Abstract
Meristems require a myriad of intercellular signaling pathways for coordination of cell division within and between functional zones and clonal cell layers. This control of cell division ensures a constant availability of stem cells throughout the life span of the meristem while limiting overproliferation of meristematic cells and maintaining the meristem structure. We have undertaken a genetic screen to identify additional components of meristem signaling pathways. We identified pluripetala (plp) mutants based on their dramatically larger meristems and increased floral organ number. PLURIPETALA encodes the α-subunit shared between protein farnesyltransferase and protein geranylgeranyltransferase-I. plp mutants also have altered abscisic acid responses and overall much slower growth rate. plp is epistatic to mutations in the β-subunit of farnesyltransferase and shows a synergistic interaction with clavata3 mutants. plp mutants lead to insights into the mechanism of meristem homeostasis and provide a unique in vivo system for studying the functional role of prenylation in eukaryotes.
Meristems are small groups of pluripotent plant cells that are the ultimate source of all adult plant structures. Meristems act by integrating external environmental signals with genetic cues to initiate leaves, branches, flowers, stems, and roots in a manner characteristic of the species but also adapted to environmental circumstances. Meristems also have a maintenance function that allows them to replenish cells lost to initiation events and to preserve their overall integrity throughout the life of the plant.
Shoot meristems consist of a small central zone composed of slowly dividing cells that replenish cells in the surrounding peripheral zone lost to the initiation of leaves, branches, and flowers (1, 2). Superimposed on these zones is an additional histological character, that of clonally distinct cell layers (3). In most dicotyledonous plants, there are three layers: L1, or the epidermal layer; L2, or the subepidermal layer; and L3, or the corpus. Because each layer is clonal, cell growth and proliferation must be coordinated between the different cell layers while the meristem carries out its maintenance and primordia initiation functions. This coordination presumably occurs via cell–cell communication between and within the different functional zones and cell layers.
The best-characterized meristem signaling pathway is the CLAVATA (CLV) pathway (4). Plants with mutations in any of three loci, CLV1, CLV2, or CLV3, show a progressive increase in meristem size beginning in the embryo and continuing throughout life, indicating a loss of cell division restriction (5–8). Available evidence indicates that CLV3 encodes a small secreted peptide expressed in outer cell layers (9, 10) and likely binds to the leucine-rich repeat receptor kinase CLV1 and its putative dimerization partner CLV2, which are expressed in inner cell layers (11–13). Activation of the CLV complex results in negative regulation of cell proliferation (14). The CLV complex also includes a kinase-associated protein phosphatase and a Rhorelated GTPase, which may participate in downstream signaling (15). Genetic evidence indicates that much of the CLV response is mediated by WUSCHEL (WUS), a homeodomain-containing putative transcription factor (16–18). wus mutants have the opposite phenotype of clv mutants in that they have reduced or absent meristems (16). wus is also epistatic to the clv mutants, indicating that wus acts downstream and is a putative target of CLV action (18).
Several other mutants that affect meristem cell proliferation control have been identified; however, their relation neither to each other nor to the pathways in which they are involved is clear. Mutations in the SHOOTMERISTEMLESS (STM) gene, another homeodomain gene, cause a phenotype similar to wus mutants, with greatly reduced or absent meristems (19, 20); however, genetic analysis indicates that STM acts in a pathway separate from that of the CLV genes (21). Other mutants that show increased apical meristem size include fasciata1 (fas1), fasciata2 (fas2), mgoun1 (mgo1), mgoun2 (mgo2), and enhanced response to abscissic acid (era1) (22–28). In each of these mutants, the meristem shows a greater increase in width instead of the height increase seen in clv mutants, a possible indication that they play a role in limiting peripheral zone cell proliferation. To identify additional genes involved in meristem proliferation control, we took advantage of the fact that overexpression of the homeobox gene BREVIPEDICELLUS (BP, also called KNAT1) induces lobing and ectopic meristem formation in leaves (29). We reasoned that mutations in genes that are required to limit meristem cell division would lead to increased meristem activity and an enhancement of the 35S::BP phenotype.
Here we describe the isolation and characterization of pluripetala (plp) mutants. plp single mutants are very slow growing and have enlarged wider meristems and extra flower organs, particularly petals. plp mutants show similar but much more severe defects in meristem function and flower development compared to era1. We show that PLP encodes a key component of the protein prenylation mechanism.
Materials and Methods
Isolation of plp-1 and Sources of Other Plant Material. 35S::BP plants (29) in ecotype Nossen-0 were mutagenized via Agrobacterium-mediated DNA insertion as described (30) by using the vector developed for vacuum infiltration (31). Approximately 15,000 resistant T1 seeds were collected in pools of five, with plp-1 isolated once in the population. plp-1 was subsequently back-crossed to Columbia-0 for analysis. plp-2 segregates in the Syngenta Arabidopsis insertion library line 519 D08 from Syngenta/Torrey Mesa Research Institute (Syngenta Biotechnology, Research Triangle Park, NC) and is in a Col-0 background (32). era1–4, originally called wig-1, and clv3–2 have been described (6, 25, 28).
Identification of PLP. plp-1 was crossed to Col-0 and Landsberg erecta (L-er), and linkage to simple sequence length polymorphism markers was analyzed (33). plp-1 was found to map between nga707 and AthFus6. We PCR-amplified and sequenced the leading candidate gene in this region, protein farnesyltransferase (PFT)/protein geranylgeranyltransferase (PGGT)-Iα, from No-0 and plp-1 genomic DNA and identified an 11-bp deletion in plp-1. This result was confirmed by sequencing of the plp-1 RT-PCR product.
Genetic Analysis. The F1 progeny of a cross between plp-1 and era1–4 resembled wild type. F2 progeny of selfed F1 plants included plants with wild type, era1–4, and plp-1 phenotypes, and no novel phenotypes. The era1–4 and plp-1 plants were allowed to self. The progeny of two-thirds of the era1–4 F2 plants segregated the plp-1 phenotype in a 3:1 ratio, whereas the progeny of all of the plp-1 plants showed the plp-1 phenotype, suggesting that the mutants are unlinked and that plp-1 is epistatic to era1–4. The identity of the plp-1 era1–4 double was confirmed by test crossing: F3 plants that were derived from era1–4 F2 plants and showed the plp-1 phenotype were crossed to both era1–4 and plp-1 single mutants. This resulted in F1 plants that all showed the respective mutant phenotype. F2 progeny of a cross between plp-1 and clv3–2 segregated wild type, plp-1, and clv3–2 phenotypes and a previously undescribed phenotype, the putative double mutant phenotype. This phenotype reappeared in selfed progeny of F2 plants with single mutant phenotypes, confirming the identity of the double mutant.
Additional Methods. The following methods were carried our essentially as described: phenotypic analysis and microscopy (8, 27), in vitro and in vivo prenylation assays (34–37), immunoblots (27, 34), and abscisic acid (ABA) germination assays (38). Detailed descriptions can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
The Role of PLP in Meristem Homeostasis. The plp-1 mutant was isolated in a T-DNA insertion mutagenesis in a screen for modifiers of the of 35S::BP phenotype (29) and was named for its flower phenotype of many extra petals. Crosses of plp-1 to wild-type strains demonstrated that plp mutants are recessive and are inherited in simple Mendelian fashion. The plp mutants display defective shoot and floral meristem functions, with the most striking phenotypes in the flower. The floral organ number in plp-1 is variable and significantly increased, with flowers developing 4–7 (average 5.1) sepals, 5–12 (8.7) petals, 5–8 (6.8) stamens, and 2–4 (3.1) carpels instead of the 4 sepals, 4 petals, 6 stamens, and 2 carpels typically found in wild-type flowers (Fig. 1 A and B).
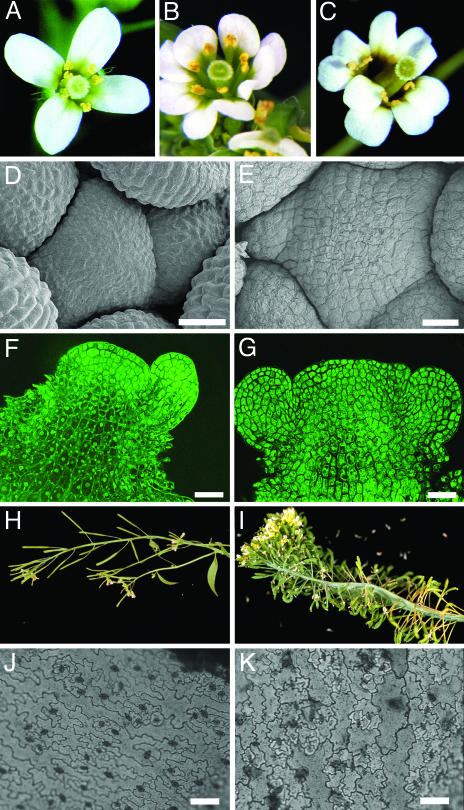
Fig. 1.
Developmental phenotypes of plp mutants. (A) Wild-type Arabidopsis flower. (B) plp-1 flowers have extra organs. (C) plp-2 flowers show the same phenotype as plp-1. (D–G) Inflorescence meristems of wild-type and plp-1 plants. (D) Scanning electron micrograph of a wild-type shoot apex. (E) The plp-1 shoot meristem is enlarged. (F) Confocal optical section of a wild-type shoot apex. (G) The plp-1 shoot apex is wider and flatter and shows some disruption of cell layering. [Bars = 25 μm (D and G).] (H) Wild-type stem. (I) plp-1 plants can show extreme fasciation and severe phyllotaxy defects. (J) Light micrograph of a wild-type leaf epidermis. (K) plp-1 leaf epidermal cells have reduced lobing and variable sizes. [Bars = 100 μm(J and K).]
The structural changes in plp flowers suggested that the shoot apical meristem of the mutant plant may be enlarged. Indeed, plp-1 plants have significantly enlarged shoot apical meristems compared to wild-type plants (92 ± 10 vs. 65 ± 8 μm in diameter), and 22% of the plants (26/120) develop a fasciated phenotype (Fig. 1 D–I). Confocal images indicate that plp-1 shoot meristems are wider and flatter than wild type and show some disruption of cell pattern in the subepidermal layer (Fig. 1 F and G). Interestingly, epidermal leaf cells of plp plants have reduced lobing typical of wild-type epidermal leaf cells (Fig. 1 J and K), indicating that PLP functions in regulation of cell shape.
plp plants show a number of pleiotropic phenotypes in addition to shoot meristem and flower organ number defects. plp plants show severely retarded growth (Fig. 2), with most plants remaining extremely small throughout their life. plp-1 plants have shorter stems with an internode length of 4.3 ± 2.4 mm for the first 10 internodes compared to 10.4 ± 1.6 mm in wild type. plp-1 plants flower late in long days, at 31 ± 5 days after stratification vs. 19 ± 1 days for wild type (Fig. 2). This delay in flowering is mostly due to a slower overall growth rate, because leaf initiation rate is slower in plp. plp mutants produce 11 ± 2 rosette leaves before flowering compared to 8 ± 1 in wild type. plp-1 mutants are self-fertile but produce markedly fewer seeds than wild type.
Fig. 2.
plp whole plant phenotype. Wild-type Arabidopsis plants (Left) and plp-1 plants (Right) 38 days after stratification.
The enlarged shoot apical meristems (SAMs) and fasciation of plp-1 are reminiscent of clavata1 (clv1), clv3, and enhanced response to aba-1 (era1) mutants. Mutations in the CLAVATA (CLV) genes also cause larger shoot and floral meristems and extra organs in the flower, particularly stamens and carpels (5–7). era1 plants have an enlarged SAM, resulting in increased numbers of flowers on the inflorescence stem. Flowers, in turn, have an increased organ number in three of the four whorls with more sepals, petals and sometime carpels, but not stamens (25–28). ERA1 encodes the β-subunit of protein farnesyltransferase (38).
Double mutants of plp-1 with both clv3–2 and era1–4 were created to determine whether they act in a common pathway with plp-1 to regulate meristem function (Fig. 3). clv3–2 single mutants have larger shoot and floral meristems, but, like in plp-1, clv3–2 shoot meristems maintain overall integrity and initiate organs throughout the life of the plant. The plp-1 clv3–2 double mutant shows a synergistic interaction, with massive overproliferation of the shoot meristem and early cessation of floral meristem initiation (Fig. 3 C and D). This finding indicates that the role of PLP in shoot apical meristem function is at least partly independent of the CLV pathway. era1–4 mutants are much less severe than plp-1, showing only slightly delayed flowering, slightly reduced fertility, and a small increase in floral organ number. plp-1 era1–4 double mutant plants are indistinguishable from plp (Fig. 3 A and B), showing markedly delayed growth, reduced fertility, and higher petal number, demonstrating that plp-1 is epistatic to era1–4 and likely acts in the prenylation pathway.
Fig. 3.
Interactions of plp with other mutants affecting meristem function. (A) era1–4 mutants have a phenotype similar to but less severe than plp-1.(B) plp-1 era1–4 double mutants are indistinguishable from plp-1 single mutants. (C) clv3–2 mutants have larger shoot meristems and extra stamens and carpels. (D) clv3–2 plp double mutants show massive shoot meristem overproliferation and cessation of flower meristem initiation.
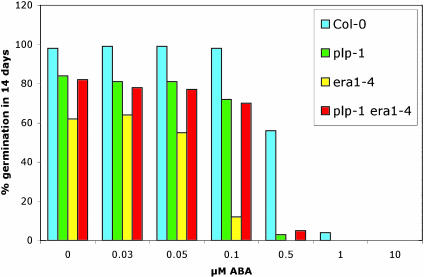
Regulation of ABA Response by PLP. era1 mutant plants are hyper-sensitive to ABA, resulting in highly reduced seed germination at 0.03 μM ABA (38) and enhanced stomatal closure (39). Because plp is epistatic to era1, we decided to examine the response of plp-1 and plp-1 era1 double mutants to ABA during seed germination. Most of the plp-1 seeds (72%) germinated on filter paper saturated with 0.1 μM ABA 14 days after stratification, whereas nearly 100% of all wild-type seeds but only 12% of era1–4 seeds germinated at this ABA concentration (Fig. 4). At 0.5 μM ABA, 56% of wild-type seeds germinated, whereas no era1–4 seeds but still 3% of plp-1 seeds germinated. The germination rate of plp-1 era1–4 plants was similar to plp-1 (Fig. 4). The reduced sensitivity of plp and plp era1 mutants to ABA compared to era1 suggests that PLP may act antagonistically to ERA1 in the maintenance of seed dormancy. The reduced sensitivity of plp to ABA compared to era1 presented an enigma, because during development, plp plants were much more affected compared to era1. To solve the enigma and elucidate the function of plp, we mapped plp-1.
Fig. 4.
Sensitivity of plp mutants to ABA during germination. Germination assays. Wild-type, plp-1, era1–4, and plp-1 era1–4 seeds were germinated on increasing concentrations of ABA. plp-1 seeds show a stronger response to exogenous ABA than wild type but weaker than era1–4. Germination was scored as positive when a radicle tip had fully penetrated the seed coat. Assays were repeated several times, and similar results were obtained in all cases. Percent germination was determined by dividing the number of seeds that germinated by the total number of seed on the plate.
PLP Encodes the α-Subunit Shared by PFT and PGGT-I. The plp-1 mutant was unlinked to the T-DNA selection marker. We mapped the plp-1 mutation to chromosome 3 between simple sequence length polymorphism markers nga707 and AthFus6. Because plp-1 is epistatic to era1, in which the PFT β-subunit (PFTβ) gene is mutated, we investigated the possibility that plp-1 is mutated in the α-subunit shared between PFT and the PGGT (PFT/PGGT-Iα) gene (At3g59380.1), located in the same region as plp-1. Sequence analysis revealed a deletion of 11 nucleotides in the third exon of the PFT/PGGT-Iα gene in plp-1. The deletion removes a conserved four amino acid motif (DAKH), which includes a putative substrate-binding site (40) and creates a stop codon predicted to result in a truncated protein of 146 amino acids (17.4 kDa). Sequencing of a PFT/PGGT-Iα-specific RT-PCR product from plp-1 confirmed this deletion. PLP encodes a protein of 326 amino acids (38 kDa) and is a single copy gene in Arabidopsis. To confirm that the plp phenotype results from the disruption of the PFT/PGGT-Iα gene, we identified an additional allele in line 519 D08 from the Syngenta Arabidopsis insertion library collection (32). This allele, plp-2, has a T-DNA insertion in the third intron of PLP (see Fig. 6) and shows the same phenotype as plp-1 during development (Fig. 1C) and ABA response (data not shown).
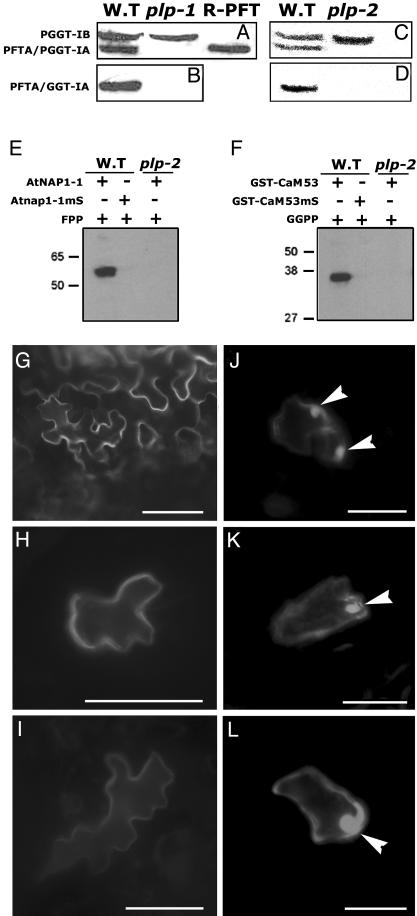
plp Mutants Lack Prenylation Activity. Immunoblots with PFT/PGGT-Iα and PGGT-Iβ polyclonal antibodies confirmed the absence of the α-subunit in plp-1 and plp-2 protein extracts (Fig. 5 A–D). Interestingly, the levels of PGGT-Iβ are similar in wild-type, plp-1, and plp-2 protein extracts, suggesting that PGGT-Iβ is not destabilized by the absence of the α-subunit. To test whether plp mutants lack prenyltransferase activity, we carried out in vitro prenylation assays (Fig. 5 E and F). Soluble protein extracts prepared from young flower tissue of either wild-type Col-0 or plp plants were used as a source for PFT and PGGT-I activities. The Arabidopsis PFT substrate AtNAP1–1 was farnesylated by a protein extract prepared from wild-type Col-0 but not by a protein extract prepared from plp plants (Fig. 5E). Similarly, the petunia calmodulin (CaM)53, which is primarily prenylated by PGGT-I but can also be prenylated by PFT (34, 41), was prenylated by a protein extract prepared from Col-0 but not by a plp protein extract (Fig. 5F). Prenylation deficient mutants of both AtNAP1–1 and CaM53, Atnap1–1mS and CaM53mS, in which the prenyl acceptor cysteines were mutated to serines, were used as additional negative controls for both reactions (Fig. 5 E and F). To further establish the lack of prenyltransferase activity in plp, we examined the subcellular location of the CaM53. Prenylated GFP-CaM53 localizes to the plasma membrane (Fig. 5 G–I) but a mutation of the CaaX-box or inhibition of prenylation results in nuclear accumulation of the protein (34, 41). In epidermal cells of plp leaves, GFP-CaM53 accumulates in the nuclei, indicating that the protein was not prenylated (Fig. 5 J–L). In contrast, fluorescing nuclei were not observed in epidermal cells of wild-type Col-0 plants (Fig. 5 G–I). The results shown in Fig. 5 G–L are of representative cells. The same results were repeated in injection experiments with different plants and leaves as described in Materials and Methods. These results confirm that both PFT and PGGT-I activities are absent in plp plants, and that neither Rab-GGT nor other unidentified prenyltransferases can prenylate CaaX box proteins in plp.
Fig. 5.
Loss of PFT/PGGT-Iα and protein prenylation activity in plp. (A–D) Immunoblots of crude protein extracts from wild-type, plp-1, and plp-2 plants and purified recombinant PFT (R-PFT) decorated with either polyclonal antibodies against PFT/PGGT-Iα and PGGT-Iβ (A and C) or affinity-purified anti-PFT/PGGT-Iα antibodies (B and D). (E and F) In vitro prenylation assays with protein extracts prepared from either Col-0 (WT) or mutant (plp-2) plants. (E) Farnesylation assay with AtNAP1–1 and prenyl acceptor mutant Atnap1–1mS as protein substrates and farnesyl pyrophosphate as prenyl group donor. (F) Gernaylgeranylation assays using GST-CaM53 and prenyl acceptor mutant GST-CaM53mS as protein substrates and geranylgeranylpyrophosphate as prenyl group donor. (G–I) GFP-CaM53 fusion protein localizes to the plasma membrane and absent from nuclei in epidermal cells of wild-type Arabidopsis leaves. (J–L) In plp-1 leaf epidermal cells, GFP-CaM53 localizes to nuclei (arrowheads), signifying the lack of prenylation (41). [Bars = 20 μm(G–L).]
Discussion
The Role of Prenylation in Plant Development. We have isolated Arabidopsis plp mutants that contain lesions in the α-subunit of both PFT and PGGT. Protein prenylation is conserved in fungi, plants, and animals and is mediated by PFT, PGGT-I, and Rab-GGT (42, 43). Protein prenylation is required for the function of >100 proteins important for cell division, growth, differentiation, morphogenesis, and environmental signaling. Protein prenyltransferases catalyze the covalent attachment of a 15-carbon farnesyl or 20-carbon geranylgeranyl isoprenoid to C-terminal cysteines of selected proteins (44). Addition of prenyl groups facilitates membrane association and protein–protein interactions of the prenylated proteins (42, 45, 46).
Substrate specificity of PFT and PGGT-I is determined by the β-subunits of each enzyme through sequence-specific recognition of a C-terminal CaaX box motif on target proteins, where C is cysteine, the a residues are often aliphatic, and X is usually alanine, cysteine, glutamine, methionine, or serine in the case of PFT but is almost exclusively a leucine when prenylated by PGGT-I (44, 47–51). PFT and PGGT-I are somewhat promiscuous for their protein substrates (47, 49, 51). PGGT-I can use farnesyl pyrophosphate (FPP) as well as geranylgeranylpyrophosphate (GGPP) as a prenyl group donor at low efficiency (44, 51–53). However, PFT cannot prenylate CaaL box proteins with GGPP, and PGGT-I cannot prenylate CaaX box proteins with FPP (47, 49, 51).
Mutations in the PGGT-Iβ gene are lethal in Saccharomyces cerevisiae (47, 49) and Drosophila (54), as well as in Schizosac-charomyces pombe when grown at a restrictive temperature of 37°C (55). S. cerevisiae cdc4/cal1 (PGGT-Iβ mutants) can be rescued by Ca2+ (47) or by overexpression of the PGGT-I substrates Rho1 and CDC42, suggesting a role of prenylated proteins in regulating cell division (49). S. pombe cwg2– (PGGT-Iβ mutant) cells are defective in β-d-glucan synthesis and can be rescued when grown at 37°C in high osmotic pressure media (55). In contrast, PFTβ loss-of-function mutants are viable in yeast and Arabidopsis, possibly due to partial compensation by PGGT-I. However, these mutants show defects in growth, mating, and development (25–28, 38, 56).
Arabidopsis enhanced response to abscisic acid1 (era1) mutants lack PFTβ-subunits. era1 plants have an enlarged shoot apical meristem, resulting in increased numbers of flowers on the inflorescence stem. Flowers, in turn, have an increased organ number in three of the four whorls with more sepals, petals, and sometime carpels, but not stamens (25–28). era1 mutants are also hypersensitive to ABA during germination (38), show enhanced stomatal closure under noninductive concentrations of ABA, and display concomitant drought resistance (39). Mutants have increased cytoplasmic CA2+ concentrations in response to ABA treatments at concentrations that are noninductive to wild-type stomata, suggesting that protein farnesylation negatively regulates ABA-induced Ca2+ release (57).
era1 mutants are not likely to reveal all of the roles for prenylation in plant biology, due to possible compensation of PFT by PGGT-I. Loss of PFT/PGGT-Iα function, which eliminates both PFT and PGGT-I activity, is lethal in yeast (58), and no mutations in PFT/PGGT-Iα have been reported from other organisms. plp mutants thus represent the first viable prenylation deficient mutants, although they are viable only under ideal growth conditions.
The Relationship of PLP to ERA1 and CLV3. PLP, ERA1, and PGGT-Iβ exist as single copy genes in Arabidopsis. That plp is epistatic to era1 lends further support to the idea that the general mechanism of protein prenylation is conserved in plants and suggests that no other prenylation mechanism compensates for the lack of plp or era1 function. That plp shows some phenotypes similar to but more severe than era1, as well as additional phenotypes, is consistent with its role in both farnesyltransferase and geranylgeranyltransferase-I. plp mutant flowers display an increase in organ number in all four whorls, including stamens, in contrast to era1 flowers in which stamen number remains six and carpel numbers increase to three in only 50% of the flowers (27). These results suggest that PGGT-I may assume PFT function in the center of the floral meristem. Another marked difference between era1 and plp meristems is in cell division patterns, which remain normal in era1 mutants. In plp mutants, cell division planes are altered with periclinal divisions occurring in L2 and some divisions in L1 that are not at right angles to the surface (Fig. 1). Thus, protein prenylation affects both the patterns of cell divisions and cellular differentiation in meristems.
Further insight into the function of protein prenylation in cellular differentiation was obtained by analysis of the plp1 clv3 double mutants (Fig. 3). These double mutants show a synergistic interaction, indicating that in addition to any potential loss of CLV function, other pathways affecting meristem function are disrupted in plp mutants. The CLV pathway acts by restricting the expression of the WUSCHEL (WUS) gene in the meristem (18). It would now be interesting to examine whether protein prenylation functions by activating an additional pathway, which regulates WUS, or through another mechanism.
The complete loss of protein prenylation in plp strongly decreases growth rates, as judged by the extremely small plant size (Fig. 2). In contrast, era1 mutant alleles are typically larger than wild-type plants having bigger rosette and cauline leaves and floral organs, because of their larger meristems (27) and additional cell division cycles (A.G., G. Beemster, and W.G., unpublished data). It is likely, therefore, that the size differences between plp and era1 result from compromised activity of PGGT-I in plp. Future analysis of PGGT-Iβ-subunit mutant will be required for assessing how PGGT-I activity affects growth.
The partial loss of ABA hypersensitivity of plp compared to era1 suggests that PGGT-I may have an opposite function to PFT in ABA signaling. An alternative explanation could be that certain farnesylated proteins are geranylgeranylated in era1, thereby inhibiting their activity, as have been shown for mammalian RhoB (59–61). In this case, ERA1 function (protein fanesylation) might not be suppression of the ABA response, as has been deduced from the phenotype of era1 mutant plants (38, 39, 57). Future analysis will be required to assess protein prenylation patterns in era1, plp, and pggt-Ib mutant backgrounds.
PLP Target Proteins and Their Potential Role in the plp Phenotype. Prenylation by PFT and PGGT-I has been confirmed in vivo for only a few proteins in plants, including APETALA1 and CaM53 (41, 62), although several more have been identified in vitro. Prenylation is required for the function of all members of the eukaryotic Ras superfamily of small GTP-binding proteins (Ras1p, Ras2p, Rho1, and CDC42 in yeast; and K-Ras, H-Ras N-Ras, Rhos, CDC42, and Racs in animals), which are essential for cell viability. Organization of the animal nuclear lamina requires prenylation of lamin A and lamin B3 (45). Phosphoinositide signaling depends on prenylation of type I inositol triphosphate 5′-phosphatase (63) and G2/M cell cycle transition requires prenylation of two kinetochore-binding proteins (64). Neither Ras proteins, Rhos, CDC42, nor prenylated isoforms of IP3 5′-phosphatase and nuclear lamins have been identified in plants. Thus, the viability of plp mutants may be attributed to the lack of these proteins in plants. In contrast, plants have a large family of Rac-like GTPases (ROPs or RACs), which have been implicated in different signaling cascades, including induction of cell polarity (65). The decreased lobing of leaf epidermal cells may be due to loss of RAC function. An as-yet-unidentified Rop/RAC is found in association with the CLV complex (15). Most type I RACs are potential substrates of PGGT-I, but membrane attachment of type II RACs is facilitated by palmitoylation (37). Thus, partial or complete loss of type I RAC function due to lack of prenylation may not entirely compromise shoot and root apical meristem function during development.
Relevance to Other Systems and Future Prospects. PFT and PGGT-I inhibitors are becoming increasingly important in medicine for treating cancer and other human diseases (66–68). The biology of protein prenylation is far less well understood. The degree of promiscuity between PFT and PGGT-I is limited in vitro (51), but until now could not be fully evaluated in vivo. Due to the lethality of PGGT-I mutants in other organisms, plp and other plant prenylation-deficient mutants now provide an unprecedented opportunity to study the involvement of prenylation in developmental processes and the relations between the prenylation enzymes in vivo. In addition, the plp mutants provide a system to study the relation between prenylation and other protein lipid modifications such as palmitoylation.
Supplementary Material
Acknowledgments
We dedicate this paper to the memory of our colleague, Dr. Gethyn Allen, in recognition of his contributions to the study of protein prenylation. We thank the Syngenta Torrey Mesa Research Institute for the plp-2 T-DNA mutant. This research was supported by grants from the U.S.–Israel Binational Science Foundation (no. 1999423) and the U.S.–Israel Binational Agricultural Research and Development Fund (no. IS-3215-01, to S.Y.), the Israel Science Foundation (no. 495/01, to N.O.), the National Science Foundation (MCB-9727611, to S.H.), and the Swiss Federal Institute of Technology (to W.G.). M.P.R was a fellow of the Damon Runyon Cancer Research fund. M.L. is a recipient of the Israeli Ministry of Science Eshkol Fellowship for Ph.D. students.
Abbreviations: ABA, abscisic acid; PFT, protein farnesyltransferase; PGGT-I, protein geranylgeranyltransferase-I; CaM, calmodulin; Ln, layer n.
References
- 1.Meyerowitz, E. M. (1997) Cell 88, 299-308. [DOI] [PubMed] [Google Scholar]
- 2.Steeves, T. A. & Sussex, I. M. (1989) Patterns in Plant Development (Cambridge Univ. Press, New York).
- 3.Tilney-Bassett, R. A. E. (1986) Plant Chimeras (E. Arnold, London).
- 4.Clark, S. E. (2001) Curr. Opin. Plant Biol. 4, 28-32. [DOI] [PubMed] [Google Scholar]
- 5.Clark, S. E., Running, M. P. & Meyerowitz, E. M. (1993) Development (Cambridge, U.K.) 119, 397-418. [DOI] [PubMed] [Google Scholar]
- 6.Clark, S. E., Running, M. P. & Meyerowitz, E. M. (1995) Development (Cambridge, U.K.) 121, 2057-2067. [Google Scholar]
- 7.Kayes, J. M. & Clark, S. E. (1998) Development (Cambridge, U.K.) 125, 3843-3851. [DOI] [PubMed] [Google Scholar]
- 8.Running, M. P., Clark, S. E. & Meyerowitz, E. M. (1995) Methods Cell Biol. 49, 217-229. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, J. C., Brand, U., Running, M. P., Simon, R. & Meyerowitz, E. M. (1999) Science 283, 1911-1914. [DOI] [PubMed] [Google Scholar]
- 10.Rojo, E., Sharma, V. K., Kovaleva, V., Raikhel, N. V. & Fletcher, J. C. (2002) Plant Cell 14, 969-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, S. E., Williams, R. W. & Meyerowitz, E. M. (1997) Cell 89, 575-585. [DOI] [PubMed] [Google Scholar]
- 12.Jeong, S., Trotochaud, A. E. & Clark, S. E. (1999) Plant Cell 11, 1925-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenhard, M. & Laux, T. (2003) Development (Cambridge, U.K.) 130, 3163-3173. [DOI] [PubMed] [Google Scholar]
- 14.Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. (2000) Science 289, 617-619. [DOI] [PubMed] [Google Scholar]
- 15.Trotochaud, A. E., Hao, T., Wu, G., Yang, Z. & Clark, S. E. (1999) Plant Cell 11, 393-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laux, T., Mayer, K. F., Berger, J. & Jurgens, G. (1996) Development (Cambridge, U.K.) 122, 87-96. [DOI] [PubMed] [Google Scholar]
- 17.Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G. & Laux, T. (1998) Cell 95, 805-815. [DOI] [PubMed] [Google Scholar]
- 18.Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jurgens, G. & Laux, T. (2000) Cell 100, 635-644. [DOI] [PubMed] [Google Scholar]
- 19.Barton, M. K. & Poethig, R. S. (1993) Development (Cambridge, U.K.) 119, 823-831. [Google Scholar]
- 20.Long, J. A., Moan, E. I., Medford, J. I. & Barton, M. K. (1996) Nature 379, 66-69. [DOI] [PubMed] [Google Scholar]
- 21.Clark, S. E., Jacobsen, S. E., Levin, J. Z. & Meyerowitz, E. M. (1996) Development (Cambridge, U.K.) 122, 1567-1575. [DOI] [PubMed] [Google Scholar]
- 22.Leyser, H. M. O. & Furner, I. J. (1992) Development (Cambridge, U.K.) 116, 397-&. [Google Scholar]
- 23.Kaya, H., Shibahara, K. I., Taoka, K. I., Iwabuchi, M., Stillman, B. & Araki, T. (2001) Cell 104, 131-142. [DOI] [PubMed] [Google Scholar]
- 24.Laufs, P., Dockx, J., Kronenberger, J. & Traas, J. (1998) Development (Cambridge, U.K.) 125, 1253-1260. [DOI] [PubMed] [Google Scholar]
- 25.Running, M. P., Fletcher, J. C. & Meyerowitz, E. M. (1998) Development (Cambridge, U.K.) 125, 2545-2553. [DOI] [PubMed] [Google Scholar]
- 26.Bonetta, D., Bayliss, P., Sun, S., Sage, T. & McCourt, P. (2000) Planta 211, 182-190. [DOI] [PubMed] [Google Scholar]
- 27.Yalovsky, S., Kulukian, A., Rodriguez-Concepcion, M., Young, C. A. & Gruissem, W. (2000b) Plant Cell 12, 1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziegelhoffer, E. C., Medrano, L. J. & Meyerowitz, E. M. (2000) Proc. Natl. Acad. Sci. USA 97, 7633-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuck, G., Lincoln, C. & Hake, S. (1996) Plant Cell 8, 1277-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 31.Bouchez, D., Camilleri, C. & Caboche, M. (1993) C. R. Acad. Sci. 316, 1188-1193. [Google Scholar]
- 32.Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., et al. (2002) Plant Cell 14, 2985-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell, C. J. & Ecker, J. R. (1994) Genomics 19, 137-144. [DOI] [PubMed] [Google Scholar]
- 34.Caldelari, D., Sternberg, H., Rodriguez-Concepcion, M., Gruissem, W. & Yalovsky, S. (2001) Plant Physiol. 126, 1416-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yalovsky, S., Loraine, A. E. & Gruissem, W. (1996) Plant Physiol. 110, 1349-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracha, K., Lavy, M. & Yalovsky, S. (2002) J. Biol. Chem. 277, 29856-29864. [DOI] [PubMed] [Google Scholar]
- 37.Lavy, M., Bracha-Drori, K., Sternberg, H. & Yalovsky, S. (2002) Plant Cell 14, 2431-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S. & McCourt, P. (1996) Science 273, 1239-1241. [DOI] [PubMed] [Google Scholar]
- 39.Pei, Z. M., Ghassemian, M., Kwak, C. M., McCourt, P. & Schroeder, J. I. (1998) Science 282, 287-290. [DOI] [PubMed] [Google Scholar]
- 40.Strickland, C. L., Windsor, W. T., Syto, R., Wang, L., Bond, R., Wu, Z., Schwartz, J., Le Hung, V., Beese, L. S. & Weber, P. C. (1998) Biochemistry 37, 16601-16611. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Concepcion, M., Yalovsky, S., Zik, M., Fromm, H. & Gruissem, W. (1999) EMBO J. 18, 1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yalovsky, S., Rodriguez-Concepcion, M. & Gruissem, W. (1999) Trends Plant Sci. 4, 429-438. [DOI] [PubMed] [Google Scholar]
- 43.Maurer-Stroh, S., Washietl, S. & Eisenhaber, F. (2003) Genome Biol. 4, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, F. L. & Casey, P. J. (1996) Annu. Rev. Biochem. 65, 241-269. [DOI] [PubMed] [Google Scholar]
- 45.Sinensky, M. (2000) Biochim. Biophys. Acta 1529, 203-209. [DOI] [PubMed] [Google Scholar]
- 46.Ramamurthy, V., Roberts, M., van den Akker, F., Niemi, G., Reh, T. A. & Hurley, J. B. (2003) Proc. Natl. Acad. Sci. USA 100, 12630-12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohya, Y., Goebl, M., Goodman, L. E., Peterson-Bjorn, S., Friesen, J. D., Tamanoi, F. & Anraku, Y. (1991) J. Biol. Chem. 266, 12356-12360. [PubMed] [Google Scholar]
- 48.Seabra, M. C., Reiss, Y., Casey, P., Brown, M. S. & Goldstein, J. L. (1991) Cell 65, 429-434. [DOI] [PubMed] [Google Scholar]
- 49.Trueblood, C. E., Ohya, Y. & Rine, J. (1993) Mol. Cell. Biol. 13, 4260-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spence, A. & Casey, P. J. (2001) in The Enzymes, eds. Tamanoi, F. & Sigman, D. S. (Academic, San Diego), pp. 1-18.
- 51.Taylor, J. S., Reid, T. S., Terry, K. L., Casey, P. J. & Beese, L. S. (2003) EMBO J. 22, 5963-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokoyama, K., Zimmerman, K., Scholten, J. & Gelb, M. H. (1997) J. Biol. Chem. 272, 3944-3952. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama, K. & Gelb, M. H. (2001) in The Enzymes, eds. Tamanoi, F. & Sigman, D. S. (Academic, San Diego), pp. 105-130.
- 54.Therrien, M., Chang, H. C., Solomon, N. M., Karim, F. D., Wassarman, D. A. & Rubin, G. M. (1995) Cell 83, 879-888. [DOI] [PubMed] [Google Scholar]
- 55.Diaz, M., Sanchez, Y., Bennet, T., Sun, C. R., Godoy, C., Tamanoi, F., Duran, A. & Perez, P. (1993) EMBO J. 12, 5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schafer, W. R., Trueblood, C. E., Yang, C.-C., Mayer, M. P., Rosenberg, S., Poulter, C. D., Kim, S.-H. & Rine, J. (1990) Science 249, 1133-1139. [DOI] [PubMed] [Google Scholar]
- 57.Allen, G. J., Murata, Y., Chu, S. P., Nafisi, M. & Schroeder, J. I. (2002) Plant Cell 14, 1649-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He, B., Chen, P., Chen, S.-Y., Vancura, K. L., Michaelis, S. & Powers, S. (1991) Proc. Natl. Acad. Sci. USA 88, 11373-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lebowitz, P. F., Casey, P. J., Prendergast, G. C. & Thissen, J. A. (1997) J. Biol. Chem. 272, 15591-15594. [DOI] [PubMed] [Google Scholar]
- 60.Du, W., Lebowitz, P. F. & Prendergast, G. C. (1999) Mol. Cell. Biol. 19, 1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu, A. & Prendergast, G. C. (2000) FEBS Lett. 481, 205-208. [DOI] [PubMed] [Google Scholar]
- 62.Yalovsky, S., Rodriguez-Concepcion, M., Bracha, K., Toledo-Ortiz, G. & Gruissem, W. (2000a) Plant Cell 12, 1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Smedt, F., Boom, A., Pesesse, X., Schiffmann, S. N. & Erneux, C. (1996) J. Biol. Chem. 271, 10419-10424. [DOI] [PubMed] [Google Scholar]
- 64.Ashar, H. R., James, L., Gray, K., Carr, D., Black, S., Armstrong, L., Bishop, W. R. & Kirschmeier, P. (2000) J. Biol. Chem. 275, 30451-30457. [DOI] [PubMed] [Google Scholar]
- 65.Fu, Y., Wu, G. & Yang, Z. (2001) J. Cell Biol. 152, 1019-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stark, W. W., Jr., Blaskovich, M. A., Johnson, B. A., Qian, Y., Vasudevan, A., Pitt, B., Hamilton, A. D., Sebti, S. M. & Davies, P. (1998) Am. J. Physiol. 275, L55-63. [DOI] [PubMed] [Google Scholar]
- 67.Chakrabarti, D., Da Silva, T., Barger, J., Paquette, S., Patel, H., Patterson, S. & Allen, C. M. (2002) J. Biol. Chem. 277, 42066-42073. [DOI] [PubMed] [Google Scholar]
- 68.Walters, C. E., Pryce, G., Hankey, D. J., Sebti, S. M., Hamilton, A. D., Baker, D., Greenwood, J. & Adamson, P. (2002) J. Immunol. 168, 4087-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.