Abstract
DNA gyrase is the bacterial DNA topoisomerase (topo) that supercoils DNA by using the free energy of ATP hydrolysis. The enzyme, an A2B2 tetramer encoded by the gyrA and gyrB genes, catalyses topological changes in DNA during replication and transcription, and is the only topo that is able to introduce negative supercoils. Gyrase is essential in bacteria and apparently absent from eukaryotes and is, consequently, an important target for antibacterial agents (e.g., quinolones and coumarins). We have identified four putative gyrase genes in the model plant Arabidopsis thaliana; one gyrA and three gyrB homologues. DNA gyrase protein A (GyrA) has a dual translational initiation site targeting the mature protein to both chloroplasts and mitochondria, and there are individual targeting sequences for two of the DNA gyrase protein B (GyrB) homologues. N-terminal fusions of the organellar targeting sequences to GFPs support the hypothesis that one enzyme is targeted to the chloroplast and another to the mitochondrion, which correlates with supercoiling activity in isolated organelles. Treatment of seedlings and cultured cells with gyrase-specific drugs leads to growth inhibition. Knockout of A. thaliana gyrA is embryo-lethal whereas knockouts in the gyrB genes lead to seedling-lethal phenotypes or severely stunted growth and development. The A. thaliana genes have been cloned in Escherichia coli and found to complement gyrase temperature-sensitive strains. This report confirms the existence of DNA gyrase in eukaryotes and has important implications for drug targeting, organelle replication, and the evolution of topos in plants.
DNA topoisomerases (topos) are key enzymes present in all cells that control the topological state of DNA (1). There are two types, topos I and II, distinguished by whether they transiently break one or both strands of the DNA. DNA gyrase is a type II enzyme that is essential for the processes of replication and transcription in prokaryotes. It is the only enzyme of this type that is able to catalyze ATP-dependent DNA supercoiling (2). The best-studied gyrase is that from Escherichia coli, which consists of two subunits, DNA gyrase protein A (GyrA; 97 kDa) and DNA gyrase protein B (GyrB; 90 kDa), which form an A2B2 complex. Due to its essential role in prokaryotes and its apparent absence from eukaryotes, gyrase is the target of a number of antibacterial agents, including quinolones, e.g., nalidixic acid (NAL) and ciprofloxacin (CFX), and coumarins, e.g., novobiocin (NOV) and coumermycin A1 (3).
Although thought to be a uniquely bacterial enzyme, there have been previous indications that there may be a gyrase in plants. Thompson and Mosig (4) found that Chlamydomonas reinhardtii contains an ATP-dependent topo activity that can supercoil DNA in vitro. The supercoiling activity was weak and, although unable to purify the enzyme, they found that gyrase-specific drugs inhibited chloroplast transcription. Further work showed that NOV could inhibit chloroplast DNA replication in vivo (5). In Nicotiana tabacum, NAL was shown to have a greater inhibitory effect on plastid than nuclear DNA synthesis at low drug concentrations (6); high concentrations affected both plastid and nuclear DNA synthesis. NAL was also found to inhibit DNA synthesis in both the chloroplast and mitochondrion of the unicellular alga Cyanidioschyzon merolae (7). Similarly, Mills et al. (8) found that NAL and NOV inhibited thymidine incorporation in pea chloroplasts; Lam and Chua (9) found that NOV affected transcription in pea chloroplasts. Ebringer et al. (10) reported that, in Euglena gracilis, the second-generation quinolone ofloxacin caused mass aberrations and subsequent loss of chloroplasts and ultrastructural changes in mitochondria. Pyke et al. (11) reported the presence of two proteins in wheat chloroplasts with molecular masses similar to gyrase, which crossreacted to yeast topo II antibodies.
These results suggest that there might be DNA gyrase activity in the chloroplasts and mitochondria of plant cells. In principle, eukaryotes do not require gyrase because negative supercoiling can be established by the wrapping of DNA around histones and the relaxation of the internucleosomal DNA by topos I and II; topo II is evolutionarily related to gyrase but lacks the ability to supercoil DNA (1). However, chloroplasts and mitochondria lack histones and their genomes resemble those of their bacterial ancestors in a number of respects, raising the possibility that these organelles may organize their DNA differently from nuclear DNA and could both require DNA gyrase activity.
With these considerations in mind, we have examined the genome sequence of Arabidopsis thaliana (12) and found four putative gyrase genes. By using a combination of in vivo and in vitro experiments we show that there is gyrase activity in A. thaliana cells that is targeted to chloroplasts and mitochondria. We find that the A. thaliana gyrase subunits are able to complement their E. coli counterparts and we speculate on the role of gyrase in organelle replication.
Materials and Methods
Sequence and Phylogenetic Analyses of A. thaliana Gyrase Genes. The putative gyrase genes were identified by a blast search of the Arabidopsis Gene Index in The Institute for Genomic Research (Rockville, MD) database. Sequence comparisons and phylogenetic tree analyses were performed by using the clustal w multiple alignment program (13). Potential organelle targeting sequences were identified by using the protein signal prediction programs targetp (14) and predotar version 0.5, which can be accessed at http://genoplante-info.infobiogen.fr/predotar/index.html.
Plant Material. A. thaliana ecotype Landsberg cell cultures were grown in darkness or under a 9-h short day photoperiod (120 μE m-2·s-1), with shaking at 120 rpm at 23°C in Murashige and Skoog ordinary media. Cultured cells were tested for drug sensitivity by the addition of appropriate concentrations of CFX (Sigma) to the liquid culture. MitoTracker Green FM (Molecular Probes) was added to the cell cultures in localization experiments to a final concentration of 200 nM. Cells were scanned with an Argon laser (488 nm) and the autofluorescence of chlorophyll (610 nm) was measured by using a Leica TCS SP confocal microscope.
Seeds of A. thaliana (L.) Heynh. ecotype Columbia were surface-sterilized and germinated on MS basal media (Sigma) containing 1% glucose or soil grown and maintained at 23°C under a 16-h long day photoperiod (120 μE m-2·s-1). CFX was added directly to solid germination media. Coumarin sensitivity assays used liquid based root suspension cultures with 20 sterile seeds per 50 ml of initiation media under a 9-h short day photoperiod with shaking at 70 rpm at 23°C.
Searches of Arabidopsis T-DNA insertion databases identified several lines with insertions in the putative gyrase gene regions. T-DNA insertion lines 077A10 and 290E11 (termed AtgyrA-1 and AtgyrA-2, respectively) were generated by using the gabikat program (B. Weisshaar, Max Planck Institute for Plant Breeding Research, Koln, Germany) (15). SAIL T-DNA mutant lines SAIL_390D05 (AtntgyrB-1), SAIL_559H08 (AtmtgyrB-1), SAIL_681G04 (AtcpgyrB-1) and SAIL_876B04 (AtmtgyrB-3) were provided by Syngenta (San Diego) (16). The Salk Institute Genomic Analysis Laboratory (La Jolla, CA) provided the sequence-indexed T-DNA insertion line SALK_002367 (Atmt-gyrB-2), which was obtained through the Nottingham Arabidopsis Stock Centre (Nottingham, UK) (17). PCR-based identification of T-DNA insertions was performed according to published protocols (18) by using primers listed in Supporting Text, which is published as supporting information on the PNAS web site.
Transgene Expression (GFP Targeting). In-frame fusions of the putative transit peptides to the GFP (GFP2.5) were generated by isolation of the transit peptides by PCR and ligation to the N terminus of GFP2.5 in p57TFIIDGFP. Each transit peptide fusion was subcloned into the binary vector pAOV downstream of the 35S CaMV promoter.
The constructs were transformed into Agrobacterium tumefaciens GV3101::pMP90 by using a freeze-thaw method (19). A. thaliana ecotype Landsberg cell cultures were transformed (20) and assayed for transient GFP expression within 72 h by light microscopy with an epifluorescent Nikon Eclipse 600 microscope.
Cloning and Expression of A. thaliana Gyrase Subunits. Total cellular RNA was prepared from 4-day-old A. thaliana ecotype Landsberg cell cultures (1 g of wet cells) by using TRIzol (Invitrogen). Complementary DNAs were synthesized by using Moloney murine leukemia virus reverse transcriptase (Promega). RT-PCR was performed by using primers corresponding to the 5′ and 3′ ends of the genes. The PCR products were cloned into pGEM-T Easy vector (Promega) except for AtcpgyrB, which was generated from two partial cDNAs. Each gene was subcloned into pET17b (Novagen) for expression. XL10-Gold (Stratagene) and DH10β strains of E. coli were used as hosts for routine manipulations and were grown at 37°C with antibiotic selection.
For in vivo complementation assays, individual constructs were transformed into E. coli ts strains N4177 (GyrBts) and KNK453 (GyrAts) (21, 22) by electroporation with a 1-h expression lag at 30°C. Complementation was performed by using single colonies streaked across replica isopropyl β-d-thiogalactoside gradient induction plates (0–50 μM) with growth at either 30°C or 42°C.
Enzyme Assays. All steps were performed at 0–4°C. A. thaliana ecotype Landsberg cell cultures (100 g of wet cells) were lysed in a total volume of 300 ml of 50 mM Mes (pH 5.8), 300 mM mannitol, 5 mM DTT, and the homogenate was filtered through four layers of Miracloth (Calbiochem). The eluate was centrifuged for 5 min at 200 × g to remove cell debris and nuclei. Chloroplasts were harvested by centrifugation at 2,500 × g for 10 min. Mitochondria were recovered by a further centrifugation of the cell free extracts at 10,000 × g for 15 min. The residual supernatant was retained and designated cytosolic cell-free extract. Organelles were further purified by Percoll gradients and assessed for purity by microscopy (18).
Purified organelles were resuspended and lysed to create chloroplast and mitochondrial extracts. All cell-free extracts were dialyzed against 4 liters of TGED buffer (50 mM Tris·HCl, pH 7.5/10% glycerol/1 mM EDTA/1 mM DTT), containing 20 mM NaCl and 0.3 mM PMSF. Extracts were applied to 5 ml NOV-Sepharose columns (23) and fractions were eluted with TGED buffer containing 0.5 M NaCl followed by 4 M urea, dialyzed overnight against 4 liters of TGED buffer containing 50 mM NaCl and were assayed for DNA supercoiling activity (24).
Results
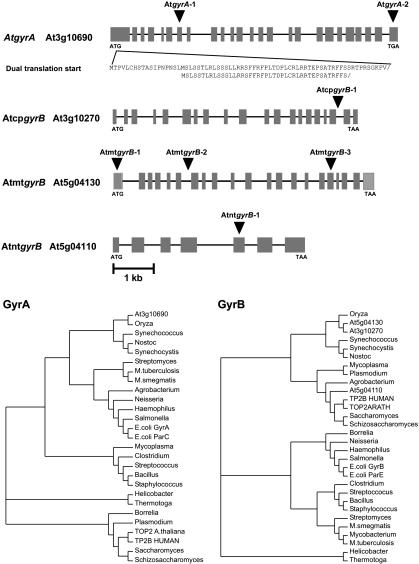
One gyrA and Three gyrB Genes in A. thaliana. The publication of the complete nuclear genome sequence of A. thaliana (12) revealed the presence of homologues of topos, including eukaryotic topo I (25) and II (26), archaeal topo VI (27), and, surprisingly, bacterial gyrA and gyrB. There is a putative gyrA gene on chromosome 3 (AtgyrA), with two translation initiation sites, and three putative gyrB genes on chromosome 3 (AtcpgyrB) and chromosome 5 (AtmtgyrB and AtntgyrB; Table 1). Originally in the A. thaliana databases, At5g04110 and the immediately adjacent sequence At5g04100, were annotated as separate genes. However, sequence alignments to other gyrase-like proteins as well as EST data (RIKEN R18897 and RT-PCR), strongly suggest that they are a single gene, referred to hereafter as At5g04110.
Table 1. DNA gyrase genes in A.thaliana.
| Gene | Locus | Chromosome | Gene product/molecular mass, Da | Transit peptide/molecular mass, Da | Mature protein/molecular mass, Da | Mature protein location |
|---|---|---|---|---|---|---|
| AtgyrA | At3g10690 | 3 | 101,413 | 8,017 | 93,395 | Chloroplast |
| 99,548 | 6,513 | 93,395 | Mitochondria | |||
| AtcpgyrB | At3g10270 | 3 | 81,192 | 8,700 | 72,492 | Chloroplast |
| AtmtgyrB | At5g04130 | 5 | 84,268 | 8,465 | 75,803 | Mitochondria |
| AtntgyrB | At5g04110 | 5 | 60,936 | None | 60,936 | Nuclear/cytosol? |
Key residues required for catalytic activity and drug interactions in both subunits of the prokaryotic enzyme are identical in the A. thaliana homologues. These residues include the activesite tyrosine for DNA breakage-reunion, residues associated with quinolone interaction, and the active-site residues for ATPase activity. AtgyrA, AtcpgyrB, and AtmtgyrB encode additional putative N-terminal transit peptides (Table 1) as identified by both targetp and predotar; the dual translation initiation sites of GyrA potentially allow targeting to plastids and mitochondria (Fig. 1), previously demonstrated for several proteins including RNA polymerase (28). Phylogenetic analyses clearly support the proposed cyanobacterial origins of AtGyrA and both the chloroplast and mitochondrial GyrB proteins with further evolution of the genes subsequent to the acquisition of the endosymbiotic organelles (Fig. 1). Surprisingly, the cladistic analysis of AtntgyrB aligned the sequence with the eukaryotic type II topos, even though the cloned enzyme is able to support DNA supercoiling. More investigation is required to resolve whether this gene encodes a functional gyrase subunit as some of the conserved topo regions are truncated in this gene.
Fig. 1.
(A) Organization of the A. thaliana DNA gyrase genes. Labels refer to our nomenclature followed by the AGI gene loci. The filled boxes represent exons, and the lines between boxes represent introns and noncoding flanking sequences, as identified by in silico analyses using defined A. thaliana consensus sequences. The amino acid sequences below AtgyrA represent the transit peptides encoded by the dual translation initiation sites, enabling targeting the gene products to both organelles. The positions of the T-DNA insertions are indicated above each gene (▾). (B) Phylogenetic relationship of the A. thaliana gyrase protein sequences to other gyrases and type II topos. The GenBank accession nos. for the protein sequences are given in Supporting Text.
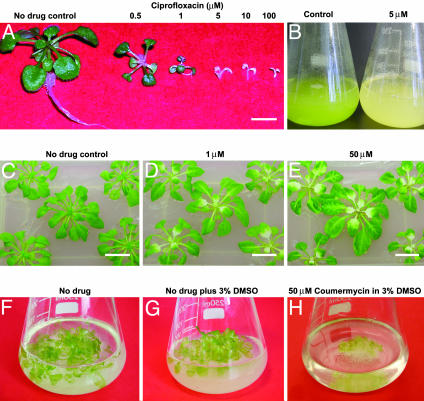
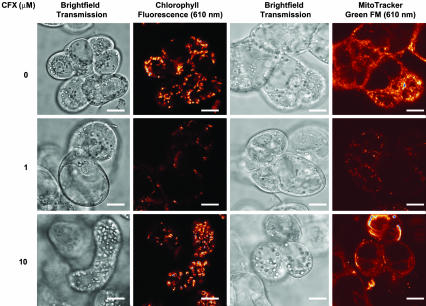
A. thaliana Seedlings and Cells in Culture Affected by Gyrase-Specific Drugs. To investigate gyrase activity in plants, we treated A. thaliana seedlings and cell cultures with gyrase-specific drugs (Figs. 2 and 3). A. thaliana ecotype Columbia seeds were germinated at a range of CFX concentrations under sterile conditions, and significant effects of the drug were observed (Fig. 2 A). Growth was inhibited immediately after germination, on uptake of the drug by the root tip, with concurrent etiolation of the hypocotyl and cotyledons. Etiolation also occurs from the base of the petiole of 6-week-old seedlings when the plants are transferred to CFX media, at a concentration of 1 μM (Fig. 2 C–E) and after venations at 50 μM CFX. These results are indicative of an effect on chloroplasts, which replicate their DNA in the basal 10 mm of the emerging leaf (29), but could also be due to an effect on mitochondria. The effects cannot be reversed by the subsequent removal of the drug from the media. When A. thaliana ecotype Landsberg cell cultures were treated with up to 5 μM CFX, the chloroplasts were eliminated and mitochondrial numbers markedly reduced (Fig. 2B and 3), with cells surviving and replicating by the addition of an exogenous energy source to the medium; the varied response to the quinolone is probably a result of differential uptake of the drug into the two organelles. At higher drug concentrations, the cells were no longer viable and plastid morphology was visibly affected (Fig. 3). Treatment of seedlings in liquid culture with the coumarin drug coumermycin A1 also showed profound effects on cell growth and total biomass (Fig. 2 F–H). Although coumarins have a much greater affinity for gyrase than quinolones (3), their relative insolubility in water appears to affect their uptake and transport to their target in planta, and the final concentration of drug internally is likely to be much less than the dose. These data suggest that gyrase-specific agents act on A. thaliana, targeting the organelles.
Fig. 2.
Effects of gyrase-specific drugs on A. thaliana. (A) Sterile seeds (ecotype Columbia) were germinated on germination media containing the indicated concentrations of CFX. (B) Effects of CFX on light-grown cell suspension cultures. Chloroplasts have been eliminated from the cells but growth is maintained due to the exogenous energy source. (C–E) Effects of CFX on 6-week-old plants. Seeds were germinated on drug-free germination media before transfer and incubation on GM media. (F–H) Effects of coumermycin A1 on A. thaliana. (F) Control. (G) Three percent DMSO (H). A total of 50 μM coumermycin A1 in 3% DMSO. (Scale bars, 10 mm.)
Fig. 3.
Effects of CFX on A. thaliana ecotype Landsberg cell cultures. Light-grown cultures (5 ml) were subcultured into 100 ml of fresh Murashige and Skoog ordinary media 4 days before visualization. Columns 1 and 3, transmission images; column 2, false-color images of chlorophyll autofluorescence in the chloroplasts after addition of stated concentrations of CFX; column 4, false-color images after treatment with MitoTracker Green FM. Samples were illuminated with a green HeNe laser by using 543 nm for chlorophyll autofluorescence and stain. Red represents the lowest level of autofluorescence, increasing through a gradient to yellow, white, and blue (highest level). (Scale bars, 10 μm.)
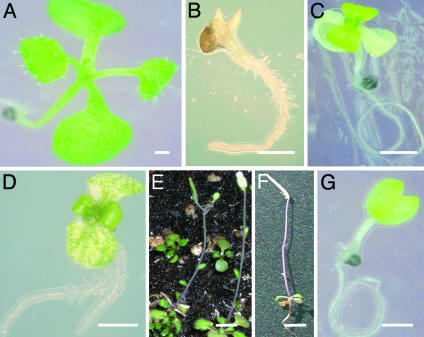
Gyrase Gene Knockouts Produce Deleterious Effects. To investigate the role played by the putative Arabidopsis gyrases during plant growth and development, we used independent mutants with T-DNA insertions in each gene. The AtgyrA-1 insertion (Fig. 1) led to an embryo-lethal phenotype (Fig. 4). No change in phenotype was apparent for the AtgyrA-2 allele, suggesting that the insertion of the T-DNA into the 3′ UTR did not affect the synthesis and processing of the protein (Fig. 1). Identical seedling lethal phenotypes were obtained from independent alleles for insertions in the plastid targeted GyrB (AtcpgyrB-1) and mitochondrial targeted GyrB (AtmtgyrB-1). The cotyledons were etiolated, the tissue being significantly more yellow than wild-type plants, with limited root growth and no further leaf or tissue development. A second phenotype in 10% of the plants in the AtmtgyrB-2 line was observed, where the cotyledons were etiolated and the primary and secondary leaf sets were green, with sporadic, aberrant trichome development on the emergent leaves and poor root growth. These plants completed a life cycle, albeit slowly and in miniature, eventually forming no more than eight leaves, taking 3 months to form no more than two flowers, setting seed in a single silique and only attaining a height of 2 cm, including the length of the silique, which was comparatively similar in size to wild-type siliques at maturation (Fig. 4G). Insertion into AtntgyrB-1 led to a seedling lethal phenotype with green cotyledons, but no further growth after emergence of the first leaves. Initial root development was apparently normal, although none of the plants grew >0.5 cm in length.
Fig. 4.
Phenotypes of plants with T-DNA insertions in the gyrase genes from the SALK, SAIL, and GABI-KAT collections. (A) Three-week-old wild-type A. thaliana ecotype Columbia seedling maintained in tissue culture. (B) Three-week-old AtgyrA-1, demonstrating the etiolated, embryo-lethal phenotype (C) AtntgyrB-1. (D) AtmtgyrB-2, displaying the seedling-lethal phenotype: etiolated cotyledons, green leaf primordia, and sporadic trichome development. A similar phenotype was also obtained in AtcpgyrB-1, AtmtgyrB-1, and AtmtgyrB-3. (E–G) Phenotype of the leaky AtmtgyrB-2 plants, which were able to complete a life cycle. (E) Two-week-old plant maintained in tissue culture before transfer to soil. (F) Ten-week-old plants after transfer to soil. (G) A 14-week-old plant after setting seed in a single silique. (Scale bars, 10 mm in A–E;25mmin F and G.)
These data therefore suggest that AtgyrA is essential for viability and no other topo is able to substitute for the enzyme in vivo. The three gyrB homologues are also essential for plant viability and for organellar replication. However, the presence of multiple gyrB sequences in the A. thaliana genome makes this result less clear than that for AtgyrA because it appears that in some cases, functional enzyme complexes are assembled with alternate GyrBs substituting in the knockout lines. Similarities of the AtcpgyrB and AtmtgyrB sequences makes this feasible, but how the targeted enzymes are transported to alternate organelles is unclear. Additional experiments involving complementation of the knockout lines with wild-type genes are warranted.
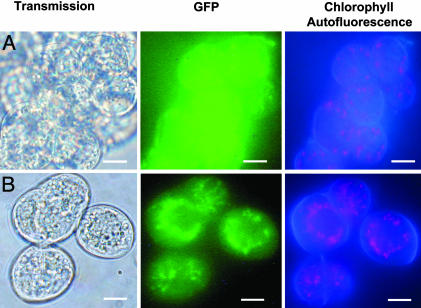
Role of the Transit Peptides in Organellar Targeting. Nuclear-encoded organellar proteins are localized to the appropriate cellular location by methods including N-terminal and internal signal sequences. N-terminal transit peptides were identified in the predicted amino acid sequences of A. thaliana GyrA and two of the GyrBs (Table 1). Transgene constructs fusing the transit peptide to the GFP reporter gene under the control of a constitutive promoter were transformed into Arabidopsis cells by Agrobacterium transformation for transient expression assays. Transformants were identified readily by epifluorescent microscopy, and dark grown cell cultures avoided significant amounts of chlorophyll autofluorescence and confirmed transgene expression and GFP localization to plastids (Fig. 5) and mitochondria (data not shown), as predicted by the transit peptide sequences (Table 1). GFP was localized randomly in the cytosol in nontargeted controls (Fig. 5). Untransformed cells and empty vector controls did not emit any fluorescence. These results confirm the in silico analysis of the genes and are further evidence of an organellar role for the gyrase gene products.
Fig. 5.
Organelle targeting of the 35S::pAOVGFP constructs in A. thaliana ecotype Landsberg cultured cells. The constructs were introduced by Agrobacterium transformation and transient GFP emission was observed by epifluorescence. (A) The 35S::pAOVGFP control transformation. Transient expression of GFP under the control of the nontargeted 35S constitutive promoter is spread through the nucleus and cytosol. (B) Transient expression of the transit peptide construct 35S::cpTPGyrB. The GFP is localized to the chloroplasts, as confirmed by the autofluorescence of the chlorophyll in Right. (Scale bars, 10 μm.)
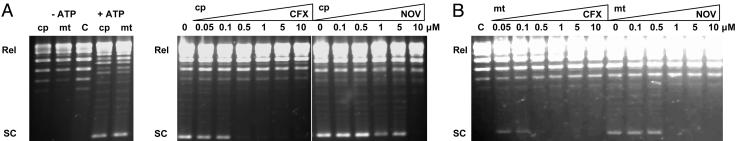
Supercoiling Activity Is Found in Chloroplast and Mitochondrial Extracts but Not in the Cytoplasm. The presence of gyrase genes in the A. thaliana genome, the deleterious effects of gyrase-specific antibacterial agents on plants and cells in culture, and the lethal effects of the gene knockouts, strongly suggest that plant cells should contain DNA supercoiling activity. To ascertain this outcome, we have grown sterile plant cell cultures and fractionated them into plastid, mitochondrial, and cytoplasmic fractions. The gyrase subunits were partially purified by using a NOV-affinity column. GyrA coeluted with weakly bound GyrB in the 0.5 M NaCl wash (with tightly bound GyrB eluting in 4 M urea fractions), which were then assayed for supercoiling activity (Fig. 6). We found that only the organellar fractions contain detectable supercoiling activity, which is ATP-dependent (Fig. 6A) and is inhibitable by the gyrase-specific drugs CFX and NOV (Fig. 6B).
Fig. 6.
(A) DNA supercoiling activity of plant gyrase. Purified total protein extracts from isolated chloroplasts (cp) and mitochondria (mt) after binding to and elution from a NOV-Sepharose column, were incubated with 400 ng of relaxed pBR322 DNA for 30 min at 37°C, with or without ATP as indicated. (B) Inhibition of supercoiling activity by drugs. Partially purified cp and mt proteins were assayed for supercoiling activity in the presence of the stated concentrations of CFX or NOV. Lane C contains no enzyme or drug.
A. thaliana Gyrase Genes Complement E. coli ts Strains. To further characterize the A. thaliana gyrase proteins, we have cloned the four gyrase genes. When each of the genes was expressed in the E. coli temperature-sensitive mutant strains (KNK453 for gyrA and N4177 for gyrB), we found that three of the cloned plant genes could complement the bacteria at the nonpermissive temperature, albeit weakly in some cases; AtcpgyrB was unable to complement N4177 (Table 2). This finding suggests that not only do the A. thaliana gyrase genes encode functional gyrase proteins but that they are able to form an active supercoiling complex (A2B2) with their partner subunit from E. coli. Supercoiling activity was obtained in vitro when the complementary purified E. coli subunit is added to the overexpressed and partially purified A. thaliana subunits (data not shown). However, despite extensive efforts, we have thus far been unable to detect supercoiling activities by using purified mixes of recombinant A. thaliana proteins expressed in E. coli (M.K.W. and L.A.M., unpublished results). The A. thaliana enzymes may require other factors that are not required for the E. coli enzyme; further work will be required to establish the optimal conditions for activity.
Table 2. Complementation of E. coli temperature-sensitive strains.
| Complementation in E. coli strain
|
||
|---|---|---|
| Gene | KNK453 (gyrAts) | N4177 (gyrBts) |
| AtgyrA | ++ | NA |
| AtcpgyrB | NA | - |
| AtmtgyrB | NA | +++ |
| AtntgyrB | NA | ++ |
| E. coli gyrA | +++ | NA |
| E. coli gyrB | NA | +++ |
cp, chloroplast; mt, mitochondrial; nt, not targeted; NA, not applicable; +++, strong complementation; +, weak complementation; -, no complementation.
Discussion
Previous work has presented circumstantial evidence for the existence of DNA gyrase in plants. In this paper, we describe the identification of four A. thaliana genes encoding five potential gene products, and demonstrate the existence of at least two gyrase enzymes, targeted to chloroplasts and mitochondria, which support DNA supercoiling.
Phylogenetic analysis of AtGyrA (Fig. 1) suggests that it is most closely related to the Oryza sativa homologue, demonstrating gene conservation from dicots to monocots. The close relationship of AtGyrA, AtcpGyrB, and AtmtGyrB to cyanobacterial gyrase subunits suggests that the genes were assimilated through the acquisition of endosymbiotic bacteria by the plant. However, AtntGyrB clusters with eukaryotic topo IIs, suggestive of an alternative origin.
Chloroplasts and mitochondria are remnants of free-living prokaryotes that lost their autonomy during evolution by establishing an endosymbiotic relationship with their host cells, and it is reasonable to expect that certain mechanisms of replication and cell division have been retained throughout evolution (30). It is therefore not unexpected that a bacterial-like gyrase activity is present in these organelles. However, gyrase has not been identified in the genomes of other eukaryotes, although there is evidence for the existence of the enzyme in unicellular parasites.
The malarial parasite, Plasmodium falciparum, contains a relict chloroplast, the apicoplast (31), which contains distinct metabolic pathways. Treatment of P. falciparum with CFX inhibits apicoplast but not nuclear replication (32). The presence of gyrase in this organism is supported by the recent publication of the P. falciparum genome (33). Therefore, the apparent absence of gyrase in other eukaryotes may be explained by the absence of a plastid-derived organelle, with secondary acquisition by plant mitochondria.
Plant organelles have maintained many of the bacterial proteins involved in DNA metabolism. Many genes for mitochondrial and chloroplast RNAs and proteins are nuclear-encoded, including all enzymes involved in DNA replication and most proteins participating in gene expression, allowing nuclear control of organellar development (12, 34). Targeting of the gyrase subunits to the chloroplast and mitochondria is consistent with the lack of any DNA replication genes found on the organelle genomes (35, 36). The situation of A. thaliana gyrase is analogous to that of RecA, where there appears to be four RecA homologues, one targeted to mitochondria, one to chloroplasts, and two others where the targeting is unclear (37).
Many plant genes encode dual-targeted proteins, enabling transfer to both chloroplasts and mitochondria, by using mechanisms including ambiguous targeting, dual targeting sequences, and multiple transcription or translation starts (38). AtGyrA apparently has two translation initiation sites that allow dual targeting to both organelles, whereas the AtcpGyrB and Atmt-GyrB proteins have unambiguous targeting sequences that determine chloroplast or mitochondrial localization, respectively (Fig. 5).
The functional significance of the plant gyrase proteins was supported by effects of gyrase-specific drugs on plant seedlings and cultured cells (Figs. 2 and 3). Quinolones, such as CFX, specifically inhibit bacterial gyrase but do not substantially affect its eukaryotic counterpart, topo II, or other eukaryotic enzymes (39). The efficacy of CFX against plants in these experiments is consistent with the concentration required to inhibit the bacterial enzyme (IC50 <1 μM) and is significantly less than the IC50 of 300 μM reported for CFX against eukaryotic topo II (40).
The full complement of chloroplasts in A. thaliana ecotype Landsberg is attained after three complete rounds of chloroplast DNA replication and division (41). It is this process that the gyrase-targeting drugs disrupt. Elimination of the organelles and subsequent cell stasis strongly suggest that gyrase is an essential plant enzyme. The biphasic effect of CFX on the organelles (Fig. 3) is reminiscent of the effects of this drug on the morphology of E. coli cells (42), suggesting the possibility of a similar mechanism of action involving arrest of the gyrase-cleavable complex and blocking of replication and transcription (43). This finding raises the possibility of compounds of the quinolone or coumarin class being developed as novel herbicides.
Further evidence for the requirement of plant gyrase came from studies with knockout mutants (Fig. 4). Disruption of AtgyrA produced an embryo lethal phenotype, whereas disruption of the AtgyrB genes led primarily to seedling lethal phenotypes, although some plants displayed a less severe phenotype, which is likely to be due to functional gyrase complexes being formed by one of the other unaffected AtGyrB homologues with AtGyrA. Partial redundancy of gene function has been observed in other A. thaliana gene families, particularly where gene divergence exists. We are currently making inducible knockouts of each of the A. thaliana genes to further investigate the biological roles of these enzymes.
Assays of fractionated extracts from plant cell cultures demonstrated that there is ATP-dependent supercoiling activity in chloroplast and mitochondrial fractions, but not in cytoplasmic fractions. Moreover, expression of the AtgyrA, AtmtgyrB, and AtntgyrB genes in the corresponding E. coli ts mutant (Table 2) shows that, like mtRecA, the A. thaliana proteins can function with their bacterial counterparts (37). The only exception is AtcpgyrB, which was unable to complement the gyrB ts strain. Partially purified A. thaliana gyrase proteins were able to support supercoiling activity when the complementary E. coli gyrase subunit was added; however, no activity was observed when GyrA and GyrB subunits from A. thaliana were mixed together. It is likely that other factors are required for the functional interaction of A. thaliana GyrA and GyrB proteins, or that one or other of the proteins is alternatively spliced or requires posttranslational modifications. More detailed investigations are warranted.
The data in this paper, coupled with earlier work, support the existence of gyrase in A. thaliana, with activities being targeted to chloroplasts and mitochondria. It seems that these organelles have retained a bacterial-like mechanism of DNA replication, which is likely to involve gyrase in relaxing positive supercoils ahead of the replication fork and in maintenance of supercoiling in the organellar DNA. Whether the plant organellar gyrases can perform other functions, such as decatenation, which are normally carried out by topo II or topo IV, remains to be elucidated.
Supplementary Material
Acknowledgments
We thank K. Roberts and K. Sugimoto for helpful suggestions and J. Mylne and F. Corke for reagents. This work was supported by grants from the Wellcome Trust (U.K.) and the Biotechnology and Biological Sciences Research Council (U.K.). M.K.W. is a Biotechnology and Biological Sciences Research Council-funded Postdoctoral Researcher. Funding for the Salk Institute Genomic Analysis Laboratory (La Jolla, CA) indexed insertion mutant collection was provided by the National Science Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CFX, ciprofloxacin; GyrA, DNA gyrase protein A; GyrB, DNA gyrase protein B; NAL, nalidixic acid; NOV, novobiocin; topo, topoisomerase.
References
- 1.Champoux, J. J. (2001) Annu. Rev. Biochem. 70, 369-413. [DOI] [PubMed] [Google Scholar]
- 2.Gellert, M., Mizuuchi, K., O'Dea, M. H. & Nash, H. A. (1976) Proc. Natl. Acad. Sci. USA 73, 3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maxwell, A. (1997) Trends Microbiol. 5, 102-109. [DOI] [PubMed] [Google Scholar]
- 4.Thompson, R. J. & Mosig, G. (1985) Nucleic Acids Res. 13, 873-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woelfle, M. A., Thompson, R. J. & Mosig, G. (1993) Nucleic Acids Res. 21, 4231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinhorst, S., Cannon, G. & Weissbach, A. (1985) Arch. Biochem. Biophys. 239, 475-479. [DOI] [PubMed] [Google Scholar]
- 7.Itoh, R., Takahashi, H., Toda, K., Kuroiwa, H. & Kuroiwa, T. (1997) Eur. J. Cell Biol. 73, 252-258. [PubMed] [Google Scholar]
- 8.Mills, W. R., Reeves, M., Fowler, D. L. & Capo, S. F. (1989) J. Exp. Bot. 40, 425-429. [Google Scholar]
- 9.Lam, E. & Chua, N.-H. (1987) Plant Mol. Biol. 8, 415-424. [DOI] [PubMed] [Google Scholar]
- 10.Ebringer, L., Polonyi, J. & Krajcovic, J. (1993) Arzneim.-Forsch. 43, 777-781. [PubMed] [Google Scholar]
- 11.Pyke, K. A., Marrison, J. & Leech, R. M. (1989) FEBS Lett. 242, 305-308. [Google Scholar]
- 12.The Arabidopsis Genome Initiative. (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 13.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emanuelsson, O., Nielsen, H. & von Heijne, G. (1999) Protein Sci. 8, 978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Y., Rosso, M. G., Strizhov, N., Viehoever, P. & Weisshaar, B. (2003) Bioinformatics 19, 1441-1442. [DOI] [PubMed] [Google Scholar]
- 16.Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., et al. (2002) Plant Cell 14, 2985-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 18.Weigel, D. & Glazebrook, J. (2002) Arabidopsis: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 19.Hofgen, R. & Willmitzer, L. (1988) Nucleic Acids Res. 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forreiter, C., Kirschner, M. & Nover, L. (1997) Plant Cell 9, 2171-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreuzer, K. N. & Cozzarelli, N. R. (1979) J. Bacteriol. 140, 424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menzel, R. & Gellert, M. (1983) Cell 34, 105-113. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell, A. & Howells, A. J. (1999) in DNA Topoisomerase Protocols I. DNA Topology and Enzymes, eds. Bjornsti, M.-A. & Osheroff, N. (Humana, Totowa, NJ), pp. 135-144.
- 24.Reece, R. J. & Maxwell, A. (1989) J. Biol. Chem. 264, 19648-19653. [PubMed] [Google Scholar]
- 25.Kieber, J. J., Tissier, A. F. & Signer, E. R. (1991) Plant Physiol. 99, 1493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie, S. & Lam, E. (1994) Plant Physiol. 106, 1701-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartung, F. & Puchta, H. (2001) Gene 271, 81-86. [DOI] [PubMed] [Google Scholar]
- 28.Hedtke, B., Borner, T. & Weihe, A. (2000) EMBO Rep. 1, 435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamura, S., Kuroiwa, T. & Nagata, T. (1990) Plant Cell Physiol. 31, 597-602. [Google Scholar]
- 30.Gray, M. W. (1993) Curr. Opin. Genet. Dev. 3, 884-890. [DOI] [PubMed] [Google Scholar]
- 31.Ralph, S. A., D'Ombrain, M. C. & McFadden, G. I. (2001) Drug Resist. Updat. 4, 145-151. [DOI] [PubMed] [Google Scholar]
- 32.Fichera, M. E. & Roos, D. S. (1997) Nature 390, 407-409. [DOI] [PubMed] [Google Scholar]
- 33.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419, 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tewari, K. K. (1988) in DNA Replication in Plants, eds. Bryant, J. A. & Dunham, V. L. (CRC Press, Boca Raton), pp. 69-116.
- 35.Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E. & Tabata, S. (1999) DNA Res. 6, 283-290. [DOI] [PubMed] [Google Scholar]
- 36.Unseld, M., Marienfeld, J. R., Brandt, P. & Brennicke, A. (1997) Nat. Genet. 15, 57-61. [DOI] [PubMed] [Google Scholar]
- 37.Khazi, F. R., Edmondson, A. C. & Nielsen, B. L. (2003) Mol. Genet. Genomics 269, 454-463. [DOI] [PubMed] [Google Scholar]
- 38.Silva-Filho, M. C. (2003) Curr. Opin. Plant Biol. 6, 589-595. [DOI] [PubMed] [Google Scholar]
- 39.Gootz, T. D., Barrett, J. F. & Sutcliffe, J. A. (1990) Antimicrob. Agents Chemother. 34, 8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammonds, T. R., Foster, S. R. & Maxwell, A. (2000) J. Mol. Biol. 300, 481-491. [DOI] [PubMed] [Google Scholar]
- 41.Pyke, K. A. & Leech, R. M. (1992) Plant Physiol. 99, 1005-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diver, J. M. & Wise, R. (1986) J. Antimicrob. Chemother. 18, Suppl., D31-D41. [DOI] [PubMed] [Google Scholar]
- 43.Drlica, K. & Malik, M. (2003) Curr. Top Med. Chem. 3, 249-282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.