Abstract
Hedgehog (Hh) signaling plays important roles in various development processes. This signaling is necessary for osteoblast formation during endochondral ossification. In contrast to the established roles of Hh signaling in embryonic bone formation, evidence of its roles in adult bone homeostasis is not complete. Here we report the involvement of Gli1, a transcriptional activator induced by Hh signaling activation, in postnatal bone homeostasis under physiological and pathological conditions. Skeletal analyses of Gli1 +/− adult mice revealed that Gli1 haploinsufficiency caused decreased bone mass with reduced bone formation and accelerated bone resorption, suggesting an uncoupling of bone metabolism. Hh-mediated osteoblast differentiation was largely impaired in cultures of Gli1 +/− precursors, and the impairment was rescued by Gli1 expression via adenoviral transduction. In addition, Gli1 +/− precursors showed premature differentiation into osteocytes and increased ability to support osteoclastogenesis. When we compared fracture healing between wild-type and Gli1 +/− adult mice, we found that the Gli1 +/− mice exhibited impaired fracture healing with insufficient soft callus formation. These data suggest that Gli1, acting downstream of Hh signaling, contributes to adult bone metabolism, in which this molecule not only promotes osteoblast differentiation but also represses osteoblast maturation toward osteocytes to maintain normal bone homeostasis.
Introduction
Hedgehog (Hh) signaling is a highly conserved pathway that plays important roles in various development processes. Hh signaling is indispensable for osteoblast formation in endochondral ossification, one of the two ossification processes in mammals, which forms bones in limbs, the trunk, and some head structures [1]. In this context, Indian hedgehog (Ihh) expressed in prehypertrophic chondrocytes is thought to act directly on progenitors in the perichondrium and the bone marrow to induce their differentiation into bone-forming osteoblasts [2]–[4]. The deletion of Ihh or smoothened (Smo), a transmembrane signaling transducer for Hh, caused no bone collar or primary spongiosa in mice [2], [4]. These mutant mice lacked the expression of runt-related transcription factor 2 (Runx2), a key determinant for osteoblasts [5], [6] and bone gamma-carboxyglutamate (gra) protein (osteocalcin; official gene symbol, Bglap), a bona fide marker for osteoblasts [1], in the perichondrium.
Gli transcription factors mediate the transcription of target genes downstream of Hh signaling. Gli1, a target gene of Hh signaling, acts as a transcriptional activator, whereas Gli2 and Gli3 can act as both activators and repressors [7]. The activator function of Gli2 and the repressor function of Gli3 were reported to mediate substantial aspects of the action of Ihh on bone development [8], [9]. In addition, we found that Gli1 participated in the Hh-mediated osteoblast formation collectively with Gli2 and Gli3 [10], [11].
Regarding postnatal roles of Hh signaling in bone metabolism, we reported that the haploinsufficiency of patched 1 (Ptch1), a transmembrane Hh receptor that represses signaling activity without Hh input, led to high bone mass in adult mice and humans [12]. Mice in which Ptch1 was deleted in Bglap-positive mature osteoblasts showed low bone mass [13]. Although both of the mutants mentioned above had high bone turnover, where both osteoblastogenesis and osteoclastogenesis were enhanced, the balance shifted toward the opposite phenotypes. When an activator form of Gli2 was forcibly expressed in osteoblast precursors expressing Sp7 (osterix-Osx), another key determinant of osteoblasts, the skeleton was not affected at the fetal stage in mice. However, the mutants postnatally showed severe osteopenia with a decrease in osteoblast number, although the number of osteoclasts was not changed [14]. Thus, in contrast to the established roles of Hh signaling in embryonic bone formation, the evidence of its roles in adult bone homeostasis is not adequate. In particular, little is known about the roles of Gli1, a potent transcriptional activator induced by Hh signaling, in postnatal bone metabolism.
In the present study, we attempted to examine Hh signaling in the postnatal skeletal system, focusing on the functions of Gli1 in the bone metabolism. We analyzed skeletal phenotypes of Gli1 heterozygous knockout mice postnatally in vivo and in vitro. We also compared fracture healing between mutant and wild-type (WT) mice. We report that Gli1 haploinsufficiency affects adult bone metabolism, at least partly through the uncoupling of bone formation and bone resorption, leading to a decrease in bone mass and a delay in fracture healing in postnatal skeletons.
Materials and Methods
Animal Experiments
Wild-type C57BL/6J mice were obtained from Charles River Japan, and Gli1 +/− mice were generated as previously described [15]. All experiments were performed in accord with the protocol approved by the Animal Care and Use Committee of The University of Tokyo (#KA13-5). Mice were kept in individual cages under controlled temperature and humidity with a 12-hr circadian rhythm. They were given ad libitum access to food and water. All efforts were made to minimize the suffering of the mice. Euthanasia of mice was performed with an overdose of barbiturates.
Reagents and Vectors
Smoothened agonist (SAG) was purchased from Calbiochem (San Diego, CA; 566660). Receptor activator of NF-kappa-B ligand (RANKL) was purchased from Pepro Tech (Rocky Hill, NJ; 184-01791). Sonic hedgehog (Shh) was purchased from R&D Systems (Minneapolis, MN; 1845-SH). Cyclopamine was purchased from Enzo Life Sciences (Farmingdale, NY; BML-GR3334). Plasmids expressing human GLI1 were constructed as previously described [12]. The adenoviral vector expressing human GLI1-Biotin-3xFLAG-IRES-dsRed was constructed using the pAd/PL-DEST vector and ViraPower Adnoviral Expression System (Life Technologies, Carlsbad, CA), according to the manufacturer's instructions. In brief, human GLI1 cDNA carrying Biotin-3xFLAG tag [16] was initially cloned into pCID vectors [17]; GLI1-Biotin-3xFLAG-IRES-dsRed was then transferred into the pENTR1A vector in conjunction with the CAGGS promoter and subjected to adenoviral vector construction using the Gateway system. The pAd/PL-DEST expressing the CAGGS promoter-driven GLI1-Biotin-3xFLAG-IRES-dsRed was linearized with Pac I and transfected into 293A cells. After amplification, the virus was stored at −80°C. The viral titer was determined by an end-point titer assay using 293A cells.
Cell Culture
C3H10T1/2 cells were obtained from the RIKEN Cell Bank (Ibaraki, Japan). Mouse primary bone marrow stromal cells were isolated from the long bones of 8-week-old male mice. Mouse primary osteoblast precursors were isolated from calvarias of mouse neonates. Cells were cultured in high-glucose Dulbecco's modified Eagle Medium (DMEM; Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. For osteogenic cultures, the cells were cultured in osteogenic media [18], [19] supplemented with Smoothened agonist (SAG). Co-culture experiments using mouse primary bone marrow stromal cells and bone marrow macrophages were performed as previously described [18]. The in vitro osteoclast differentiation of RAW264.7 cells was performed as previously described [20]. Adenoviruses expressing GLI1-Biotin-3xFLAG-IRES-dsRed (Ax-Gli1) or GFP (Ax-GFP) were used to infect cells at MOI (multiplicity of infection) 10. Alkaline phosphatase (ALP), von Kossa, and tartrate-resistant acid phosphatase (TRAP) staining were performed as previously described [12], [18].
Real-Time RT-PCR
Total RNA extraction and real-time reverse transcription-polymerase chain reaction (RT-PCR) were performed as previously described [21]. All reactions were run in triplicate. The primer sequences are as follows: β-actin, AGATGTGGATCAGCAAGCAG (forward) and GCGCAAGTTAGGTTTTGTCA (reverse); Alp, GCTGATCATTCCCACGTTTT (forward) and CTGGGCCTGGTAGTTGTTGT (reverse); Ibsp, CAGAGGAGGCAAGCGTCACT (forward) and CTGTCTGGGTGCCAACACTG (reverse); Bglap, AAGCAGGAGGGCAATAAGGT (forward) and TTTGTAGGCGGTCTTCAAGC (reverse); Gli1, GCACCACATCAACAGTGAGC (forward) and GCGTCTTGAGGTTTTCAAGG (reverse); Gli2, CTGAAGGATTCCTGCTCGTG (forward) and ACAGTGTAGGCCGAGCTCAT (reverse); Runx2, CCGCACGACAACCGCACCAT (forward) and CGCTCCGGCCCACAAATCTC (reverse); Sp7, ACTCATCCCTATGGCTCGTG (forward) and GGTAGGGAGCTGGGTTAAGG (reverse); Tnfsf11, AGCCATTTGCACACCTCAC (forward) and CGTGGTACCAAGAGGACAGAGT (reverse); Tbfrsf11b, GTTTCCCGAGGACCACAAT (forward) and CCATTCAATGATGTCCAGGAG (reverse); Dmp1, CAGTGAGGATGAGGCAGACA (forward) and TCGATCGCTCCTGGTACTCT (reverse); Sost, AAGCCGGTCACCGAGTTGGT (forward) and GTGAGGCGCTTGCACTTGCA (reverse); Ptch1, CTGGACTCTGGCTCCTTGTC (forward) and CAACAGTCACCGAAGCAGAA (reverse); Ctsk, ACGGAGGCATCGACTCTGAA (forward) and GATGCCAAGCTTGCGTCGAT (reverse); and Nfatc1, CCTTCGGAAGGGTGCCTTTT (forward) and AGGCGTGGGGCCTCAGCAGG (reverse).
Luciferase Assay
Cells were plated onto 24-well plates and transfected with 0.4 µg of DNA in a mixture containing the reporter plasmids (8×3′-Gli BS-luc) [22], the control reporter plasmids encoding Renilla luciferase, and effector plasmids (pCMV-myc-GLI1) or adenoviral vectors expressing GLI1. A dual-luciferase assay was performed as previously described [12].
Immunoblot
Whole-cell lysates were isolated using RIPA buffer as previously described [23]. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were performed using anti-Gli1 (sc-20687, 1∶1000, Santa Cruz Biotechnology, Santa Cruz, CA) and HRP-conjugated anti-FLAG M2 antibodies (A8592-2MG, 1∶500, Sigma-Aldrich) as previously described [12].
Radiological analysis
X-ray photographs of the left tibiae of WT and Gli1 +/− mice (n = 10 each) were taken using a soft x-ray system (M-60; Softex Co., Tokyo). Micro-computed tomography (CT) scanning of the harvested femurs was performed using a microfocus X-ray CT system SMX-90CT (Shimadzu, Kyoto, Japan) under the following conditions: tube voltage, 90 kV; tube current, 110 µA; layer thickness, 5.3 mm; and field of view (XY), 10.4 mm. The resolution of one CT slice was 512×512 pixels. The three-dimensional construction software package TRI/3D-BON (Ratoc System Engineering, Tokyo) was used for quantitative analysis.
Histological analysis
We intraperitoneally injected calcein (0.16 mg per 10 g of body weight; Sigma) into mice 4 days and 1 day before sacrifice. We stained the undecalcified sections of the femur with toluidine blue, von Kossa, and TRAP as previously described [12]. Images were taken using an Axio Imager A1 (Carl Zeiss, Jena, Germany) and processed using AxioVision (Carl Zeiss). Histomorphometric analyses were performed using the HistometryRT CAMERA system. We analyzed five mice for each group.
Fracture model
Ten 8-week-old male mice were used in each group. Under general anesthesia with isofluorane in O2, the left hind limb was shaved and sterilized for surgery. A 15-mm incision was made longitudinally, and a blunt dissection of the muscle was made to expose the tibia. A transverse osteotomy was performed using disk-shaped dental steel bars at the mid point of the tibia. The fracture was repositioned, and then the full-length of the bone marrow cavity was internally stabilized as previously described [24]. After irrigation with saline, the skin was closed with 4-0 nylon sutures. Fourteen days after surgery, the tibias were harvested from the euthanized mice.
Statistical analysis
The means of groups were compared by an analysis of variance (ANOVA), and the significances of the differences were determined by Student's t-test. P-values <0.05 were considered significant.
Results
Gli1 haploinsufficiency causes decreased bone mass in adult mice
As reported, we found that Gli1 −/− mice had no gross abnormalities at birth [11], [15]. Although Gli1 −/− pups were born in Mendelian ratios, the number of Gli1 −/− adults turned out to be approximately 10% of the total pups obtained because of their reduced survival rates during the first 10 days after birth (Table 1). In addition, the growth rate of the surviving Gli1 −/− mice was significantly lower than those of both the wild-type (WT) or Gli1 +/− mice, and the body weights of the 8-week-old Gli1 −/− mice were approximately 20% lower than those of the WT and Gli1 +/− mice (Figure S1).
Table 1. Survival rate of Gli1 mutant mice during postnatal 10 days.
| Genotype | Postnatal day | |
| 1 | 10 | |
| Gli1 +/+ | 11 (30.6) | 37 (28.5) |
| Gli1 +/− | 16 (44.4) | 81 (62.3) |
| Gli1 −/− | 9 (25.0) | 12 (9.2) |
| Total | 36 | 130 |
Percentages are in parentheses.
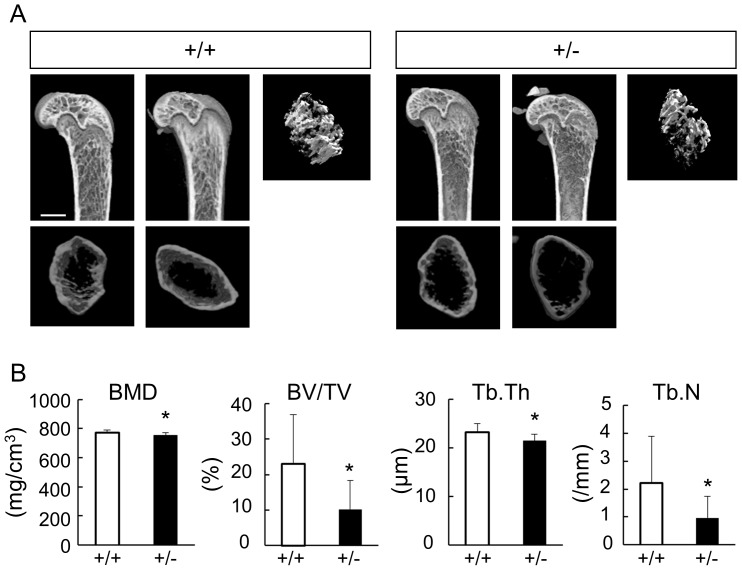
These findings suggest that the Gli1 −/− mice were largely affected by systemic abnormalities due to complete loss of Gli1 gene. Therefore, to investigate the roles of Gli1 in adult bone metabolism as directly as possible, we analyzed the skeletal system in Gli1 +/− male mice, which had weights and gross appearance comparable to those of the WT male mice. Micro-computed tomography (µ-CT) analyses of distal femurs revealed that the trabecular density of the 8-week-old Gli1 +/− mice was reduced compared to that of WT mice (Figure 1A). The bone morphometric analysis using the micro-CT data supported the finding, as the Gli1 +/− mice showed less bone mineral density (BMD) along with decreased parameters for bone formation (Figure 1B). These skeletal phenotypes were also observed in the Gli1 +/− female mice (Figure S2). In contrast, there was no significant difference in the cortical bone between WT and Gli1 +/− mice (Figure S3), suggesting differences between trabecular and cortical bones with regard to the contribution of Gli1.
Figure 1. Radiological findings of long bones in wild-type (WT) and Gli1 +/− mice.
(A) Three-dimensional micro-computed tomography (3D-micro-CT) images of the distal femurs of representative 8-week-old WT and Gli1 +/− male mice. Sagittal sections, transverse sections, and 3D reconstruction images of the primary spongiosa are shown for each genotype. Bar, 1 mm. (B) Histomorphometric analyses of 3D-micro-CT data. BMD, bone mineral density; BV/TV, bone volume per tissue volume; Tb.Th, trabecular thickness; Tb.N trabecular number parameters. Data are means ± SDs of eight male mice per genotype. *p<0.05 vs. WT.
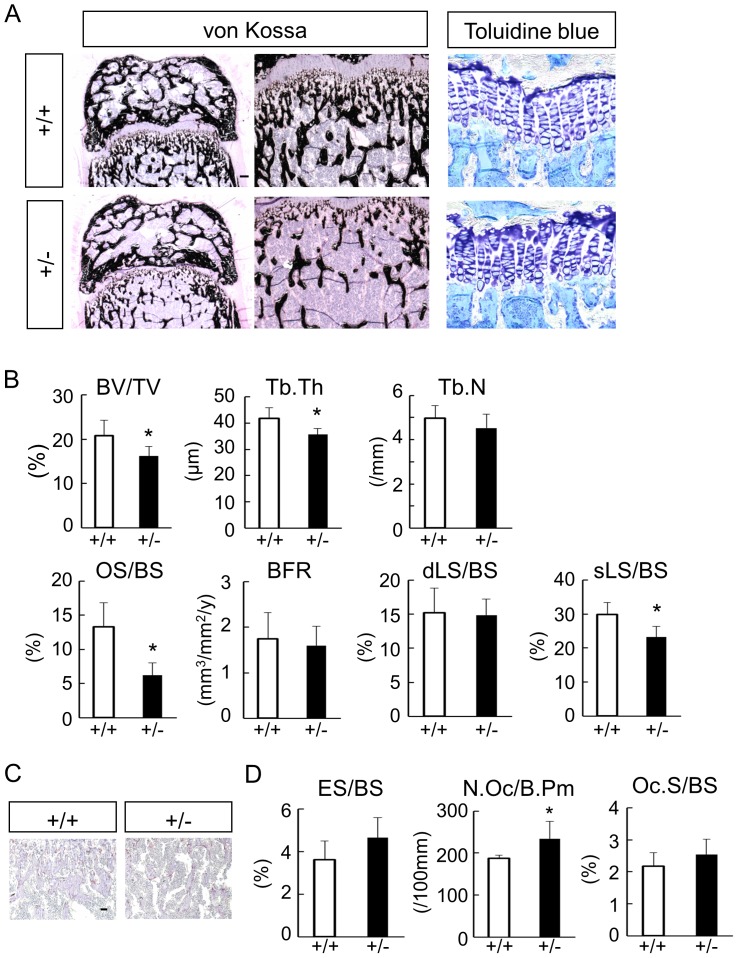
The histological analyses of the distal femurs of 8-week-old mice showed that Gli1 +/− mice had reduced trabecular bones compared to the WT mice (Figure 2A, see von Kossa), whereas the growth plate appeared normal (Figure 2A, see toluidine blue). Indeed, the bone volume/tissue volume (BV/TV) and trabecular thickness (Tb.Th) values were significantly decreased in Gli1 +/− mice compared to WT mice, as observed in micro-CT-based analyses (Figure 2B). Gli1 +/− mice also had significantly lower values of osteoid surface/bone surface (OS/BS) and single-labeled surface/bone surface (sLS/BS), which are parameters of the osteogenic capacity (Figure 2B).
Figure 2. Histological findings of adult WT and Gli1 +/− mice.
(A) von Kossa staining and toluidine blue staining of the distal femur sections of representative 8-week-old WT and Gli1 +/− male mice. Bar, 100 µm. (B) Histomorphometric analyses of bone volume and bone formation parameters in distal femurs from 8-week-old WT and Gli1 +/− male mice. OS/BS, osteoid surface per bone surface; MAR, mineral apposition rate; BFR, bone formation rate per bone surface; dLS/BS, double-labeled surface per bone surface; sLS/BS, single-labeled surface per bone surface. Data are means ± SDs of five male mice per genotype. *p<0.05 vs. WT. (C) TRAP staining of the distal femur sections of representative 8-week-old WT and Gli1 +/− male mice. Bar, 100 µm. (D) Histomorphometric analyses of bone resorption parameters in the distal femurs of 8-week-old WT and Gli1+/− male mice. ES/BS, eroded surface per bone surface; N. Oc/B. Pm, number of osteoclasts per 100 mm of bone perimeter; Oc. S/BS, osteoclast surface per bone surface. In (B) and (D), data are means ± SDs of five mice per genotype. *p<0.05 vs. WT.
Regarding bone resorption, the numbers of TRAP-positive osteoclasts were significantly higher in the Gli1 +/− mice compared to the WT mice (Figure 2C and D, see N.Oc/B.Pm). Consistent with this finding, the Gli1 +/− mice showed a trend toward increased bone resorption capacity parameters, compared to WT mice (Figure 2D, see ES/BS and Oc.S/BS). These data suggest that Gli1 haploinsufficiency causes an uncoupling of bone turnover in adult mice, which leads to decreased bone mass.
Osteoblast differentiation is impaired by Gli1 haploinsufficiency
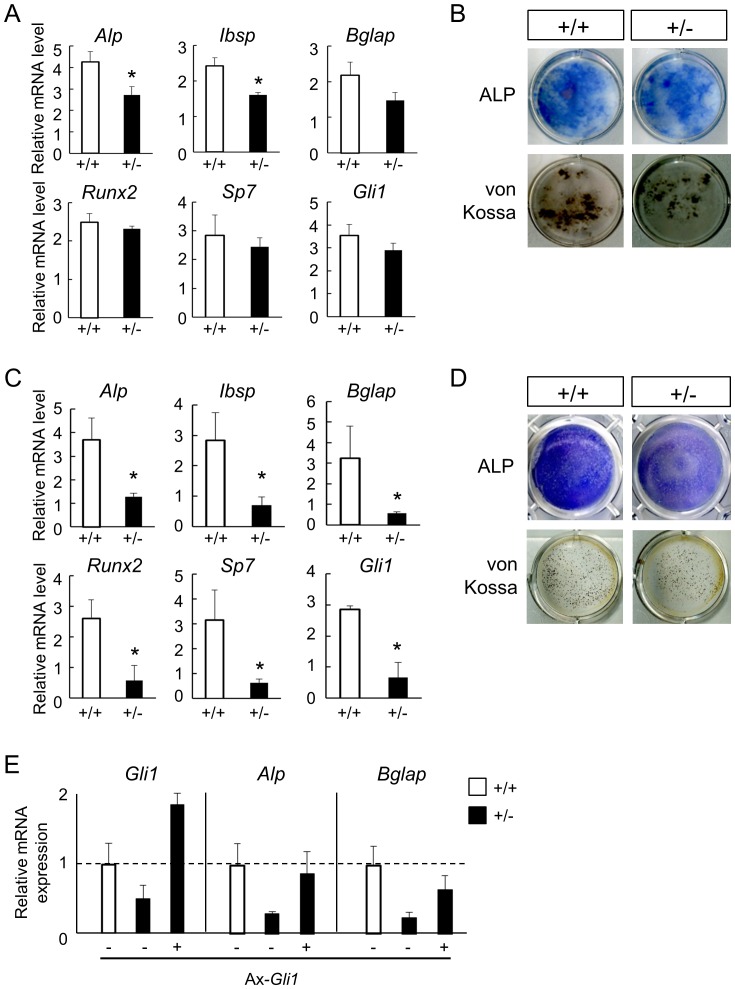
To investigate whether Gli1 haploinsufficiency caused decreased osteogenic capacity through the impairment of osteoblast differentiation from precursors, we examined osteoblast differentiation in ex vivo cultures of primary bone marrow stromal cells (BMSCs) harvested from 8-week-old WT and Gli1 +/− mice (Figure 3A and B) and primary osteoblast precursors (OPs) from neonates of each genotype (Figure 3C and D). Given that Gli1 expression is induced upon Hh signaling, we suspected that any difference resulting from the loss of one allele of Gli1 would be observed under Hh signaling-activated conditions. We therefore performed the experiments in the presence of Smoothened agonist (SAG), a small molecule that activates Hh signaling. The mRNA expression levels of osteoblast-related genes were lower in Gli1 +/− BMSCs than in WT BMSCs. In particular, alkaline phosphatase (Alp) and integrin-binding sialoprotein (Ibsp) expression levels were significantly reduced in the Gli1 +/− BMSCs (Figure 3A), which was consistent with our previous finding that Gli1 directly induced the expression of these two genes [11]. ALP activity and matrix calcification, key features of osteoblasts, were also suppressed in the Gli1 +/− BMSCs as evidenced by ALP and von Kossa staining (Figure 3B). The impaired osteoblast differentiation due to loss of one allele of Gli1 was more prominent in the OPs. All of the genes tested here showed significantly lower expression levels in the Gli1 +/− OPs compared to WT (Figure. 3C), and ALP activity and calcification were impaired in the Gli1 +/− OPs (Figure 3D). Lastly, Gli1 was reduced via the loss of one allele of Gli1 in both cell types, although a significant reduction was observed only in the OPs (Figure 3A and C, see Gli1).
Figure 3. Osteoblast differentiation in cultures of precursor cells from WT and Gli1 +/− mice.
(A) mRNA expression of osteoblast marker genes in 14-day osteogenic cultures of BMSCs in the presence of the Smoothened agonist. mRNA expression was analyzed by real-time RT-PCR. (B) ALP and von Kossa stainings in 14-day osteogenic cultures of BMSCs in the presence of the Smoothened agonist. (C) mRNA expression of osteoblast marker genes in 7-day osteogenic cultures of osteoblast precursors (OPs) isolated from neonatal calvarias, in the presence of the Smoothened agonist. (D) ALP and von Kossa staining in 7-day osteogenic cultures of OPs in the presence of the Smoothened agonist. (E) Rescue of the expression levels of osteoblast marker genes in Gli1 +/− OPs by the adenoviral overexpression of Gli1 in the presence of the Smoothened agonist. WT or Gli1 +/− OPs were infected with Ax-GFP (−) or Ax-Gli1 (+) at MOI 10.
The data above suggest that Gli1 haploinsufficiency affects osteoblast differentiation in a cell-autonomous manner. We then attempted to further verify the hypothesis by testing whether the recovery of Gli1 expression would rescue the osteoblast phenotypes in Gli1 +/− cells. We prepared an adenoviral vector expressing Gli1 (Ax-Gli1) (Figure S4A). Gli1 protein expression induced by Ax-Gli1 and its function were confirmed by western blotting using a specific antibody, luciferase reporter assays using the Gli-responsive element, and mRNA expression analyses of Alp and Ibsp in C3H10T1/2 cells (Figures S4B–D). As shown in Figure 3E, the introduction of Gli1 into Gli1 +/− OPs induced the expression of Alp, an early osteoblast marker, at a level comparable to that in WT OPs. The expression of Bglap, an osteoblast marker that is expressed at a later stage, was also upregulated but not fully recovered by Gli1 introduction. Thus, Gli1 haploinsufficiency in osteoblast precursors is likely to cause impairment of their differentiation, which underlies reduced bone formation in Gli1 +/− mice.
The Hh-Gli1 axis is involved in the capacity of osteoblasts/osteocytes to support osteoclastogenesis, but not in osteoclastogenesis itself
Bone homeostasis is achieved by the coupling of bone formation and bone resorption via cross-talk between osteoblasts/osteocytes and osteoclasts. Mature osteoblasts and osteocytes are known to support osteoclastogenesis by expressing the receptor activator of NF-kappa-B ligand (RANKL; official symbol, Tnfsf11 - tumor necrosis factor (ligand) superfamily, member 11) and osteoprotegerin (OPG; official symbol, Tnfrsf11b - tumor necrosis factor receptor superfamily, member 11b), a stimulator and an inhibitor of osteoclastogenesis, respectively [25]–[28]. However, Gli1 +/− mice exhibited impaired bone formation and accelerated bone resorption, suggesting an uncoupling state of bone metabolism.
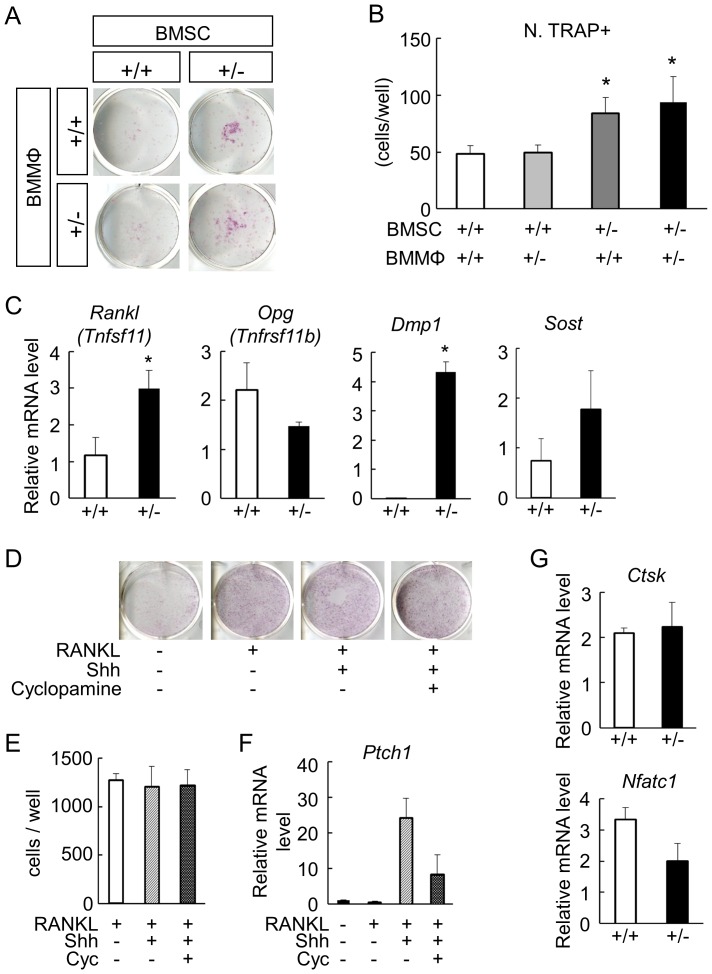
To identify the cellular mechanism underlying the aberrant state in Gli1 +/− bones, we conducted co-culture experiments using BMSC and bone marrow macrophages (BMMΦ) derived from either WT or Gli1 +/− mice. The number of TRAP-positive multinucleated osteoclasts co-cultured with Gli1 +/− BMMCs was significantly higher than those with WT BMMCs, regardless of the Gli1 genotypes in the BMMΦ (Figure 4A and B). Thus, bone-forming cells and their progenitors, not osteoclastic cells, were likely to be responsible for the abnormalities in osteoclastogenesis of the Gli1 +/− mice. This led us to analyze BMSCs in terms of their ability to support the osteoclastogenesis.
Figure 4. Osteoclast differentiation in cultures of bone marrow cells from WT and Gli1 +/− mice.
(A) Formation of TRAP-positive multinucleated osteoclasts by the co-culture of BMSCs and BMMΦ derived from WT or Gli1 +/− mice. (B) The numbers of osteoclasts expressed as means ± SDs of 4 wells per group in (A). *p<0.05 vs. the control group (WT BMSC × WT BMMΦ). (C) mRNA expression of Rankl, Opg, Dmp1, and Sost in 14-day osteogenic cultures of BMSCs. The mRNA expression was analyzed by real-time RT-PCR. Rankl, receptor activator of nuclear factor-κB ligand; Opg, osteoprotegerin; Dmp1, dentin matrix acidic phosphoprotein 1; Sost, sclerostin. *p<0.05 vs. WT. (D) Formation of TRAP-positive multinucleated osteoclasts in 5-day cultures of RAW cells in the presence or absence of sonic hedgehog (Shh) and cyclopamine (Cyc). (E) The numbers of osteoclasts in (D) expressed as means ± SDs of 4 wells per group. (F) mRNA expression of Ptch1 in cultured RAW cells in the presence or absence of Shh and cyclopamine (Cyc). Data are means ± SDs of 4 wells per group. (G) mRNA expression of cathepsin K (Ctsk) and NFATc1 in osteoclasts derived from WT or Gli1 +/− BMMΦ. Cells were cultured with recombinant M-CSF (10 ng/mL), RANKL (100 ng/mL), and Shh (25 nM). The mRNA expression was analyzed by real-time RT-PCR.
In BMSCs harvested from the femurs of 8-week-old Gli1 +/− mice, the mRNA expression of RANKL (Tnfsf11) was significantly upregulated, whereas that of OPG (Tnfrsf11b) showed a trend toward downregulation compared to WT BMSCs (Figure 4C). In addition, expression levels of dentin matrix acidic phosphoprotein 1 (Dmp1) and sclerostin (Sost), markers for osteocytes, were higher in the Gli1 +/− BMSCs than in the WT (Figure 4C). This trend was also observed in OPs (Figure S5). Taken together with the downregulation of osteoblast marker genes in Gli1 +/− BMSCs, these findings indicate that Gli1 haploinsufficiency may promote a premature differentiation of osteoblast precursors into osteocytes, which have been reported as a major source of RANKL [25], and these results may indicate that the premature differentiation not only affects bone formation, but also stimulates bone resorption by inducing RANKL expression at a supraphysiological level. Given that the recovery of Gli1 expression negated the upregulation of RANKL mRNA expression in Gli1 +/− BMSCs (Figure S6), it is also possible that Gli1 not only suppresses the differentiation of osteoblast precursors into osteocytes, but also negatively acts on the transcription of RANKL.
We next investigated the cell-autonomous effects of Hh signaling on osteoclastogenesis using RAW264.7 cells, which have been shown to differentiate into TRAP-positive osteoclasts in the presence of RANKL [20], [29]. Treatment with neither Shh nor cyclopamine, an inhibitor of hedgehog signaling, affected the RANKL-induced osteoclast differentiation of RAW264.7 cells (Figures 4D and E) although the cells were responsive to Hh signaling, as indicated by the expression change of Ptch1, a readout of the Hh signaling, upon Shh and cyclopamine (Figure 4F). Finally, we analyzed osteoclastogenesis of primary BMMΦ harvested from 8-week-old WT or Gli1 +/− mice. Expression levels of cathepsin K (Ctsk) and Nfatc1, markers for osteoclasts, were not affected by loss of Gli1 in cultures of BMMΦ with M-CSF and RANKL (Figure 4G).
Thus, the Hh-Gli1 axis is unlikely to mediate osteoclastogenesis in a cell-autonomous manner, which further suggests that the accelerated bone resorption in Gli1 +/− mice is not caused by defects in osteoclastic cells. Abnormalities in osteoblast precursors due to Gli1 haploinsufficiency may mediate all the aspects of the disruption of bone homeostasis in Gli1 +/− bones, which explains why Gli1 +/− mice have a low bone mass phenotype.
Fracture healing was impaired by Gli1 haploinsufficiency
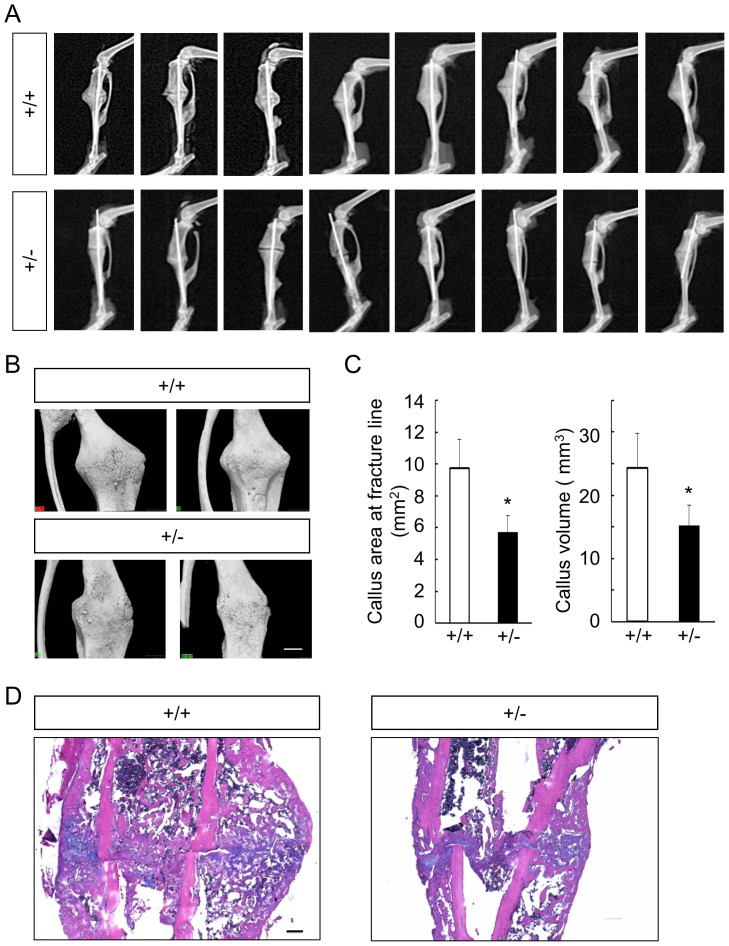
We next set out to examine the involvement of Gli1 in postnatal bones under a pathological condition, comparing fracture healing between WT and Gli1 +/− mice in a model that was surgically created in the tibias of 8-week-old males. During the fracture healing process, both intramembranous ossification and endochondral ossification are observed, and osteo-chondroprogenitor cells from the periosteum adjacent to fracture sites are major source of cells that contribute to the healing process [30], [31]. Fracture healing was evaluated 2 weeks after the surgery, given that bony bridging at the fracture site was typically observed as early as the point [30], [31]. In soft X-ray analyses, we observed impairment of callus formation and bone union in Gli1 +/− mice compared to WT mice, as well as the variability and reproducibility of the fracture model itself (Figure 5A). Using 3D-micro-CT analyses, we evaluated the areas of the calluses on horizontal cross-sections at the fracture lines and the volume of the callus. Both the areas and the volumes of the callus in the Gli1 +/− mice were significantly lower than those in WT mice (Figures 5B and C).
Figure 5. Comparison of bone fracture healing between 8-week-old WT and Gli1 +/− mice.
(A) Soft X-ray pictures of the fracture sites of all WT (n = 8) and Gli1 +/− (n = 8) male mice tested at 2 weeks after the fracture. (B) Representative micro-CT images of the callus in WT and Gli1 +/− male mice. Bar, 1 mm. (C) The areas of horizontal cross-sections at fracture lines (left) and the volume of the calluses (right) of WT and Gli1 +/− mice, calculated using 3D-micro-CT data. Data are means ± SDs of eight mice per genotype. *p<0.05 vs. WT. (D) H&E and alcian blue double staining of the calluses 2 weeks after the fracture. Bar, 200 µm.
We then performed histological analyses on sections stained with the hematoxylin-eosin (H–E) and alcian blue to identify differences in the healing process between WT and Gli1 +/− mice (Figure 5D). In WT mice, large volumes of soft callus surrounded by hard calluses were observed around the fracture sites, as previously reported [32] (Figure 5D). However, in Gli1 +/− mice, both the soft and hard calluses were reduced compared to those in the WT mice (Figure 5D), suggesting that both endochondral ossification and intramembranous ossification were affected by Gli1 haploinsufficiency during the fracture healing process.
Discussion
The present study had six major findings. (1) Adult bone mass was affected by Gli1 haploinsufficiency in mice, although body length and body weight were not. (2) The low bone mass phenotypes were accompanied by impaired bone formation and accelerated bone resorption, that is, an uncoupling of bone metabolism. (3) Gli1 haploinsufficiency had a negative impact on Hh-mediated osteoblast differentiation in cultures of precursors. (4) Despite the impairment of osteoblast differentiation, the expression levels of Dmp1and Rankl were upregulated in cultures of Gli1 +/− precursors, suggesting that Gli1 haploinsufficiency induced the premature differentiation of osteoblasts into osteocytes, which have a greater ability to promote osteoclastogenesis than do osteoblasts. (5) Hh-Gli1 was not involved in osteoclastogenesis in a cell-autonomous manner in vitro. (6) Gli1 haploinsufficiency affected fracture healing in adult mice. Based on these findings, we propose that Gli1, acting downstream of Hh signaling, not only promotes osteoblast differentiation but also acts as a repressor of osteoblast maturation toward osteocytes to maintain normal bone homeostasis in adult mice.
There are two possible mechanisms underlying the aberrant upregulation of osteocyte marker genes in Gli1 +/− precursors. The first possibility is that the overall differentiation of osteoblast precursors is accelerated by Gli1 haploinsufficiency, although the differentiation program is kept normal. The second possibility is that the program itself is disturbed by Gli1 haploinsufficiency, and the disturbance may induce the premature differentiation of precursors into osteocytes by skipping proper phases of osteoblast differentiation. The second possibility is more likely, because osteocyte marker genes (Dmp1 and Sost) were highly induced in Gli1 +/− cells, despite downregulation of both early and late osteoblast marker genes (Alp, Ibsp, Runx2, Sp7, and Bglap). If the first possibility was the case, Gli1 +/− precursors would show upregulation of both osteoblast and osteocyte marker genes.
It remains to be elucidated how Gli1 haploinsufficiency disturbs the osteoblast differentiation program, which results in accelerated bone resorption as well as decreased bone formation, i.e., an uncoupling of bone metabolism in development. Gli1, collectively with Gli2 and Gli3, is involved in the specification of osteo-chondroprogenitor cells in the perichondrium into an osteoblast lineage, and it promotes early osteoblast differentiation in a Runx2-independent manner [11]. The removal of Smo after the specification of Sp7-positive osteoblast precursors showed normal osteoblast development, suggesting that Hh signaling is not required for the differentiation of Sp7-positive osteoblast precursors into osteoblasts [33]. In contrast, the phenotypes of Gli1 +/− mice suggest that Gli1 acts to repress the maturation of osteoblasts into osteocytes in postnatal bones directly or indirectly via a negative feedback loop-like mechanism, in addition to its specifier function. In addition to the repressive effects of Gli1 on the terminal differentiation of osteoblasts, Gli1-mediated negative regulation of Rankl transcription may also explain the enhanced osteoclastogenesis in Gli1 +/− mice. An extensive search for Gli binding regions in the osteoblast genome will be useful for understanding the precise roles of Gli1 in bone metabolism.
Given that one copy of Gli1 allele was removed in all the cells of the mice used in the present study, cells other than skeletal lineages may also be involved in the osteoblast phenotypes seen in the mutants via systemic factors or direct contact with osteoblastic cells. Indeed, the body weights of the 8-week-old Gli1 −/− mice were about 20% less than those of the WT and Gli1 +/− mice, suggesting systemic abnormalities upon complete loss of Gli1. Stage- and tissue-specific manipulation of Gli1 using a floxed Gli1 allele would clarify the distinct function of Gli1 at different stages of osteoblast development, although such mutant mice are not yet available.
The involvement of Hh signaling in the regulation of bone metabolism has been debated. We previously found that Ptch1 +/− mice and patients with nevoid basal cell carcinoma syndrome, in which Hh signaling was activated by the loss of one copy of the Ptch1 allele, demonstrated high bone mass phenotypes [12], whereas Mak et al. reported that the disruption of the gene in Bglap-positive cells caused low bone mass [13]. At cellular levels, a common phenomenon underlies the contradictory phenotypes. Both osteoblastogenesis and osteoclastogenesis were enhanced in both mutants. Therefore, those different outcomes might be due to an alteration of the balance between enhanced bone formation and bone resorption. These studies and the present investigation have provided consistent evidence of the direct promotion of osteoblastogenesis by Hh signaling in adult mice.
In contrast, there are some arguments with respect to the involvement of Hh signaling in osteoclastogenesis. The studies mentioned above [12], [13] and the present one support the concept that Hh signaling stimulates osteoclastogenesis indirectly via the augmentation of osteoblasts or osteocytes. Conversely, Heller et al. described that the inhibition of Smo suppressed osteoclastogenesis in a cell-autonomous manner [34]. The reasons for these conflicting findings should be elucidated with regard to cell type and culture conditions. Joeng et al. recently reported postnatal skeletal phenotypes in mice expressing a constitutively active form of Gli2 in Sp7-positive cells. The mutants showed osteopenia with decreased bone formation and unaltered bone resorption, although osteoblast differentiation was enhanced in cultures of precursor cells isolated from the mutants [14]. Although Gli1 is thought to be upregulated in the mutants as Ptch1 upregulation was confirmed [14], their skeletal phenotypes are seemingly inconsistent with those in the Gli1 +/− mice. The discrepancy may be caused by the difference in populations in which Hh signaling was manipulated or the difference in Gli factors that were manipulated between the studies. Overall, these results suggest that as Joeng et al. mentioned [14], the roles of Hh signaling in bone metabolism depend on its target population, the timing of its activation, and complex regulation by Gli factors downstream of the signaling; the roles are possibly modulated by non-skeletal cells or factors.
We demonstrated roles of Gli1 in the fracture healing process as well as in mouse adult bone homeostasis. A previous report on defects of bone healing in Smo-deleted mice [35] supports the impairment of callus formation in the Gli1 +/− mice although the tested models are different between that study and our present investigation. Given that cartilage development was not largely affected by the complete loss of Gli1 [11], the reduced size of soft calluses during fracture healing of the Gli1 +/− mice was unexpected. In development, Gli3 repressor, rather than Gli activators, was shown to play a major role in the Hh-mediated control of cartilage formation [8]. Thus, it is possible that the involvement of Gli1 in chondrogenesis and/or cartilage metabolism is different between embryos and adults or physiological conditions and pathological ones. Hh signaling may require a contribution from the Gli activators during fracture healing, where the rapid growth of cartilaginous tissues is likely to depend more on cellular proliferation than on increases in cellular volume or matrix deposition [32]. The promotion of proliferation is known to be a direct action of Hh signaling on chondrocytes [36].
When discussing the regulation of the adult skeleton by Hh-Gli signaling under physiological and pathological conditions, we should consider target cell types and sources of Hh ligands. Maeda et al. demonstrated that Ihh secreted from chondrocytes in the growth plate is required for the maintenance of trabecular bones in postnatal mice [37]. Type I collagen-positive cells in bone lining were shown to express Ihh in humans [38]. Both Ihh expressed in soft calluses [39] and sonic hedgehog (Shh) expressed in the periosteum [40] or osteoblasts/osteocytes [41] have been implicated in fracture healing. These findings imply that Gli1 is likely to contribute to adult bone metabolism upon the inputs of Ihh and Shh. In addition, given that Hh signaling acts to maintain the skeleton in adults, and some species close the growth plate after puberty, skeletal components other than growth plate chondrocytes may produce Hh ligands in this context.
In conclusion, the results of the present study lead us to propose the involvement of Gli1 in postnatal bone homeostasis under physiological and pathological conditions although further studies are necessary to obtain an integrative understanding of the roles of all Gli family members in this context. Collectively with previous studies, the present study also indicates the importance of tissue- and stage-specific manipulation of Hh signaling for the treatment of bone-related disease, as well as the need for a greater understanding of all the actions of Hh signaling on the skeletal tissue throughout life.
Supporting Information
Gross appearance of WT and Gli1 mutant male mice. (A) Gross appearance of WT, Gli1 +/−, and Gli1 −/− male mice at 8 weeks of age. (B) Comparison of postnatal growth between WT, Gli1 +/−, and Gli1 −/− male mice. Body weight was measured on the indicated dates after birth. *p<0.05 vs. WT or Gli1 +/−.
(PDF)
Radiological analyses of long bones in female WT and Gli1 +/− mice. (A) 3D-micro-CT images of the distal femurs of representative 8-week-old WT and Gli1 +/− female mice. Sagittal sections, transverse sections, and 3D reconstruction images of the primary spongiosa are shown for each genotype. Bar, 1 mm. (B) Histomorphometric analyses of the 3D-micro-CT data in (A). BMD, bone mineral density; BV/TV, bone volume per tissue volume; Tb.Th, trabecular thickness; Tb.N, trabecular number parameters. Data are means ± SDs of five female mice per genotype. *p<0.05 vs. WT.
(PDF)
Radiological analyses of cortical bones in WT and Gli1 +/− mice. (A) 3D-micro-CT images of distal femurs of representative 8-week-old WT and Gli1 +/− male mice. Transverse sections of the primary spongiosa are shown for each genotype. Bar, 500 µm. (B) Histomorphometric analyses of 3D-micro-CT data in (A). Cv/Av, cortical bone volume per all bone volume; Cvt, cortical bone thickness; BMD, bone mineral density. Data are means ± SDs of ten male mice per genotype.
(PDF)
Construction of adenoviral vector expressing GLI1 . (A) Schematic representation of pAd/PL-DEST vectors expressing GLI1. (B) Luciferase reporter assay using the 8×3′-Gli BS-luc in combination with dsRed, Myc-GLI1, and the constructed adenoviral vector (GLI1-IRES-deRed). The luciferase assay was performed 48 hours after transfection in C3H10T1/2 cells. (C) Protein expression of GLI1 in C3H10T1/2 cells transfected with Myc-GLI1 or adenovirally transduced with GLI1-IRES-dsRed. (D) mRNA expression of Alp and Ibsp in C3H10T1/2 cells transfected with Myc-GLI1 or adenovirally transduced with GLI1-IRES-dsRed.
(PDF)
mRNA expression of Rankl , Opg , Dmp1 , and Sos t in 7-day osteogenic cultures of OPs. The mRNA expression was analyzed by real-time RT-PCR. Rankl, receptor activator of nuclear factor-κB ligand; Opg, osteoprotegerin; Dmp1, dentin matrix acidic phosphoprotein 1; Sost, sclerostin. *p<0.05 vs. WT.
(PDF)
Suppression of Rankl expression in response to the recovery of Gli1 expression in Gli1 +/− BMSCs. (A) Scheme of the experiment. Gli1+/− BMSCs were cultured in osteogenic media supplemented with Smoothened agonist (SAG). Cells were infected with either Ax-GFP (control) or Ax-GLI1-IRES-dsRed on Day 4 and cultured for another 7 days. (B) mRNA expression of Rankl on days 0, 4, and 11. The mRNA expression was analyzed by real-time RT-PCR analyses. *p<0.05 vs. Ax-GFP.
(PDF)
Acknowledgments
We thank Drs. Alexandra L. Joyner, Chi-chung Hui, Gen Yamada, Andrew P. McMahon, and Hiroshi Sasaki for providing experimental materials. We are also grateful to Katsue Morii, Harumi Kobayashi, and Nozomi Nagumo for providing technical assistance.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a Grant-in-Aid for Young Scientists (#23689079), the Center for Medical System Innovation, the Graduate Program for Leaders in Life Innovation, Core-to-Core Program A (Advanced Research Networks), the Funding Program for World-Leading Innovative R&D on Science and Technology, the Center for NanoBio Integration, and the S-innovation Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kronenberg HM (2003) Developmental regulation of the growth plate. Nature 423: 332–336. [DOI] [PubMed] [Google Scholar]
- 2. St-Jacques B, Hammerschmidt M, McMahon AP (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13: 2072–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chung UI, Schipani E, McMahon AP, Kronenberg HM (2001) Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J Clin Invest 107: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, et al. (2004) Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131: 1309–1318. [DOI] [PubMed] [Google Scholar]
- 5. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- 6. Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, et al. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771. [DOI] [PubMed] [Google Scholar]
- 7. Ingham PW, McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- 8. Hilton MJ, Tu X, Cook J, Hu H, Long F (2005) Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 132: 4339–4351. [DOI] [PubMed] [Google Scholar]
- 9. Joeng KS, Long FX (2009) The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development 136: 4177–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hojo H, Ohba S, Taniguchi K, Shirai M, Yano F, et al. (2013) Hedgehog-Gli Activators Direct Osteo-chondrogenic Function of Bone Morphogenetic Protein toward Osteogenesis in the Perichondrium. J Biol Chem 288: 9924–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hojo H, Ohba S, Yano F, Saito T, Ikeda T, et al. (2012) Gli1 Protein Participates in Hedgehog-mediated Specification of Osteoblast Lineage during Endochondral Ossification. Journal of Biological Chemistry 287: 17860–17869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohba S, Kawaguchi H, Kugimiya F, Ogasawara T, Kawamura N, et al. (2008) Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev Cell 14: 689–699. [DOI] [PubMed] [Google Scholar]
- 13. Mak KK, Bi Y, Wan C, Chuang PT, Clemens T, et al. (2008) Hedgehog signaling in mature osteoblasts regulates bone formation and resorption by controlling PTHrP and RANKL expression. Dev Cell 14: 674–688. [DOI] [PubMed] [Google Scholar]
- 14. Joeng KS, Long F (2013) Constitutive activation of Gli2 impairs bone formation in postnatal growing mice. PLoS ONE 8: e55134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park HL, Bai C, Platt KA, Matise MP, Beeghly A, et al. (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127: 1593–1605. [DOI] [PubMed] [Google Scholar]
- 16. Zhang X, Peterson KA, Liu XS, McMahon AP, Ohba S (2013) Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells 31: 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, et al. (2006) The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell 10: 647–656. [DOI] [PubMed] [Google Scholar]
- 18. Ogata N, Chikazu D, Kubota N, Terauchi Y, Tobe K, et al. (2000) Insulin receptor substrate-1 in osteoblast is indispensable for maintaining bone turnover. J Clin Invest 105: 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akune T, Ogata N, Hoshi K, Kubota N, Terauchi Y, et al. (2002) Insulin receptor substrate-2 maintains predominance of anabolic function over catabolic function of osteoblasts. J Cell Biol 159: 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogasawara T, Katagiri M, Yamamoto A, Hoshi K, Takato T, et al. (2004) Osteoclast differentiation by RANKL requires NF-kappaB-mediated downregulation of cyclin-dependent kinase 6 (Cdk6). J Bone Miner Res 19: 1128–1136. [DOI] [PubMed] [Google Scholar]
- 21. Ohba S, Ikeda T, Kugimiya F, Yano F, Lichtler AC, et al. (2007) Identification of a potent combination of osteogenic genes for bone regeneration using embryonic stem (ES) cell-based sensor. Faseb J 21: 1777–1787. [DOI] [PubMed] [Google Scholar]
- 22. Sasaki H, Hui C, Nakafuku M, Kondoh H (1997) A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124: 1313–1322. [DOI] [PubMed] [Google Scholar]
- 23. Ogasawara T, Kawaguchi H, Jinno S, Hoshi K, Itaka K, et al. (2004) Bone morphogenetic protein 2-induced osteoblast differentiation requires Smad-mediated down-regulation of Cdk6. Mol Cell Biol 24: 6560–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kugimiya F, Kawaguchi H, Kamekura S, Chikuda H, Ohba S, et al. (2005) Involvement of endogenous bone morphogenetic protein (BMP) 2 and BMP6 in bone formation. J Biol Chem 280: 35704–35712. [DOI] [PubMed] [Google Scholar]
- 25. Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, et al. (2011) Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat Med 17: 1231–1234. [DOI] [PubMed] [Google Scholar]
- 26. Takahashi N, Akatsu T, Udagawa N, Sasaki T, Yamaguchi A, et al. (1988) Osteoblastic cells are involved in osteoclast formation. Endocrinology 123: 2600–2602. [DOI] [PubMed] [Google Scholar]
- 27. Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, et al. (1999) Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev 20: 345–357. [DOI] [PubMed] [Google Scholar]
- 28. Udagawa N, Takahashi N, Yasuda H, Mizuno A, Itoh K, et al. (2000) Osteoprotegerin produced by osteoblasts is an important regulator in osteoclast development and function. Endocrinology 141: 3478–3484. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto A, Miyazaki T, Kadono Y, Takayanagi H, Miura T, et al. (2002) Possible involvement of IkappaB kinase 2 and MKK7 in osteoclastogenesis induced by receptor activator of nuclear factor kappaB ligand. J Bone Miner Res 17: 612–621. [DOI] [PubMed] [Google Scholar]
- 30. Shimoaka T, Kamekura S, Chikuda H, Hoshi K, Chung UI, et al. (2004) Impairment of bone healing by insulin receptor substrate-1 deficiency. J Biol Chem 279: 15314–15322. [DOI] [PubMed] [Google Scholar]
- 31. Murao H, Yamamoto K, Matsuda S, Akiyama H (2013) Periosteal cells are a major source of soft callus in bone fracture. J Bone Miner Metab 31: 390–398. [DOI] [PubMed] [Google Scholar]
- 32. Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA (2003) Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J Cell Biochem 88: 873–884. [DOI] [PubMed] [Google Scholar]
- 33. Rodda SJ, McMahon AP (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133: 3231–3244. [DOI] [PubMed] [Google Scholar]
- 34. Heller E, Hurchla MA, Xiang J, Su X, Chen S, et al. (2012) Hedgehog signaling inhibition blocks growth of resistant tumors through effects on tumor microenvironment. Cancer Res 72: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Q, Huang C, Zeng F, Xue M, Zhang X (2010) Activation of the Hh pathway in periosteum-derived mesenchymal stem cells induces bone formation in vivo: implication for postnatal bone repair. Am J Pathol 177: 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Long F, Zhang XM, Karp S, Yang Y, McMahon AP (2001) Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128: 5099–5108. [DOI] [PubMed] [Google Scholar]
- 37. Maeda Y, Nakamura E, Nguyen MT, Suva LJ, Swain FL, et al. (2007) Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proc Natl Acad Sci U S A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakase T, Miyaji T, Kuriyama K, Tamai N, Horiki M, et al. (2001) Immunohistochemical detection of parathyroid hormone-related peptide, Indian hedgehog, and patched in the process of endochondral ossification in the human. Histochem Cell Biol 116: 277–284. [DOI] [PubMed] [Google Scholar]
- 39. Vortkamp A, Pathi S, Peretti GM, Caruso EM, Zaleske DJ, et al. (1998) Recapitulation of signals regulating embryonic bone formation during postnatal growth and in fracture repair. Mech Dev 71: 65–76. [DOI] [PubMed] [Google Scholar]
- 40. Miyaji T, Nakase T, Iwasaki M, Kuriyama K, Tamai N, et al. (2003) Expression and distribution of transcripts for sonic hedgehog in the early phase of fracture repair. Histochem Cell Biol 119: 233–237. [DOI] [PubMed] [Google Scholar]
- 41. Horikiri Y, Shimo T, Kurio N, Okui T, Matsumoto K, et al. (2013) Sonic hedgehog regulates osteoblast function by focal adhesion kinase signaling in the process of fracture healing. PLoS One 8: e76785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gross appearance of WT and Gli1 mutant male mice. (A) Gross appearance of WT, Gli1 +/−, and Gli1 −/− male mice at 8 weeks of age. (B) Comparison of postnatal growth between WT, Gli1 +/−, and Gli1 −/− male mice. Body weight was measured on the indicated dates after birth. *p<0.05 vs. WT or Gli1 +/−.
(PDF)
Radiological analyses of long bones in female WT and Gli1 +/− mice. (A) 3D-micro-CT images of the distal femurs of representative 8-week-old WT and Gli1 +/− female mice. Sagittal sections, transverse sections, and 3D reconstruction images of the primary spongiosa are shown for each genotype. Bar, 1 mm. (B) Histomorphometric analyses of the 3D-micro-CT data in (A). BMD, bone mineral density; BV/TV, bone volume per tissue volume; Tb.Th, trabecular thickness; Tb.N, trabecular number parameters. Data are means ± SDs of five female mice per genotype. *p<0.05 vs. WT.
(PDF)
Radiological analyses of cortical bones in WT and Gli1 +/− mice. (A) 3D-micro-CT images of distal femurs of representative 8-week-old WT and Gli1 +/− male mice. Transverse sections of the primary spongiosa are shown for each genotype. Bar, 500 µm. (B) Histomorphometric analyses of 3D-micro-CT data in (A). Cv/Av, cortical bone volume per all bone volume; Cvt, cortical bone thickness; BMD, bone mineral density. Data are means ± SDs of ten male mice per genotype.
(PDF)
Construction of adenoviral vector expressing GLI1 . (A) Schematic representation of pAd/PL-DEST vectors expressing GLI1. (B) Luciferase reporter assay using the 8×3′-Gli BS-luc in combination with dsRed, Myc-GLI1, and the constructed adenoviral vector (GLI1-IRES-deRed). The luciferase assay was performed 48 hours after transfection in C3H10T1/2 cells. (C) Protein expression of GLI1 in C3H10T1/2 cells transfected with Myc-GLI1 or adenovirally transduced with GLI1-IRES-dsRed. (D) mRNA expression of Alp and Ibsp in C3H10T1/2 cells transfected with Myc-GLI1 or adenovirally transduced with GLI1-IRES-dsRed.
(PDF)
mRNA expression of Rankl , Opg , Dmp1 , and Sos t in 7-day osteogenic cultures of OPs. The mRNA expression was analyzed by real-time RT-PCR. Rankl, receptor activator of nuclear factor-κB ligand; Opg, osteoprotegerin; Dmp1, dentin matrix acidic phosphoprotein 1; Sost, sclerostin. *p<0.05 vs. WT.
(PDF)
Suppression of Rankl expression in response to the recovery of Gli1 expression in Gli1 +/− BMSCs. (A) Scheme of the experiment. Gli1+/− BMSCs were cultured in osteogenic media supplemented with Smoothened agonist (SAG). Cells were infected with either Ax-GFP (control) or Ax-GLI1-IRES-dsRed on Day 4 and cultured for another 7 days. (B) mRNA expression of Rankl on days 0, 4, and 11. The mRNA expression was analyzed by real-time RT-PCR analyses. *p<0.05 vs. Ax-GFP.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.