Abstract
The high effectiveness of cyclic oligosaccharides like cyclodextrins in the production of trans-resveratrol in Vitis vinifera cell cultures is enhanced in the presence of methyl jasmonate. In order to dissect the basis of the interactions among the elicitation responses triggered by these two compounds, a transcriptional analysis of grapevine cell cultures treated with cyclodextrins and methyl jasmonate separately or in combination was carried out. The results showed that the activation of genes encoding enzymes from phenylpropanoid and stilbene biosynthesis induced by cyclodextrins alone was partially enhanced in the presence of methyl jasmonate, which correlated with their effects on trans-resveratrol production. In addition, protein translation and cell cycle regulation were more highly repressed in cells treated with cyclodextrins than in those treated with methyl jasmonate, and this response was enhanced in the combined treatment. Ethylene signalling was activated by all treatments, while jasmonate signalling and salicylic acid conjugation were activated only in the presence of methyl jasmonate and cyclodextrins, respectively. Moreover, the combined treatment resulted in a crosstalk between the signalling cascades activated by cyclodextrins and methyl jasmonate, which, in turn, provoked the activation of additional regulatory pathways involving the up-regulation of MYB15, NAC and WRKY transcription factors, protein kinases and calcium signal transducers. All these results suggest that both elicitors cause an activation of the secondary metabolism in detriment of basic cell processes like the primary metabolism or cell division. Crosstalk between cyclodextrins and methyl jasmonate-induced signalling provokes an intensification of these responses resulting in a greater trans-resveratrol production.
Introduction
Vitis vinifera produces stilbenes, which are a small group of compounds characterized by a 1,2-diphenylethylene backbone. Most plant stilbenes are derivatives of the monomeric unit trans-resveratrol (trans-R; 3,5,4′-trihydroxystilbene). The formation of stilbenes is considered to be a part of the general defense mechanism since they display strong antifungal and antimicrobial activities [1]–[3]. In fact, trans-R is produced in both grapevine vegetative tissues and berries as well as in cell cultures in response to abiotic and biotic stress [1], [4]–[5]. Moreover, hundreds of studies have reported the beneficial effects of trans-R on neurological system [6], cardiovascular diseases [7], preventing carcinogenesis [8]–[9] and as an antiaging agent in the treatment of age-related human diseases [10].
Biosynthesis of trans-R starts from phenylalanine which is a key intermediate linking the primary metabolism and the secondary metabolism. Thus, the first step in the stilbene biosynthesis pathway consists in the transformation of phenylalanine into cinnamic acid in a reaction catalyzed by the enzyme phenylalanine ammonia lyase (PAL). The consecutive action of cinnamate 4-hydroxylase (C4H) and 4-coumarate-CoA ligase (4CL) transforms cinnamic acid into 4-coumaroyl-CoA, which is the common precursor of most of the phenolic compounds found in plants: lignins, flavonoids and stilbenoids. Then, one molecule of 4-coumaroyl-CoA is condensed with three malonyl-CoA units to produce either trans-R, through the action of stilbene synthase (STS), or naringenin chalcone by the action of chalcone synthase ([11] and references therein). Nowadays, different transcription factors (TFs) involved in the regulation of lignin, flavonol and anthocyanin metabolism are known [12]–[14]. The regulation of stilbene biosynthesis remains largely unknown, and only a role of TFs MYB14 and MYB15 has been recently reported in grapevine [15].
Stilbene biosynthesis is induced in response to a wide range of biotic and abiotic elicitors, which in turn, activate the expression of genes encoding stilbene biosynthesis pathway enzymes [16]–[17]. The activation of stilbene biosynthesis by particular elicitors is well-documented in grapevine, where the expression of STS genes and the production of stilbenes are induced upon elicitation with different fungal pathogens [18]–[19]. Stilbene biosynthesis is also triggered by signalling molecules such as methyl jasmonate (MJ) [20]. In this way, upon perception of jasmonate signals, the plant cell activates several defense mechanisms, reflected in a massive reprogramming of gene expression which leads to both the activation of stilbene biosynthesis and the expression of pathogenesis related-proteins (PR-proteins) [17], [21]. Moreover, the addition of MJ induces both the repression of cell cycle progression and the induction of phenylpropanoid metabolism in Arabidopsis thaliana cell cultures [22].
On the other hand, cyclodextrins (CD) are cyclic oligosaccharides that chemically resemble to the pectic oligosaccharides naturally released from the cell walls during a fungal attack [2]. They act as true elicitors since they provoke stilbene accumulation and induce the accumulation of new gene products like peroxidases, β-1,3-glucanases and chitinases [23]–[24]. Martínez-Esteso et al. [25] observed that enzymes from the trans-R biosynthesis pathway like STS were up-regulated by CD in grapevine (V. vinifera cv Gamay) cell cultures. In addition, CD are not only inducers of trans-R biosynthesis but also are promoters of adducts that remove trans-R from the culture medium, reducing the feedback inhibition and trans-R degradation, and allowing its accumulation at high concentrations [26]. Interestingly, Lijavetzky et al. [27] demonstrated that the combined use of CD and MJ enhanced the production of trans-R, which was strongly associated to an increased expression of STS, PAL, C4H, 4CL genes in grapevine (V. vinifera cv Monastrell) cell cultures.
Global transcriptomic approaches can provide new clues on both the transcriptional cascade activated by elicitors and the possible interactions taking place when they are applied in combination. Considering the synergistic interaction of CD and MJ on trans-R production, we have transcriptionally analyzed the responses of grapevine cell cultures to the treatments with CD and MJ separately and in combination using the GrapeGen GeneChip. In this work, we identified regulators which are specifically activated in each treatment and discussed their possible involvement in the control of different physiological processes as well as the putative interaction of physiological and cellular responses giving rise to the synergistic accumulation of trans-R in the combined treatment.
Results
Cyclodextrins and methyl jasmonate synergistically induce trans-resveratrol production
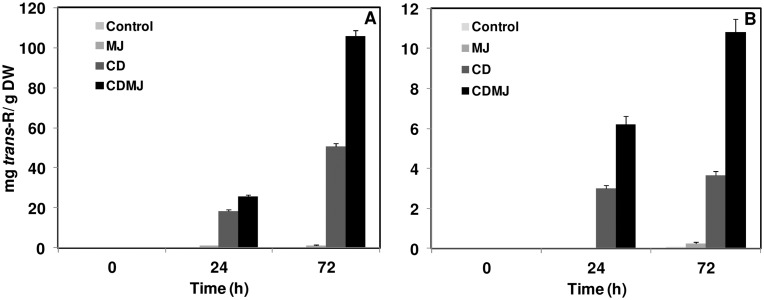
In order to compare the effects of the different treatments on the production of trans-R in the cell cultures in which the transcriptomic analyses were carried out, the level of this compound was analyzed both in cells and extracellular medium at 0, 24 and 72 h. As shown in Figure 1A, a negligible amount of trans-R was detected in the extracellular medium when cell cultures were elicited with MJ (0.019 mg g−1 dry weight (DW) that means 0.19 mg L−1) in comparison with CD-treated cells. In contrast, the level of extracellular trans-R increased linearly up to 72 h in CD-treated cells (50.4 mg DW that means 504 mg L−1) and, when cells were simultaneously elicited with CD and MJ, the accumulation of trans-R in the medium increased more than twice (105.9 mg g−1 DW that means 1059 mg L−1) (Figure 1A). Similarly, the highest trans-R levels detected in the cells were reached in the presence of CD and MJ after 72 h of treatment (10.8 mg g−1 DW that means 108 mg L−1) (Figure 1B) although it was of one order of magnitude lower than the final concentration obtained in the medium.
Figure 1. Effect of elicitation time course on trans-R (trans-resveratrol) production in grapevine cells treated with CD (cyclodextrins) and/or MJ (methyl jasmonate).
(A) trans-R content in the extracellular medium. (B) trans-R content in the cells. Experiments were repeated three times. Data are the mean ± SD of the replicates.
Global gene expression is differentially modulated by cyclodextrins and methyl jasmonate
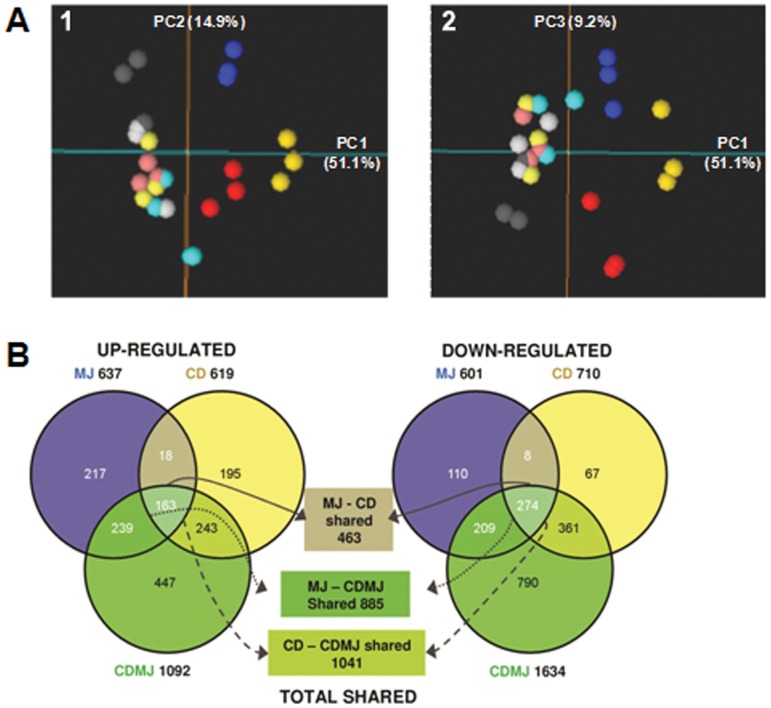
RNA samples obtained from the four treatments (C, MJ, CD and CDMJ) at 0 and 24 h were used for the gene expression analyses. In order to validate the results obtained with the microarray analysis, a quantitative real time RT-PCR (qRT-PCR) assay on 7 differentially expressed transcripts was carried out using gene-specific primers (Table S1) based on the corresponding GrapeGen GeneChip probe set sequences. Linear regression analysis displayed highly significant correlations (average R2 = 0.96±0.02) for the 7 evaluated genes (Figure S1). Altogether, the experiment was considered highly reliable to support further transcriptomic analyses. Moreover, the results of a Principal Component Analysis (PCA) plot showed a strong consistency across biological replicas, and at 0 h, no apparent effect of elicitor addition was observed, since all the samples were grouped together (Figure 2A). Moreover, a consistent biological interpretation can be extracted from the three PCs, indicating that expression differences amongst samples were dominated by the elicitation response. The principal component 1 (PC1) explained 51.1% of the variation and displayed a similar trend in the response of the cells to all treatments (CD, MJ and CDMJ), although the combined treatment presented an intensified global expression response (Figure 2A). PC2 (14.9%) pointed to the specific effect of MJ, while PC3 (9.2%) showed the individual effect of CD and MJ which in turn, resulted to be opposite (Figures 2A.1 and 2A.2).
Figure 2. A, PCA over the expression dataset after the average of the redundant probesets expression values.
1, PC1 vs PC2; 2, PC1 vs PC3. Grey, Control; Blue, MJ (methyl jasmonate); Red, CD (cyclodextrins); Yellow, CD and MJ (cyclodextrins and methyl jasmonate); Light colors, just before cell treatments; Dark colors, 24 h cell treatments. B, Genes significantly regulated by elicitor treatments. C, control; MJ, methyl jasmonate; CD, cyclodextrins; CDMJ, cyclodextrins and methyl jasmonate. Venn diagram summary of significantly regulated transcripts. Non-redundant significant transcripts were obtained from probesets showing an at least 2-fold change and 5% FDR and P-value <0.05 for model variable in the corresponding control versus treatment 24 h series maSigPro comparison. Left side, up-regulated transcripts in each treatment; Right side, down-regulated transcripts in each treatment; Center, summary of significantly regulated transcripts shared by any two treatments.
Expression changes over the whole dataset (Table S2) were analyzed by means of maSigPro time series comparisons with the control (C). In this way, we identified 3,659 differential probesets (5% FDR and two-fold change, Table S3), representing 3,306 non-redundant transcripts. Few differences were observed in the control between 0 and 24 h, while no significant differences were identified within the four samples at 0 h, even using a loose threshold (Bonferroni and Hochberg adjusted P-value <0.1; data not shown). These results demonstrate the homogeneity of the starting cell cultures for the four treatments, as it was previously shown by the PCA analysis (Figure 2A). Also consistent with the PCA results, most transcripts regulated in treatments with either CD or MJ were similarly regulated in the combined treatment, as it can be seen in the Venn diagrams comparing differentially expressed genes in each treatment (Figure 2B). In addition, a high proportion of significant transcripts were exclusively regulated by the joint action of CD and MJ.
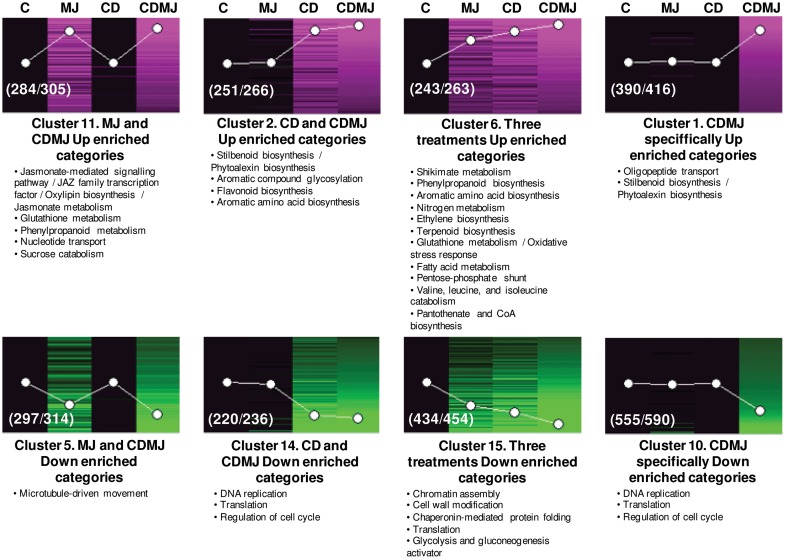
With the aim of establishing the relationship amongst the particular responses to the treatments (MJ, CD, CDMJ), significant probesets were grouped according to their expression profiles in every treatment normalized to the control. According to a ‘gap’ analysis, 15 clusters were generated in a SOM analysis that allowed the identification of specific responses for each treatment as well as those shared by them (Figure S2; Table S4). According to the differentially expressed genes clustering, the most abundant expression profiles corresponded to the combined treatment-specific response (clusters 1 and 10), followed by responses shared by all three treatments (clusters 6 and 15) (Figure S2). Self-organizing maps (SOM) clustering analysis also identified a large proportion of transcripts co-regulated by one of the individual treatments as well as by CD and MJ, the proportion corresponding to CD separately or in combination with MJ (clusters 2, 9 and 14, Figure S2) was greater than that of MJ alone and in combination with CD (clusters 5 and 11, Figure S2). As it is shown in the PCA plot, the SOM analysis also revealed that the MJ-specific response (clusters 4 and 12) was stronger than that of CD (clusters 3 and 13) (Figure S2).
Functional analysis of elicitor transcriptional responses
To understand the putative biological meaning of the different expression profiles detected, a FatiGO functional enrichment analysis (Figure 3) was performed in the transcripts grouped in each of the 15 SOM clusters (Figure S2). Significant functional enrichment (Bonferroni-Hochberg adjusted P-value <0.05 in a Fisher’s exact test) was only detected in the largest clusters (1, 2, 5, 6, 10, 11, 14 and 15; Figure 3) that corresponded to the transcript groups regulated by all treatments (clusters 6 and 15), only by CDMJ (1 and 10), by CDMJ and MJ (5 and 11) or by CDMJ and CD (2 and 14). Considering the functional categories enriched in cluster 6, the three treatments shared the activation of several pathways related with stilbene biosynthesis, including both the initial steps of the pathway and those of their precursors at different levels (i.e., nitrogen metabolism, shikimate biosynthesis, aromatic amino acid and phenylpropanoid biosynthesis; Figure 3 and Table S4). Other enriched categories in cluster 6 were ethylene and terpenoid biosynthesis, glutathione metabolism and the oxidative stress response, fatty acid metabolism, pentose-phosphate shunt, branched-chain amino acid catabolism, and pantothenate and CoA biosynthesis. In view of the expression trend in cluster 6, the magnitude of the activation of the mentioned processes was greater for CD than for MJ, and even greater for the combined treatment (Table S3).
Figure 3. Functional categories enriched in clusters of transcripts sharing the same expression response to the treatments.
Significant enrichment was only found in clusters 1, 2, 5, 6, 10, 11, 12, 14, and 15 from Figure S2. All non-redundant over-represented categories are listed below the corresponding cluster in order of significance except for related categories that are grouped in the same line. Significant enrichment according to Bonferroni-Hochberg adjusted P-value <0.05 in a Fisher’s exact test. Magenta and green intensity, level of up-regulated or down-regulated expression respectively, 24 h after treatment and normalized to the control; Black, transcripts not significantly regulated by the treatment. C, control; MJ, methyl jasmonate; CD, cyclodextrins; CDMJ; cyclodextrins and methyl jasmonate. The numbers in brackets indicate non-redundant transcripts/total probesets included in the cluster.
On the other hand, jasmonate metabolism and signalling mediated by jasmonate were the most significantly enriched categories in the cluster of transcripts up-regulated by MJ as well as by the combined treatment (cluster 11, Figure 3). Within the same profile, a significant enrichment of other categories like the glutathione and phenylpropanoid metabolism, nucleotide transport and sucrose catabolism was found. As regards transcripts up-regulated only by CD and CDMJ (cluster 2, Figure 3), the functional enrichment analysis pointed to the activation of stilbenoid and flavonoid biosynthesis by CD as well as aromatic compound glycosylation and aromatic amino acid biosynthesis. The concurrent presence of CD and MJ synergistically activated stilbenoid biosynthesis, since this category was also over-represented in the CDMJ specifically up-regulated transcripts (cluster 1, Figure 3). Moreover, the expression of oligopeptide transporters was also specifically activated by the combined treatment.
The enriched functions within the transcripts repressed by the three treatments included basic cellular processes like chromatin assembly, translation, chaperonin-mediated protein folding and cell wall modification (cluster 15, Figure 3). As regards the genes repressed by MJ and CDMJ, an over-representation of the cytoskeleton organization (cluster 5, Figure 3) was observed. Finally, CD and CDMJ repressed DNA replication and the regulation of cell cycle-related processes, while some transcripts involved in those processes were repressed only by the combined treatment (cluster 14 and 10, respectively, Figure 3).
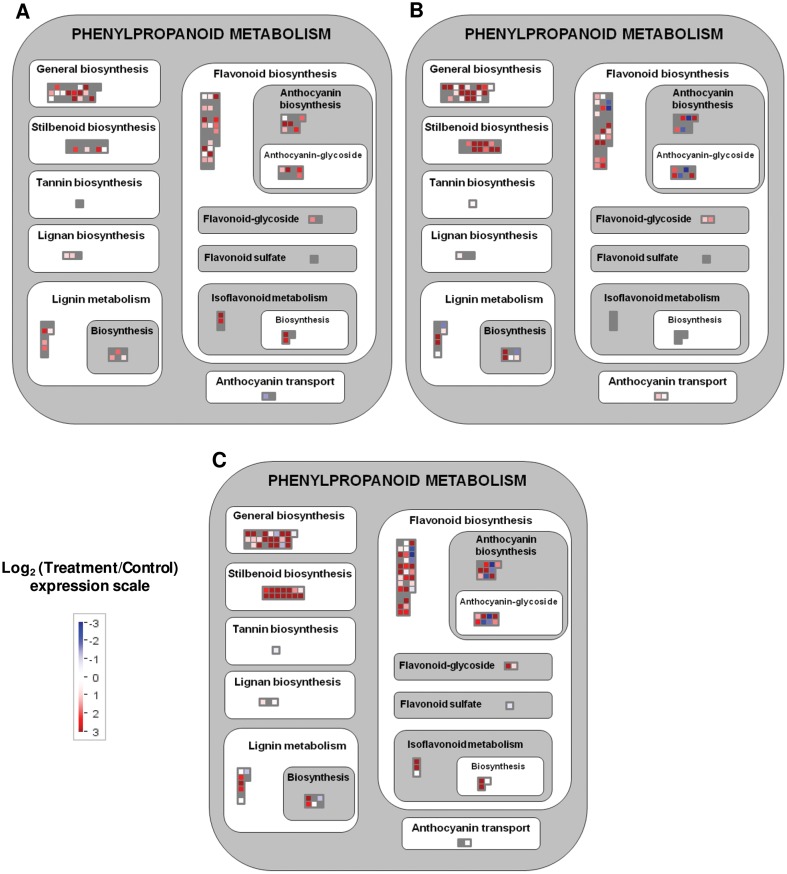
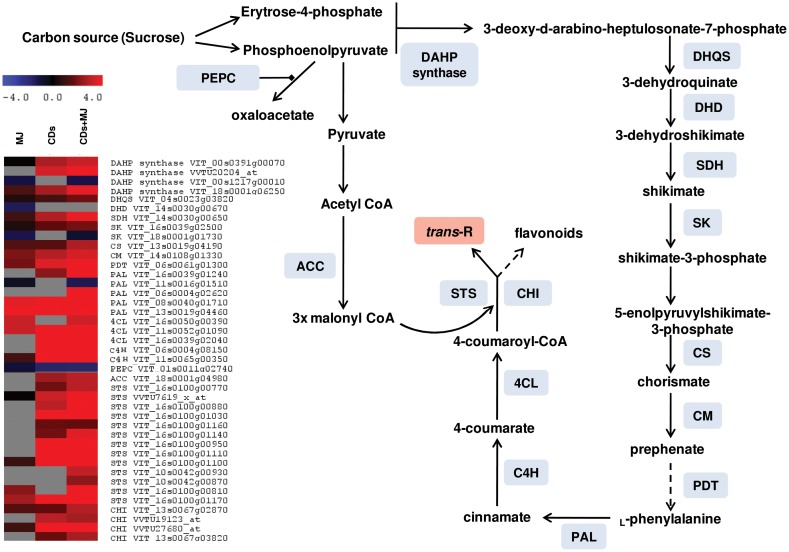
A graphical overview of the main pathways altered by the different treatments was depicted using the MapMan software. Several components of the phenylpropanoid pathway were widely activated by the presence of elicitors (Figures 4 and 5 and Table S3). More particularly, CD separately or in combination with MJ activated a greater number of PAL, 4CL, C4H and STS genes than MJ alone (Figure 5). Moreover, the activation of some phenylpropanoid biosynthesis precursors, like shikimate and aromatic amino acids, was significantly higher in cells treated with CD (separately or in combination with MJ) than in MJ-treated cells (Figure S3). In fact, CD and CDMJ strongly increased the expression of genes like 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAHP), 3-dehydroquinate synthase (DHQS), dehydroquinate dehydratase (DHD), shikimate dehydrogenase (SDH), shikimate kinase (SK), chorismate synthase (CS), chorismate mutase (CM), and prephenate dehydratase (PDT) (Figure 5). Interestingly, most of these genes were more highly induced by CDMJ than by CD. In contrast, MJ alone only provoked a slight increase in the expression of DAHP, SDH, CS, CM and PDT genes (Figure 5). Moreover, it is important to highlight that CD and MJ provoked the down-regulation of one gene encoding one phosphoenolpyruvate carboxylase (PEPC), whereas one acetyl-CoA carboxylase (ACC) involved in malonyl-CoA biosynthesis was only up-regulated in the presence of CD (Figure 5).
Figure 4. Mapman visualization of the significant genes in the ‘Phenylpropanoid metabolism’ pathway.
A, MJ (methyl jasmonate); B, CD (cyclodextrins); C, CDMJ (cyclodextrins and methyl jasmonate). Resultant transcripts were considered after the average of significant redundant probesets expression values. Significant probesets according to a 5% FDR and P-value <0.05 for model variable in the corresponding control versus treatment 24 h series maSigPro comparison. Expression changes in the treatment normalized to these in the control are shown. Red, treatment up-regulated transcripts; blue, treatment down-regulated transcripts; grey, transcripts not significant in the treatment.
Figure 5. Modulation of trans-resveratrol biosynthesis pathway encoding enzymes gene expression by CD (cyclodextrins) and/or MJ (methyl jasmonate).
Enzyme abbreviations: 4CL, 4-coumarate CoA ligase; ACC, acetyl-CoA carboxylase; C4H, cinnamate-4-hydroxylase; CHI, chalcone isomerase; CM, chorismate mutase; CS, chorismate synthase; DAHP synthase, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase; DHD, dehydroquinate dehydratase; DHQS, 3-dehydroquinate synthase; PAL, phenylalanine ammonia lyase; PDT, prephenate dehydratase; PEPC, phosphoenolpyruvate carboxylase; SDH, shikimate dehydrogenase; SK, shikimate kinase; STS, stilbene synthase.
In relation to transport overview, the joint action of CD and MJ enhanced the expression of transcripts related with the transport of amino acids, oligopeptides and nucleotides (Figure S4). The activation of multidrug transporters was considerably higher in cells treated with MJ separately or in combination with CD than in CD-treated cells.
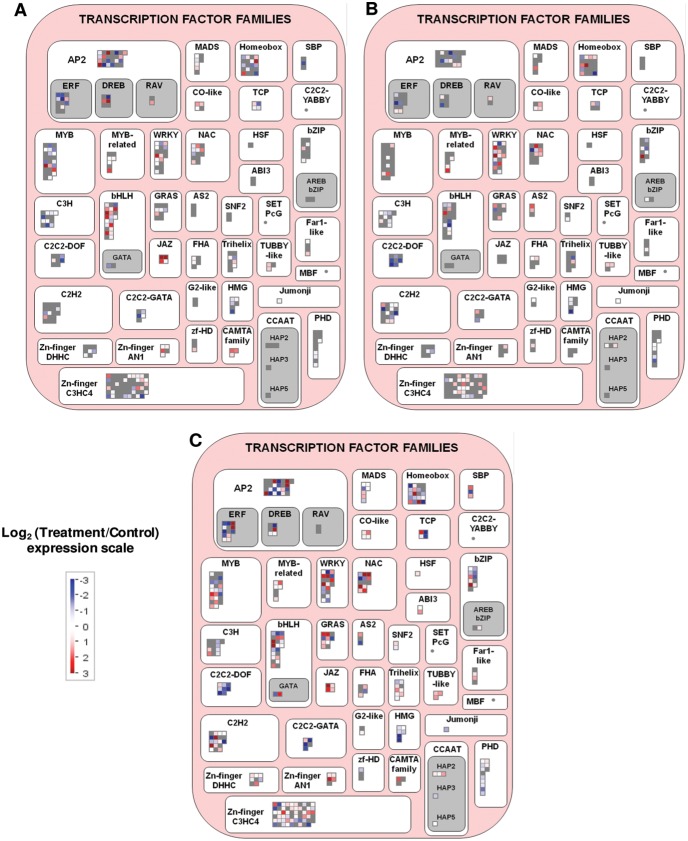
On the other hand, MJ induced both JAZ type and bHLH TFs, while CD significantly induced WRKY and NAC TFs, and repressed the C2C2-DOF (Figure 6 and Table S3). Moreover, CD and MJ applied together enhanced the induction of some WRKY, NAC, AP2 and JAZ type TFs (Figure 6). In relation to JAZ expression, the activation of jasmonic acid (JA) metabolism and signalling-related transcripts was higher in MJ-treated cells (Figure S5).
Figure 6. Mapman visualization of the significant genes in the ‘Transcription factor families’ functional category.
A, MJ (methyl jasmonate); B, CD (cyclodextrins); C, CDMJ (cyclodextrins and methyl jasmonate). Resultant transcripts were considered after the average of significant redundant probesets expression values. Significant probesets according to a 5% FDR and P-value <0.05 for model variable in the corresponding control versus treatment 24 h series maSigPro comparison. Expression changes in the treatment normalized to these in the control are shown. Red, treatment up-regulated transcripts; blue, treatment down-regulated transcripts; grey, transcripts not significant in the treatment.
As regards hormonal regulation, all the treatments activated ethylene biosynthesis genes and repressed salicylic responsive genes (Figure S5). Finally, the CDMJ treatment also activated the expression of transcripts related with brassinosteroid signalling, and metabolism and transport of auxins (Figure S5).
Discussion
Cyclodextrins and/or methyl jasmonate induced the biosynthesis of defense-related proteins
The use of CD and MJ, separately or in combination as elicitors, has been proved to be very effective in cell reinforcing, stimulating the defensive arsenal in the apoplast-like extracellular medium of grapevine cell cultures through the accumulation of trans-R, and PR-proteins [2], [21], [24], [25]. In fact, functional categories enriched in clusters of transcripts from stilbenoid biosynthesis were found in CD and CD+MJ treatments (clusters 1 and 2, Figure 3). In addition, individual genes encoding PR-proteins were up-regulated in all treatments, more specifically two class IV chitinases (VIT_05s0094g00340 and VIT_05s0094g00330) and one class I endochitinase (VIT_03s0038g03400). In addition, one acidic class III chitinases (VIT_15s0046g01590) and one thaumatin-like protein (VIT_02s0025g04330) were also up-regulated in the MJ and CDMJ treatments (Table S3), and a protease inhibitor cystatin (VIT_00s0187g00040) in MJ-treated cells (Table S3). Moreover, defense responses related to oxidative stress were also induced by all treatments (Figure 3). These results are in accordance with those shown by Belchí-Navarro et al. [28] since the CD-mediated trans-R accumulation in V. vinifera cv Monastrell cell cultures was dependent on H2O2 production, and H2O2 levels increased significantly in the combined treatment with MJ after 24 h. In this sense, several peroxidases were down-regulated in CDMJ-treated cells (Table S3), what is consistent with higher H2O2 levels and higher trans-R accumulation in the combined treatment.
On the other hand, proteins that participate in the regulation of cellular redox that protect from the oxidative stress were also up-regulated in all treatments (cluster 6, Table S3). Amongst them, a METHIONINE SULFOXIDE REDUCTASE (VIT_19s0014g00170, Table S3) that reduces methionine sulfoxide in a thioredoxin-dependent reaction, provides a pathway to repair proteins damaged by reactive oxygen species in cells instead of having them be degraded [29].
The up-regulation of genes involved in the biosynthesis of trans-resveratrol by cyclodextrin is enhanced in the presence of methyl jasmonate
Grapevine cell cultures elicited with CD and/or MJ displayed a significant inductive effect on biosynthetic processes related with phenylpropanoid and stilbenoid biosynthesis (clusters 1, 2, 11 and 6, Figure 3). Altogether, the biosynthesis of stilbenoids was up-regulated from the very early steps leading to the precursors, which involved genes coding for synthesis of phosphoenolpyruvate (enolase, VIT_17s0000g04540, Table S3) and D-erythrose 4-phosphate (functional category pentose-phosphate shunt, cluster 6, Figure 3), both responsible for feeding the shikimate pathway (Figure 5). In addition, the over-expression of genes encoding enzymes involved in the shikimate metabolism, aromatic amino acid, phenylpropanoid and stilbenoid biosynthesis was observed principally in the presence of CD either alone or in combination with MJ (clusters 1, 2, 11 and 6, Figure 3), thereby promoting the carbon flux toward these pathways (Figure 5) what is well-correlated with the high trans-R production found in these treatments (Figure 1). The simultaneous enrichment of functional categories corresponding to nitrogen metabolism, pantothenate and CoA biosynthesis and fatty acid metabolism within the transcripts up-regulated in all treatments (cluster 6, Figure 3) partly resulted from the presence in this cluster of genes encoding enzymes involved in the biosynthesis of acetyl-CoA and malonyl-CoA, such as ATP citrate lyase (VIT_05s0077g00950) and acetyl-CoA carboxylase (ACC, VIT_18s0001g04980), respectively (Table S3). Such elicitor response whose magnitude was greater in CD-treated cells and enhanced in CDMJ-treated cultures could be directed towards the production of phenylalanine and precursors of carbon skeletons for the biosynthesis of stilbenes and other phenylpropanoid compounds. Moreover, it is important to highlight a down-regulation of one gene encoding PEPC (VIT_01s0011g02740, Figure 5 and Table S3) in all treatments. Phosphoenolpyruvate is a key precursor in the production of oxaloacetate via PEPC, 4-coumaroyl CoA, via the shikimate-phenylpropanoid pathway, and also a precursor of malonyl-CoA, via pyruvate kinase, pyruvate dehydrogenase and ACC. Therefore, the down-regulation of PEPC might lead to a depletion of oxaloacetate in favor of phosphoenolpyruvate, which would provoke a critical change in the partition of carbon flux between primary and secondary metabolism since the excess of acetyl-CoA produced from pyruvate in CD- and CDMJ-treated cells would be used for biosynthesis of malonyl-CoA, which is extensively used for the production of stilbenes and flavonoids (Figure 5, [30]). Considering that up to a 20% of the carbon supplied to the cells as sucrose (20 g·l−1) is recovered as trans-R in the culture medium (around 4 g·l−1) after several days of elicitation [2], [31], the role of PEPC as a point to decide the fate of the incoming carbon could be of great relevance.
PAL, C4H and 4CL genes encoding enzymes of the general phenylpropanoid biosynthetic pathway, and particularly these together with STS corresponding to stilbene biosynthesis were largely up-regulated by CD, and this response was intensified in the presence of MJ (Figure 4B and 4C and Table S3), and correlated with a high production of trans-R (Figure 1). Similar results have been found by Martínez-Esteso et al. [25] who observed that the addition of CD and CDMJ to V. vinifera cv Gamay cell cultures provoked a large increase in trans-R production which was correlated with an increase in STS protein abundance. Overall, the activation of both, phenylpropanoid precursors and stilbene gene expression by CD together with the enhancement of this activation in the presence of MJ, was highly correlated with the high levels of trans-R induced in grapevine cell cultures elicited with CD and MJ (Figure 1). Additionally, many transcripts coding for enzymes in other branches of the phenylpropanoid metabolism, including flavonoid and monolignol biosynthesis, were strongly up-regulated by the presence of CD and/or MJ (Table S3). These findings are in agreement with other studies performed in Arabidopsis [22] and grapevine [32]–[33] cell cultures.
On the other hand, the pathways for transport of trans-R to the extracellular medium in grapevine cells, being of utmost biotechnological relevance, are mostly unknown. In this sense, an increase in the transcript levels related to MATE and ABC transporters, which was more intense in cells treated with MJ alone or combined with CD than in CD-treated cells, was observed (Figure S4 and Table S3). In addition, some glutathione transferases, which are involved in the trafficking and accumulation of secondary metabolites such as anthocyanins [34]–[36] and in the sequestration of xenobiotics [37], were specifically up-regulated by MJ or CDMJ (cluster 11 in Figure 3 and Table S4). In agreement with our results, Martinez-Esteso et al. [25] observed that CD and MJ provoked an increase of glutathione transferases in V. vinifera cv Gamay cell cultures, with abundance profiles similar to those found for STS gene expression, and suggested the existence of a coordinated action between the biosynthesis and transport of trans-R in grapevine cells treated with CD and MJ. Future targeted studies are required to determine whether ABC or MATE transporters and glutathione transferases induced by CD and/or MJ (Table S3) would play a role in trans-R mobilization or sequestration.
Regulatory cascades activated by cyclodextrins and/or methyl jasmonate
Consideration of the putative regulatory genes showing an altered expression in grapevine cell cultures elicited with CD and/or MJ, can help understanding the different responses that they trigger. The elicitation response triggered by MJ involves the JA signalling cascade, as it was shown in the functional analysis (Figure 3 and Table S4). Although jasmonate responses are controlled at the protein level, via ubiquitin-dependent proteolysis of JASMONATE ZIM-DOMAIN (JAZ) TF [38]–[39], MJ up-regulated MYC2 and MYC3 grapevine homologues (VIT_15s0046g00320 and VIT_02s0012g01320 in Table S2), which are the main factors triggering JA responses that directly interact with JAZ proteins in Arabidopsis [40]–[41]. Concurrently, three JAZ TF encoding genes (VIT_01s0146g00480, VIT_11s0016g00710 and VIT_17s0000g02230; Table S2), which are the main repressors of JA responses [38]–[39], were also up-regulated in response to MJ, suggesting a mechanism which is activated as a result of an increased sensitivity of the signalling pathway in response to the treatment. In fact, a positive self-regulating loop activating oxylipin and JA biosynthesis also seems to occur in the presence of MJ (Figure 3 and Table S4). This agrees with other studies showing that jasmonate biosynthesis and its signalling are interlinked by a positive feedback loop whereby jasmonates stimulate their own biosynthesis [42]. Besides, genes encoding other TFs like WRKY (VIT_11s0052g00450 and VIT_14s0108g01280), NAC (VIT_13s0019g05240), ARF2 (VIT_17s0000g00320) and MYB (VIT_08s0007g07230) (Figure 6 and Table S3) were significantly activated only in the presence of MJ. Therefore, the cell responses transcriptionally activated by MJ may operate by inducing these TFs.
Bearing in mind that CD and CDMJ-mediated trans-R accumulation is held for several days, it is interesting to note that transcripts encoding proteins involved in putative early signalling events were up-regulated. For instance, the presence of CD up-regulated nine transcripts coding for protein receptor-like kinases and three transcripts for calmodulins, while the joint addition of CD and MJ up-regulated nineteen kinases and receptor-like kinases, and four calcium sensors (Table S3). Our studies on early signalling events using pharmacological approaches have pointed out the central role of phosphorylation/dephosphorylation cascades and calcium signalling in the production of trans-R in grapevine cell cultures [28]. Additionally, other TFs such as MYB15 (VIT_05s0049g01020), three NACs (VIT_08s0007g07670, VIT_18s0001g02300 and VIT_19s0014g03290) and a WRKY (VIT_09s0018g00240) recently annotated as VvWRKY30 [43], were up-regulated only in the presence of both elicitors (Figure 6 and Table S3), indicating that they could participate in the enhancement of the elicitation response when both compounds are added together. Indeed, MYB15 and MYB14 TFs specifically activate the promoter of STS genes in grapevine [15], indicating that the enhanced expression of STS genes in CDMJ-treated cells may result, at least in part, from the up-regulation of MYB15. Although the significant GrapeGen GeneChip VVTU9342_was previously annotated as MYB14 (Table S2; [44]), the blast alignment of the probeset confirmed that it actually corresponds to the MYB15 grapevine gene [45]. Unfortunately probesets representing the current MYB14 gene (VIT_07s0005g03340) are missing from the GrapeGen GeneChip and therefore, its expression was not tested in our work. In the same way, VvWRKY30 might be another STS expression-promoting TF in view that it was also co-induced with STS genes in grapevine leaves exposed to UV light, as well as in response to circadian rhythms that occur in grapevine fruits [46]–[47]. Furthermore, the identification of MYB15, VvWRKY30 and NAC TFs and protein kinases that are specifically induced by the combined treatment suggests the synergistic activation of additional regulatory pathways due to crosstalk between the signalling cascades activated by CD and MJ. In this way, the induction of genes encoding protein kinases by CD might result in an enhancement of the elicitation responses activated by the two WRKY genes (cited above) induced only in the presence of MJ. Also, the gibberellin signalling repressor VvGAI1 might be another node of crosstalk between CD and MJ signalling, resulting in increased jasmonate responses, since this DELLA gene (VIT_01s0011g05260) was specifically up-regulated in the presence of CD and MJ. In Arabidopsis, DELLA proteins bind to MYC2 preventing its interaction to JAZ repressors and allowing the activation of JA responses [48]–[49].
In parallel, a putative MAPKKK encoding gene (VIT_07s0031g00530) which was up-regulated in a similar extent in grapevine cell cultures treated with MJ alone or combined with CD, might participate in the enhancement of phosphorylation cascades triggered by CD.
The antagonisms and synergisms between JA and SA signals in the activation of pathogen responses are well documented [50]. These compounds coordinate the activation of a large set of defense responses and, when applied exogenously, they are able to induce resistance in both plants and cell cultures [11]. It is well known that most of the SA produced in planta is converted into SA glucoside by a pathogen-inducible SA glucosyltransferase [51]. This SA glucoside is actively transported from the cytosol to the vacuole in soybean and tobacco cells, where this inactive form of SA is stored [52]–[53]. Noticeably, we observed over-expression of genes encoding enzymes like UDP-glucose glucosyltransferase and UDP-glucose:SA glucosyltransferase, only in the presence of CD (Table S3). Moreover, SA-responsive transcripts were down-regulated upon elicitor treatment, and especially in the combined treatment (Table S3). These observations are consistent with the decrease in trans-R levels provoked by the addition of SA to grapevine cell cultures [21], suggesting that MJ and CD might increase trans-R biosynthesis through the attenuation of SA signalling induced by SA conjugation.
Finally, as ethylene biosynthesis is activated in all treatments, this molecule or its signalling may participate in the regulation of responses triggered by CD and/or MJ (Cluster 6, Figure 3). In fact, a great number of transcripts involved in ethylene signalling, like an EIN4 putative ethylene receptor encoding gene (VIT_14s0081g00630), several ethylene response factors (ERF, VIT_07s0141g00690, VIT_10s0003g00580 and VIT_15s0021g01630; Table S3), and those from ethylene biosynthetic pathway were up-regulated in grapevine cells treated with CDMJ. In agreement with our results, Zamboni et al. [32] also observed the induction of a 1-aminocyclopropane-1-carboxylate oxidase in V. riparia × V. berlandieri cell cultures elicited with CD after 6 h of treatment.
Growth-related gene expression is repressed upon elicitor treatment
Microarray analysis of grapevine cell cultures elicited with CD and/or MJ showed a specific down-regulation of genes involved in the control of cell wall modification (cluster 15, Figure 3 and Table S3). These results paralleled those of Pontin et al. [46], who showed a reduction in cell wall loosening-related gene expression concurrently to STS genes induction as part of the defense mechanism against UV-B in V. vinifera cv Malbec leaves. Our results also showed that genes encoding enzymes involved in microtubule-driven movement were down-regulated in all treatments (Table S3) although this effect was stronger in the presence of MJ (cluster 5, Figure 3 and Tables S3 and S4). It is well-documented that the plant cytoskeleton is a highly dynamic and versatile scaffold composed of microtubules and actin microfilaments that plays an important role in many aspects of cell growth, including cell division and expansion, and intracellular organization [54]–[55]. The down-regulation of genes coding for α- and β-tubulin, villin, and kinesin in cells treated with CD and/or MJ (Cluster 5, Figure 3 and Tables S3 and S4) might indicate an arrest in both cell division and expansion. In fact, Ebel et al. [56] observed that mRNA degradation of a β-tubulin in soybean cells elicited with Phytophthora sojae provoked a repression of cell growth, and a regulation of cell expansion controlled by the cytoskeleton has also been shown in grapevine cell cultures [57]. Also, a down-regulation of genes encoding enzymes involved in both microtubule-driven movement and vesicle-mediated trafficking (Table S3) was observed in MJ treatments. This result agrees with the functional role of cytoskeleton in cellular trafficking mechanisms, that regulates the movement of the vesicles and endosomes on actin filaments or microtubules [58]. Moreover, the presence of CD provoked a significant repression of genes related to DNA replication, translation and the regulation of the cell cycle, responses that were even more intensively repressed when grapevine cells were elicited with CDMJ. These results suggest that the presence of CD predominantly decreased cell growth and division processes. As regards to the regulation of the cell cycle, it is important to highlight that a cell division cycle protein 48 encoding gene (CDC48, VIT_10s0071g00680) was specifically down-regulated in the presence of CD (Table S3). In fact, CDC48 protein is an important regulator of the cell cycle, and is required for the cell-cycle commitment point via degradation of the G1-cyclin-dependent kinase inhibitor [59]. In the same way, CDC48 protein promotes cell proliferation in maize roots [60] and it is also down-regulated upon elicitation in other species like tobacco [61]. Taking into account these results, CD could reduce cell proliferation through the down-regulation of CDC48. In addition, a probable explanation of the strong repression of cell division triggered by CD and MJ might be the need of the cell to redirect all the available resources to the activation of a defense-related metabolism. CD and MJ also strongly down-regulated translation-related transcripts like a set of ribosomal proteins (Table S3) as it has also been reported in soybean and rice cell cultures treated with fungal pathogens [62]–[63]. Therefore, the arrest of the cell cycle and the decrease of the expression of a set of ribosomal proteins in the grapevine cells treated with both elicitors would lead to the suppression of protein translation, thus contributing to saving energy. Finally, the level of repression of these basic processes is well-correlated with the high levels of trans-R observed in the elicited grapevine cell cultures (Figure 1 and Table S4), indicated that CD and MJ activated the secondary metabolism in detriment of basic cell processes like primary metabolism and cell division.
Experimental Procedures
Plant material
V. vinifera cv Monastrell calli were established in 1990 as described by Calderón et al. [64]. Grapevine cell cultures derived from them have been routinely maintained by periodical subcultures as described by Belchí-Navarro et al. [21].
Elicitor treatments
Elicitation experiments were performed in triplicate using 12 day old grapevine cell cultures. At this stage of cell development, 20 g of fresh weight of cells were transferred into 250 mL flask and suspended in 100 mL of fresh culture medium described by Belchí-Navarro et al. [21]. Then, cell cultures were maintained (110 rpm, 25°C) during 10 h at 25°C in darkness in a rotary shaker (110 rpm) and then, they were supplemented with 50 mM CD (Wacker Chemie, Spain) and/or 100 µM MJ (Duchefa, Spain). Control treatments without elicitors (C) were always run in parallel. All cell cultures were incubated up to 72 h under the same conditions described above. After elicitation, cells were separated from the culture medium by filtration, rapidly washed with cold distilled water, weighted and frozen at −80°C until use. The elicited culture medium was used for trans-R quantification.
Quantification of trans-resveratrol in both culture medium and cells
For this, 20 µL of the spent medium were analyzed in a HPLC-DAD (Waters 600E, Waters 996) as described by Belchí-Navarro et al. (2012). In addition, 50 mg of freeze-dried cells were extracted overnight in 4 mL methanol at 4°C. The cell extract was diluted with water to a final concentration of 80% (v/v) methanol. Then, 20 µl of the diluted extracts was filtered (Anopore 0.2 µm) and analyzed in a HPLC-DAD (Waters 600E, Waters 996) as described by Bru et al. (2006) using a Spherisorb ODS2 C-18 column (250×4.6 mm, 5 µm). trans-R was identified at 304 nm and quantified by comparison with authentic standard of >99% purity (Sigma-Aldrich, Spain).
Gene expression analyses
RNA isolation, cDNA synthesis and real-time quantitative PCR
To validate the microarray expression data were performed qRT-PCR experiments. For this purpose, Total RNA was isolated from frozen cells (0.5 g fresh weight) using the TRIZOL reagent (INVITROGEN, Spain) following the manufacture’s recommendations. The concentration of each RNA sample was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Only the RNA samples with a 260/280 ratio between 1.9 and 2.1 were used for the analysis. The integrity of RNA samples was also assessed by agarose gel electrophoresis. One µg of total RNA from each sample was reverse-transcribed by first-strand cDNA synthesis using MMLV RT (Invitrogen), according to the manufacturer’s instructions. SYBR Green PCR Core Reagents (Applied Biosystems) were used for qRT-PCR in an ABI PRISM 7500 instrument (Applied Biosystems). Gene specific primers (Table S1) were designed using the OligoAnalyzer 3.1 software (IDT, Integrated DNA Technologies) based on the corresponding probeset sequence from the custom GrapeGen GeneChip. For each primer pair, reaction efficiency estimates were derived from a standard curve generated from a serial dilution of pooled cDNA. For each gene, the expression levels were normalized with respect to grapevine EFα1 gene (Forward primer: GAACTGGGTGCTTGATAGGC; Reverse primer: AACCAAAATATCCGGAGTAAAAGA) used as reference control as described Lijavetzky et al. [27] and Reid et al. [65]. The same RNA samples used for the microarray hybridizations were used for the synthesis of the cDNA analyzed by qRT-PCR: three biological replicates of C, CD, MJ and the combined treatment (CDMJ) at 0 and 24 h. Correlation coefficients of 0 to 24 h expression ratios on each treatment normalized to the same expression temporal ratio in the control between microarray and qRT-PCR data were calculated for validation.
GeneChip hybridization
Samples of 0 and 24 h from each treatment were hybridized at the Genomics Unit of the National Biotechnology Centre (CNB-CSIC, Madrid). RNA integrity analysis was performed with an Agilents Bioanalyzer 2100. The custom GrapeGen Affymetrix GeneChip (A-AFFY-162 and GPL11004 ArrayExpress and GEO accession numbers, respectively), which contained 23,096 probesets, corresponding to 18,711 non-redundant grapevine transcripts [66] was processed as previously described [46].
Expression data preprocessing and Principal Component Analysis plot
The full GeneChip raw expression dataset is available on PlexDB [67] under the accession number VV44. Probeset signal values from all the microarray hybridizations were normalized together using Robust Microarray Average (RMA) [68] by RMA Express (http://rmaexpress.bmbolstad.com). A PCA [69] was directed over the full dataset after the average of redundant probesets expression values. Probeset redundancy was reduced according to the GrapeGen GeneChip 12Xv1 annotations version [44] using Babelomics preprocessing tools [70]. The three first Principal Components were analyzed using Acuity 4.0 (Axon Molecular Devices, http://www.moleculardevices.com).
Identification and clustering of differentially accumulated transcripts
Each treatment-time series was compared to the control in a maSigPro analysis conducted in Babelomics suite from RMA normalized expression data [70]–[71]. A 0.05 significance level after Benjamini and Hochberg multiple test adjustment and P-value <0.05 for model variable were applied. Besides, probesets were considered significant only when the expression difference normalized to the control was more than 2-fold in at least one treatment. RMA normalized data, maSigPro significance level and expression ratio for all probesets can be found in Table S2. The significant transcripts identified were grouped according to shared responses normalized to the control. SOM clustering [72] with Euclidean squared and Scale rows metrics was applied in Acuity 4.0 for the clustering. Fifteen groups were generated in a 5×3 clustering as estimated by gap statistics [73] in Acuity 4.0. Venn diagrams comparing up-regulated and down-regulated non-redundant transcripts among treatments were performed in Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Functional analysis
Non-redundant GrapeGen 12Xv1 transcripts found within each expression profile cluster were analyzed in FatiGO [74] to search for significant enrichment of GrapeGen 12Xv1 functional categories [44]. Fisher’s exact test was carried out in FatiGO to compare each study list with the list of total non-redundant transcripts housed in the GrapeGen GeneChip. Significant enrichment was considered in case of P-value <0.05 after Benjamini and Hochberg correction for multiple testing. GrapeGen 12Xv1 MapMan pathways [44] were selected, taking into account significantly over-represented functional categories for gene expression responses to treatments, to be depicted in MapMan software [75]. An expression heatmap including differentially expressed genes in the biosynthetic pathway leading to stilbene production was performed in MeV version 4.8 [76].
Supporting Information
qRT-PCR expression validation of the microarray hybridization experiments.
(DOCX)
Treatments significantly regulated gene expression profiling summary.
(DOCX)
Mapman visualization of the significant genes in the ‘Amino acid metabolism’ pathway.
(DOCX)
Mapman visualization of the significant genes in the ‘Transport overview’ pathway.
(DOCX)
Mapman visualization of the significant genes in the ‘Hormone signalling’ pathway.
(DOCX)
PCR primers used to amplify gene-specific regions for expression analyses.
(XLS)
RMA normalized expression dataset. Log2 RMA GrapeGen GeneChip probesets signal in each sample.
(XLSX)
Probesets significantly regulated 24 h after any treatment.
(XLS)
Functional categories over-represented in each expression cluster of significantly regulated genes.
(XLS)
Acknowledgments
We are grateful to the European Cooperation in the field of Scientific and Technical Research (Cost Actions FA1006 and FA1106).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Fundación Séneca, Agencia de Ciencia y Tecnología de la Región de Murcia en el marco de II PCTRM 2007-10 (08799/PI/08) and Ministerio de Economía y Competitividad-Fondo Europeo de Desarrollo Regional (BIO2008-03892 and BIO2011-29856-C02-02); Agencia Nacional de Promoción Científica y Tecnológica-MINCYT-Argentina (PAE-PICT 2360). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pezet R, Gindro K, Viret O, Richter H (2004) Effects of resveratrol and pterostilbene on Plasmopara viticola zoospore mobility and disease development. Vitis 43: 145–148. [Google Scholar]
- 2. Bru R, Selles S, Casado-Vela J, Belchí-Navarro S, Pedreño MA (2006) Modified cyclodextrins are chemically defined glucan inducers of defense responses in grapevine cell cultures. J Agri Food Chem 54: 65–71. [DOI] [PubMed] [Google Scholar]
- 3. Adrian M, Jeandet P (2012) Effects of resveratrol on the ultrastructure of Botrytis cinerea conidia and biological significance in plant/pathogen interactions. Fitoterapia 83: 1345–1350. [DOI] [PubMed] [Google Scholar]
- 4. Cantos E, Espín JC, Fernández MJ, Oliva J, Tomás-Barberán FA (2003) Postharvest UV-C irradiated grapes as potential source for producing stilbene-enriched red wines. J Agri Food Chem 51: 1208–1214. [DOI] [PubMed] [Google Scholar]
- 5. Wang W, Tang K, Yang HR, Wen PF, Zhang P, et al. (2010) Distribution of resveratrol and stilbene synthase in young grape plants (Vitis vinifera L. cv. cabernet sauvignon) and the effect of UV-C on its accumulation. Plant Physiology and Biochemistry 48: 142–152. [DOI] [PubMed] [Google Scholar]
- 6. Okawara M, Katsuki H, Kurimoto E, Shibata H, Kume T, et al. (2007) Resveratrol protects dopaminergic neurons in midbrain slice culture from multiple insults. Biochem Pharmacol 73: 550–560. [DOI] [PubMed] [Google Scholar]
- 7. Bradamante S, Barenghi L, Villa A (2004) Cardiovascular protective effects of resveratrol. Cardiovasc Drug Rev 22: 169–188. [DOI] [PubMed] [Google Scholar]
- 8. Vang O, Ahmad N, Baile CA, Baur JA, Brown K, et al. (2011) What is new for an old molecule?. Systematic review and recommendations on the use of resveratrol. PLoS ONE 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernández-Pérez F, Belchí-Navarro S, Almagro L, Bru R, Pedreño MA, et al. (2012) Cytotoxic effect of natural trans-Resveratrol obtained from elicited Vitis vinifera cell cultures on three cancer cell lines. Plant Foods Hum. Nutr 67: 422–429. [DOI] [PubMed] [Google Scholar]
- 10. De la Lastra CA, Villegas I (2005) Resveratrol as an anti-inflammatory and anti-aging agent: mechanisms and clinical implications. Mol Nutr Food Res 49: 405–430. [DOI] [PubMed] [Google Scholar]
- 11.Almagro L, Belchí-Navarro S, Sabater-Jara AB, Vera-Urbina JC, Selles-Marchart S, et al. (2013) Bioproduction of trans-resveratrol from grapevine cell cultures. In: Ramawat KG and Merillon JM, editors. Handbook of Natural Products. Heidelberg, Springer. 1683–1713. [Google Scholar]
- 12. Chong JL, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177: 143–155. [Google Scholar]
- 13. Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, et al. (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483. [DOI] [PubMed] [Google Scholar]
- 14. Czemmel S, Heppel SC, Bogs J (2012) R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249: 109–118. [DOI] [PubMed] [Google Scholar]
- 15. Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, et al. (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera . Plant Cell 25: 4135–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferri M, Tassoni A, Franceschetti M, Righetti L, Naldrett MJ, et al. (2009) Chitosan treatment induces changes of protein expression profile and stilbene distribution in Vitis vinifera cell suspensions. Proteomics 9: 610–624. [DOI] [PubMed] [Google Scholar]
- 17. Tassoni A, Fornalè S, Franceschetti M, Musiani F, Michael AJ, et al. (2005) Jasmonates and Na-orthovanadate promote resveratrol production in V. vinifera cv. Barbera cell cultures. New Phytol 166: 895–905. [DOI] [PubMed] [Google Scholar]
- 18. Donnez D, Kim KH, Antoine S, Conreux A, De Luca V, et al. (2011) Bioproduction of resveratrol and viniferins by an elicited grapevine cell culture in a 2L stirred bioreactor; Process Biochemistry. 46: 1056–1062. [Google Scholar]
- 19. Santamaria AR, Mulinacci N, Valletta A, Innocenti M, Pasqua G (2011) Effects of elicitors on the production of resveratrol and viniferins in cell cultures of Vitis vinifera L. cv Italia. J Agri Food Chem 59: 9094–9101. [DOI] [PubMed] [Google Scholar]
- 20. Kiselev KV (2011) Perspectives for production and application of resveratrol. Appl Microbiol Biotechnol 90: 417–425. [DOI] [PubMed] [Google Scholar]
- 22. Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, et al. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morales M, Bru R, García-Carmona F, Ros Barceló A, Pedreño MA (1998) Effect of dimethyl-β-cyclodextrins on resveratrol metabolism in Gamay grapevine cell cultures before and after inoculation with shape Xylophilus ampelinus . Plant Cell Tiss Org Cult 53: 179–187. [Google Scholar]
- 24. Martínez-Esteso MJ, Sellés-Marchart S, Vera-Urbina JC, Pedreño MA, Bru-Martinez R (2009) Changes of defence proteins in the extracellular proteome of grapevine (Vitis vinifera cv. Gamay) cell cultures in response to elicitors. J Proteomics 73: 331–341. [DOI] [PubMed] [Google Scholar]
- 25. Martínez-Esteso MJ, Selles-Marchart S, Vera-Urbina JC, Pedreño MA, Bru-Martinez R (2011) DIGE analysis of proteome changes accompanying large resveratrol production by grapevine (Vitis vinifera cv. Gamay) cell cultures in response to methyl-β-cyclodextrin and methyl jasmonate elicitors. J Proteomics 74: 1421–1436. [DOI] [PubMed] [Google Scholar]
- 26.Almagro L, Sabater-Jara AB, Belchí-Navarro S, Fernández-Perez F, Bru R, et al. (2011) Effect of UV light on secondary metabolite biosynthesis in plant cell cultures elicited with cyclodextrins and methyljasmonate. In: Hemanth KN and Vasanthaiah MCA, editors. Plants and Environment. Intech. 115–136. [Google Scholar]
- 27. Lijavetzky D, Almagro L, Belchí-Navarro S, Martínez-Zapater JM, Bru R, et al. (2008) Synergistic effect of methyljasmonate and cyclodextrin on stilbene biosynthesis pathway gene expression and resveratrol production in Monastrell grapevine cell cultures. BMC Res Not 1: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weissbach HL, Resnick N (2005) Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta 1703: 203–212. [DOI] [PubMed] [Google Scholar]
- 30. Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, et al. (2013) The Flavonoid Biosynthetic Pathway in Arabidopsis: Structural and Genetic Diversity. Plant Physiol. Biochem 72: 21–34. [DOI] [PubMed] [Google Scholar]
- 31.Pedreño MA, Belchí-Navarro S, Almagro L, Bru R (2009) Uso combinado de metil jasmonato y ciclodextrinas para la producción de resveratrol. PCT Patent WO/2009/106662.
- 32. Zamboni A, Gatto P, Cestaro A, Pilati S, Viola R, et al. (2009) Grapevine cell early activation of specific responses to DIMEB, a resveratrol elicitor. BMC Genomics 10: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Belhadj A, Telef N, Cluzet S, Bouscaut J, Corio-Costet MF, et al. (2008) Ethephon elicits protection against Erysiphe necator in grapevine. J Agri Food Chem 56: 5781–5787. [DOI] [PubMed] [Google Scholar]
- 34. Kitamura S, Shikazono N, Tanaka A (2004) Transparent testa 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114. [DOI] [PubMed] [Google Scholar]
- 35. Conn S, Curtin C, Bézier A, Franco C, Zhang W (2008) Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J Exp Bot 59: 3621–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, et al. (2011) In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J 67: 960–970. [DOI] [PubMed] [Google Scholar]
- 37. Edwards R, Dixon DP, Walbot V (2000) Plant glutathione S-transferases: Enzymes with multiple functions in sickness and health. Trends Plant Sci 5: 193–198. [DOI] [PubMed] [Google Scholar]
- 38. Chini A, Fonseca S, Fernández G, Adie B, Chico JM, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- 39. Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, et al. (2007) JAZ repressor proteins are targets of the SCF (COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- 40. Gfeller A, Liechti R, Farmer EE (2010) Arabidopsis jasmonate signaling pathway. Sci Signal 3: cm4. [DOI] [PubMed] [Google Scholar]
- 41. Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, et al. (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 8: 153–161. [DOI] [PubMed] [Google Scholar]
- 43. Guo C, Guo R, Xu X, Gao M, Li X, et al. (2014) Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot 65: 1513–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lijavetzky D, Carbonell-Bejerano P, Grimplet J, Bravo G, Flores P, et al. (2012) Berry Flesh and Skin Ripening Features in Vitis vinifera as Assessed by Transcriptional Profiling. PloS one 7: e39547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pontin MA, Piccoli PN, Francisco R, Bottini R, Martínez-Zapater JM, et al. (2010) Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol 10: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carbonell-Bejerano P, Rodríguez V, Royo C, Hernáiz S, Moro-González LC, et al. (2014) Circadian oscillatory transcriptional programs in grapevine ripening fruits. BMC Plant Biol 14: 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- 49. Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, et al. (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beckers GJ, Spoel SH (2006) Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol 8: 1–10. [DOI] [PubMed] [Google Scholar]
- 51. Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu Rev Phytopathol 47: 177–206. [DOI] [PubMed] [Google Scholar]
- 52. Dean JV, Mills JD (2004) Uptake of salicylic acid 2-O-beta-D-glucose into soybean tonoplast vesicles by an ATP-binding cassette transporter-type mechanism. Physiol Plant 120: 603–612. [DOI] [PubMed] [Google Scholar]
- 53. Dean JV, Mohammed LA, Fitzpatrick T (2005) The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221: 287–296. [DOI] [PubMed] [Google Scholar]
- 54. Staiger CJ (2000) Signalling to the actin cytoskeleton in plants. Annu Rev Plant Physiol Plant Mol Biol, 51, 257–288. [DOI] [PubMed] [Google Scholar]
- 55. Wasteneys GO, Galway ME (2003) Remodelling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Physiol 54: 691–722. [DOI] [PubMed] [Google Scholar]
- 56. Ebel C, Gomez LG, Schmit AC, Neuhaus-Url G, Boller T (2001) Differential mRNA degradation of two β-tubulin isoforms correlates with cytosolic Ca2+ changes in glucan-elicited soybean cells. Plant Physiol 126: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu Q, Qiao F, Ismail A, Chang X, Nick P (2013) The plant cytoskeleton controls regulatory volume increase. Biochim Biophys Acta 1828: 2111–2120. [DOI] [PubMed] [Google Scholar]
- 58.Peer WA (2011) Plasma membrane protein trafficking. The Plant Plasma Membrane. In: Murphy AS, Peer WA, Schulz, B, editors. Plant Cell Monographs. Springer. 31–56. [Google Scholar]
- 59. Jiang Q, Lu Z, Zhang C (2004) Role of Ran GTPase in cell cycle regulation. Chin Sci Bull 49: 535–541. [Google Scholar]
- 60. Li K, Xu C, Li Z, Zhang K, Yang A, et al. (2008) Comparative proteome analyses of phosphorus response in maize (Zea mays L.) roots of wild-type and a low-P-tolerant mutant reveal root characteristics associated with phosphorus deficiency. Plant J 55: 927–939. [DOI] [PubMed] [Google Scholar]
- 61. Guo M, Chena K, Zhanga P (2012) Transcriptome profile analysis of resistance induced by burdock fructooligosaccharide in tobacco. J Plant Physiol 169: 1511–1519. [DOI] [PubMed] [Google Scholar]
- 62. Ludwig A, Tenhaken R (2001) Suppression of the ribosomal L2 gene reveals a novel mechanism for stress adaptation in soybean. Planta 212: 792–798. [DOI] [PubMed] [Google Scholar]
- 63. Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, et al. (2003) Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J 33: 425–434. [DOI] [PubMed] [Google Scholar]
- 64. Calderón AA, Zapata JM, Muñoz R, Pedreño MA, Ros Barceló A (1993) Resveratrol production as a part of the hypersensitive-like response of grapevine cells to an elicitor from Trichoderma viride . New Phytol 124: 455–463. [Google Scholar]
- 65. Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 14: 6–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grimplet J, Van Hemert J, Carbonell-Bejerano P, Diaz-Riquelme J, Dickerson J, et al. (2012) Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res Not 5: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res 40: 1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. (2003) Summaries of AffymetrixGeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Raychaudhuri S, Stuart JM, Altman RB (2000) Principal components analysis to summarize microarray experiments: application to sporulation time series. Pacific Symposium on Biocomputing 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, et al. (2010) Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Research 38 (Web Server issue): W210–213. [DOI] [PMC free article] [PubMed]
- 71. Conesa A, Nueda MJ, Ferrer A, Talon M (2006) maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 22: 1096–1102. [DOI] [PubMed] [Google Scholar]
- 72. Toronen P, Kolehmainen M, Wong G, Castren E (1999) Analysis of gene expression data using self-organizing maps. FEBS Let 451: 142–146. [DOI] [PubMed] [Google Scholar]
- 73. Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Series B 63: 411–423. [Google Scholar]
- 74. Al-Shahrour F, Díaz-Uriarte R, Dopazo J (2004) FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20: 578–580. [DOI] [PubMed] [Google Scholar]
- 75. Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, et al. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939. [DOI] [PubMed] [Google Scholar]
- 76. Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, et al. (2006) TM4 Microarray Software Suite. Meth Enzymol 411: 134–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR expression validation of the microarray hybridization experiments.
(DOCX)
Treatments significantly regulated gene expression profiling summary.
(DOCX)
Mapman visualization of the significant genes in the ‘Amino acid metabolism’ pathway.
(DOCX)
Mapman visualization of the significant genes in the ‘Transport overview’ pathway.
(DOCX)
Mapman visualization of the significant genes in the ‘Hormone signalling’ pathway.
(DOCX)
PCR primers used to amplify gene-specific regions for expression analyses.
(XLS)
RMA normalized expression dataset. Log2 RMA GrapeGen GeneChip probesets signal in each sample.
(XLSX)
Probesets significantly regulated 24 h after any treatment.
(XLS)
Functional categories over-represented in each expression cluster of significantly regulated genes.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.