Summary
Hedgehog (Hh) signaling during development and in postembryonic tissues requires activation of the 7TM oncoprotein Smoothened (Smo), by mechanisms that may involve endogenous lipidic modulators. Exogenous Smo ligands previously identified include the plant sterol cyclopamine (and its therapeutically useful synthetic mimics) and hydroxylated cholesterol derivatives (oxysterols); Smo is also highly sensitive to cellular sterol levels. The relationships between these effects are unclear because the relevant Smo structural determinants are unknown. We identify the conserved extracellular cysteine rich domain (CRD) as the site of action for oxysterols on Smo, involving residues structurally analogous to those contacting the Wnt lipid adduct in the homologous Frizzled CRD; this modulatory effect is distinct from that of cyclopamine mimics, from Hh-mediated regulation, and from the permissive action of cellular sterol pools. These results imply that Hh pathway activity is sensitive to lipid binding at several Smo sites, suggesting mechanisms for tuning by multiple physiological inputs.
Introduction
The Hedgehog (Hh) signal transduction pathway controls numerous aspects of embryonic patterning and regeneration of postembryonic tissues (Beachy et al., 2004; Varjosalo and Taipale, 2008). Insufficient Hh signaling can result in a range of birth defects including holoprosencephaly (Beachy et al., 2004; Muenke and Beachy, 2000; Varjosalo and Taipale, 2008), while inappropriate pathway activity is linked to human malignancies such as basal cell carcinoma and medulloblastoma (Teglund and Toftgård, 2010). Despite intensive study, the mechanisms that ensure proper reception and transduction of the Hh signal remain largely mysterious (Lum and Beachy, 2004; Rohatgi and Scott, 2007). Pathway activation is initiated by binding of the cholesterol-modified Hh morphogen (Mann and Beachy, 2004) to its receptor, the transporter-like tumor suppressor Patched1 (Ptch1). Hh-induced inactivation of Ptch1 releases the inhibition of Smoothened (Smo), a seven-transmembrane (7TM) oncoprotein with extended extracellular and cytoplasmic termini (Lum and Beachy, 2004; Rohatgi and Scott, 2007). Activated Smo then accumulates in the vertebrate primary cilium, ultimately triggering transcription of Hh target genes via activation of Gli transcription factors (Lum and Beachy, 2004; Rohatgi et al., 2007).
A key question in the study of Hh signal transduction concerns the physiological mechanisms that influence Smo activity in pathway-responsive cells. One leading model hypothesizes that Smo is regulated by an endogenous lipidic modulator whose availability is controlled by Ptch1 transport activity (Eaton, 2008; Hausmann et al., 2009; Taipale et al., 2002). Indeed, a remarkable array of exogenous small molecules impinges on vertebrate Smo, including the plant-derived sterol cyclopamine and its synthetic mimics (Chen et al., 2002a; 2002b), which are finding therapeutic uses as pathway antagonists (Hoff et al., 2009; Rudin et al., 2009; Tang et al., 2012; Teglund and Toftgård, 2010). Smo can also be activated by non-cellular hydroxylated cholesterol derivatives such as 20(S)-hydroxycholesterol (20(S)-OHC) (Corcoran and Scott, 2006; Dwyer et al., 2007; Nachtergaele et al., 2012). Furthermore, Smo-mediated signal transduction is highly sensitive to genetic or pharmacological depletion of endogenous cellular sterols (Cooper et al., 2003; Corcoran and Scott, 2006) (Fig. 1A). It is tempting to postulate that all of these sterols act on Smo via shared structural mechanisms that might also be employed by endogenous Ptch-regulated ligands. However, the mechanisms of small molecule action and the relationship of these mechanisms to the modulatory effects of Ptch1 activity remain unclear, and in most cases the relevant Smo structural determinants have not been precisely delineated.
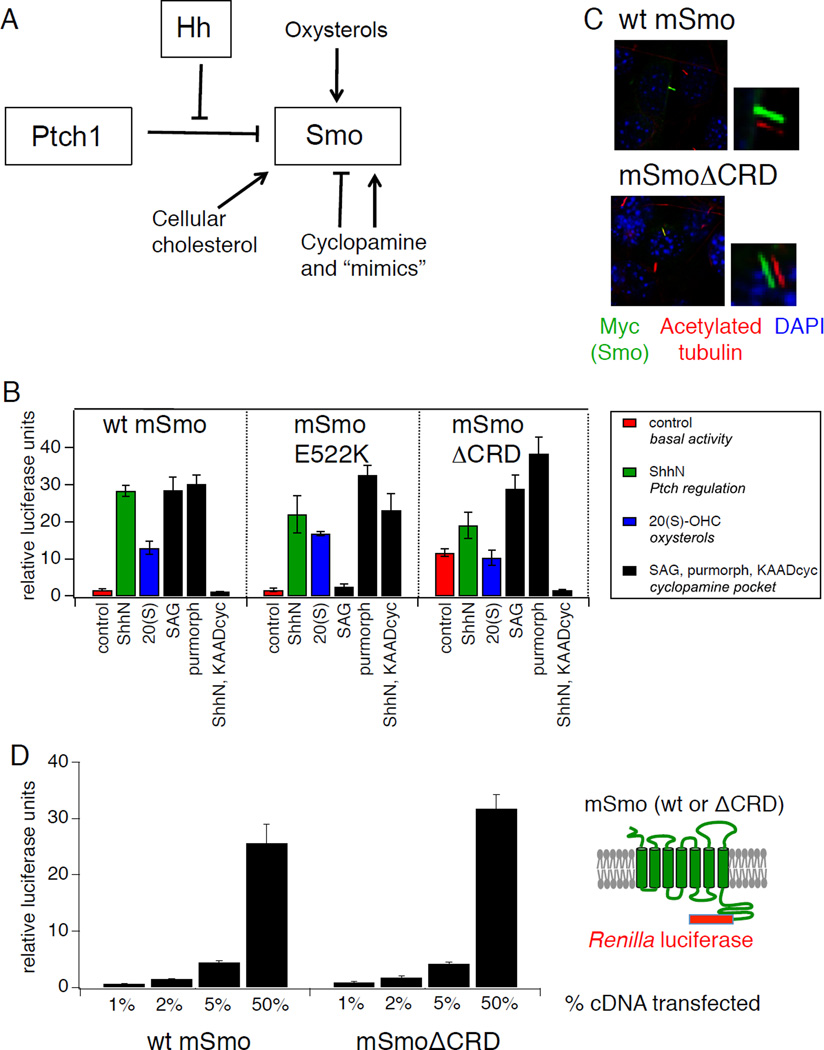
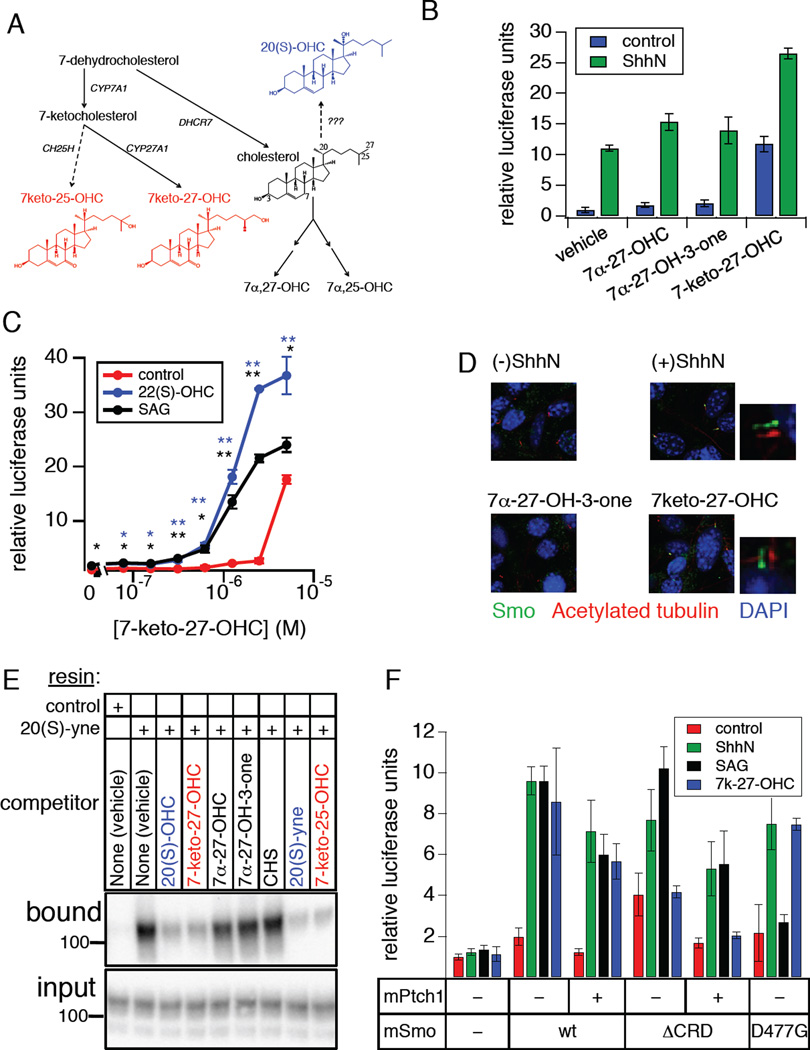
Figure 1.
Distinct structural determinants required for regulation of Smo by Ptch and by endogenous and exogenous ligands. (A) Schematic diagram of Hh pathway components showing multiple regulatory mechanisms that impinge on Smo, including Hh antagonism of Ptch-mediated suppression, direct modulation by cyclopamine and related small molecule agonists and antagonists, activation by oxysterols, and a requirement for cholesterol. (B) Luciferase activity in Smo−/− mouse embryonic fibroblasts (MEFs) cotransfected with Gli-luciferase reporter plasmids and cDNAs encoding wild-type (wt) mSmo or the E522K or ΔCRD mutants following treatment with: control or ShhN conditioned medium (red and green bars, respectively); 20(S)-OHC (10 µM, blue bars); SAG (200 nM), purmorphamine (2.5 µM), or ShhN conditioned medium with KAAD-cyclopamine (KAADcyc, 300 nM) (black bars). Error bars represent standard deviations (n = 3 independent transfections per data point). A summary of the experiment is shown; complete data set is presented in Fig. S1A. (C) Ciliary localization of Myc-tagged wt or ΔCRD Smo constructs was examined in fixed NIH3T3 cells stained with antibodies against acetylated tubulin (red) or the Myc epitope (green); nuclei were visualized with DAPI (blue). Small panels to the right of selected images display shifted overlays of the acetylated tubulin and Myc channels. See Fig. S1B for quantification. (D) Quantification of wt Smo and Smo ΔCRD protein in transfected Smo−/− MEFs over a range of expression levels using Cterminal fusions to Renilla luciferase. X-axis indicates % of Smo cDNA transfected into each well, while Y-axis indicates Renilla measurement, normalized to a control secreted alkaline phosphatase (SEAP) construct; replicates and error bars are as in panel (B). See also Figure S1 and Table S1.
Although the involvement of heterotrimeric G proteins in Smo-mediated transduction is debated (Ayers and Thérond, 2010), Smo and the related group of Frizzled (Fz) Wnt receptors clearly constitute a GPCR-like subfamily within the larger group of 7TM proteins. A recent crystal structure of the human Smo heptahelical domain in complex with a cyclopamine mimic confirmed its seven-pass topology and revealed a ligand binding site enclosed by a cage of transmembrane helices and the interior surfaces of extracellular loops, and connected to the aqueous extracellular environment via a narrow tunnel (Wang et al., 2013). This membrane-bounded pocket, a canonical location for ligand-binding in GPCRs (Lagerström and Schiöth, 2008; Venkatakrishnan et al., 2013)., was initially defined in Smo by studies with cyclopamine (Chen et al., 2002a; 2002b) and is the apparent site of action for pathway agonists or antagonists that compete for cyclopamine binding to Smo (cyclopamine mimics). Apart from the presence of a canonical 7TM domain, several structural features distinguish Smo and Fz from the GPCRs typically found in physiologic signaling systems and, despite numerous apparent differences between the modes of ligand reception and intracellular signal transduction in their respective developmental pathways, the existence of common Smo and Fz structural elements implies that some aspects of their activation mechanisms may be fundamentally related. One hallmark of the Smo and Fz subfamily is the N-terminal cysteine rich domain (CRD), a three-helix bundle stabilized by five stereotyped disulfide bonds, which is biochemically, crystallographically, and genetically established in Fz receptors as its site of interaction with the diffusible Wnt ligand (Bhanot et al., 1996; Cadigan et al., 1998; Janda et al., 2012; Povelones and Nusse, 2005). The Smo CRD is also highly conserved (Ayers and Thérond, 2010; Huangfu and Anderson, 2006; Ingham et al., 2011), and missense mutations in this domain reduce Hh signaling during development (Aanstad et al., 2009; Chen and Struhl, 1998; Nakano et al., 2004). In contrast to Fz, however, Smo plays no known role as a site for morphogen reception, as this function is performed in the Hh pathway by Ptch1 and co-receptors (Beachy et al., 2010). Thus the biochemical function of the Smo CRD remains unknown and any potential connection with the corresponding domain in Fz remains unexplored.
In this study we identify the Smo CRD as the major binding site for action of several oxysterols, including 20(S)-OHC and two newly identified cellular modulators, 7-keto-25- and 7-keto-27-hydroxycholesterol (7-keto-25-OHC, 7-keto-27-OHC). Oxysterol binding to Smo involves residues structurally analogous to those contacting the Wnt lipid adduct in the homologous Fz CRD (Janda et al., 2012). Oxysterol activation furthermore is distinct from modulation by cyclopamine or its mimics, from Ptch1-mediated regulation, and from the permissive action of endogenous sterol pools in Hh-responsive cells. These results suggest that Smo activation involves a conformational rearrangement modulated by lipidic molecules acting at multiple distinct sites within the extracellular and heptahelical domains, and further suggest the possibility that Smo activity is tunable by multiple physiological inputs.
An intact CRD is required for Smo activation by oxysterols
Oxysterol activation of EBI2/GPR183 (Epstein Barr Virus-induced Molecule 2), a class A GPCR (G-protein coupled receptor), is triggered in immune cells by binding of 7α-25- or 7α-27- hydroxycholesterol (7α-25-OHC, 7α-27-OHC) to a pocket located within the external half of the membrane-spanning heptahelical bundle (Benned-Jensen et al., 2012; Zhang et al., 2012). A similar ligand-binding pocket exists in Smo, and mutations in this region disrupt the binding of cyclopamine, of its synthetic mimics, and of certain agonists (Buonamici et al., 2010; Dijkgraaf et al., 2011). We examined the pharmacological properties of various Smo sequence alterations using a transcriptional reporter assay of Hh pathway activation in Smo−/− mouse embryonic fibroblasts; in these cells, Smo-dependent pathway activation depends entirely on Smo exogenously introduced by transfection ((Varjosalo et al., 2006); see also Fig. S1A), permitting experimental analysis of Smo structure-function relationships. We found that responsiveness to 20(S)-OHC either alone or in synergistic combination with 22(S)-OHC was not affected by “cyclopamine pocket” mutations D477G and E522K, previously shown to block the effects of one or more cyclopamine-competitive Smo agonists and antagonists (Fig. 1B; Fig. S1A). These results suggest that the heptahelical bundle might not contain the binding site for modulatory oxysterols, consistent with earlier studies showing that oxysterols and cyclopamine do not compete with each other for binding to Smo (Dwyer et al., 2007; Nachtergaele et al., 2012).
Bioinformatic and structural modeling studies have revealed an unexpected parallel between the N-terminal cysteine-rich domain (CRD) of Fz receptors and the cholesterol- or riboflavin-binding modules of Niemann-Pick C1 and RBP proteins, respectively (Bazan and de Sauvage, 2009), suggesting that binding of small lipidic molecules may be a property of some CRD folds. We therefore tested whether Smo oxysterol responsiveness was altered upon deletion of the CRD (SmoΔCRD). We found that SmoΔCRD exhibited significant constitutive activity relative to wild-type Smo or the “cyclopamine pocket” mutants. (Fig. 1B, Fig. S1A). However, the resulting SmoΔCRD basal activity was wholly resistant to induction by 20(S)-OHC and partially resistant to induction by ShhN (Fig. 1B). Despite loss of oxysterol modulation, SmoΔCRD remained fully responsive to agonists (SAG and purmorphamine) and antagonists (KAAD-cyclopamine and GDC-0449) that bind within the transmembrane domain (Fig. 1B; Fig. S1A). SmoΔCRD thus displays a selective defect in stimulation by ShhN or oxysterols that is nearly opposite to the pharmacological profile of the D477G and E522K mutants. We noted no significant differences between wild type (wt) and SmoΔCRD in their accumulation in primary cilia upon transient overexpression in NIH3T3 cells (Fig. 1C; Fig. S1B), consistent with previous measurements of the ciliary localization of CRD-deleted forms of Smo in cell lines or in embryos (Aanstad et al., 2009; Dorn et al., 2012). In quantitative enzyme fusion-based titration assays of Smo expression, we noted no dramatic effects of CRD deletion on expression level; while a modest (~30%) increase in SmoΔCRD protein was detected at the lowest level of plasmid transfection relative to its wild type counterpart, this effect was not consistent across a range of transfection ratios and is not sufficient to account for the dramatic functional effects of CRD deletion on basal- or oxysterol-induced activity (Fig. 1D, Table S1). The functional defect of SmoΔCRD in response to ShhN or oxysterol stimulation thus is not due to alterations in Smo protein levels, defects in ciliary entry, nor in the ability of SmoΔCRD to engage downstream transduction machinery.
The CRD is required for oxysterol stimulation but exerts indirect effects on basal and Hhinduced Smo activity states
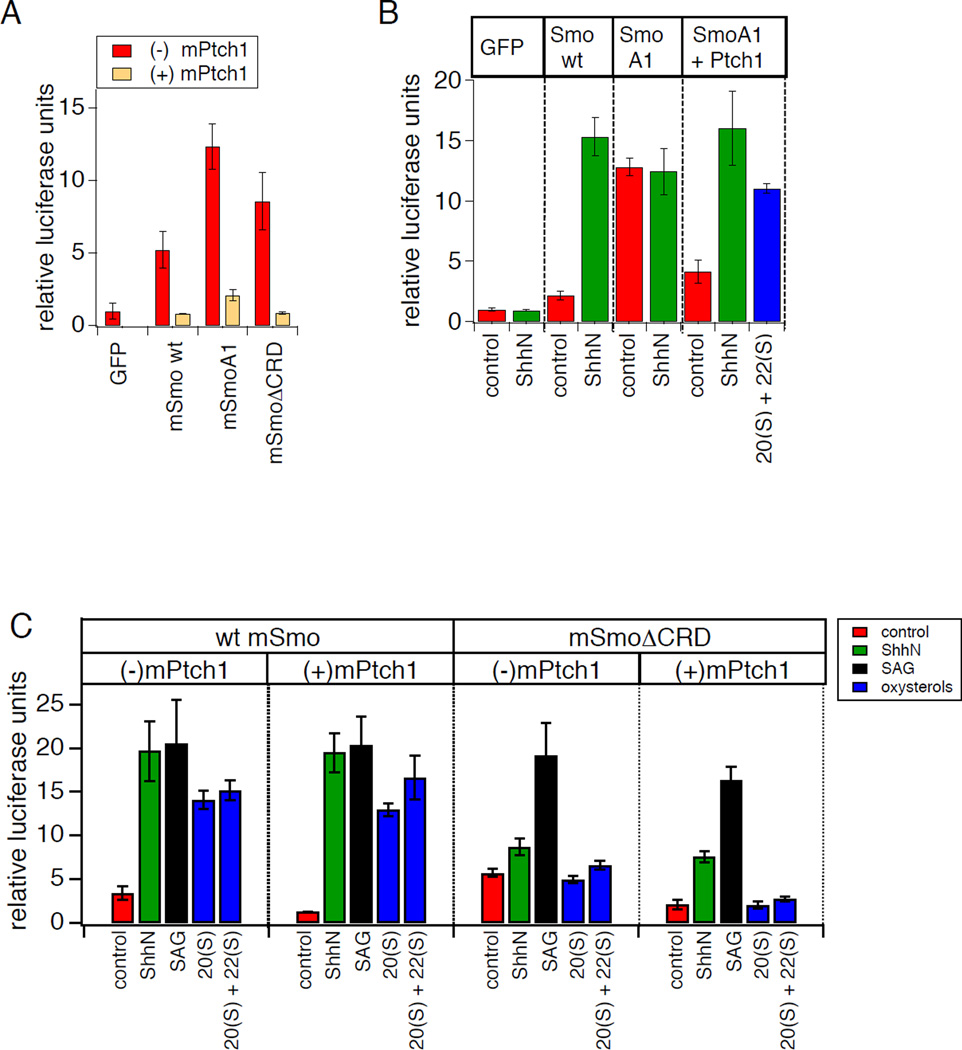
The phenotype of SmoΔCRD raises the possibility that an intact CRD is critical for modulation by oxysterols as well as by Hh-mediated Ptch1 inactivation. However, comparison between wt and CRD-deleted Smo is complicated by the latter’s abnormally high basal activity, which could in principle mask the activating effects of ShhN or oxysterols. To more conclusively test whether the CRD mediates the regulatory action of Ptch on Smo, as suggested by the partial loss of SmoΔCRD response to ShhN stimulation, we examined the effect of elevated levels of Ptch1 on basal and Hh-induced activation of this Smo deletion mutant. Ptch1 over-expression has previously been shown to reduce the basal activity of wt Smo or of the fully constitutively active oncogenic mutant SmoA1 (corresponding to the SmoM2 allele in human basal cell carcinoma) (Taipale et al., 2002; 2000). In each of these cases, Smo constitutive activity is strongly suppressed by substoichiometric amounts of Ptch1, and Smo activity is restored by treatment with Hh protein (Taipale et al., 2002), illustrating that the underlying inhibitory effects are dependent on Ptch1 activity rather than nonspecific consequences of protein over-expression. An insensitivity of SmoΔCRD basal activity to Ptch1 co-expression would be consistent with a direct role for the CRD in mediating Ptch1 effects on Smo. In contrast, if the basal activity of SmoΔCRD diminishes upon Ptch coexpression, then the Ptch1 regulatory mechanism must be intact in this mutant and any observed reduction in ShhN-induced activation is indirect, likely allosteric in nature. We observed in transfected Smo−/− MEFs that like wt Smo or SmoA1, the basal activity of SmoΔCRD was potently suppressed by Ptch1 coexpression, and its activity was restored in the presence of ShhN or SAG (Fig. 2B,C). To confirm this result in an independent cell line, we tested whether Ptch1 coexpression could reduce the basal activity of SmoΔCRD transiently expressed in NIH3T3 cells; although these cells express endogenous wt Smo, their low basal Hh pathway activity in transcriptional reporter assays permits measurements of constitutive activity in cDNA transfection experiments (Taipale et al., 2000; 2002). We indeed observed suppression of SmoΔCRD basal activity in NIH3T3 cells (Fig. 2A), similar to our results in Smo−/− MEFs. Thus, our Ptch1 coexpression studies show that although CRD deletion compromises full activation of Smo upon Hh-mediated release of Ptch1 inhibition, this domain is not the major site on Smo for Hh-sensitive regulation by Ptch1.
Figure 2.
The Smo CRD is required for modulation by oxysterols and indirectly affects Ptch1-mediated regulation. (A) Basal (non-ShhN-stimulated) Gli-luciferase activity in NIH3T3 cells transfected with i the indicated cDNA constructs in the absence (red) or presence (orange) of cotransfected Ptch1 (25% DNA for Smo wt and ΔCRD; 5% DNA for the constitutively active SmoA1 construct). (B) Gli-luciferase assay in Smo−/− MEFs revealed that SmoA1 activity is induced by oxysterols (20(S)-OHC + 22(S)-OHC, 5 μM each) upon suppression of its basal activity by co-expression of Ptch1. (C) 8xGli-luciferase assay in Smo−/− MEFs transfected with wt or ΔCRD Smo, in the presence or absence of overexpressed mPtch1, following treatment with control or ShhN conditioned medium, SAG (200 nM), 20(S)-OHC (10 µM), or 20(S)-OHC + 22(S)-OHC (5 µM each). Comparable Ptch1-mediated suppression was demonstrated by transfecting increased wt Smo cDNA to compensate for the higher ΔCRD Smo basal activity. Error bars represent standard deviations (n = 3 independent transfections per data point). See also Table S1.
Although SmoΔCRD remains sensitive to Ptch1 regulatory effects, the ability of Ptch1 coexpression to reduce the basal activity of SmoΔCRD provides an opportunity to test whether the observed loss of oxysterol sensitivity (Fig. 1B) could be a secondary consequence of the mutant’s enhanced basal activity. Unlike the effect of ShhN or SAG treatment, however, suppression of SmoΔCRD activity by Ptch1 was not reversed by 20(S)-OHC either alone or in the presence of 22(S)-OHC (Fig 2C). This result illustrates that an intact CRD is absolutely required for Smo activation by oxysterols. In contrast, wt Smo or oncogenic SmoA1 clearly retain oxysterol sensitivity even in the presence of elevated Ptch1 levels (Fig. 2B,C). Our Ptch1 coexpression experiments thus confirm that the Smo CRD is essential for the effects of oxysterols on Smo. Because the phenotype of CRD deletion with regard to basal and ShhN-induced activity can be partially reversed under experimental conditions of Ptch1 over-expression, any role for the CRD in basal or ShhN-induced activity probably stems from indirect, allosteric effects on other parts of Smo. In contrast, the same experimental conditions fail to restore oxysterol sensitivity to SmoΔCRD, arguing that the CRD must play a direct and indispensable role in the oxysterol activation process.
The Smo CRD binds oxysterols via residues structurally analogous to the Wnt lipidbinding groove on the Fz CRD
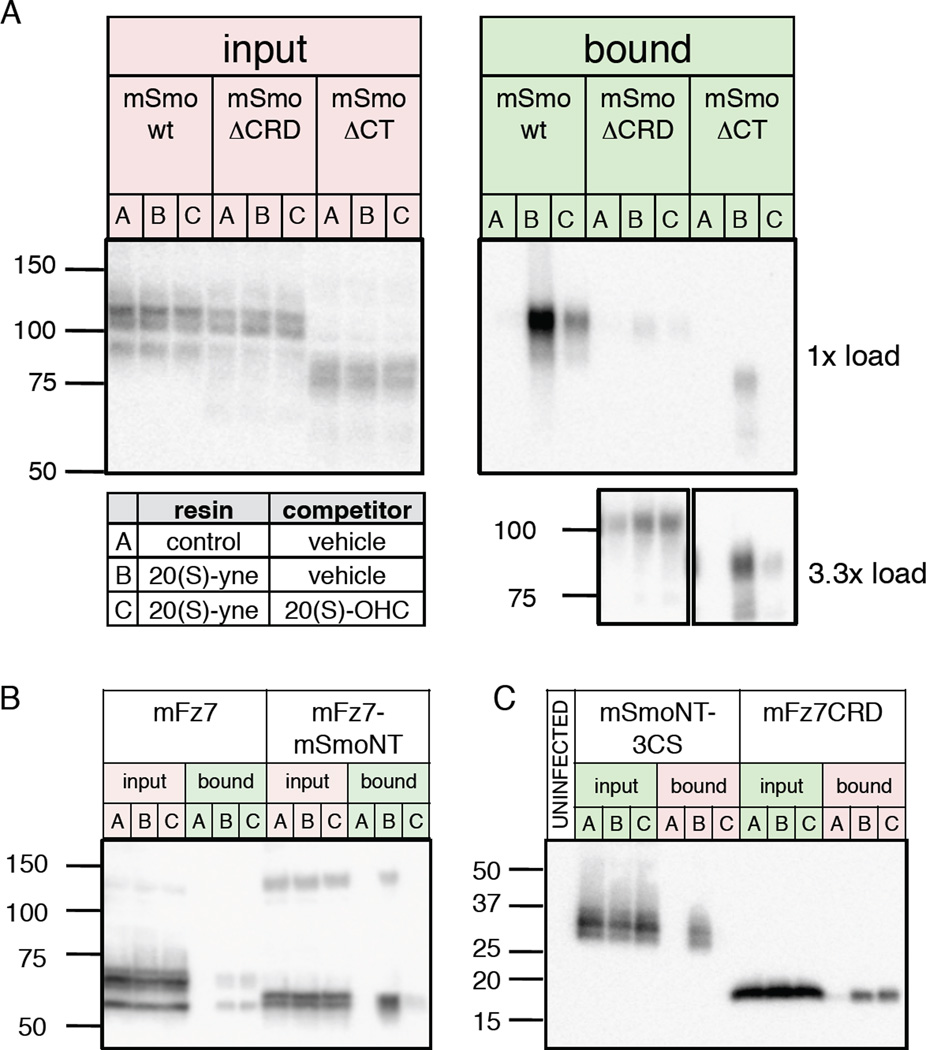
To test whether the CRD serves as the physical site of interaction for 20(S)-OHC, we employed a binding assay that utilizes a 20(S)-OHC alkyne derivative (20(S)-yne) immobilized on magnetic beads. In this assay, consistent with previous results (Nachtergaele et al., 2012), we found that wild type Smo expressed in HEK293 cells and solubilized in n-dodecyl-β-D-maltoside (DDM) robustly binds to the 20(S)-yne affinity resin, and that the interaction is specific as it is competed by excess 20(S)-OHC (Fig. 3A). In contrast, the level of binding to mSmoΔCRD was dramatically reduced, and no competition was observed with free 20(S)-OHC, indicating that any residual binding is nonspecific (Fig. 3A). We also tested binding of a C-terminal truncation (SmoΔCT) and found, as previously noted for binding of cyclopamine to this mutant protein (Chen et al., 2002a), that binding was less efficient as compared to wt Smo; binding to immobilized 20(S)-yne was nevertheless potently inhibited by 20(S)-OHC (Fig. 3A), indicating that deletion of the Smo CRD, but not the Smo cytoplasmic region, eliminates specific binding to 20(S)-OHC in vitro.
Figure 3.
The Smo CRD directly binds oxysterols. (A) Detergent-solubilized membranes from HEK293FT cells transfected with Myc-tagged wt Smo, ΔCRD, or ΔCT deletion mutants were incubated with control or 20(S)-yne affinity resin in the presence of 50 µM 20(S)-OHC competitor or vehicle, as indicated in the table. After washing, bound protein was eluted and analyzed by immunoblotting (“bound”). The ΔCRD and ΔCT mutants bound less efficiently than wt Smo to 20(S)-yne matrix; analysis of 3.3× equivalents of bound material nevertheless revealed specific binding for ΔCT, but not ΔCRD. (B) A similar experiment as in (A) with Myc-tagged wt mFz7 or a mFz7-mSmoNT chimera. (C) Experiments as above, but using concentrated conditioned medium (no added detergent) from 293S-GnT1− cells infected with BacMam virus encoding Protein C-tagged mSmoNT-3CS or mFz7 CRD constructs. In this and all subsequent blots the migration of molecular weight markers (in kDa) is indicated to the left. Results are representative of multiple independent experiments. See also Figure S2.
To determine whether the Smo CRD suffices to confer specific 20(S)-OHC binding, we examined a chimera in which the mouse Fz7 (mFz7) N-terminal domain (including its CRD) was replaced with that of mouse Smo (mFz7-mSmoNT). We observed a low level of nonspecific binding to wt mFz7 but substantially greater binding to mFz7-mSmoNT, which appears specific as it was dramatically reduced in the presence of free 20(S)-OHC (Fig. 3B). We also examined binding to a secreted protein comprising the Smo CRD and the short linker sequence just prior to the start of TM1, with three linker region cysteines altered to prevent adventitious aggregation (Fig. S2). The resulting protein, mSmoNT-3CS bound specifically to 20(S)-OHC, whereas a similar soluble CRD construct from mFz7 displayed only nonspecific binding (Fig. 3C). These experiments indicate that the core determinants of specific 20(S)-OHC binding are contained within the N-terminal module of Smo rather than in its transmembrane or cytoplasmic portions.
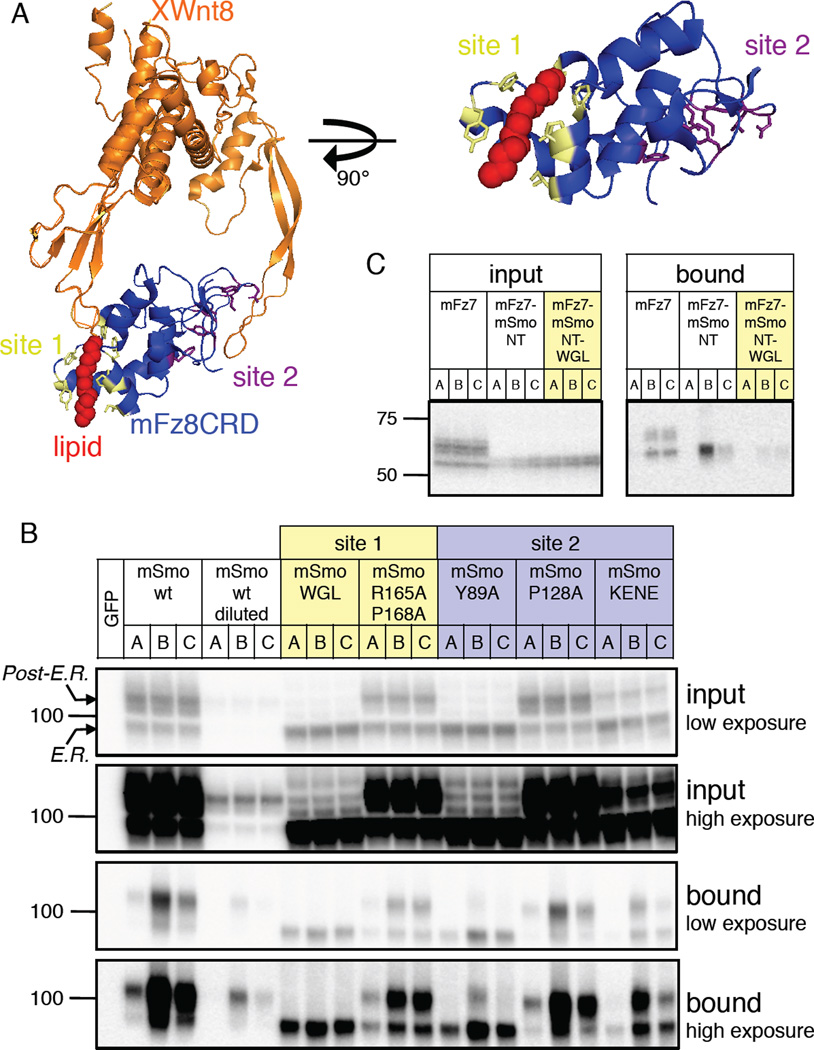
Recent crystallographic studies of the mFz8 CRD in complex with its acylated Wnt ligand revealed an unexpected mode of Wnt binding that resembles a hand grasping two distinct sites on opposite surfaces of the CRD (Janda et al., 2012) (Fig. 4A). Site 1 of the CRD comprises a hydrophobic pocket formed by side chains from CRD α-helices that engulf the Wnt palmitoleyl “thumb”, with little contribution to binding from Wnt amino acid side chains. Site 2 in contrast is a shallow depression that engages amino acid side chains on the Wnt protein’s “forefinger” (Fig. 4A). We hypothesized that the Smo CRD might bind to oxysterols in a manner resembling the interaction between the Fz8 CRD and the Wnt palmitoleoyl adduct. Accordingly, we tested the effects of alterations in CRD site 1 and site 2 residues (alignment in Fig. S3) on specific binding to 20(S)-OHC, focusing our analysis on the slower-migrating fully glycosylated post-ER forms (Chen et al., 2002a) of each altered protein. Alanine substitutions of CRD site 2 residues Y89, P128, and K137 E139 N140 E144 (KENE) had no qualitative effect on specific binding of 20(S)-OHC to Smo (Fig. 4B). In contrast, the site 1 substitutions R165A P168A and W113A G115A L116A (WGL) blocked specific 20(S)-OHC binding, whether in the context of the Smo protein (Fig. 4B) or in the mFz7-mSmoNT chimera (Fig. 4C). We conclude that Smo CRD site 1 residues are essential for interaction with 20(S)-OHC, whereas site 2 residues are largely dispensable.
Figure 4.
A conserved CRD lipid-binding interface in Smo and Fz CRDs. (A) Structure of a complex between XWnt8 and the mFz8 CRD as reported by Garcia and colleagues (Janda et al., 2012) (orange and blue, respectively; PDB code 4F0A) showing the Wnt lipid adduct (red) and the location of CRD site 1 (yellow) and site 2 (purple) residues. In the image at right, a magnified view of the Fz8 CRD site 1 is presented to highlight residues that contact the lipid. See sequence alignment in Fig. S3. (B) In vitro binding assay as in Fig. 2A and 2B with the indicated Myc-tagged Smo constructs transfected into HEK293FT cells. Site 1 and 2 mutants are indicated in yellow and purple to correspond to the structural model. Low and high exposures of the same input and bound fractions are also provided, and arrows above and below the 100-kDa marker indicate the positions of the immature ER-resident and fully glycosylated post-ER forms of wt Smo. Note that this assay sensitively detects specific 20(S)-OHC binding to low amounts of post-ER Smo, as seen for wt Smo diluted 33-fold in a control membrane extract and for the site 2 mutant Y89A; no specific binding, however is observed for the site 1 WGL mutant, which exhibits similar post-ER levels (see “input” panel). (C) Effect of the site 1 WGL mutation on oxysterol binding to Myc-tagged mFz7-mSmoNT chimera. See also Figure S3.
Activation of Smo by naturally occurring cellular oxysterols
Although 20(S)-OHC is capable of activating Smo when supplied exogenously to Hh-responsive cells, the physiological relevance of this lipid in the context of Hh pathway stimulation is not clear, as no sterol hydroxylase has been reported to produce 20(S)-OHC in cells. Furthermore, although 20(S)-OHC could easily be identified when added to NIH3T3 cell cultures (Fig. S4A–C, Table S1), we and others have been unable to detect endogenous 20(S)-OHC in these cells (Roberg-Larsen et al., 2012). These observations prompted a search for naturally occurring oxysterols that modulate Smo activity. 7-keto-27-OHC has been identified as a metabolic intermediate in the elimination of 7-ketocholesterol from tissues, which in turn is formed via oxidation of an immediate cholesterol precursor by cytochrome P450 (CYP) 7A1. 7-keto-27- OHC is detectable in extracts from retinal pigment epithelial (RPE) cells and can also be formed 10 in vitro by incubating 7-ketocholesterol with RPE homogenates or recombinant sterol 27- hydroxylase (CYP27A1) (Heo et al., 2011) (Fig. 5A). In experiments with Hh-pathway responsive NIH3T3 cells, we found that 7-keto-27-OHC could activate the Hh transcriptional reporter to approximately the same level as ShhN, while the structurally related oxysterols 7α-27-dihydroxycholesterol (7α-27-OHC) and 7α-27-dihydroxycholestenone (7α-27-OH-3-one) were substantially less active (Fig. 5B). 7-keto-27-OHC also produced additive activation of the reporter when combined with ShhN (Fig. 5B), and like 20(S)-OHC it synergized with SAG or with 22(S)-OHC (Fig. 4C). In a survey of other oxysterols for Smo-modulatory capacity, we found that 7-keto-25-OHC, another potential metabolite of 7-ketocholesterol, also produced substantial Gli-luciferase reporter activation when combined with 22(S)-OHC (Fig. S4D,E). As with other Hh pathway modulators acting directly on Smo, 7-keto-27-OHC induced the accumulation of endogenous Smo in the primary cilia of NIH3T3 cells (Fig. 5D; Fig. S4F), and its activity in transcriptional reporter assays was abolished by cyclopamine (Fig. S4G). We also found that 7-keto-27-OHC and 7-keto-25-OHC specifically block binding of Smo to immobilized 20(S)-yne in vitro (Fig. 5E). Using Smo−/− MEFs to test the effect of mutations in the Smo extracellular domain or “cyclopamine pocket” on activation by 7-keto-27-OHC, we found that the 7-keto oxysterol failed to activate SmoΔCRD (Fig. 5F). Taken together, these results suggest that 7-keto-27-OHC and 7-keto-25-OHC, like 20(S)-OHC, act on Smo via direct binding to the CRD.
Figure 5.
Naturally occurring oxysterols modulate Smo activity via CRD engagement. (A) Schematic diagram of sterol biosynthetic pathways leading to the formation of 7-keto-25-OHC and 7-keto-27-OHC (in red). Enzyme names are indicated in black italics; solid and dashed arrows represent experimentally verified or proposed biosynthetic steps, respectively. Abbreviations: cholesterol 7α-hydroxylase (CYP7A1), sterol 27-hydroxylase (CYP27A1), cholesterol 25-hydroxylase (CH25H). For comparison, the structure of the non-cellular 20(S)-OHC is shown in blue. (B) Gli-luciferase activity in Hh pathway-responsive NIH3T3 Shh-LIGHT2 cells was measured following treatment with the indicated sterols either alone (blue) or in combination with ShhN conditioned medium (green). (C) Concentration-response analysis of Gli-luciferase activity in Shh-LIGHT2 cells treated with 7-keto-27-OHC alone (red), in combination with 22(S)-OHC (5 µM, blue), or with a threshold concentration of SAG (1 nM, black). Statistical significance (Student’s t-test): p<0.01 (*); p<0.001 (**). (D) Ciliary accumulation of endogenous Smo in NIH3T3 cells treated with control or ShhN conditioned medium, or 7-keto-27-OHC; the inactive oxysterol 7α-27-OH-3-one serves as a negative control. Fixed cells were analyzed by immunofluorescence using antibodies against Smo (green) or acetylated tubulin (red) with DAPI counterstain (blue). Shifted overlays of Smo and acetylated tubulin stains are displayed as small panels to the right of selected images. See Fig. S4F for image quantification. (E) Binding of wt Smo to immobilized 20(S)-yne was tested as in Fig. 2A, using the indicated sterols as competitors (all at 50 µM except 20(S)-yne, at 100 µM). Sterols newly identified in this study as Smo modulators are shown in red, while inactive analogs are shown in black. For comparison, 20(S)-OHC and related derivatives are shown in blue. (F) Gliluciferase assay in Smo−/− MEFs transfected with the indicated Smo and Ptch1 constructs and stimulated with control (red) or ShhN conditioned medium (green), 100 nM SAG (black), or 5 µM 7-keto-27-OHC (blue). Error bars for A, B, C, and F represent standard deviations (n = 3 independent culture wells or transfections per data point). See also Figure S4 and Tables S1–S2.
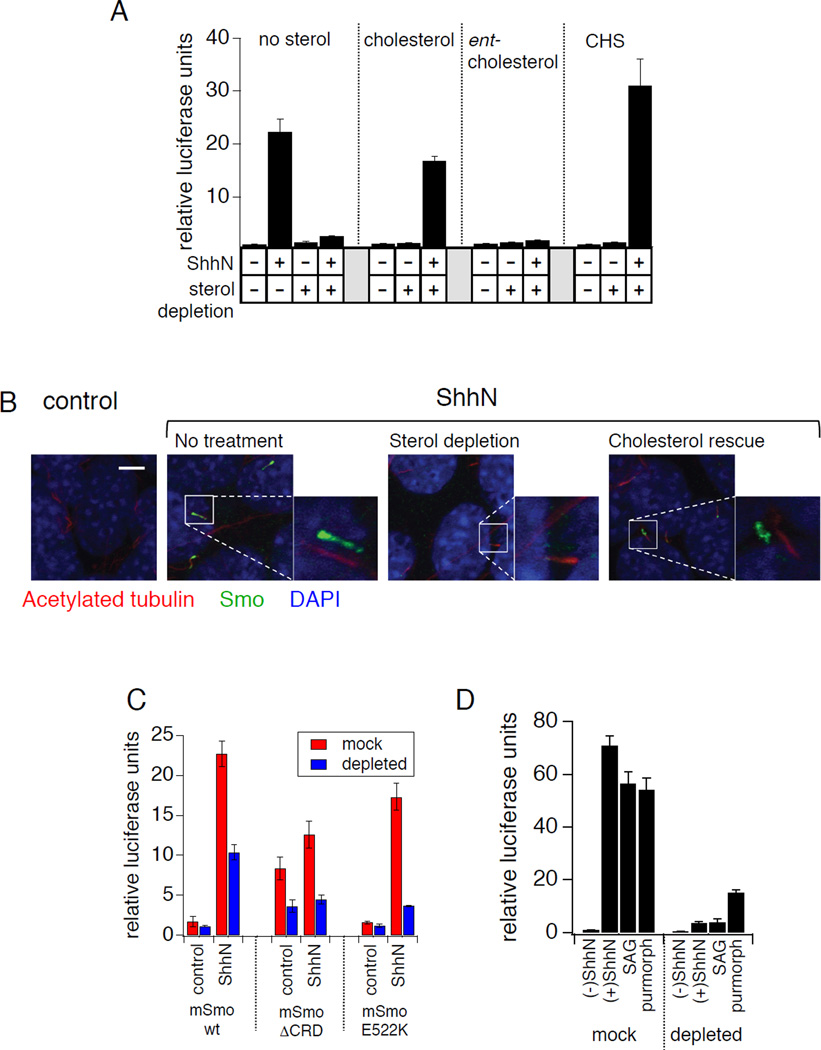
Multiple distinct sterol effects converge on the Hh pathway at the level of Smo
Human cholesterol synthesis disorders, such as Smith-Lemli-Opitz Syndrome (SLOS), combined with cultured cell studies have established a link between cellular sterols and Smo activation in response to Hh stimulation (Cooper et al., 2003). This requirement for sterols maps genetically to Smo (Cooper et al., 2003) and a relationship to the modulatory effects of oxysterols has been suggested (Corcoran and Scott, 2006; Nachtergaele et al., 2012), but the molecular basis of this role for sterols is not known. This sterol requirement can be experimentally demonstrated by β-cyclodextrin- and statin-mediated sterol depletion and consequent loss of transcriptional reporter activation (Fig 6A). We also observed that sterol depletion blocks ShhN-induced ciliary Smo accumulation (Fig. 6B; Fig. S5). The effects of sterol depletion are reversed by replenishment of cells with cholesterol or the more water-soluble cholesteryl hemisuccinate (CHS), but not by a synthetic enantiomer of cholesterol (ent-cholesterol) (Belani and Rychnovsky, 2008) whose stereochemistry is reversed at every chiral center (Fig. 6A). The failure of ent-cholesterol to rescue Hh response indicates a stereospecific site of action for cholesterol and is consistent with a direct Smo-cholesterol interaction rather than an effect on membrane structure and fluidity. However, we note that this result does not rule out the possibility of relevant physical interactions between cholesterol and chiral membrane lipids.
Figure 6.
The Smo activation state is regulated by multiple distinct sterol effects. (A) NIH3T3 cells were transfected with Gli-luciferase reporter plasmids and stimulated with ShhN conditioned medium in combination with the indicated exogenous sterols (at 100 µM) following endogenous sterol depletion (with methyl-β-CD plus lovastatin as described in the methods section). (B) Ciliary accumulation of endogenous Smo in ShhN-stimulated NIH3T3 cells following depletion of endogenous sterols (as described in panel A) and subsequent rescue with exogenous cholesterol. See Fig. S5 for image quantification. (C) In Smo−/− MEFs transfected with the indicated Smo constructs, Gli-luciferase activity induced by ShhN stimulation was measured under mock-treated (red) or sterol-depleted (blue) conditions as in (A). (D) In NIH3T3 cells, endogenous sterol depletion (“depleted”) reduced Gli-luciferase activity induced by treatment with ShhN conditioned medium, SAG (500 nM), or purmorphamine (2.5 µM), as compared to mock-depleted controls (“mock”). Error bars in all luciferase assays represent standard deviations (n = 3 independent transfections per data point). See also Figure S5 and Table S1.
Our identification of the CRD as the major site of action on Smo for activating oxysterols provides an opportunity to ask whether the requirement of an intact sterol synthesis pathway for Smo function derives from an analogous CRD-lipid interaction. Despite its unresponsiveness to activating oxysterols, we found that SmoΔ CRD remained sensitive to sterol depletion (Fig. 6C). Sensitivity to sterol depletion is also retained with the E522K alteration of the cyclopamine pocket (Fig. 6C), and in the context of stimulation by the agonists SAG and purmorphamine (Fig. 6D). In testing whether this role for cholesterol is linked to oxysterol modulatory effects, we also found that CHS failed to compete for specific binding to immobilized 20(S)-OHC (Fig. 5E). These data together show that the permissive effect of cholesterol on Smo is mediated by structural determinants that lie outside the CRD oxysterol binding site and are likely distinct from the cyclopamine pocket (Fig. 7). Furthermore, the requirement of intact cellular cholesterol pools for efficient Smo function is mechanistically independent of any Smo activation that occurs via oxysterol-CRD interactions.
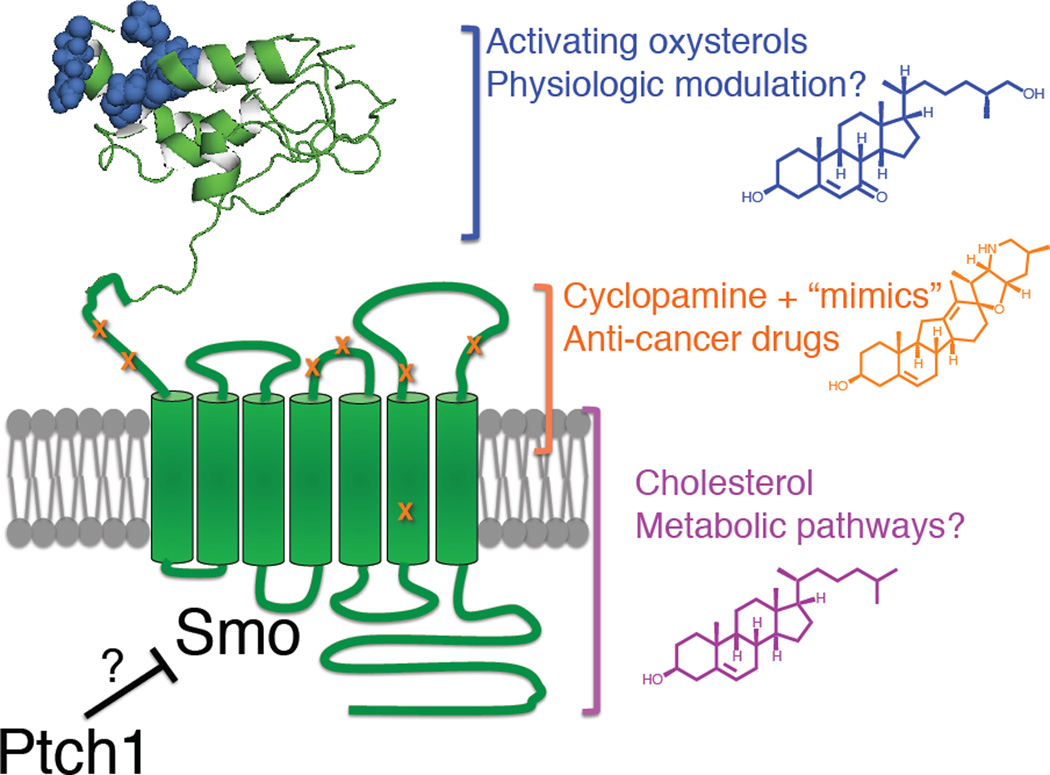
Figure 7.
Summary model indicating multiple sterol modulatory effects in Hh signal transduction at the level of mSmo as well as their binding determinants. Oxysterols (blue) activate Smo via CRD engagement (hypothetical structural model of human Smo CRD, with mutations affecting oxysterol binding indicated in blue), while cyclopamine and related anticancer drugs (orange) bind to a pocket in the extracellular half of the heptahelical domain. Mutations that affect binding and/or signaling inhibition by cyclopamine and its mimics GDC-0449 and NVP LDE-225 are displayed in orange; cholesterol (purple) may bind directly to an allosteric site facing the membrane, as has been shown for other GPCRs. See also Figure S6.
Discussion
A direct role for cholesterol in Smo modulation echoes the structurally defined interactions of sterols with other 7TM proteins (Burger and Gimpl, 2000). Modulatory ligand binding near the extracellular portion of the heptahelical bundle is also a common feature of class A GPCRs (Venkatakrishnan et al., 2013), analogous to the Smo cyclopamine pocket, and several metabotropic neurotransmitter receptors bind their ligands through structured extracellular domains (Lagerström and Schiöth, 2008), reminiscent of our findings with the Smo CRD. Strikingly, each of these three Smo modulatory sites can be occupied by distinct types of steroidal ligands. The identification of these multiple sites and their modulators should facilitate structural elucidation of active and inactive full-length Smo conformations through ligand-mediated stabilization strategies that have accelerated the crystallographic analysis of GPCR activation (Venkatakrishnan et al., 2013). Within the Smo/Fz subfamily, it is intriguing that the Wnt lipid adduct is recognized by site 1 of the Fz CRD, and that a corresponding portion of the Smo CRD mediates oxysterol binding and regulation. Although sterols and fatty acids are structurally unrelated, a predicted swap in the Smo CRD disulfide bond pattern with respect to Fz CRDs (J.F.B., data not shown; Fig. S3) suggests possible alterations in the dimensions of the Smo CRD hydrophobic groove. Photoaffinity labeling and crystallographic experiments of fulllength Smo or its isolated CRD will more precisely define the structural determinants of oxysterol interaction in the Smo extracellular domain, and may also identify additional endogenous lipids that bind to this region.
How does Ptch1 regulate Smo activity? Although an intact Smo CRD is required for activation by oxysterols, our functional analysis shows that Ptch1 coexpression restores basal-and Hh-induced activation to SmoΔCRD, thereby indicating that this domain is not the major site of regulation by Ptch1. By similar reasoning, the ability to dissociate oxysterol action on Smo from Ptch regulatory effects argues that oxysterols like 20(S)-OHC or the 7-ketocholesterol derivatives studied here are unlikely to subserve the regulatory functions of Ptch1 (Fig. 2C). The failure of SmoΔCRD to become fully activated in response to Hh may point instead to a more indirect facilitative CRD function in stabilizing an active Smo conformation induced by loss of Ptch1-mediated repression. The structural basis for such stabilization could be a physical interaction between the CRD and the extracellular surface of the Smo transmembrane domain (discussed in more detail below) that could permit the CRD to influence the conformational equilibrium of the heptahelical bundle, ensuring low basal activity and high Hh-induced responsiveness. This stabilizing effect of the CRD could be independent of its lipid-binding capacity but still involve an interaction with the transmembrane domain. Alternatively, the ligand-binding function of the CRD may be intimately related to its ability to regulate Smo conformation, under which circumstances one or more bound lipids (such as oxysterols) would likely serve as cofactors required for proper basal and Hh-induced Smo activation but whose availability is not directly controlled by Ptch1 activity. As another possible, albeit more complex regulatory mechanism, Ptch1 conceivably could act on both the Smo transmembrane domain and on the CRD lipid binding pocket as well. Future structural and functional investigations, as well as identification of endogenous Smo ligands, will be required to distinguish between these potential mechanisms of Smo regulation.
Ptch1 regulation of Smo involves sequences outside of the CRD, but the major site of action for Ptch1 on Smo has yet to be delineated. The Smo transmembrane ligand-binding cavity, which is the site of interaction for cyclopamine and its mimics, has been proposed to bind an endogenous ligand whose availability is controlled by Ptch1. In this regard, however, it is interesting that point mutants in the Smo transmembrane and extracellular loop (ECL) domains, defined by their ability to disrupt action of one or more cyclopamine mimics (some developed as anticancer drugs), display essentially normal basal and Hh-induced activity (Fig 1C, Fig. S6, (Buonamici et al., 2010; Dijkgraaf et al., 2011; Kim et al., 2013; Yauch et al., 2009)), raising the possibility that Ptch1 acts on Smo via mechanisms that do not involve modulation of small molecule ligand binding within the cyclopamine pocket. Such a mechanism could involve lipids binding to additional sites on Smo that are yet to be identified. Another plausible scenario is that endogenous Smo ligands regulated by Ptch1 activity engage the cyclopamine pocket in a manner that is insensitive to these previously described mutations, although we note from examination of the recent X-ray structure of the Smo 7TM domain in complex with an anticancer drug (Fig. S6A,B) that the residues highlighted by drug-resistant mutations collectively cover a significant portion of the ligand binding surface. Delineation of the Ptch1 site of action on Smo ultimately awaits identification of Ptch-regulated bioactive small molecules (steroidal or otherwise) as well as more extensive mutagenesis of the crystallographically resolved heptahelical ligand binding site.
In any case, the similar roles for lipid interaction in these CRDs defined here and recently confirmed by another study (Nedelcu et al., 2013) raise the possibility that Smo and Fz activation may share some features, despite divergent modes of protein ligand recognition and intracellular signal transduction of the biologically distinct Hh and Wnt signaling pathways. In activation of its canonical pathway, the Wnt protein ligand nucleates a ternary complex with Fz and low-density lipoprotein receptor related protein 5/6, and transduction of this binding event results from recruitment of cytoplasmic effectors to a newly generated intracellular surface (Clevers and Nusse, 2012). Oxysterol activation of Smo may similarly proceed through lipid-mediated changes in protein association, such as the Smo dimerization that has been reported to depend on CRD integrity (Zhao et al., 2007). Alternatively, the oxysterol-bound Smo CRD might influence the conformation and activity state of the heptahelical bundle by docking with the large Smo ECLs, as occurs with the N-terminal tethered ligand exposed by thrombin cleavage in protease activated receptor 1 (PAR 1) (Coughlin, 2000). In support of this idea, point mutations in of ECL cysteines lead to increased basal activation and decreased Hh sensitivity in Drosophila Smo (Carroll et al., 2012),; in fact, one such cysteine substitution is predicted to break a disulfide bond that tethers ECL1 to the structured linker region joining the CRD to TM1. This phenotype is remarkably similar to that resulting from deletion of the mouse Smo CRD, consistent with the hypothesis that each of these sequence alterations affects the same regulatory process. Although the CRD is not present in the recently published crystal structure of the Smo heptahelical bundle in an inactive conformation (Wang et al., 2013), it is intriguing that the ECLs form a highly structured “tower” whose external surface could scaffold an interaction with the CRD, and the observed rigid extension of the sixth transmembrane helix into ECL3 (see Fig. S6B) might provide a lever arm to translate changes in CRD-ECL interactions to the Smo heptahelical and intracellular domains.
The Smo CRD is highly conserved, and missense mutations in this domain lead to defects in Hh signaling in vivo (Aanstad et al., 2009; Chen and Struhl, 1998; Nakano et al., 2004), but the biochemical function of the CRD in Smo regulation has remained a mystery. The ability of the Smo CRD to bind oxysterols raises the possibility that this domain may mediate Smo-modulatory effects of physiologically relevant small molecules; this possibility is supported by oxysterol response for a range of vertebrate Smo proteins (data not shown). Indeed, the functional defect resulting from removal of the Smo CRD (Fig. 1C) suggests that the CRD can strongly affect basal or Hh-stimulated Smo activity, and future biochemical and genetic studies may delineate roles for 7-keto-25-OHC, 7-keto-27-OHC or other CRD-binding lipids in influencing Smo under normal or pathological conditions. Of particular interest for this mechanism of regulation, the cell surface accessibility of the Smo N-terminus (Denef et al., 2000; Milenkovic et al., 2009) may permit the CRD to sample the extracellular milieu, potentially rendering Smo susceptible to modulation by diffusible messengers such as secreted hormones and metabolites. In contrast, the influence of Ptch on Smo is strictly cell-autonomous (Briscoe et al., 2001; Chen and Struhl, 1996), suggestive of a membrane-delimited biochemical regulatory event. These dual modes of Smo regulation may serve to couple a systemic input with the inherently local Hh response, tuning Hh pathway output based on feedback from metabolic circuits (Teperino et al., 2012) or other aspects of organismal physiology.
Experimental Procedures
Cell culture
4C20 Smo−/− MEFs (Varjosalo et al., 2006), HEK293FT cells (Life Technologies), and stably transfected HEK293-ShhN cells (Maity et al., 2005) were maintained in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (Omega Scientific) and 1% penicillin/streptomycin/glutamine. NIH3T3 cells and stable Shh-LIGHT2 cells were maintained in DMEM with 1% penicillin/streptomycin/glutamine and 10% bovine calf serum (HyClone) as previously described (Kim et al., 2010). 293-Freestyle suspension cells were maintained in 293-Freestyle medium (Invitrogen). 293S-GnT1- cells were cultured in pro-293(S)-CDM (Lonza) with 0.1% FBS (BenchMark, Gemini), 10 mM GlutaMAX, and 0.01% penicillin/streptomycin as previously described(Dukkipati et al., 2008). Spodoptera frugiperda (Sf9) cells were cultured in Sf900-III (Life Technologies) with 10% FBS (BenchMark, Gemini) and 10 µg/ml gentamicin as previously described (Dukkipati et al., 2008).
Antibodies, chemicals, and small molecules
The following antibodies were used in this study: anti-acetylated tubulin (mouse monoclonal, Sigma, T6793, 1:1000), anti-c-Myc (rabbit polyclonal, Santa Cruz Biotechnology, A14, 1:1000), anti-protein C (mouse monoclonal, Roche, HPC4, 1:400). A previously described polyclonal antibody against mouse Smo (Kim et al., 2009) was used at 1:1000. For immunoblotting, HRP-conjugated goat anti-rabbit and anti-mouse secondary antibodies (1:20,000) were from Promega. For immunofluorescence microscopy, Alexa Fluor-488 and -594-conjugated secondary antibodies (Life Technologies) were used at 1:1000. Control and ShhN conditioned medium were produced from HEK293-ShhN stable cells as described (Maity et al., 2005) and diluted 10–20-fold for cell treatments. SAG was purchased from Enzo Life Science. Purmorphamine was purchased from VWR Scientific. 20(S)-OHC, 22(S)-OHC, and CHS were purchased from Sigma. Azide-PEG4-NHS ester was purchased from Click Chemistry Tools. 7-keto-27-OHC and its analogs were purchased from Avanti Polar Lipids. All other sterols were purchased from Steraloids. 20(S)-yne was purchased from Cayman Chemical. SiMAG amine magnetic beads were purchased from Chemicell. Ent-cholesterol was synthesized as previously described (Belani and Rychnovsky, 2008).
Transfections and reporter assays
Shh-LIGHT2, NIH3T3, and 4C20 Smo−/− cells were seeded into 24 or 96 well plates and transfected with various cDNAs (TransIT 2020, Mirus) along with a mixture of 8xGli-luciferase and SV40-Renilla plasmids as previously described (Kim et al., 2009). Transfection mixtures were supplemented with a GFP expression plasmid to normalize the amount of DNA delivered to each well. For expression in Smo−/− MEFs, transfection mixtures generally contained 1–2% (w/w) of each Smo cDNA except in figures 1D or 2C where cells received 1–50% (w/w) or 1–10 (w/w) of Smo cDNA as described in the respective legends. Upon reaching confluency, cells were shifted to DMEM with 0.5% serum containing ShhN conditioned medium, agonists or antagonists (or appropriate vehicle controls) where indicated and incubated for 24–48 hours, at which point luciferase activity was measured (Dual Luciferase, Promega) using a Berthold Centro XS3 luminometer. Depletion of cellular sterols was performed with 45 minutes of 8 mM methyl-β–CD extraction followed by sustained lovastatin (5 µM) treatment as previously described (Kim et al., 2010). For quantificaiton of Smo-Renilla expression a secreted alkaline phosphatase plasmid (pSEAP-CT) was cotransfected as a normalization control and the assay (Phospha-Light, Applied Biosystems) was performed as previously described. HEK293FT cells in 10 cm dishes were transfected with Lipofectamine 2000 (3.75–7.5 µg DNA per dish) according to the manufacturer’s instructions. For BacMam-mediated viral expression in HEK293 suspension cell lines, recombinant baculovirus was produced in Sf9 cells via cotransfection with linearized baculovirus DNA (Sapphire, Allele Biotechnology) using Cellfectin II and amplified as previously described (Dukkipati et al., 2008); high-titer baculovirus stocks were used to infect log-phase HEK293 cells for 2–3 days in the presence of 10 mM sodium butyrate.
Immunofluorescence
Indirect immunofluorescence was performed essentially as previously described. Briefly, NIH3T3 cells were grown to confluency on coverslips, at which point they were shifted to low serum medium and incubated overnight with various treatments as outlined in the main text. The next day, cells were fixed in 4% paraformaldehyde/PBS (Fig. 1) or ice-cold methanol (Fig. 5, 6) and stained with the indicated primary and secondary antibodies along with DAPI counterstain. After mounting, coverslips were imaged on a Zeiss LSM 510 laser scanning confocal microscope (Fig. 6) or a Leica SD6000 spinning disc confocal microscope with three blind iterative cycles of computerized Z-stack deconvolution using LAS AF software as previously described (Kim et al., 2009) (Fig. 1, 5).
Preparation of 20(S)-yne affinity matrix
20(S)-yne affinity matrix was prepared by a modification of a previous method (Nachtergaele et al., 2012) using a two-step coupling procedure. 4 mg of SiMAG amine beads were washed into 0.8 ml of high-quality dimethylformamide (DMF) and reacted with 525 nmol azide-PEG4-NHS ester crosslinker in the prsence of 1.05 µmol of triethylamine for 2 hr. After extensive washing in DMF, the azide-derivatized matrix was used for alkyne-azide cycloaddition with 350 nmol of 20(S)-yne (stock solution at 10 mM in ethanol) in the presence of 250 mmol CuSO4-5H2O and 5 mM freshly prepared sodium ascorbate, The reaction proceeded for 2 hours, after which point the beads were incubated in acetic anhydride/DMF (2:8 (v/v)) for several hours to block any free amine groups. Beads were subsequently washed in DMF and stored in ethanol (0.8 ml volume) until ready for use. Control resin was prepared in the same manner but the azide-PEG4-NHS and 20(S)-yne were omitted. Coupling efficiency was assessed at the azide-alkyne cycloaddition step by thin layer chromatography (TLC) of reaction supernatants essentially as described (Nachtergaele et al., 2012), except that methanol/dichloromethane 1:9 (v/v) was used as solvent and the TLC plate was visualized by staining in Hanessian’s reagent. Each binding reaction typically employed 20 µl of affinity resin per 400 µl of solubilized membranes or conditioned medium.
Oxysterol ligand affinity chromatography
Transfected HEK293FT cells were pelleted by low-speed centrifugation and subjected to hypotonic lysis, membrane isolation, and solubilization in 0.1% DDM-containing extraction buffer as previously described (Nachtergaele et al., 2012). 5% of each binding reaction was typically reserved an an “input” fraction, and competitor ligands (or an equivalent amount of vehicle, either DMSO or ethanol) were preincubated for 1 hour with the solubilized membranes prior to addition of equilibrated control or 20(S)-yne affinity resin. Binding was allowed to proceed overnight, after which point the beads were washed three times in extraction buffer and eluted with SDS-PAGE sample buffer for 20 minutes at 37°C. For binding studies with secreted extracellular domains, 20 ml of 0.45 µm-filtered conditioned medium from BacMam-infected HEK293S-GnT1- cells was concentrated ~10-fold (Amicon 3 kDa MWCO centrifugal concentrator) and subjected to ligand affinity chromatography as described above, except that cell culture medium (pro-293(S) CDM, Lonza) with no added detergent was used for the equilibration and wash steps.
Immunoblotting
Protein samples were separated on Criterion TGX SDS-PAGE gels (BioRad), transferred to polyvinylidene difluoride (PVDF) membranes, blocked in PBS + 0.1% Tween-20 (PBST) with 5% milk and blotted with the relevant primary and secondary antibodies. For experiments with protein C-tagged constructs, Tris-buffered saline (TBS) with 1 mM CaCl2 was used in place of PBS owing to the Ca2+-dependent nature of the antibody-epitope interaction. Membranes were visualized using an electronic CCD-equipped Chemidoc XRS+ imager (BioRad).
Supplementary Material
Highlights.
Select oxysterols that activate Smo bind its extracellular cysteine-rich domain (CRD)
CRD binding determinants are analogous to those in Frizzled that bind the Wnt lipid
Candidate endogenous modulators that act via the Smo CRD include 7-keto oxysterols
Diverse sterol modulatory effects act via distinct Smo structural determinants
Acknowledgments
We thank S. Nachtergaele and R. Rohatgi for providing 20(S)-yne, K. Krasinska for assistance with mass spectrometry, and K. Shin, R. Mann, A. Creanga, K.C. Garcia, D. Leahy, and J. K. Chen for comments on the manuscript. We thank D. Waghray and K.C. Garcia for providing HEK293SGnT1- cells, Sf9 cells, and BacMam vectors, and E. Piccione for providing HEK293- Freestyle cells. This research was supported in part funding from NIH. B.R.M. and N.S. were fellows of the Damon Runyon Cancer Research Foundation. Y.C. is a fellow of the Agency for Science, Technology, and Research (A*STAR). P.A.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
Supplemental Information
Supplemental information includes six figures, two tables, experimental procedures, and references, and can be found online at_____.
References
- Aanstad P, Santos N, Corbit KC, Scherz PJ, Trinh LA, Salvenmoser W, Huisken J, Reiter JF, Stainier DYR. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr Biol. 2009;19:1034–1039. doi: 10.1016/j.cub.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers KL, Thérond PP. Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends in Cell Biology. 2010;20:287–298. doi: 10.1016/j.tcb.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Bazan JF, de Sauvage FJ. Structural ties between cholesterol transport and morphogen signaling. Cell. 2009;138:1055–1056. doi: 10.1016/j.cell.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes & Development. 2010;24:2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- Belani JD, Rychnovsky SD. A concise synthesis of ent-Cholesterol. J. Org. Chem. 2008;73:2768–2773. doi: 10.1021/jo702694g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benned-Jensen T, Norn C, Laurent S, Madsen CM, Larsen HM, Arfelt KN, Wolf RM, Frimurer T, Sailer AW, Rosenkilde MM. Molecular characterization of oxysterol binding to the Epstein-Barr virus-induced gene 2 (GPR183) Journal of Biological Chemistry. 2012;287:35470–35483. doi: 10.1074/jbc.M112.387894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Chen Y, Jessell TM, Struhl G. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol Cell. 2001;7:1279–1291. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Gimpl G. Regulation of receptor function by cholesterol. Cellular and Molecular Life Sciences. 2000 doi: 10.1007/PL00000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93:767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- Carroll CE, Marada S, Stewart DP, Ouyang JX, Ogden SK. The extracellular loops of Smoothened play a regulatory role in control of Hedgehog pathway activation. Development. 2012;139:612–621. doi: 10.1242/dev.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes & Development. 2002a;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002b;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Cooper MK, Wassif CA, Krakowiak PA, Taipale J, Gong R, Kelley RI, Porter FD, Beachy PA. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Scott MP. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci USA. 2006;103:8408–8413. doi: 10.1073/pnas.0602852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubüser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf GJP, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, Burdick D, Goldsmith R, Robarge K, Sutherlin D, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71:435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 Complex Transduces the Hedgehog Signal at Primary Cilia. Developmental Cell. 2012:1–13. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, Parhami F. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J Biol Chem. 2007;282:8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9:437–445. doi: 10.1038/nrm2414. [DOI] [PubMed] [Google Scholar]
- Hausmann G, Mering von C, Basler K. The Hedgehog Signaling Pathway: Where Did It Come From? PLoS Biol. 2009;7:e1000146. doi: 10.1371/journal.pbio.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo G-Y, Bederman I, Mast N, Liao W-L, Turko IV, Pikuleva IA. Conversion of 7-ketocholesterol to oxysterol metabolites by recombinant CYP27A1 and retinal pigment epithelial cells. J Lipid Res. 2011;52:1117–1127. doi: 10.1194/jlr.M014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff Von DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011 doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy P. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Kim J, Chen B, King EM, Borodovsky A, et al. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell. 2013;23:23–34. doi: 10.1016/j.ccr.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tang JY, Gong R, Kim J, Lee JJ, Clemons KV, Chong CR, Chang KS, Fereshteh M, Gardner D, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17:388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors 21 and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Lum L, Beachy P. The Hedgehog Response Network: Sensors, Switches, and Routers. Science. 2004 doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci USA. 2005;102:17026–17031. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Genetics of ventral forebrain development and holoprosencephaly. Curr. Opin. Genet. Dev. 2000;10:262–269. doi: 10.1016/s0959-437x(00)00084-8. [DOI] [PubMed] [Google Scholar]
- Nachtergaele S, Mydock LK, Krishnan K, Rammohan J, Schlesinger PH, Covey DF, Rohatgi R. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat Chem Biol. 2012;8:211–220. doi: 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Nystedt S, Shivdasani AA, Strutt H, Thomas C, Ingham PW. Functional domains and sub-cellular distribution of the Hedgehog transducing protein Smoothened in Drosophila. Mech Dev. 2004;121:507–518. doi: 10.1016/j.mod.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat Chem Biol. 2013 doi: 10.1038/nchembio.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M, Nusse R. The role of the cysteine-rich domain of Frizzled in Wingless-Armadillo signaling. Embo J. 2005;24:3493–3503. doi: 10.1038/sj.emboj.7600817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg-Larsen H, Strand MF, Grimsmo A, Olsen PA, Dembinski JL, Rise F, Lundanes E, Greibrokk T, Krauss S, Wilson SR. High sensitivity measurements of active oxysterols with automated filtration/filter backflush-solid phase extraction-liquid chromatography-mass spectrometry. J Chromatogr A. 2012;1255:291–297. doi: 10.1016/j.chroma.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9:1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be 22 reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K, Coppola C, Chanana AM, Marji J, Bickers DR, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366:2180–2188. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglund S, Toftgård R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Teperino R, Amann S, Bayer M, McGee SL, Loipetzberger A, Connor T, Jaeger C, Kammerer B, Winter L, Wiche G, et al. Hedgehog Partial Agonism Drives Warburg-like Metabolism in Muscle and Brown Fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes & Development. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Li S-P, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Developmental Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- Wang C, Wu H, Katritch V, Han GW, Huang X-P, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013:1–20. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJP, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shih AY, Yang XV, Kuei C, Wu J, Deng X, Mani NS, Mirzadegan T, Sun S, Lovenberg TW, et al. Identification of structural motifs critical for epstein-barr virus-induced molecule 2 function and homology modeling of the ligand docking site. Mol Pharmacol. 2012;82:1094–1103. doi: 10.1124/mol.112.080275. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.