, we are still uncovering filopodial roles in vivo (Sanders et al., 2013; Roy et al., 2014). Thus defining mechanisms by which filopodia form within a single cell type and how they differ among different cell types during development remains a key challenge.
Filopodia formation is believed to rely on actin elongation factors such as Diaphanous-related formins (DRFs) and Enabled (Ena)/VASP proteins (Michelot and Drubin, 2011). Both protein families bind actin filament barbed ends to promote polymerization and block capping, and members of both protein families can, when overexpressed/activated in cultured cells or neurons, promote formation of filopodia (Bachmann et al., 1999; Lanier et al., 1999; Rottner et al., 1999; Bear et al., 2002; Svitkina et al., 2003; Lebrand et al., 2004; Barzik et al., 2005; Applewhite et al., 2007; Pasic et al., 2008; Chesarone and Goode, 2009; Hansen and Mullins, 2010). Cell protrusive behavior is crucial in many developmental events (Rørth, 2009), and current models suggest that Ena/VASP proteins and DRFs should play key roles in regulating this (Gupton and Gertler, 2007). One puzzle has been why cells have two different families of actin elongation factors. The roles of Ena/VASP proteins in filopodia formation and protrusive behavior were confirmed in cell culture by loss-of-function approaches (Bear et al., 2000; Loureiro et al., 2002; Schirenbeck et al., 2006; Gonçalves-Pimentel et al., 2011). In contrast, whereas activated DRFs clearly can induce filopodia in cultured cells and localize to their tips (Yang et al., 2007; Block et al., 2008), assessing whether DRFs are essential for protrusive behavior and filopodia during development has been more challenging due to their conserved roles in cytokinesis and the partial redundancy of mammalian DRFs.
DRFs play a wide variety of roles in vivo. They were first identified via their conserved roles in cytokinesis (Castrillon and Wasserman, 1994; Chang et al., 1997; Imamura et al., 1997; Swan et al., 1998; Tominaga et al., 2000; Tolliday et al., 2002; Peng et al., 2003; Echard et al., 2004; Ingouff et al., 2005). Subsequent work in cultured mammalian cells identified a diverse array of functions, ranging from stress fiber assembly, to coordinating microtubules and actin, to targeted secretion and organelle dynamics, to modulating cell adhesion and phagocytosis (Faix and Grosse, 2006). Knockdown studies clearly implicate DRFs in directional cell migration (Yamana et al., 2006; Gupton et al., 2007; Lai et al., 2008; Shi et al., 2009; Dong et al., 2013; Daou et al., 2014), but, with the exception of work in Dictyostelium and Drosophila epidermal cells (Schirenbeck et al., 2005; Homem and Peifer, 2009), their roles in protrusive behavior and filopodia formation in vivo remain less well established. In cultured cells, mDia1 knockdown reduces filopodia formation induced by IRSp53 or Rif (Goh et al., 2011; Goh and Ahmed, 2012), whereas mDia2 antibody blockade or knockdown in B16F1 cells reduces both filopodia and lamellipodia (Peng et al., 2003; Yang et al., 2007). The roles of mammalian DRFs in protrusive behavior during development are even less clear. Mammals have three DRFs, complicating analysis. mDia1 mutants have immune system defects (Tanizaki et al., 2010), whereas compound mDia1; mDia3 mutants have defects in neuronal migration (Thumkeo et al., 2011), and mDia2-knockout mice have multinucleate erythroblasts (Watanabe et al., 2013). However, in none of these cases were clear effects on protrusive behavior defined.
Drosophila provides a simple system for studying these proteins, as there is only one Ena/VASP protein, Ena, and one DRF, Diaphanous (Dia). In Drosophila, Dia's early role in the modified form of cytokinesis known as cellularization (Afshar et al., 2000; Grosshans et al., 2005) obscured the search for later functions. However, use of RNA interference or zygotic mutants in which maternal protein had run down revealed roles in apical protein secretion in tracheae (Massarwa et al., 2009), cytoskeletal regulation during wound healing (Antunes et al., 2013; Abreu-Blanco et al., 2014), segmental groove formation (Mulinari et al., 2008), germband retraction (Homem and Peifer, 2008) and synapse growth (Pawson et al., 2008). However, once again, in vivo roles for Dia in protrusive behavior proved more elusive.
Recent work revealed that these two processive actin elongators, Ena and Dia, are biochemically distinct. Dia is a more efficient elongation factor than Ena, being seven times more processive and elongating twice as fast (Bilancia et al., 2014). These differences in biochemical properties are reflected in the filopodia each stimulates in cell culture, with Dia-driven filopodia having longer lifetimes and Ena-driven filopodia being more dynamic. Further, Ena and Dia directly interact via their EVH1 and FH1 domains (Bilancia et al., 2014; Barzik et al., 2014), and through this interaction, Ena can negatively regulate Dia, most likely at filament initiation (Bilancia et al., 2014). These data suggest that there are distinct roles for each of these elongation factors in filopodia. However, we still lack a clear understanding of how actin-elongation proteins work together or separately to initiate and elongate protrusions in vivo and how these protrusions contribute to major developmental processes. Our desire to define the roles of Dia in protrusive behavior was further sharpened by the recent discovery of a likely role for Dia in generating cytonemes, “signaling filopodia,” whose roles in paracrine signaling are becoming increasingly apparent (Roy et al., 2014).
To study the regulation and roles of protrusive behavior in embryonic development, we use Drosophila dorsal closure as a model (Jacinto et al., 2002b; Heisenberg, 2009). During this process, two lateral sheets of epidermal cells move dorsally to meet at the dorsal midline, displacing the central amnioserosal (AS) cells and enclosing the embryo in skin (Supplemental Movie S1; Figure 1, A and B). As closure proceeds, lateral epidermal cells elongate along their dorsal-ventral axes and move toward the midline. Closure is partially powered by an actin-based supracellular cable at the leading edge (LE) of the epidermis, which acts as a purse string to draw the two sheets together (Young et al., 1991; Kiehart et al., 2000; Jacinto et al., 2002a), while the central AS cells undergo waves of apical constriction (Kiehart et al., 2000; Gorfinkiel et al., 2009; Solon et al., 2009), drastically reducing their area. Actin-based protrusions from LE cells play key roles in closure, aligning the two contralateral sheets as they meet at the anterior and posterior canthi (Jacinto et al., 2002a; Millard and Martin, 2008).
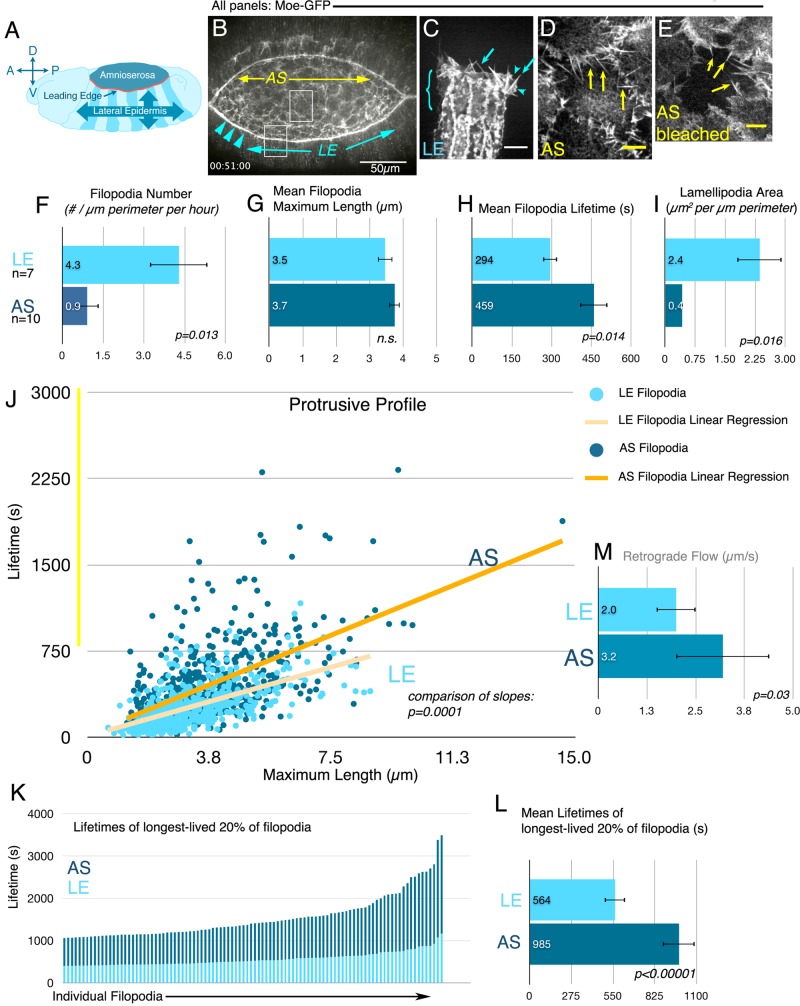
FIGURE 1:
The AS and LE cells as a model for differential regulation of protrusive behavior during normal development. For all embryo images. anterior is left. (A) Schematic, stage 14 embryo during dorsal closure. (B) Maximum intensity projection movie still. Dorsal closure in an embryo expressing the moesin actin-binding domain fused to GFP driven in both AS and LE via spaghetti squash promoter. LE cells (blue arrows), LE actin cable (arrowheads), AS cells (yellow region). Boxes indicate regions of embryo like those shown in C vs. D and E. (C–E) Scale bars, 5 μm. (C) Lateral epidermal cells expressing Moe-GFP in engrailed stripes. LE cells (bracket) produce filopodia (arrows) that often arise from lamellipodia (arrowheads). (D, E) AS cells (D, before bleaching; E, after bleaching central cell). AS cells produce filopodia (arrows) and fewer lamellipodia. (F–I) Quantitation of protrusive parameters of LE vs. AS cells. Error bars in all graphs equal SD; n ≥ 5 embryos. (F) LE cells produce more filopodia per micrometer perimeter than AS cells. (G) Filopodia mean maximum length is the same. (H) AS cell filopodia have a longer mean lifetime than LE filopodia. (I) LE cells produce more lamellipodial area per micrometer of perimeter. (J) Protrusive profiles of AS and LE cells, or plots of filopodium maximum length vs. lifetime. Slopes of linear regression illustrate that AS filopodia are longer lived per unit length. Individual distributions are shown in Supplemental Figure S2. Longer-lived subset of AS cell filopodia indicated by yellow bar. (K) Top 20% of filopodia by lifetime from each tissue. (L) The longest-lived 20% AS cell filopodia have a lifetime almost double that of the longest-lived LE filopodia. (M) AS cell filopodia have a higher rate of retrograde flow.
Although LE cells are not classical migrating cells, their filopodia and lamellipodia are likely to serve a similar purpose, helping sense the cellular environment and provide protrusive force, matching the epidermal sheets on the two sides and zippering them closed, akin to the role of filopodia in junction formation in primary keratinocytes, endothelial cells, or Caenorhabditis elegans ventral enclosure (Raich et al., 1999; Vasioukhin et al., 2000; Woolner et al., 2005; Millard and Martin, 2008; Hoelzle and Svitkina, 2012). Surprisingly, the nonmigratory cells of the amnioserosa also make actin-based protrusions, although they have not been characterized nor their roles defined.
We thus built on previous work by our lab and others in vitro and in cultured cells, using the LE and AS to uncover whether and how Ena and Dia regulate protrusive behavior during normal development, assessing whether roles these proteins can have in stimulating filopodial behavior after overexpression or activation play out in important roles during normal development. Combining detailed quantitative analysis of cell behavior with both loss-of-function and gain-of function genetic tools helped reveal the mechanisms by which regulated Ena and Dia activity shape protrusive behavior in these two cell types—one migratory and one not—and to assess their contributions to the tissue level process of dorsal closure.
RESULTS
AS and LE cells provide a model for differential regulation of protrusive behavior during normal development
We have learned much about how actin regulators modulate actin polymerization in vitro and how they regulate protrusive behavior in cultured cells, but their roles in vivo during normal development are less clear. Protrusions produced by LE cells during dorsal closure provide a model for how different actin regulators shape protrusive activity within a single cell type during normal development (Figure 1, A–C; Jacinto et al., 2000; Woolner et al., 2005; Gates et al., 2007; Millard and Martin, 2008; Homem and Peifer, 2009; Bilancia et al., 2014). During dorsal closure, LE cells elongate and migrate to meet contralateral partners, producing protrusions that are a mix of broad lamellipodia (Figure 1C, arrowheads; Supplemental Movie S2, right) and filopodia (Figure 1C, arrows; we used the en-GAL4 driver to express our filopodial marker—moesin–green fluorescent protein [GFP] or actin-GFP—in alternating stripes of cells in the lateral epidermis). Their nearest neighbors, the AS cells (Figure 1, A and B), also use the actin cytoskeleton to drive oscillatory behavior, resulting in net apical constriction (Kiehart et al., 2000; Jacinto et al., 2002b; Solon et al., 2009). Previous work suggested that AS cells, although they are nonmigratory, also produce filopodia (Figure 1D; Jacinto et al., 2000), but their characteristics and dynamics have not been assessed. These two cell types provide a setting to test the hypothesis that Ena and Dia act separately and together to help shape the distinctive protrusive behaviors of different cell types in normal development. Our first task was to determine whether filopodial dynamics differ in these two cell types, which undergo very different cell shape changes, by characterizing AS cell protrusions and comparing them to those in the LE.
We visualized protrusions by labeling f-actin structures by expressing the moesin actin-binding domain fused to GFP (Moe-GFP) via the myosin light chain promoter (spaghetti squash [sqh]), driving expression both in the AS and the lateral epidermis, or using the UAS-GAL4 system to drive Moe-GFP along the LE in stripes (Table 1). To facilitate comparison with our and other previous work, we defined filopodia as any thin protrusion (width, <1.25 μm; length, >1.15 μm) extending beyond the lamellipodium or leading edge, and lamellipodia were defined as protrusions >1.25 μm in width. We then used live confocal imaging to visualize filopodia dynamics in both tissues.
TABLE 1:
Fly stocks, antibodies, and probes.
| Stock | Source | |
|---|---|---|
| Sqh moesin-GFP | D. Kiehart (Duke University, Durham, NC) | |
| UAS-GFP-actin | P. Martin (University of Bristol, Bristol, United Kingdom) | |
| UAS moesin-GFP | D. Kiehart | |
| UASGFPDia∆DAD | P. Rorth (EMBL, Heidelberg, Germany) | |
| UASHADia∆DAD | Homem and Peifer (2009) | |
| GAL4 drivers | Expression pattern | |
| C381-GAL4 | Expression starts at stage 12 in the amnioserosa | |
| engrailed-GAL4 | Expression starting at stage 12 in engrailed-expressing epidermal cells in the posterior of each segment; occasionally a few amnioserosa cells | |
| Antibody | Dilution | Source |
| Anti-Enabled | 1:100 | Developmental Studies Hybridoma Bank (DSHB; University of Iowa, Iowa, City, IA) |
| Anti-Diaphanous | 1:5000 | S. Wasserman (University of California, San Diego, La Jolla, CA) |
| Anti-DE-CAD2 | 1:100 | DSHB |
| Anti-neurotactin | 1:100 | DSHB |
| Anti-phosphotyrosine | 1:1000 | Upstate Biotechnology (Lake Placid, NY) |
| Anti-lamin | 1:500 | DSHB |
| Tetramethylrhodamine isothiocyanate–phalloidin | 1:500 | Molecular Probes (Eugene, OR) |
| Secondary antibodies: Alexa 488, 568, 647 | 1:500 | Molecular Probes |
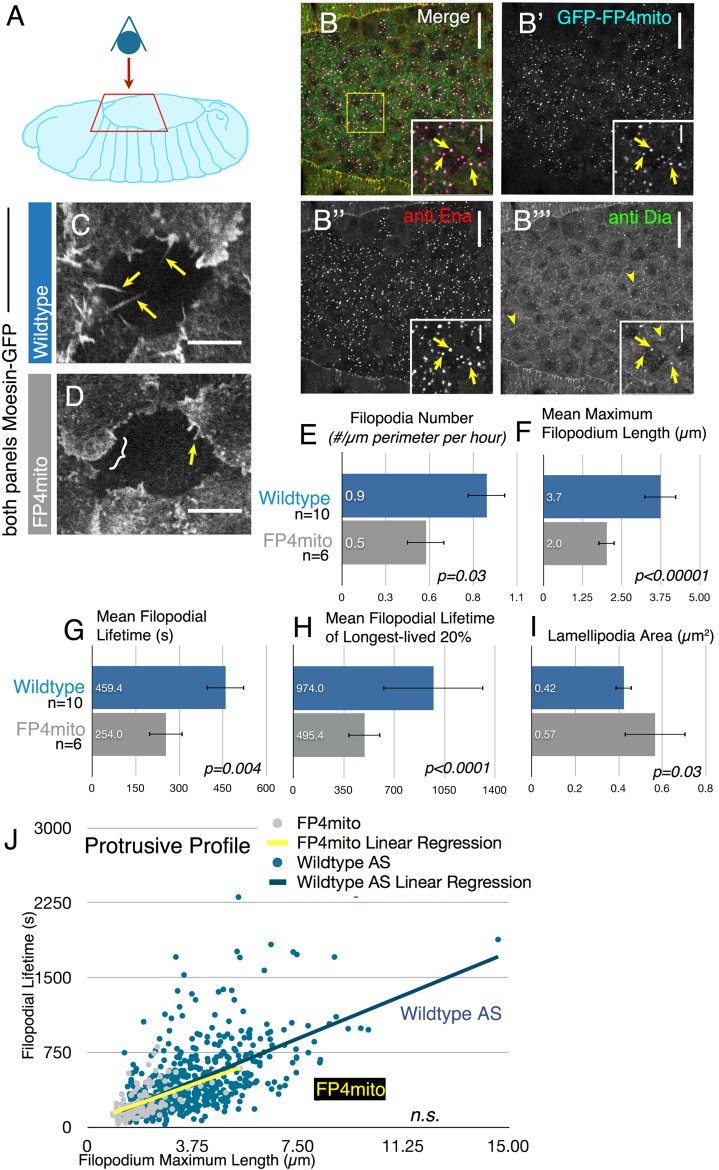
This revealed that AS cells produced thin, actin-based filopodia (Figure 1D, arrows; Supplemental Movie S2, left). However, because AS cells protrude over one another (Figure 1D), quantifying AS filopodial number, length, and lifetime was obstructed by lack of contrast against which to accurately measure individual filopodia. To better visualize AS filopodia, we developed a bleaching assay, photobleaching individual AS cells (Figure 1E). Bleaching allowed us to correctly assign protrusions to neighboring cells and reliably discern spatial and temporal properties of individual protrusions, facilitating quantitative comparison of AS and LE. It is important to note that the bleached cells were not those whose protrusions we measured—they simply provided the darkened background against which to more easily visualize neighbors. To ensure that bleaching did not adversely affect cell behavior or viability, we verified that it did not obviously alter filopodial behavior (by following filopodia before and after bleaching), that embryos with bleached cells completed dorsal closure in a timely manner, and that bleached cells were no more likely than unbleached neighbors to be among the fraction of AS cells to undergo early apoptosis (5% observed in our combined experiments vs. 10–30% of all AS cells/every closure event normally; Kiehart et al., 2000; Toyama et al., 2008).
Visual inspection suggested that the protrusive behavior of LE and AS cells was qualitatively distinct, with the LE producing filopodia emerging from lamellipodia and the AS protrusions emerging more often from the cell body (Figure 1, C vs. E, and Supplemental Movie S2). To refine this comparison, we quantitated different aspects of protrusive behavior in the two tissues. We did this manually, as the complex motion of both LE and AS filopodia in the x-y and z axes (Supplemental Movie S3) prevented automated quantitation. This revealed a number of striking differences. When we quantitated the number of filopodia formed per micrometer of cell perimeter per hour, we found that AS cells produce four times fewer filopodia than LE cells (Figure 1F; 0.9 ± 0.04 vs. 4.3 ± 1.0, p = 0.013). In contrast, the mean maximum lengths of filopodia in each tissue were similar (Figure 1G; 3.7 ± 0.5 vs. 3.5 ± 0.6 μm), as was the distribution of maximum filopodial lengths (Supplemental Figure S1B). Two other parameters were also strikingly different in the two tissues—lamellipodia area and filopodia lifetime. Filopodia along the LE most often arise from broad lamellipodia (Figure 1C, arrow; Gates et al., 2007; Homem and Peifer, 2009), but the actin-based structures associated with AS filopodia remained largely unexplored. To test the hypothesis that differences in the filopodia/lamellipodial balance might help account for the visual difference between the two tissues, we quantified lamellipodial area. There was significantly less lamellipodial area/cell perimeter in AS versus LE cells (Figure 1I; AS, 0.4 ± 0.04 μm2/μm perimeter, n = 6 embryos, vs. LE, 2.4 ± 1.3 μm2/μm perimeter, n = 5 embryos). Thus AS filopodia largely emerge directly from the cell cortex, whereas LE filopodia emerge from lamellipodia (Figure 1, E vs. C). The difference in lamellipodial area accounted for part of the visual difference between protrusive behaviors in the two tissues, but a second dynamic parameter also played an important role. AS cell filopodia had significantly longer mean lifetimes than their LE cell counterparts (Figure 1H; AS, 459 ± 65 s, n = 9 embryos, 567 filopodia, vs. LE, 294 ± 72 s, n = 7 embryos, 429 filopodia; p = 0.014; this was also reflected in a shift of the distributions of the lifetimes of individual filopodia; Supplemental Figure S1B).
This difference in filopodial lifetime prompted us to look closely at the relationship between filopodial length and lifetime, which could be influenced in part by barbed-end polymerization rates. To explore this, we plotted the maximum length versus lifetime for each wild-type LE or AS filopodium, generating a “protrusive profile” of individual filopodia (AS and LE distributions are superimposed in Figure 1J; the separate distributions are in Supplemental Figure S2A). We applied a linear regression fit to each data set, the slope of which revealed the relationship between filopodial length and lifetime. As expected, longer filopodia generally have longer lifetimes in all cases. However, the relationship between these two parameters differed significantly in the two tissues (p = 0.0001), with filopodia in the AS generally having longer lifetimes at the same length. This could be consistent with differences in filopodial extension rates or persistence of extension, which may reflect differences in activity of the actin regulators governing filopodial behavior in the AS and LE.
The AS filopodial protrusive profile also revealed outliers that are longer lived than any at the LE (Figure 1J, region highlighted by yellow bar). This is prominently illustrated by comparing the top 20% longest-lived filopodia in LE versus AS cells (Figure 1K)—the mean lifetimes of the longest-lived filopodia differ by almost twofold (Figure 1L; 564 ± 152 vs. 985 ± 358 s). Finally, we examined whether this increase in lifetime reflected differences in the time filopodia spent extending or retracting (Supplemental Figure 1, C and D). To do so, we measured the time each filopodium spent extending (defined as the time before maximum length was reached) and/or retracting (defined as the time from maximum length until the filopodium shortened below our minimum length). Both were increased proportionately in AS cells versus the LE, such that in both tissue types, filopodia spent roughly half of their lifetime before they reached maximum length and half after maximum length (Supplemental Figure 1, C–E). Although we were unable to measure instantaneous extension rates, due to the inability to use automated tracking, the analysis did reveal average rates of filopodia extension, which were similar in the two tissues (Supplemental Figure 1F). Together these data suggest that the difference in lifetime is primarily shaped by the longer time AS filopodia spent extending and retracting rather than a difference in rate. Thus these two tissues have distinct filopodial dynamics as revealed by differences in both the behavior of individual filopodia and their protrusion profiles.
The parameters of filopodial length and lifetime are also likely to be modulated in part by retrograde flow, which in turn depends on rates of actin polymerization at the tip and myosin-based contractility at the base of the filopodia. To determine whether retrograde flow rates in filopodia were different between the two tissues, we expressed actin-GFP in the AS or the LE and either bleached a small transverse region of a filopodia or followed fiduciary marks via kymography for a portion of the filopodia's lifetime in which it did not move laterally in x-y. From this, we found that the AS has a small but significant increase in retrograde flow compared with the LE (Figure 1M; 3.2 ± 1.3 vs. 2.0 ± 0.5 μm/s), suggesting increased elongation at the tip and/or increased myosin contractility.
Together the data revealed that AS filopodia have a higher rate of retrograde flow and are generally significantly longer lived than LE filopodia of the same length and that a subset of the AS filopodia have substantially longer lifetimes overall. These differences were intriguing, given our recent results in cultured cells in which Dia-driven filopodia had longer lifetimes and often emerged directly from the cell body, whereas Ena-driven filopodia were more dynamic and emerged from broad lamellipodia (Bilancia et al., 2014), suggesting Ena and Dia have properties that might allow them to drive filopodia with these sorts of distinct dynamic properties in vivo.
Ena and Dia have distinct localization patterns in the AS versus the LE
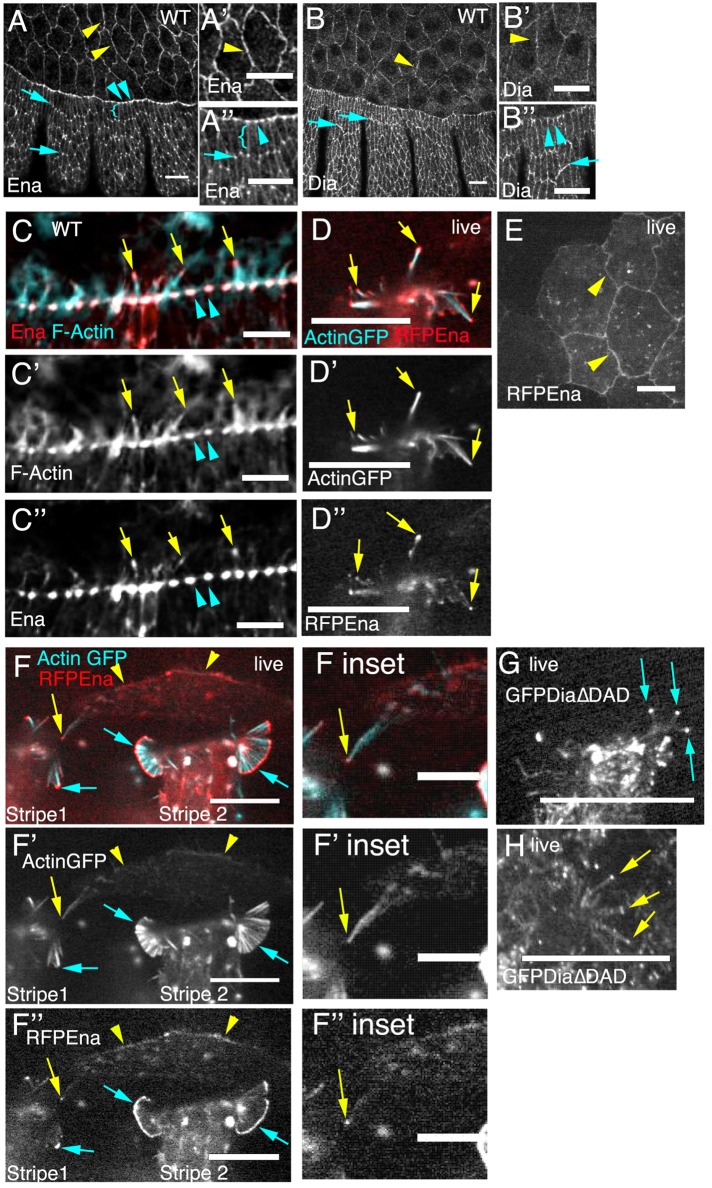
The simplest version of the hypothesis that Dia and Ena drive different types of protrusive behavior in different cell types would involve a situation in which either Ena or Dia predominates in one or the other tissue. However, in fixed embryos, both endogenous Ena and Dia are present in both cell types but localize differently in each. In LE cells, endogenous Ena localized robustly with F-actin to LE “dots” where LE cells abut one another and project over the AS (Figure 2, A, A′′, and C–C′′, cyan arrowheads), as well as to the tips of LE filopodia (in embryos where these were successfully preserved; Figure 2, C–C′′, yellow arrows). In epidermal cells ventral to the LE, endogenous Ena was planar polarized, being enriched at tricellular junctions, with lower levels at the lateral cortex (Figure 2A′′, cyan arrows vs. brackets; Bilancia et al., 2014). Endogenous Dia was cortical in all cells of the lateral epidermis (Figure 2B) and less polarized than Ena, though there was enrichment in the region where LE cells abut the AS (Figure 2B′′, cyan arrowheads) and somewhat elevated accumulation at dorsal-ventral cell borders (Figure 2, B and B′′, arrows); unfortunately the fixation conditions needed for the Dia antibody prevented preservation of the LE filopodia. Together these localization patterns suggest that there may be differential activity of these two elongation factors in the lateral epidermis versus the LE cells. In the AS, endogenous Ena localized robustly to the cell cortex (Figure 2, A and A′, yellow arrowheads), even from early stages (unpublished data), at levels roughly comparable to those for the cortex of LE cells. However, in contrast to the LE, Ena's localization in AS cells was radially symmetric. Endogenous Dia was also cortically localized in the AS, at levels slightly lower than for the LE cell cortex, and also was radially symmetric (Figure 2, B and B′). Thus both proteins are present in both the LE cells and the AS cells and are potentially poised to play roles in filopodia formation and elongation in both tissues.
FIGURE 2:
Ena and Dia localization in AS and LE cells: similarities and distinctions. Embryos, stage 13/14, anterior left and dorsal up, antigens indicated. (D–H) Expressed fluorescent protein–tagged versions of the indicated protein using engrailed (D, F, G) or c381 AS (E, H) GAL4 drivers. Fixed embryos (A–C) and live embryos (D–H). (A–A′′) Endogenous Ena localizes circumferentially to the AS cortex (yellow arrowheads) and in the lateral epidermis is enriched in LE dots in LE cells (cyan arrowheads) and at tricellular junctions (cyan arrows) in more-ventral epidermal cells relative to the lateral cell cortex (cyan brackets). (B–B′′) Endogenous Dia localizes cortically in the AS (yellow arrowheads) and is cortical in the lateral epidermis, with some enrichment just behind the LE (B′′, cyan arrowheads) and at dorsal/ventral cell borders (arrows). (C–C′′) Endogenous Ena localizes to filopodia tips along the LE (arrows) and robustly localizes to LE dots (arrowheads). (D–D′′) RFP-Ena also localizes to tips of LE cell filopodia (arrows). (E) When expressed in the AS, RFP-Ena localizes predominantly to the cell cortex (arrowheads). (F–F′′) RFP-Ena robustly localizes to tips of LE cell filopodia and filopodial fans (cyan arrows), whereas in an adjacent AS cell, it is cortical (arrowheads) and occasionally localizes to filopodial tips (yellow arrow). (G, H) Constitutively active GFPDia∆DAD can localize to LE filopodia tips (G, arrows) and localizes robustly to tips of AS cell filopodia (H, arrows). Scale bars, 20 μm, except F insets, 10 μm.
Ena is preferentially enriched in LE filopodia
This analysis of fixed embryos provided insights into localization differences of these two actin regulators in vivo but did not reliably capture the dynamic cell protrusions we study. To assess this, we used live analysis of fluorescently tagged proteins. Endogenous Ena, GFP-tagged Ena, and red fluorescent protein (RFP)–tagged Ena can all localize to filopodia tips in LE cells (Figure 2, C and D, arrows; Gates et al., 2007). Preserving endogenous Dia localization to filopodia proved impossible, as fixation conditions preserving LE filopodia fail to preserve Dia antibody signal. Full-length Dia tagged with GFP is presumably in the autoinhibited state (it has no effects on filopodial behavior), and it is largely cytoplasmic, with some cortical enrichment but no enrichment to either the edges of lamellipodia or the tips of filopodia (Homem and Peifer, 2009). Thus, to assess Dia's ability to localize to filopodia tips, we live imaged embryos expressing a GFP-tagged, constitutively active version of Dia (Dia∆DAD, lacking the DAD autoinhibitory domain). Like Ena, active Dia was also enriched at tips of LE filopodia (Figure 2G, arrows; Homem and Peifer, 2009). Thus both elongation factors can localize to filopodia tips along the LE.
Because we cannot reliably preserve AS cell protrusions after fixation, we assessed the ability of Ena and Dia to localize to AS filopodia tips by live imaging. Surprisingly, live imaging of RFP-Ena expressed in the AS using the c381-GAL4 driver revealed strong localization to the AS cortex (Figure 2E, arrowheads) but did not reveal clear enrichment in AS filopodia. When we expressed RFP-Ena using the engrailed-GAL4 driver, we saw robust localization to LE filopodia (Figure 2, D and F, cyan arrow, left) and to LE filopodial “fans” (Figure 2F, cyan arrows, right). In the rare, isolated AS cells expressing both actin and RFP-Ena via this driver, we saw strong RFP-Ena localization to the cortex (Figure 2F, yellow arrowheads) and only occasional localization to the tips of AS cell filopodia (Figure 2F, inset, arrow); thus the ratio of RFP-Ena at AS filopodial tips relative to the AS cell cortex was lower than in LE cells (Figure 2F, yellow arrowheads and arrow vs. cyan arrows). This is not solely due to reduced filopodia number, as Ena overexpression increases the number of AS filopodia (see later discussion). In contrast, GFP-tagged active Dia localized robustly to the tips of AS filopodia (Figure 2H, arrows). Together these data demonstrate that both Ena and Dia can localize to the tips of filopodia in both AS and LE cells but suggest that Ena enrichment to filopodial tips may be more robust at the LE.
Roles for Ena levels in governing filopodia number and dynamics along the LE
Our assessment of the biochemical properties of Ena and Dia and their effects on protrusive behavior in cultured Drosophila cells revealed that Dia generates longer, longer-lived filopodia emerging from the cell body, whereas Ena generates shorter-lived filopodia that emerge from lamellipodia (Bilancia et al., 2014). On this basis, we hypothesized that Ena is responsible for the more dynamic LE filopodia, whereas Dia might play a more dominant role in the AS. Consistent with this, reducing Ena function using loss-of-function mutants substantially reduced LE filopodial number (Homem and Peifer, 2009).
To further test this hypothesis, we used quantitative tools to assess the dynamic properties of filopodia driven by either Ena or activated Dia. To do so, we expressed tagged versions of Ena or active Dia (Dia∆DAD; Homem and Peifer, 2009) along with actin-GFP in epidermal stripes using engrailed-GAL4. We used actin-GFP for these experiments to allow comparison to our earlier work with LE cells. Although actin-GFP can have toxic effects, as were observed in the ovary (Spracklen et al., 2014), we did not observe any obvious toxic effects driving UAS-actin-GFP in the lateral epidermis using the en-GAL4 driver—these embryos complete dorsal closure in a time consistent with the time taken by wild-type embryos and embryos expressing UAS-Actin-GFP hatch (unpublished data).
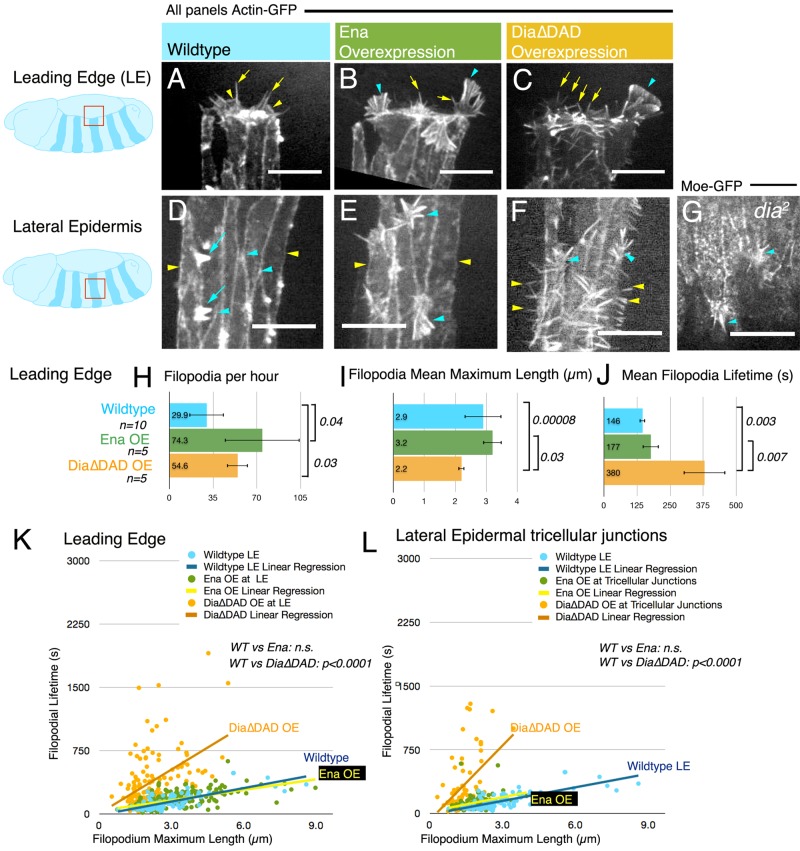
Wild-type LE protrusions are a mix of lamellipodia (Figure 3A, arrowheads; Supplemental Movie S4, left) and their associated filopodia (Figure 3A, arrows). Both Ena and activated Dia can promote formation of filopodia along the LE (our data; Gates et al., 2007; Homem and Peifer, 2009). Overexpressing RFP-Ena in the lateral epidermis altered LE protrusive behavior, promoting both excess filopodia (Figure 3B, arrows, H; Gates et al., 2007) and “filopodial fans,” which may be many closely packed “filopodia” elongating in concert within in a common membrane (Figure 3B, arrowheads, and Supplemental Movie S4 middle). Both filopodia and fan-like protrusions have RFP-Ena at the tips (Figure 2F, cyan arrows). When we quantitated LE filopodia induced by Ena overexpression (excluding fan protrusions to allow direct comparisons), we found higher filopodia number, suggesting increased filopodia initiation (Figure 3H; Gates et al., 2007). However, mean filopodial lifetime and length were not different from those for control LEs (Figure 3, I and J), nor were there striking changes in the distribution of filopodial maximum lengths or lifetimes (Supplemental Figure S1A). We next examined the protrusive profile, comparing filopodia length and lifetime. The protrusive profile of Ena-induced filopodia at the LE was not significantly different from that for wild type (Figure 3K, green vs. blue dots and lines, and Supplemental Figure S2B). The observation that elevating Ena along the LE increases filopodia initiation without changing filopodia dynamics is consistent with the idea that Ena normally initiates and governs elongation of LE filopodia, whereas the elevation of filopodia number after elevation of Ena levels suggests that Ena may be limiting for LE filopodial initiation.
FIGURE 3:
Expressing Ena or active Dia at the LE induces filopodia with distinct dynamic behaviors. Schematics indicate locations of LE cells and more-ventral lateral epidermal cells and expression pattern of engrailed driver (dark stripes). (A–G) Stage 14 embryos expressing GFP-actin in stripes in the ventral epidermis under control of en-GAL4 (A–F) or moe-GFP ubiquitously (G). Scale bars, 10 μm. (A–C) LE cells. (A) Wild-type, revealing normal filopodia (arrows) that arise from broad lamellipodia (arrowheads). (B) Overexpressing Ena yields filopodia (arrows) and morphologically distinct filopodial-fan protrusions (arrowheads). (C) Expressing constitutively active Dia∆DAD induces short filopodia (arrows), as well as fan-like structures (arrowheads). (D–G) Lateral epidermal cells. (D) Wild-type epidermal cells ventral to the LE normally do not form filopodia or lamellipodia, either laterally (yellow arrowheads) or at tricellular junctions (cyan arrowheads), but do produce actin-based dorsal hairs at the posterior borders of cells by late closure (arrows). (E) Ena overexpression yields filopodia and filopodial-fan-like protrusions at tricellular junctions (cyan arrowheads) but no lateral filopodia (yellow arrowheads). (F) Dia∆DAD overexpression induces numerous lateral filopodia (yellow arrowheads) and filopodia emerging from lamellipodia at tricellular junctions (cyan arrowheads). (G) dia2 zygotic mutants also have filopodia emerging from tricellular junctions (cyan arrowheads). (H–J) Quantitation of filopodial dynamics. n, number of embryos. (H) Mean number of filopodia produced per micrometer perimeter per hour along the LE in the genotypes indicated. Statistics, t test. (I) Mean maximum filopodia length (μm). (J) Mean filopodia lifetime (s). (K, L) Protrusive profile plots of maximum length (μm) vs. lifetime (s) with linear regression analysis. The p value is by ANCOVA for difference in slopes. (K) LE filopodia, genotypes indicated. (L) Filopodia induced at tricellular junctions.
Active Dia induces long-lived LE filopodia that mimic AS filopodia
We then contrasted filopodia induced by Ena at the LE with filopodia induced by active Dia. Overexpressing hemagglutinin (HA)-tagged Dia∆DAD had a dramatic affect, triggering filopodia-like protrusions all along the epidermal cell cortex of all lateral epidermal cells (Figure 3F and Supplemental Movie S4, right; Homem and Peifer, 2009). Like Ena, active Dia also significantly increased the number of filopodia initiated along the LE (Figure 3, C, arrows, and H) and induced filopodial fans (Figure 3C, arrowhead). However, in contrast to Ena, Dia substantially altered the dynamic properties of LE filopodia, with mean length significantly decreased and mean lifetime significantly increased (Figure 3, I and J; there were parallel changes in the distributions of individual maximum lengths and lifetimes; Supplemental Figure S1A). We thus examined the protrusive profile, plotting maximum length versus lifetime for each wild-type LE filopodium, as well as those induced by Ena overexpression or activated Dia (Figure 3K). Whereas the protrusive profile of Ena-induced filopodia was not significantly different from that of wild type (Figure 3K, green vs. blue dots, blue vs. yellow lines; individual distributions are in Supplemental Figure S2B), in contrast, the slope of the protrusive profile of Dia-induced LE filopodia was significantly shifted, with active Dia inducing generally shorter and longer-lived filopodia (Figure 3K). Thus, in contrast to Ena, Dia does not seem to be a strong candidate for playing the dominant role in normal LE filopodial dynamics. Intriguingly, Dia overexpression at the LE altered the slope of the protrusive profile such that it was more similar to that of normal AS cells.
Ena activity in the lateral epidermis is polarized
In the wild type, endogenous Ena is strongly enriched at LE dots (Figure 2, A and A′′, arrowheads) and robustly but more weakly enriched at tricellular junctions in more ventral epidermal cells (Figure 2, A and A′′, arrows). Overexpressing RFP-Ena in lateral epidermal stripes induced ectopic filopodia at the place where its levels are normally elevated—the tricellular junctions in more ventral cells (Figure 3E, cyan arrowheads, and Supplemental Movie S5, middle)—but not at the places were endogenous Ena levels are lower—the lateral cell cortices (Figure 3E, yellow arrowheads). RFP-Ena localized to the tips of the ectopic protrusions (Supplemental Figure S3A, arrows). This allowed us to ask questions about polarization of Ena activity. At the LE, protrusions are restricted to dorsal borders of the LE cells. The ectopic filopodia induced by RFP-Ena in more ventral epidermal cells also appeared to be oriented dorsally. To test directly whether these filopodia were polarized, we coexpressed RFP-Ena and GFP-actin, photobleached the GFP-actin in a subset of lateral epidermal cells, and watched over time as filopodia appeared on unbleached neighbors. These filopodia emerged from the dorsal tricellular junctions of unbleached cells (100%, n = 5 embryos; Supplemental Figure S3B, yellow arrows). Controls in the same tissue (ventral tricellular junctions abutting more-ventral bleached cells) never produced filopodia (Supplemental Figure S3B, cyan arrows). Thus the lateral epidermis is polarized toward the LE, suggesting that Ena activity at tricellular junctions is polarized.
Active Dia also induced filopodia in cells ventral to the LE—like Ena, Dia induced filopodia at tricellular junctions, but active Dia also induced filopodia along the lateral cortex (Figure 3F, cyan vs. yellow arrowheads, and Supplemental Movie S5, right). Dia-induced filopodia at the lateral cortex, where Ena levels are low, have exceptionally long lifetimes (Bilancia et al., 2014), consistent with our hypothesis that Dia promotes longer-lived filopodia. Dia-induced filopodia at tricellular junctions, where Ena levels are higher, were shorter-lived. We analyzed and compared protrusive profiles of filopodia at tricellular junctions. The occasional wild-type filopodia observed at tricellular junctions and those induced by Ena had similar protrusive profiles (Supplemental Figure S3C), and the slopes of both of these protrusive profiles resembled those of wild-type LE cells (Figure 3L). In contrast, Dia-induced filopodia at tricellular junctions had a significantly different protrusive profile, being generally longer lived with respect to length (Figure 3L), and thus are similar to wild-type AS filopodia. Therefore, even in this ectopic location, Ena-induced filopodia more resembled those at the wild-type LE, whereas Dia-induced filopodia had dynamic properties more similar to those of the AS.
Elevating Ena expression in the AS induces filopodia with LE characteristics, whereas active Dia promotes long-lived filopodia in the AS with AS-like dynamics
The foregoing data combined with our earlier work suggest that Ena may be the dominant actin elongator acting at the LE. The differences in protrusive profile between the wild-type LE and AS, along with the shift in protrusive profile induced by Dia at the LE, led us to hypothesize that Dia may be more important in the AS. To begin to test this, we elevated either Ena or Dia activity in AS cells, expressing RFP-Ena or HA-Dia∆DAD specifically in the AS using c381-GAL4, and driving sqh-Moe-GFP to visualize protrusions and compare them to wild type (Figure 4, A–C, and Supplemental Movie S6). Elevating levels of these actin regulators did not prevent completion of dorsal closure. HA-Dia∆DAD did not substantially change either the overall process of dorsal closure or its timing, as assessed by area change or canthi migration within the last 90 min of closure (Supplemental Figure S4, A and C–E). Overexpressing Ena did not alter overall closure or the change in AS area, but it did increase the rate of closure as determined by canthi distance (Supplemental Figure S4, A, B, D, and E, and Supplemental Movie S7).
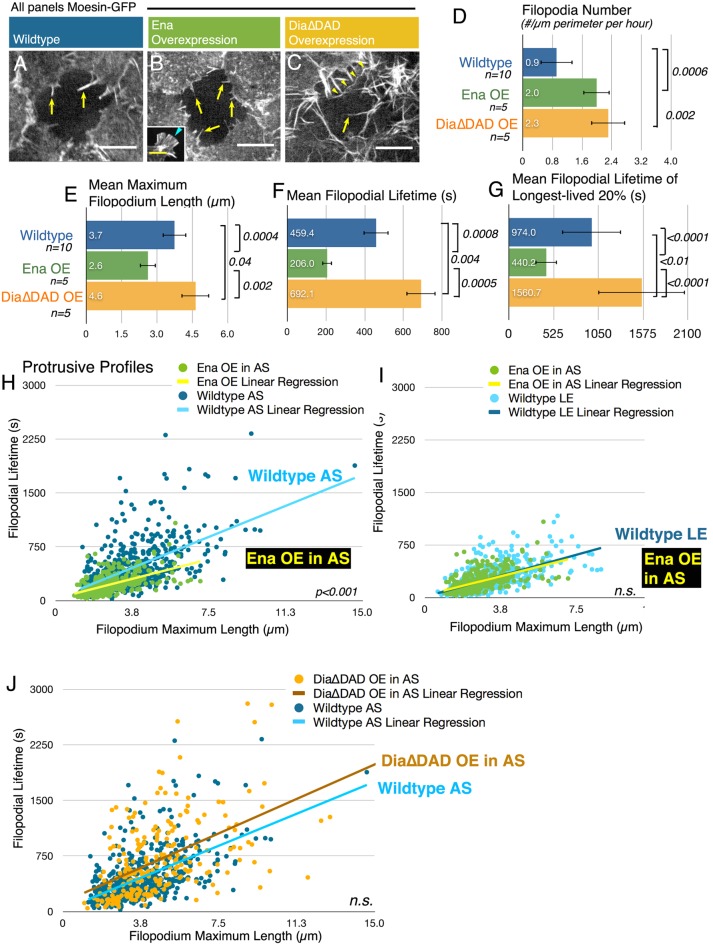
FIGURE 4:
Elevating Ena or Dia activity in the AS has effects on protrusive behavior distinct from one another and from their effects at the LE. (A–C) Representative movie stills of AS cell bleach experiments in stage 14 embryos ubiquitously expressing Moe-GFP in genotypes indicated. Yellow arrows note filopodia. Scale bars, 10 μm, except B inset, 5 μm. (A) Wild-type AS cells produce filopodia (arrows) without many lamellipodia. (B) Ena overexpression increases filopodia number (arrows) but does not induce the filopodial fans seen in LE cells (B inset, arrowhead). (C) Dia∆DAD induces long filopodia (arrow, arrowheads). (D–F) Statistics via t test. (D) Mean filopodia number per micrometer of perimeter per hour. (E) Mean maximum filopodium length (μm). (F) Mean filopodial lifetime (s). (G) Mean filopodial lifetime of longest-lived 20%. (H–J) Protrusive profiles. Statistical test, ANCOVA for difference in slopes, (H) Ena overexpression in the AS alters the protrusive profile, leading to shorter-lived filopodia when controlled for length. (I) Ena overexpression in the AS yields filopodia with a protrusive profile slope resembling that of wild-type LE cells. (J) Expressing Dia∆DAD in the AS does not significantly alter the protrusive profile slope of AS cell filopodia.
Both RFP-Ena and HA-Dia∆DAD expression increased AS filopodia number (Figure 4D and Supplemental Movie S6), as expected. Unexpectedly, Ena expression in the AS did not induce the fan-like protrusions driven by Ena overexpression along the LE (Figure 4, B vs. B, inset, and Supplemental Movie S4 vs. Supplemental Movie S6). However, elevating Ena levels significantly altered filopodial behavior. Whereas Ena overexpression increased filopodia number, as it did when overexpressed at the LE, both filopodial length and lifetime decreased (Figure 4, E and F, and Supplemental Figure S1B). Ena overexpression also decreased the lifetime of the 20% longest-lived filopodia (Figure 4G), and the effect on filopodial lifetime included parallel reductions in the time spent extending and that spent retracting, without a significant change in rate (Supplemental Figure S1, C–F). Analysis of the protrusive profile of AS filopodia after Ena expression provided an interesting insight into this difference. Ena overexpression in the AS altered the relationship between filopodial length and lifetime, with lifetime generally reduced for filopodia of the same length relative to AS controls (Figure 4H and Supplemental Figure S2C). As a result, the protrusive profile of Ena-induced AS filopodia strongly resembles that of filopodia produced by Ena expression along the LE and is statistically indistinguishable from that of wild-type LE filopodia (Figure 4I). The fact that Ena overexpression induces “LE-like” filopodia in the AS is consistent with the hypothesis that Ena governs wild-type filopodia dynamics along the LE.
To begin to test whether Dia governs the longer-lived AS filopodia, we examined effects of expressing Dia∆DAD in the AS, predicting it would increase filopodia lifetime. Indeed, both lifetime and maximum length of filopodia are significantly increased (Figure 4, E and F, and Supplemental Figure S1B; Dia∆DAD also increased the lifetime of the 20% longest-lived filopodia; Figure 4G). The increase in length is readily apparent, with occasional filopodia spanning the cortex of one to several AS cells, an event never seen in controls (Figure 4C, arrowheads, and Supplemental Movie S6, right). The increase in filopodial lifetime primarily involved more time spent extending without a significant change in rate (Supplemental Figure S1, C–F). However, despite these changes in length and lifetime, the slope of the protrusive profile of Dia∆DAD-induced filopodia in the AS was the same as that of wild-type AS cells, as length and lifetime increased in parallel (Figure 4J and Supplemental Figure S2C). These data are consistent with the model that Dia normally drives wild-type AS cell filopodial behavior, helping make them longer lived.
Ena/Dia sequestration via FP4mito expression alters and delays dorsal closure and drastically reduces filopodia number, length, and lifetime
Both elongation factors are thus poised to play roles in filopodia in each tissue, and each can influence filopodia in distinctive ways. However, the integral test for whether Ena and Dia truly regulate normal filopodial behavior during development is to ask whether and how filopodia are altered when each elongation factor is reduced or removed. We first reduced function of both Ena and Dia by sequestering Ena away from the cell cortex by expressing FP4mito, a high-affinity Ena-EVH1 domain-binding site tethered to mitochondria (Bear et al., 2000), specifically in the AS. FP4mito efficiently recruits Ena to mitochondria in Drosophila, leading to loss-of-function (Figure 5B′′, inset, arrows, Gates et al., 2007). FP4mito also recruits some Dia to mitochondria, whereas some remains at the cortex (Figure 5B′′′, inset, arrows vs. arrowhead; Homem and Peifer, 2009; FP4 also recruits Dia family members in mammalian cells; Barzik et al., 2014). FP4mito drastically reduces filopodia in LE cells (Gates et al., 2007).
FIGURE 5:
Expressing FP4mito in the AS attenuates filopodia initiation, length, and lifetime. (A–D) Stage 13/14 embryos. (A) Schematic illustrating view in B–D. (B–B′′′) FP4mito expression in the AS using c381 GAL4 recruits both Ena and some Dia to mitochondria. Arrows in insets, FP4mito punctae; arrowheads, residual cortical Dia. Scale bars, 20 μm (B), 10 μm (B insets, C, and D). (C) Representative movie still, wild-type AS cell expressing Moe-GFP. Arrows, filopodia. (D) Representative movie still, FP4mito expression in the AS. Arrow, short filopodium; bracket, lamellipodium. (E) Mean filopodia number per micrometer of perimeter per hour. (F) Mean filopodium maximum length (μm). (G) Mean filopodial lifetime (s). (H) Mean filopodial lifetime of longest-lived 20%. (I) Mean lamellipodial area. (J) FP4mito filopodia, although substantially shorter, have an unchanged protrusive profile slope.
To explore the effect of depleting both Ena and Dia in the AS, we expressed FP4mito specifically there and visualized protrusions using Moe-GFP (Figure 5, A and D, and Supplemental Movie S8). FP4mito substantially reduced AS filopodia number (Figure 5, C vs. D and E; 0.53 ± 0.03 filopodia/μm perimeter per hour vs. 0.9 ± 0.4 in wild type; p = 0.03) and dramatically reduced the maximum length and lifetime of the remaining filopodia relative to controls (Figure 5, F and G, and Supplemental Figure S1B). FP4mito also drastically decreased the lifetime of the 20% longest-lived filopodia (Figure 5H). The effect on filopodial lifetime resulted from parallel reduction in times of filopodial extension and retraction, without a significant change in rate (Supplemental Figure S1, C–F). Of interest, however, the slope of the protrusive profile of the remaining filopodia was unchanged from that for wild type (Figure 5J and Supplemental Figure S2C). In contrast, lamellipodial area increased relative to wild type (Figure 5I, 0.57 ± 0.14 μm2/μm perimeter vs. 0.42 ± 0.04 in wild type; p = 0.03). These data suggest that some combination of Ena and Dia is important for filopodia initiation in AS cells, and, because FP4mito more effectively recruits Ena relative to Dia, are consistent with the idea that the filopodia remaining after FP4mito expression may be driven by residual cortical Dia, explaining the unchanged slope of the protrusive profile.
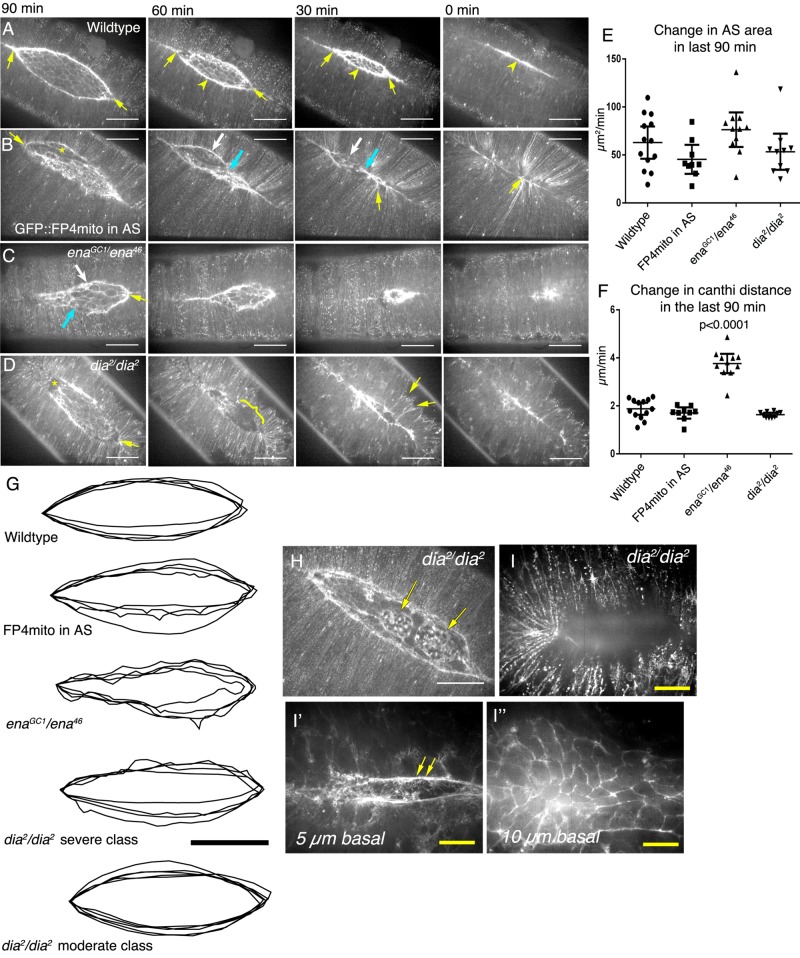
We next explored how depleting Ena and Dia from the AS affected the larger-scale process of dorsal closure. The AS provides a significant but partially redundant contribution to the forces required to ensure completion of dorsal closure (Kiehart et al., 2000; Hutson et al., 2003), resulting, at least in part, from pulsatile apical constrictions of each AS cell that reduce AS area. However, the role of AS cell protrusions in dorsal closure has remained untested. Filopodial protrusions of LE cells do play an important role in late dorsal closure, helping zipper the two sheets together and correctly matching contralateral partners (Woolner et al., 2005; Gates et al., 2007; Millard and Martin, 2008).
To explore the hypothesis that AS cell protrusions are also important for dorsal closure, we assessed morphogenetic movements and dorsal closure timing in embryos expressing FP4mito in the AS. These embryos completed dorsal closure and hatched, as lethality was negligible (96% embryonic viable, n = 327), suggesting that either the few remaining filopodia are sufficient or filopodia in the AS are not essential for dorsal closure. However, our movies revealed substantial alterations in the process. In wild type, the advancing epidermis encloses an eye-shaped opening (Figure 6A; Figure 6Gdisplays four representative examples of the LE at the same stage of closure) with zippering occurring at the canthi (Figure 6A, arrows) and a straight leading edge (Figure 6A, arrowhead, and G). In contrast, embryos expressing FP4mito had abnormally shaped openings (Figure 6, B and G), with apparent difficulties in zippering at the canthi (Figure 6B, arrow). Some regions along the leading edge progressed toward the dorsal midline more rapidly than others (Figure 6B, 60 and 30 min, white vs. blue arrows), and, as closure was completed, the epidermis was puckered (Figure 6B, 30 and 0 min, yellow arrows). In a subset of embryos, there was ripping between the AS and LE (two of eight embryos; Figure 6B, asterisk), although this was repaired and closure completed. Ripping between the AS and LE is also characteristic of mutants lacking the integrin βPS subunit in both tissues (Narasimha and Brown, 2004; Gorfinkiel et al., 2009). Finally, in a subset of embryos expressing FP4mito in the AS, there was a slight delay in dorsal closure. In normal development, dorsal closure precedes initiation of muscle constriction. If dorsal closure is delayed, muscles begin to twitch before the epidermis fully encloses the embryo. Whereas muscle contraction never precedes closure in wild type (n = 13), in embryos expressing FP4mito in the AS, muscles started to twitch just before closure in half of the embryos (three of six; Supplemental Movie S7—twitching begins at 1 h, 50 min); this delay is likely restricted to early stages, as mean rates of area change and canthi advancement in late closure were not significantly altered (Figure 6, E and F, and Supplemental Movie S7). Thus depleting Ena plus Dia substantially alters dorsal closure, but the robustness of the morphogenesis program allows closure to go to completion despite their depletion.
FIGURE 6:
Zygotic ena or dia mutants have substantial alterations in the process of dorsal closure but still complete closure in a timely manner. (A–D, H, I) Movie stills, embryos expressing sqh-driven Moe-GFP in genotypes indicated. (A–D) Embryos filmed from 90 min before closure. Scale bars, 50 μm. (A) Wild type. Note eye-shaped opening with zippering at canthi (arrows) and straight LE (arrowhead). (B) FP4mito expressed in AS. Note alteration in shape of the opening (arrow, 90 min), rip between AS and LE (asterisk), differential progress of different LE cells (white and blue arrows), and puckering of epidermis (30 and 0 min, yellow arrows). (C) ena zygotic mutant. Note alteration in shape of the opening, slowed zippering (yellow arrow), and differential progress of different LE cells (white and blue arrows). (D) dia zygotic mutant. Note alteration in shape of the opening, rip between AS and LE (asterisk), difference in z-plane of LE and AS (bracket), and severe puckering of the epidermis (yellow arrows). (E, F) Quantitation of rates of dorsal closure. Horizontal lines indicate mean and vertical bracket 95% confidence interval. Statistical test, ANOVA. (E) Rates of area change in the last 90 min of closure (μm2/min) are unaltered. (F) Rate of canthi distance change in the last 90 min of closure differs only in ena mutants. (G) Representative scale-matched outlines of LE actin cable aligned by anterior canthi for genotypes indicated, captured when canthi were ∼160 μm apart. (H) dia mutant displaying ripping of the AS (arrows). (I) The relationship between the epidermis and AS cells was often altered in dia mutants: the LE cable (I′, arrows) and AS (I′′) were both several micrometers lower in focal plane than the lateral epidermis (I), unlike what we observed in wild type.
Ena regulates AS filopodial length and lifetime and is required for proper dorsal closure
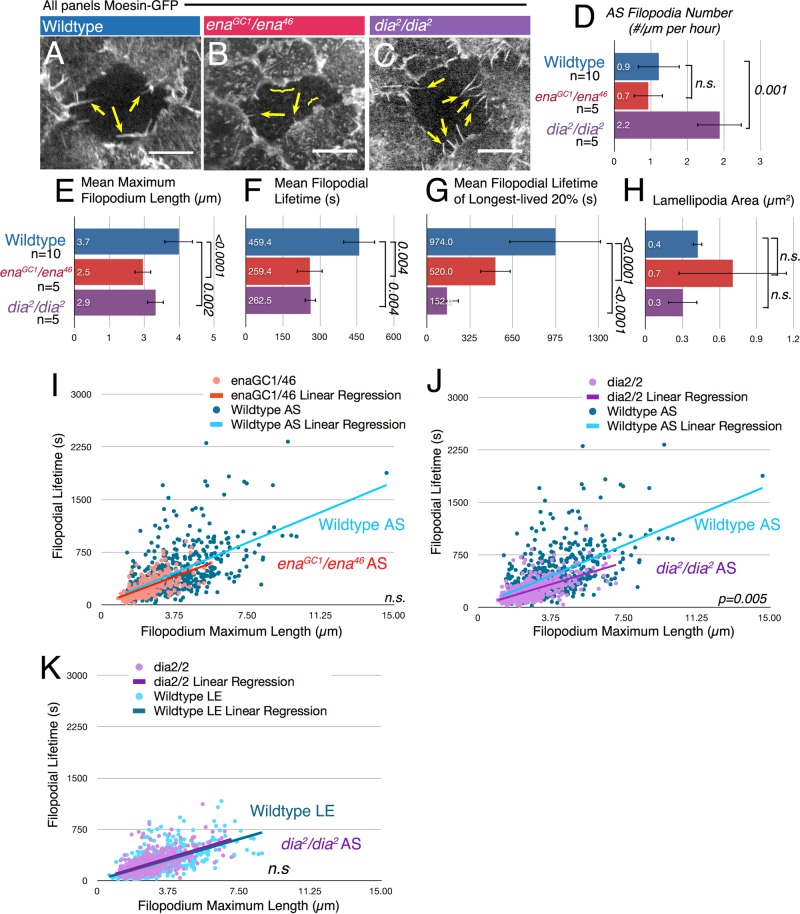
We next sought to determine which elongation factor was responsible for these changes in dorsal closure and in AS protrusions induced by expression of FP4mito, by examining embryos zygotically mutant for either ena or dia. For ena, we examined embryos transheterozygous for enaGC1, a null allele (Gertler et al., 1995), and ena46, which truncates Ena and eliminates the EVH2 domain responsible for Ena's interaction with actin (Li et al., 2005). In transheterozygotes, Ena levels and cortical localization in fixed embryos were substantially reduced by the onset of dorsal closure (Supplemental Figure S5, A and A, inset).
Ena reduction had striking qualitative effects on filopodial behavior (Figure 7, A and B, and Supplemental Movie S8). We thus quantitated different filopodial parameters. Loss of zygotic Ena significantly reduced both filopodial maximum length and lifetime (Figure 7, E and F, and Supplemental Figure S1B), including effects on time spent extending and retracting (Supplemental Figure S1, C–F). Ena reduction had a parallel effect on the lifetime of the 20% longest-lived filopodia (Figure 7G). In contrast, lamellipodial area remained statistically indistinguishable from that of wild type (Figure 7H), and Ena reduction did not significantly alter AS filopodial number relative to that of wild type (Figure 7D), indicating that either Ena is not responsible for most filopodia initiation in the AS or residual maternal Ena is sufficient. This is in marked contrast to the LE, for which reducing Ena significantly reduces filopodia number (Homem and Peifer, 2009). Thus Ena reduction partially phenocopied FP4mito expression, suggesting that Ena does, in fact, help regulate filopodial behavior in this tissue. However, analysis of the protrusive profile of filopodia in ena mutants revealed that filopodia are both shorter and shorter-lived, and thus the slope of the protrusion profile was unchanged (Figure 7I and Supplemental Figure S2C). This suggested that Dia might elongate the remaining filopodia, consistent with the idea that Dia normally regulates the wild-type AS protrusion profile.
FIGURE 7:
Reducing Dia or Ena each affects AS protrusive behavior, but only Dia reduction alters the protrusive profile. (A–C) Representative movie stills, AS cells, stage 13/14 embryos expressing Moe-GFP. Scale bars, 10 μm. (A) Wild-type AS cell. Arrows, filopodia. (B) enaGC1/ena46 zygotic mutant. Arrows, short filopodia; bracket, lamellipodia. (C) dia2 zygotic mutant. Arrows, filopodia. (D) Reducing Dia increases mean filopodia number, whereas reducing Ena does not alter it. (E) Both Ena and Dia are important for mean maximum filopodium length (μm). (F) Both Ena and Dia help maintain the mean filopodial lifetime of filopodia (s). (G) Mean filopodial lifetime of the longest-lived 20% reveals that loss of Dia drastically reduces the lifetimes of the top 20%. (H) Lamellipodia area is statistically similar in all three genotypes. (I–K) Protrusive profiles. (I) In ena mutants, length and lifetime are reduced in proportion, and thus protrusive profile slope is unchanged. (J) Reducing Dia alters the protrusive profile of filopodia toward shorter lifetimes at a given length. (K) Reducing Dia in the AS yields a filopodia protrusive profile with a slope more like that of WT LE filopodia.
We then assessed the requirement for Ena for dorsal closure, analyzing dynamics of the process via live imaging (Figure 6C). As expected from analysis of fixed embryos (Gates et al., 2007), ena zygotic mutants displayed obvious defects in dorsal closure, many of which were shared with FP4mito-expressing embryos. These varied in severity, and most prominently affected the normally “eye-shaped” dorsal closure front, with delays in zippering at the canthi (Figure 6C; 90 min, yellow arrow) and uneven progress of the leading edge (Figure 6G), with some LE cells slowed compared with neighbors (Figure 6C, 90 min, blue vs. white arrows, and Supplemental Movie S7). We next assessed whether these differences in morphogenetic movements slowed the overall rate of closure, by measuring the average rate of area change and advancement of canthi during the last 90 min of closure. Surprisingly, despite the obvious visual defects in dorsal closure shape, the change in area over time was not different from wild type (Figure 6E), and canthi migration rate actually increased (Figure 6F), perhaps because the wavy, elongated LE initially lags behind in zippering and then quickly increases its rate in last 30–60 min before closure (Figure 6C, 60 vs. 30 min). Further, in contrast to embryos expressing FP4mito, only 10% (1 of 10) of ena mutants displayed twitching before closure. Thus, although reducing Ena dramatically alters the process of dorsal closure, it is not required for either completing closure or for its correct timing, suggesting that the timing defects seen after FP4mito expression in the AS are not solely a result of Ena loss.
Dia regulates AS filopodia protrusive profile and plays a role in normal dorsal closure
Dia and its family members can induce filopodia, and cultured cell studies are consistent with a role in protrusive behavior. However, Dia's essential role in cytokinesis and the redundancy of mammalian family members have largely precluded assessment of whether Dia-class formins regulate protrusive behavior during normal development. To assess this, we reduced Dia levels by zygotic loss of function using the null allele dia2 (Castrillon and Wasserman, 1994). We found that zygotic dia2 mutants have substantially reduced cortical Dia at dorsal closure (Supplemental Figure S5B), are embryonic lethal and display significant phenotypes by the time they initiated dorsal closure, including severe head involution defects (Supplemental Figure S6, A and B). The lateral epidermis of dia2 zygotic mutants also had the characteristic defect associated with dia loss in other tissues (Castrillon and Wasserman, 1994; Afshar et al., 2000): fewer and larger cells, with correspondingly large nuclei (Supplemental Figure S6, C–E, yellow arrows), which in other cases resulted from cytokinesis failure. Of importance, because AS cells do not divide after the blastoderm stage (Frank and Rushlow, 1996; Reim et al., 2003), AS cells and nuclei were of normal size in dia2 mutants (Supplemental Figure S6E, magenta arrows). We thus used dia2 zygotic mutants to assess Dia's role in AS filopodial behavior and in the overall process of dorsal closure.
Zygotic dia mutants had significant alterations in AS filopodial behavior (Figure 7, C–H, and Supplemental Movie S8). We first assessed Dia's role in AS filopodia number. Unexpectedly, dia2 mutants had significantly more AS filopodia (Figure 7, C and D). This increase is in direct contrast to the effects of either FP4mito or loss of Ena, for which filopodia number was either reduced or remained the same (Figures 5E and 7D and Supplemental Movie S8). This is also in contrast to the LE, for which reducing Dia reduced filopodia number (Homem and Peifer, 2009). We consider possible explanations for this in the Discussion. Reducing Dia also affected other parameters of protrusive behavior. Like Ena reduction, reducing Dia reduced both the maximum length and lifetime of filopodia (Figure 7, E and F, and Supplemental Figure S1C). The effect on filopodial lifetime included parallel reductions in the time spent extending and that spent retracting, without a significant change in rate (Supplemental Figure S1, C–F). Intriguingly, Dia reduction affected the lifetime of the 20% longest-lived filopodia even more dramatically (Figure 7G). Finally, in contrast to what we observed in ena mutants, the slope of the AS protrusive profile of dia mutants significantly differed from that of wild type; dia-mutant AS filopodia had a generally lower lifetimes per length relative to either control or ena-mutant filopodia (Figure 7J and Supplemental Figure S2C). Strikingly, the protrusive profile of AS filopodia in dia mutants was shifted to resemble that of wild-type LE filopodia (Figure 7K), for which, as we discussed earlier, Ena largely drives protrusive behavior. These data are consistent with a model in which Dia activity normally shapes the protrusive profile of filopodia in AS cells, as Ena does at the LE, and when Dia is reduced, Ena confers filopodial behavior on the AS cells as it normally does at the LE.
Like Ena, Dia is not essential for completion of dorsal closure, as dia zygotic mutants close (nine of nine live embryos). However, closure was morphologically very different. The shape of the dorsal opening was substantially altered in many embryos (Figure 6, D and G), with delays in zippering both anterior and posterior (Figure 6D, 90 min, yellow arrow). Unlike ena mutants, but reminiscent of FP4mito expression in the AS, dia mutants displayed ripping along the LE (10 of 13 embryos; Figure 6H, arrows). In addition, during the last hour of closure, when the AS, the LE cable, and LE cells in wild-type embryos are normally in roughly the same position on the z-axis, in dia mutants, both the LE cable and the AS drop below the level of the LE cells (Figure 6, D, bracket, and I–I′′, and Supplemental Movie S7). This is not due to premature AS cell apoptosis, as we can still visualize both the cable and the AS cells in lower z-planes (Figure 6, I′ and I′′). Despite these morphological differences, the rate of closure as assessed by the rate of change in area or reduction in canthi distance was not affected (Figure 6, E and F), and only occasional (one of nine) dia mutants had muscle twitching before closure. Together these data are consistent with the hypothesis that the AS-LE ripping seen after FP4mito expression results from Dia reduction and suggest that the delay in dorsal closure seen after FP4mito expression is an additive result of reducing both Ena and Dia in the AS.
In recent work, we found that Ena and Dia directly bind one another and that Ena can inhibit Dia both in vitro and in vivo (Bilancia et al., 2014). In examining the lateral epidermis of dia zygotic mutants, we saw a surprising phenotype that suggested this relationship may be even more complex. Whereas protrusive behavior in wild type is essentially restricted to the dorsal surface of LE cells, in dia zygotic embryos we observed frequent ectopic protrusions at tricellular junctions of more-ventral cells (Figure 3G, arrowheads; 14 of 17 embryos), reminiscent of the effects of Ena overexpression (Figure 3E). One possible explanation is that Dia can also inhibit Ena. We consider this further in the Discussion.
DISCUSSION
Cell protrusions help drive many cell behaviors, including cell migration. We are interested in how different cells craft distinct suites of cell protrusions using the same genomic toolkit of actin regulators and how these distinct cell behaviors contribute to morphogenesis. We addressed these questions using gain- and loss-of-function approaches during embryonic morphogenesis to alter the function of two actin elongation factors with known roles in cultured cells, to assess how their biochemical properties and activities in simpler systems play out in the more complex environment in vivo.
Both Ena and Dia play important roles in regulating protrusive behavior during normal development
Studies in vitro and in cultured cells demonstrated that both Ena/VASP and DRFs bind to barbed ends and accelerate filament elongation, and gain-of-function studies revealed that both can stimulate formation of filopodia (see Introduction). However, the contributions of these proteins to protrusive behavior during normal development remain significantly less clear. Ena/VASP proteins have clearly defined roles in vivo in filopodia formation in several settings: Dictyostelium (Schirenbeck et al., 2006), Xenopus retinal ganglion cells (Dwivedy et al., 2007), and Drosophila primary neurons (Gonçalves-Pimentel et al., 2011), LE cells (Homem and Peifer, 2009), and hemocytes (Tucker et al., 2011). In contrast, functional redundancy of mammalian DRFs and their essential role in cytokinesis limit loss-of-function studies during normal development. As a result, there are only a handful of examples suggesting important roles for DRFs in protrusive behavior in vivo: these include roles in planar polarized protrusions during Drosophila bract cell induction (Peng et al., 2012), a role in LE protrusions during dorsal closure (Homem and Peifer, 2009), and a recently described role in promoting cytonemes, the long signaling filopodia in fly imaginal disk cells (Roy et al., 2014).
Our data provide new insights into these issues, allowing us to directly compare the roles of Ena and Dia in shaping the dynamics of protrusive behavior during normal development within the same cell types. Our quantitative analyses of both gain- and loss-of-function mutants reveal that Ena and Dia each induces filopodia with characteristic dynamic behaviors that are most clearly depicted by their distinct “protrusive profiles,” which relate filopodial length and lifetime and thus provide a filopodial fingerprint for each actin regulator. This parameter may be useful as we and others move forward to define the regulators of filopodial behavior in other tissues—it will be interesting to test whether it serves in other places and times as a means of distinguishing whether Ena or Dia is the dominant regulator. It is important to note that the protrusive profile is an emergent property, the precise mechanistic basis of which is unclear. In the simplest terms, the difference in protrusive profile slope means that filopodia are, for example, “more stable” in the AS versus the LE, with longer lifetimes at the same length. Understanding its mechanistic basis will require probing its possible components. Filopodia are likely to exhibit distinct phases of elongation, retraction, and pause (Schäfer et al., 2011), and rates and duration of filopodial extension and retraction reflect a complex mix of several parameters: 1) which actin elongators are present at the tips of filopodia, 2) the intrinsic extension speed and processivity of the actin elongation factor(s) at the tip and the way these parameters are modulated by other factors in the tissue, 3) the levels and activity of capping protein, and 4) the rate of retrograde flow in the tissue. It will be important moving forward to develop new tools to measure these modes reliably in tissues in vivo and to further dissect the contributions of different factors contributing to protrusive behavior.
Together our data reveal that, in general, Dia promotes filopodia with longer lifetimes, whereas Ena-driven filopodia are shorter-lived. These differences in filopodial dynamics regulated by Ena and Dia during embryogenesis parallel the biochemical differences we recently documented for these two proteins: Drosophila Dia is a more effective actin elongation factor than Ena, promoting actin elongation two times more efficiently and remaining at the barbed end seven times longer (Bilancia et al., 2014). The ability to assess the roles of Ena and Dia in parallel at all levels of complexity from purified proteins to cultured cells to tissues in vivo during morphogenesis is beginning to provide a picture of why cells retain two different proteins that both promote actin filament elongation, by suggesting that cells use them differentially to craft distinct protrusive behaviors.
These simplified “rules,” suggesting, for example, that Dia will produce longer filopodia, are not universally true in complex tissues in vivo, however; for example, activated Dia overexpression in the lateral epidermis decreased LE filopodia length. What might account for this variation? In this tissue, filopodia are normally restricted to the leading edge of the LE cells. Activated Dia not only elevates filopodia number at the leading edge, but it also induces filopodia all around the cell perimeter. We suspect this may lead to competition for limiting levels of G-actin, thus reducing the length of individual filopodia. This competition for G-actin illustrates one way in which actin regulators can interact with one another. Further, the complexity in generation of filopodia is likely to be present even in the filopodia produced by a single cell type, with different subsets of protrusions driven by different combinations of actin regulators. Although neither our filopodial profiles nor our binning of the lengths and lifetimes of filopodia revealed clearly discrete populations of filopodia within a given cell type, our previous analysis in cultured cells suggested that individual filopodia can have Ena, Dia, or both at their tips (Bilancia et al., 2014). Further, whereas Dia loss in the AS reduced the mean lifetime of all filopodia, it had its most striking effect on the longest-lived filopodia, consistent with the possibility that it may differentially regulate a filopodial subset. As we move forward, we may develop tools that allow us to distinguish these different populations in the intact animal.
Different elongation factors play predominant roles in shaping filopodia in different tissues
Dorsal closure is driven by two cell types that differ dramatically in many ways: the migratory LE cells and the apically constricting but nonmigratory AS cells. The protrusive behavior of LE cells serves as a model for understanding the role of filopodia in collective cell migration (Woolner et al., 2005; Gates et al., 2007; Millard and Martin, 2008; Homem and Peifer, 2009), but the nature and role of cell protrusions of AS cells remained mysterious. Our quantitative comparison revealed that the two cell types differ in protrusive behavior in specific ways: 1) AS cells produce significantly fewer filopodia and substantially less lamellipodia, and 2) AS filopodia had significantly longer lifetimes. Extending this analysis beyond simply comparing mean values of individual parameters further revealed that the relationship between length and lifetime of individual filopodia—the protrusive profile—highlighted differences in filopodial dynamics between these two tissues. The differences in dynamics of LE and AS filopodia led us to hypothesize that the longer-lived AS filopodia were governed by Dia, whereas the more dynamic LE filopodia were governed by Ena.
Our combination of gain- and loss-of-function approaches allowed us to test this hypothesis. Elevating Ena levels promoted dynamic, shorter-lived filopodia in both cell types, with protrusive profile slopes matching those of wild-type LE cells, whereas active Dia induced longer-lived filopodia at all locations we examined, whose protrusive profiles were more similar to those of wild-type AS cells. This is consistent with the idea that Ena is responsible for the normal protrusive profile of the LE cells. In contrast, whereas ena loss of function did not alter the slope of the protrusive profile of AS filopodia, dia loss of function converted the AS cell protrusive profile slope to one resembling that of LE cells, consistent with the idea that Dia is normally the dominant player in the AS. However, the full picture that emerged is not that simple: our present and previous (Homem and Peifer, 2009) loss-of-function analyses reveal that both actin regulators play roles in both tissues—for example, the data are consistent with a model in which Dia is important for elongation of AS filopodia, whereas Ena may play a supporting role, perhaps helping regulate filopodia initiation and working in concert with fascin-based bundling (Winkelman et al., 2014). It will be important to define the mechanistic underpinnings of these differences.
How might cells differentially deploy these two actin regulators? Our analysis ruled out the simplest model, in which one or the other cell type expressed only Ena or Dia. However, the two tissues do localize both actin regulators in distinctive fashions, with both Ena and to a lesser extent Dia having polarized localizations in the lateral epidermis, whereas both proteins are uniformly cortical in the AS. These differences in localization could reflect direct recruitment of Ena or Dia to these discrete places by distinct actin architectures or indirect recruitment via binding partners, including partners binding Ena's EVH1 domain, such as lamellipodin or zyxin (Krause et al., 2004; Drees et al., 2002). Another potential clue emerged from examination of fluorescently tagged Ena and Dia. GFP-DiaΔDad localized effectively to filopodia tips in both the AS and at the LE. However, RFP-Ena localized distinctly differently in the two tissues. In LE cells, it readily localized to both filopodia tips and to the “filopodial fans” that it induced. However, in the AS, RFP-Ena was largely cortical rather than strongly localized to filopodial tips and did not induce filopodial fans. Perhaps Ena localization and/or activity is differentially regulated in these two tissues. Defining the mechanisms differentially regulating Ena at different subcellular locations will be of interest as we move forward.
Both actin elongation factors are important for proper dorsal closure, but the process is robust to their depletion
Although AS cells are not migratory, they still produce dynamic filopodia and lamellipodia. Why do these nonmigratory cells have filopodia, and are they important for dorsal closure? FP4mito expression specifically in the AS, which sequesters Ena and some Dia, substantially reduced filopodia length and lifetime. It also strikingly altered the shape/movement of the LE, along with more subtle changes in proper timing of closure and tissue integrity. The ena and dia zygotic mutants, which each exhibited alterations in AS and LE filopodial behavior, also close in a manner morphologically quite distinct from wild type, although the timing of closure remains unaltered. Together these data are consistent with the possibility that AS filopodia may help regulate dorsal closure. However, AS cells also undergo dramatic cycles of actomyosin-based apical constriction and relaxation, which play important roles in driving closure and presumably use the same actomyosin machinery that drives protrusions (Franke et al., 2005; Solon et al., 2009; David et al., 2010). Because we lack a method for depleting cell protrusions without affecting cortical actin and the actomyosin network involved in cell constriction, it is unclear whether the altered closure process we observed is directly related to filopodia loss or occurs via effects on another structure/process altered by elongation factor loss. The ability of FP4mito-expressing embryos and ena and dia zygotic mutants to close in a time frame largely similar to wild type despite considerable larger-scale defects also highlights the robustness of the dorsal closure process.
The question of why nonmigratory AS cells have filopodia is intriguing. The dynamic LE filopodia are important for neighbor sensing and alignment as the epithelial sheets meet at the midline (Millard and Martin, 2008). One possible role for the long-lived filopodia in the nonmigratory AS cells is in cell–cell signaling over the course of the closure process. Cytonemes, long, basally associated filopodia-like structures present in the nonmigratory but shape-changing Drosophila wing disk cells, play a role in Dpp signaling, and Dia is required for proper cytoneme elongation (Roy et al., 2014). JNK and Dpp signaling regulate dorsal closure (Reed et al., 2001; Fernández et al., 2007), and thus it will be of interest to look at the relationship between AS cell filopodia and signaling. Alternatively, filopodia might play an entirely different function in the AS: because AS cells rapidly reduce their apical area in each pulse of apical constriction, these long-lived protrusions may simply function as external membrane storage to allow time for the endocytic machinery to catch up. Assessing coordination of constriction with protrusive behavior will be of interest in the future.
Integrating Ena, Dia, and other actin regulators
Cells rely on arrays of actin regulators to accomplish complex tasks. Even the seemingly simple process of antagonizing actin capping and promoting filament elongation involves Ena and Dia working together in complex ways, based in part on their direct physical interaction (Homem and Peifer, 2009; Bilancia et al., 2014). In vitro and cultured cell assays revealed that Ena can negatively regulate Dia activity, and, consistent with this, active Dia induces long-lived filopodia at lateral borders of epidermal cells, where Ena levels are low, and more dynamic filopodia at tricellular junctions, where Ena levels are high (Bilancia et al., 2014). However, Dia can also influence Ena localization and perhaps activity. Dia∆DAD expression induced fan-like protrusions along the LE; these structures are never seen in wild type but are also induced upon Ena overexpression. One possible mechanism by which active Dia could mimic Ena activation is suggested by the fact that active Dia triggers Ena localization from LE “dots” to the protrusive front (Homem and Peifer, 2009). We observed a similar relationship at tricellular junctions in the lateral epidermis: both overexpressing Ena and expressing active Dia induce similar ectopic protrusions. However, paradoxically, zygotic dia2 mutants also exhibit ectopic protrusions at tricellular junctions in the lateral epidermis and also produce more AS filopodia. Both effects closely resemble Ena overexpression, suggesting that Dia may also negatively regulate Ena at those locations. Perhaps reduced Dia levels release Ena from Ena:Dia complexes, allowing it to interact with other partners and stimulate protrusive behavior. Thus, although Ena and Dia are both important in each tissue, their relationship is not a simple one. This complex relationship between Ena and Dia, including both competition for barbed ends and profilin-actin and direct interactions, will have to be taken into account when defining mechanisms in vivo. Finally, filament elongation does not exist in a vacuum; instead, diverse actin regulators work in competition and synergy with each other, making the network plastic. Ena and Dia are likely to be heavily influenced by other actin regulators, and vice versa. For example, competition for actin monomers with the Arp2/3 complex and the roles of filament bundling and severing are likely to be critical. One potential indication of this sort of competition is the increase in AS lamellipodial area in embryos in which we used FP4-mito to sequester Ena and Dia. It will be of interest to probe integration of multiple players using quantitative tools to assess the dynamics of protrusions induced by different combinations of actin regulators.
MATERIALS AND METHODS
Fly stocks
All experiments were performed at 25°C. Mutations are described at http://flybase.org. Wild type was y w for fixed imaging and sqh promoter-driven moesin actin-binding domain fused to GFP (Moe-GFP), UAS-driven Moe-GFP, or UAS-driven actin-GFP for live imaging. Zygotic mutants were selected using the Cy twiGFP balancer. A full-length Ena transgene was cloned into a derivative of pUASp modified for Gateway cloning by T. Murphy in order to add an N-terminal mRFP tag. Males carrying UAS-transgenes were crossed to females with a GAL4 driver. See Table 1 for additional fly stock information.
Image acquisition and analysis
The following fixations were used. For Dia and neurotactin, heat-methanol treatment (Müller and Wieschaus, 1996; Laplante and Nilson, 2006), held in methanol for at least 48 h at 4°C before antibody application. All other immunohistochemistry used a 4% formaldehyde fix for 1 h. Phalloidin staining required hand-devitellinized embryos. All fixed embryos were incubated in primary antibodies overnight with agitation at 4°C and in secondary antibodies (Alexa; Molecular Probes, Eugene, OR) for 2 h at room temperature, then mounted in Aquapolymount (Polysciences, Warrington, PA) and imaged with a Zeiss 710 Confocal (Zeiss, Thornwood, NY). Antibodies and concentrations used are in Table 1.
For live imaging, embryos were bleach-dechorionated in 50% bleach and mounted in halocarbon oil (series 700; Halocarbon Products, River Edge, NJ) between a coverslip and a permeable membrane (petriPERM [Sartorius, Edgewood, NJ] or Lumox [Sarstedt, Nümbrecht, Germany]). For high-resolution movies, single-plane images or 0.5-μm z-stacks were acquired every 5 s using a 100×/1.4 numerical aperture (NA) Plan Apo VC Nikon objective on an inverted TE2000-E microscope (Nikon, Melville, NY) with a VTHawk confocal system (VisiTech, Sunderland, United Kingdom) and an Orca R2 (Hamamatsu Photonics, Hamamatsu, Japan) charge-coupled device camera. Retrograde flow data were acquired as described but with 1-s intervals. ImageJ (National Institutes of Health, Bethesda, MD) was used to quantitate filopodial lifetime and maximum length. Movies with z-stacks were compiled as maximum intensity projections for quantification. Filopodia were manually tracked and defined as any thin protrusion (width, <1.25 μm; length, >1.15 μm) extending beyond the lamellipodium or leading edge. Lamellipodia were defined as protrusions >1.25 μm in width. For leading-edge cells, quantitation was performed on one or more pairs of contralateral engrailed-expressing stripes (as the leading edge moved from 30 to 15 μm apart) per embryo. For amnioserosal cells, quantitation was performed on one to two bleached cells per embryo during the mid stage of closure. In both tissues, n is the number of embryos analyzed per genotype. For low-magnification movies, single-plane images were acquired every 15 s using a 40×/1.3NA Plan Fluor Nikon objective on a Wallac Ultraview System (PerkinElmer, Norwalk, CT). Direct comparison of top 20% of filopodia lifetimes used a random number generator function in Matlab (MathWorks, Natick, MA) to reduce the n of AS cells by 12 in a nonbiased manner to match that of LE cells in Figure 1K. Statistical significance for averaged, normally distributed data sets were determined using an unpaired Student's t test or analysis of variance (ANOVA) when comparing more than two samples, and a Mann–Whitney U test was used for the nonnormal distribution data set associated with the top 20% lifetime data using Prism for Windows, version 5.00 (GraphPad Software, San Diego, CA). When producing and analyzing the protrusive profile, analysis of covariance for linear regression (ANCOVA) was determined using Matlab. To verify that the differences in slope of the line in the protrusive profiles of LE and AS filopodia reflected differences in the relationship between filopodia length and lifetime, we examined the average lifetimes at given lengths (from the binned lengths in Supplemental Figure S1); for each bin examined that had a significant number of filopodia, differences in mean lifetime were p < 0.05. Photoshop CS4 (Adobe, San Jose, CA) was used to adjust input levels so the main range of signals spanned the entire output grayscale and to adjust brightness and contrast. In cases in which comparisons were made of levels between images, adjustments were made in parallel.
Supplementary Material
Acknowledgments
We thank all members of our lab and R. Cheney for helpful discussions. We are very grateful to T. Perdue for assistance with imaging, to T. Salmon for access to critical equipment, to J. Dale for keeping us sane while quantitating filopodia, and to Kyle Smith for help with time-lapse imaging. Additional thanks go to D. Tatomer, C. Bilancia, J. Poulton, S. Rogers, and A. Maddox for constructive comments on the manuscript. These experiments were supported by National Institutes of Health Grant R01 GM47857.
Abbreviations used:
- AS
amnioserosal
- Dia
Diaphanous
- Ena
Enabled
- LE
leading edge
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-05-0951) on August 20, 2014.
REFERENCES
- Abreu-Blanco MT, Verboon JM, Parkhurst SM. Coordination of Rho family GTPase activities to orchestrate cytoskeleton responses during cell wound repair. Curr Biol. 2014;24:144–155. doi: 10.1016/j.cub.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar K, Stuart B, Wasserman SA. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 2000;127:1887–1897. doi: 10.1242/dev.127.9.1887. [DOI] [PubMed] [Google Scholar]
- Antunes M, Pereira T, Cordeiro JV, Almeida L, Jacinto A. Coordinated waves of actomyosin flow and apical cell constriction immediately after wounding. J Cell Biol. 2013;202:365–379. doi: 10.1083/jcb.201211039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima S-I, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, McClain LM, Gupton SL, Gertler FB. Ena/VASP regulates mDia2-initiated filopodial length, dynamics, and function. Mol Biol Cell. 2014;25:2604–2619. doi: 10.1091/mbc.E14-02-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer D, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bilancia CG, Winkelman JD, Tsygankov D, Nowotarski SH, Sees JA, Comber K, Evans I, Lakhani V, Wood W, Elston TC, et al. Enabled negatively regulates diaphanous-driven actin dynamics in vitro and in vivo. Dev Cell. 2014;28:394–408. doi: 10.1016/j.devcel.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Stradal TEB, Hänisch J, Geffers R, Köstler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou P, Hasan S, Breitsprecher D, Baudelet E, Camoin L, Audebert S, Goode BL, Badache A. Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol Biol Cell. 2014;25:658–668. doi: 10.1091/mbc.E13-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DJV, Tishkina A, Harris TJC. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development. 2010;137:1645–1655. doi: 10.1242/dev.044107. [DOI] [PubMed] [Google Scholar]
- Dong B, Zhang SS, Gao W, Haichun S, Chen J, Jin F, Bhargava A, Chen X, Jorgensen L, Alberts AS, et al. Mammalian diaphanous-related formin 1 regulates GSK3β-dependent microtubule dynamics required for T cell migratory polarization. PLoS One. 2013;8:e80500. doi: 10.1371/journal.pone.0080500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and Ena/VASp family of proteins: implications for actin cytoskeleton organization. J Biol Chem. 2002;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- Dwivedy A, Gertler FB, Miller J, Holt CE, Lebrand C. Ena/VASP function in retinal axons is required for terminal arborization but not pathway navigation. Development. 2007;134:2137–2146. doi: 10.1242/dev.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A, Hickson GRX, Foley E, O'Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Fernández BG, Arias AM, Jacinto A. Dpp signalling orchestrates dorsal closure by regulating cell shape changes both in the amnioserosa and in the epidermis. Mech Dev. 2007;124:884–897. doi: 10.1016/j.mod.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Frank LH, Rushlow C. A group of genes required for maintenance of the amnioserosa tissue in Drosophila. Development. 1996;122:1343–1352. doi: 10.1242/dev.122.5.1343. [DOI] [PubMed] [Google Scholar]
- Franke JD, Montague RA, Kiehart DP. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol. 2005;15:2208–2221. doi: 10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, Van Vactor D, Gertler FB, Peifer M. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–2039. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Goh WI, Ahmed S. mDia1-3 in mammalian filopodia. Commun Integr Biol. 2012;5:340–344. doi: 10.4161/cib.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Sudhaharan T, Lim KB, Sem KP, Lau CL, Ahmed S. Rif-mDia1 interaction is involved in filopodium formation independent of Cdc42 and Rac effectors. J Biol Chem. 2011;286:13681–13694. doi: 10.1074/jbc.M110.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves-Pimentel C, Gombos R, Mihály J, Sánchez-Soriano N, Prokop A. Dissecting regulatory networks of filopodia formation in a Drosophila growth cone model. PLoS One. 2011;6:e18340. doi: 10.1371/journal.pone.0018340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Blanchard GB, Adams RJ, Martinez-Arias A. Mechanical control of global cell behaviour during dorsal closure in Drosophila. Development. 2009;136:1889–1898. doi: 10.1242/dev.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans J, Wenzl C, Herz H-M, Bartoszewski S, Schnorrer F, Vogt N, Schwarz H, Muller H-A. RhoGEF2 and the formin Dia control the formation of the furrow canal by directed actin assembly during Drosophila cellularisation. Development. 2005;132:1009–1020. doi: 10.1242/dev.01669. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci. 2007;120:3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007:re5. doi: 10.1126/stke.4002007re5. 2007. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG. The outgrowth of the nerve fiber as a mode of protoplasmic movement. J Exp Zool. 1910;9:787–848. doi: 10.1002/jez.1401420103. [DOI] [PubMed] [Google Scholar]
- Heisenberg C-P. Dorsal closure in Drosophila: cells cannot get out of the tight spot. Bioessays. 2009;31:1284–1287. doi: 10.1002/bies.200900109. [DOI] [PubMed] [Google Scholar]
- Hoelzle MK, Svitkina T. The cytoskeletal mechanisms of cell-cell junction formation in endothelial cells. Mol Biol Cell. 2012;23:310–323. doi: 10.1091/mbc.E11-08-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CCF, Peifer M. Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development. 2008;135:1005-1018. doi: 10.1242/dev.016337. [DOI] [PubMed] [Google Scholar]
- Homem CCF, Peifer M. Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell. 2009;20:5138–5155. doi: 10.1091/mbc.E09-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MS, Tokutake Y, Chang M-S, Bloor JW, Venakides S, Kiehart DP, Edwards GS. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Fitz Gerald JN, Guerin C, Robert H, Sorensen MB, Van Damme D, Geelen D, Blanchoin L, Berger F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat Cell Biol. 2005;7:374–380. doi: 10.1038/ncb1238. [DOI] [PubMed] [Google Scholar]
- Jacinto A, Wood W, Balayo T, Turmaine M, Martinez-Arias A, Martin P. Dynamic actin-based epithelial adhesion and cell matching during Drosophila dorsal closure. Curr Biol. 2000;10:1420–1426. doi: 10.1016/s0960-9822(00)00796-x. [DOI] [PubMed] [Google Scholar]
- Jacinto A, Wood W, Woolner S, Hiley C, Turner L, Wilson C, Martinez-Arias A, Martin P. Dynamic analysis of actin cable function during Drosophila dorsal closure. Curr Biol. 2002a;12:1245–1250. doi: 10.1016/s0960-9822(02)00955-7. [DOI] [PubMed] [Google Scholar]
- Jacinto A, Woolner S, Martin P. Dynamic analysis of dorsal closure in Drosophila: from genetics to cell biology. Dev Cell. 2002b;3:9–19. doi: 10.1016/s1534-5807(02)00208-3. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Lai S-L, Chan T-H, Lin M-J, Huang W-P, Lou S-W, Lee S-J. Diaphanous-related formin 2 and profilin I are required for gastrulation cell movements. PLoS One. 2008;3:e3439. doi: 10.1371/journal.pone.0003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Laplante C, Nilson LA. Differential expression of the adhesion molecule Echinoid drives epithelial morphogenesis in Drosophila. Development. 2006;133:3255–3264. doi: 10.1242/dev.02492. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- Li W, Li Y, Gao F-B. Abelson, enabled, and p120 catenin exert distinct effects on dendritic morphogenesis in Drosophila. Dev Dyn. 2005;234:512–522. doi: 10.1002/dvdy.20496. [DOI] [PubMed] [Google Scholar]
- Loureiro JJ, Rubinson DA, Bear JE, Baltus GA, Kwiatkowski AV, Gertler FB. Critical roles of phosphorylation and actin binding motifs, but not the central proline-rich region, for Ena/vasodilator-stimulated phosphoprotein (VASP) function during cell migration. Mol Biol Cell. 2002;13:2533–2546. doi: 10.1091/mbc.E01-10-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R, Schejter ED, Shilo B-Z. Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Dev Cell. 2009;16:877–888. doi: 10.1016/j.devcel.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Curr Biol. 2011;21:R560–R569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Martin P. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development. 2008;135:621–626. doi: 10.1242/dev.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulinari S, Barmchi MP, Hacker U. DRhoGEF2 and diaphanous regulate contractile force during segmental groove morphogenesis in the Drosophila embryo. Mol Biol Cell. 2008;19:1883–1892. doi: 10.1091/mbc.E07-12-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha M, Brown NH. Novel functions for integrins in epithelial morphogenesis. Curr Biol. 2004;14:381–385. doi: 10.1016/j.cub.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA. Ena/VASP proteins capture actin filament barbed ends. J Biol Chem. 2008;283:9814–9819. doi: 10.1074/jbc.M710475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson C, Eaton BA, Davis GW. Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. J Neurosci. 2008;28:11111–11123. doi: 10.1523/JNEUROSCI.0833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Han C, Axelrod JD. Planar polarized protrusions break the symmetry of EGFR signaling during Drosophila bract cell fate induction. Dev Cell. 2012;23:507–518. doi: 10.1016/j.devcel.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Raich WB, Agbunag C, Hardin J. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr Biol. 1999;9:1139–1146. doi: 10.1016/S0960-9822(00)80015-9. [DOI] [PubMed] [Google Scholar]
- Reed BH, Wilk R, Lipshitz HD. Downregulation of Jun kinase signaling in the amnioserosa is essential for dorsal closure of the Drosophila embryo. Curr Biol. 2001;11:1098–1108. doi: 10.1016/s0960-9822(01)00318-9. [DOI] [PubMed] [Google Scholar]
- Reim I, Lee H-H, Frasch M. The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development. 2003;130:3187–3204. doi: 10.1242/dev.00548. [DOI] [PubMed] [Google Scholar]
- Rørth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science. 2014;343:1244624. doi: 10.1126/science.1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497:628–632. doi: 10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer C, Faust U, Kirchgessner N, Merkel R, Hoffmann B. The filopodium: a stable structure with highly regulated repetitive cycles of elongation and persistence depending on the actin cross-linker fascin. Cell Adh Migr. 2011;5:431-8. doi: 10.4161/cam.5.5.17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TEB, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Shi Y, Zhang J, Mullin M, Dong B, Alberts AS, Siminovitch KA. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182:3837–3845. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- Spracklen AJ, Fagana TN, Lovander KE, Tootle TL. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol. 2014 doi: 10.1016/j.ydbio.2014.06.022. S0012-1606(14)00318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S-I, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, Schnabel R, Bowerman B. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111:2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- Tanizaki H, Egawa G, Inaba K, Honda T, Nakajima S, Moniaga CS, Otsuka A, Ishizaki T, Tomura M, Watanabe T, et al. Rho-mDia1 pathway is required for adhesion, migration, and T-cell stimulation in dendritic cells. Blood. 2010;116:5875–5884. doi: 10.1182/blood-2010-01-264150. [DOI] [PubMed] [Google Scholar]
- Thumkeo D, Shinohara R, Watanabe K, Takebayashi H, Toyoda Y, Tohyama K, Ishizaki T, Furuyashiki T, Narumiya S. Deficiency of mDia, an actin nucleator, disrupts integrity of neuroepithelium and causes periventricular dysplasia. PLoS One. 2011;6:e25465. doi: 10.1371/journal.pone.0025465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol. 2002;12:1864–1870. doi: 10.1016/s0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science. 2008;321:1683-1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Evans IR, Wood W. Ena drives invasive macrophage migration in Drosophila embryos. Dis Model Mech. 2011;4:126–134. doi: 10.1242/dmm.005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Watanabe S, De Zan T, Ishizaki T, Yasuda S, Kamijo H, Yamada D, Aoki T, Kiyonari H, Kaneko H, Shimizu R, et al. Loss of a Rho-regulated actin nucleator, mDia2, impairs cytokinesis during mouse fetal erythropoiesis. Cell Rep. 2013;5:926–932. doi: 10.1016/j.celrep.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci USA. 2014;111:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W, Martin P. Structures in focus—filopodia. Int J Biochem Cell Biol. 2002;34:726–730. doi: 10.1016/s1357-2725(01)00172-8. [DOI] [PubMed] [Google Scholar]
- Woolner S, Jacinto A, Martin P. The small GTPase Rac plays multiple roles in epithelial sheet fusion—dynamic studies of Drosophila dorsal closure. Dev Biol. 2005;282:163–173. doi: 10.1016/j.ydbio.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Yamana N, Arakawa Y, Nishino T, Kurokawa K, Tanji M, Itoh RE, Monypenny J, Ishizaki T, Bito H, Nozaki K, et al. The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol Cell Biol. 2006;26:6844–6858. doi: 10.1128/MCB.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S-I, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PE, Pesacreta TC, Kiehart DP. Dynamic changes in the distribution of cytoplasmic myosin during Drosophila embryogenesis. Development. 1991;111:1–14. doi: 10.1242/dev.111.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.