Abstract
Immune responses and metabolic regulation are tightly coupled in all animals, but the underlying mechanistic connections are nowhere completely clear. In flies and in humans, prolonged or excessive immune activation can drive metabolic disruption and cause loss of metabolic stores. Conversely, disruptions of metabolic homeostasis, such as periods of malnutrition, can have significant impacts on immune function. We have recently identified the transcription factor MEF2 as a critical switch between anabolic and immune function in the adult Drosophila fat body. A conserved phosphorylation determines the affinity of MEF2 for the TATA-binding protein, effecting a choice between energy storage and immune function. The goal of this review is to place this molecular event in the broader context of metabolic-immune interaction in Drosophila, exploring what is and is not known about the ties between these 2 critical physiological functions.
Keywords: metabolism, immunity, physiology, nutrient signaling, bacterial infection, antimicrobial peptides, MEF2
Drosophila Immunity and the Humoral Immune Response
Flies have several physiological mechanisms that are engaged in response to infections. Distinct suites of genes respond to infection with viruses, bacteria and fungi, parasites, and parasitoids; the activated effector mechanisms include cell-autonomous responses to intracellular pathogens, oxidative stress and melanization responses, cell-mediated responses such as phagocytosis and encapsulation, and local or systemic secretion of antimicrobial effectors such as lysozymes and antimicrobial peptides.1 Of these pathways and mechanisms, the best-studied is the humoral immune response to bacterial and fungal infection. The humoral response is characterized by the transcriptional induction of secreted antimicrobial peptides (AMPs) and lysozymes, primarily in the fat body. This response is rapid (most genes reach their transcriptional peak within 6 h of the initial infection event) and very strong (AMPs can be induced more than 1000-fold). An analogy is often drawn between the humoral immune response of the fly and the mammalian acute-phase response to infection; the acute-phase response involves similarly rapid induction of a suite of genes encoding secreted proteins and a consequent dramatic increase of the levels of these proteins in serum. In each case, the primary tissue producing these secreted proteins is one that, in uninfected animals, has key roles in metabolism—for mammals, the liver, and for flies, the fat body.
Drosophila humoral immunity has received significant experimental attention over the past 2 decades, beginning from the groundbreaking observation that the Toll and imd signaling pathways act in parallel to drive fat body expression of AMPs in response to detection of different classes of microbes.2-4 Several recent reviews have covered the current state of our knowledge.5,6 To summarize, the Toll and imd pathways are activated by distinct types of infection. Though the 2 pathways share no core components, each pathway culminates in activation of at least one NF-κB family transcription factor (for Toll, DIF and/or DORSAL; for imd, primarily RELISH). Because these factors have similar DNA recognition biases, they have overlapping repertoires of target genes, though some individual targets can be identified that are specific to one pathway or the other. The Toll and imd pathways have been held to be the primary or only signal-regulated aspects of this response in the adult fat body. However, other molecules not thought to be involved in the signal transduction process are also known to be required for normal immune function in this tissue. For example, GATA factors provide critical tissue-specific transactivation functions for targets of both pathways.7,8
MEF2 Integrates Nutrient Signaling with Immunity
Our interest in immune-metabolic interactions originated in our previous work on the consequences of Mycobacterium marinum infection in the fly.9 We showed that M. marinum infection drove a loss of triglyceride and glycogen that was associated with systemic disruption of insulin-pathway signaling (observable as a mild increase in free sugar combined with progressive reductions in AKT activity).10 The AKT-inactivated transcription factor FOXO was responsible for some aspects of pathology in this model. Similar metabolic effects have been observed more recently with other persistent pathogenic bacterial infections,11 while the non-pathogenic intracellular symbiont Wolbachia appears to exert an opposing effect,12 suggesting that these metabolic effects may be one aspect of the difference between pathogens and symbionts. Pathogenic infections in humans have broadly similar consequences.13,14
The fact that foxo mutants still exhibited triglyceride loss and hyperglycemia after M. marinum infection suggested that impaired AKT function and consequent excessive FOXO activity were not the sole drivers of infection-induced metabolic dysfunction.10 To find other metabolic-immune links, we performed a computational screen to identify transcription factor binding sites over- and under-represented on genes transcriptionally regulated by M. marinum infection, making use not only of our own expression data but also of publicly available data previously generated in Bruno Lemaitre’s laboratory.15-17 This kind of computational screen generates many false positives and false negatives; rather than trying to sort through these predictions computationally, we used our computational analysis as the basis for a targeted in vivo RNAi screen, testing infection susceptibility of fat body RNAi knockdowns for predicted regulators of metabolism and immunity. The strongest phenotype in this functional screen was given by fat-body Mef2 knockdowns, which were profoundly immunocompromised.
Mef2 was originally identified as a key myogenic transcription factor in flies and mice.18-20 In mammals, Mef2c regulates B-cell and neutrophil proliferation21-23 and myeloid cell fates,24 and is phosphorylated in response to inflammatory signaling in human monocytes.25 However, the possibility that Mef2-family proteins might be important direct activators of innate responses had not previously been examined. Similarly, Mef2-family proteins can promote expression of the glucose transporter Glut4 in muscle and adipose tissue,26 but other roles in adipose biology had not been explored.
As our computational screen had suggested MEF2 was a direct regulator of anabolic enzymes and AMPs, we went on to test the effects of this knockdown on predicted target genes. We found that nearly all tested AMPs required fat body MEF2 for normal infection-induced expression, while many enzymes of triglyceride and glycogen synthesis required fat body MEF2 for expression in uninfected flies. These 2 groups of target genes exhibited apparent counter-regulation: expression of metabolic MEF2 targets was lost in animals that were producing AMPs in response to infection. (An overall scheme of the role of MEF2 is shown in Fig. 1.)
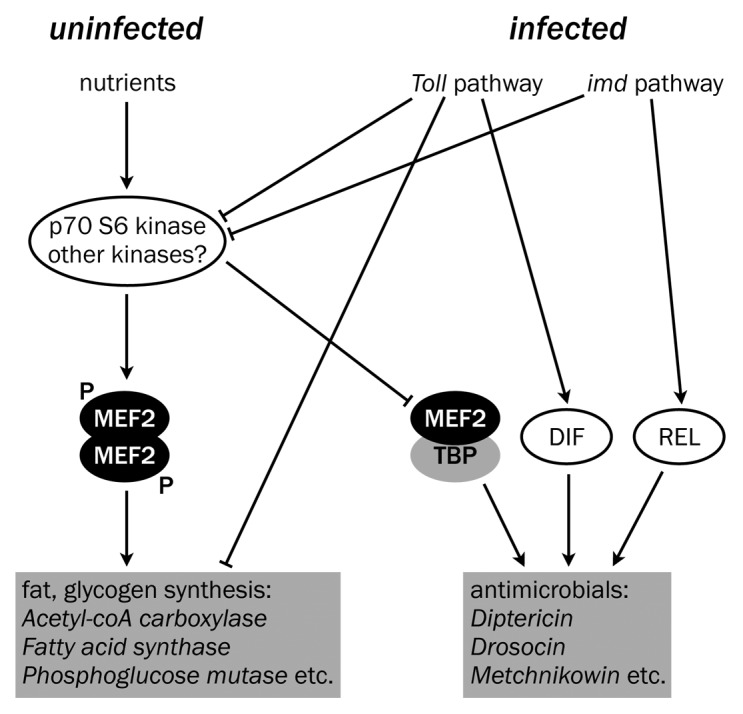
Figure 1. Model of signaling regulation of metabolic and immune function in the adult fat body in healthy flies (left) and flies experiencing an infection (right). Some aspects are speculative; for example, the stoichiometry of different MEF2-containing complexes is unknown.
We identified the relevant MEF2 sites in AMP genomic neighborhoods with reporters in which GFP was placed under the control of regulatory regions from 2 representative AMPs, Metchnikowin and Drosocin. Remarkably, in each case we found a strong requirement for a MEF2 site overlapping the TATA box. We found that the TATA-Binding Protein (TBP) and MEF2 physically interact in vivo only upon infection and that the TBP-MEF2 complex can bind the Metchnikowin TATA box sequence; this binding is dependent on residues outside the TATA-binding core that are predicted to be bound by MEF2. We then identified perfect matches to the MEF2-TATA box on 20/30 identified AMPs and AMP-like peptides. Conversely, the metabolic targets of MEF2 generally lack TATA boxes or obvious MEF2 sites in the core promoter. Instead, their predicted high-affinity MEF2 sites are more distant, and often appear in introns or 3′ flanking regions. It is still an open question whether these distant MEF2 sites are directly required for expression of these metabolic targets, though prior work suggests that many of the predicted MEF2 sites on the genes of glycogen synthesis are occupied by MEF2 in the embryo (the genes of triglyceride synthesis are not, in general, highly expressed during the embryonic stages examined).27,28
We then examined regulation of the MEF2-TBP complex. We found that MEF2 immunoprecipitated from healthy flies shows significant phosphorylation at Threonine 20. This phosphorylation is significantly reduced by infection. We showed that MEF2 T20 can be phosphorylated in vitro by p70 S6 Kinase (S6K), and that S6K activation is reduced by infection. Finally, we showed that phosphomimetic (T20E) MEF2 is unable to associate with TBP independent of infection and rescues expression of many MEF2 metabolic targets in flies with Gram-negative bacterial infection, while nonphosphorylatable (T20A) MEF2 can associate with TBP in uninfected animals and enhances AMP transcriptional induction. Together, these data suggest a model in which, in healthy animals, S6K phosphorylates fat-body MEF2 on T20 to permit expression of enzymes of triglyceride and glycogen synthesis, while upon infection, S6K activity is lost, resulting in MEF2 T20 dephosphorylation and MEF2-TBP complex formation, permitting expression of antimicrobial peptides at the cost of anabolic gene expression.
Numerous questions remain. Apparent MEF2-binding TATA boxes are found on many genes that are not antimicrobial peptides; does this reflect a requirement for the MEF2-TBP complex in other biological processes? Is S6K the only relevant in vivo T20 kinase? Are AKT and S6K shut down by a common mechanism during infection? Is T20 actively dephosphorylated during infection? Does MEF2 bind at different sites in infected and uninfected fat body? What is the relevance of more distal MEF2 sites on antimicrobial peptides?
Unanswered Questions in Immune-Metabolic Interaction
Other regulators
Importantly, while our analysis indicates that MEF2 dysregulation may be the primary event driving metabolic dysfunction due to infections that exclusively activate the imd pathway, the Toll pathway is able to inhibit expression of most anabolic enzymes by some other mechanism; we do not yet know what that mechanism is. Possibilities include, but are not limited to, direct effects of NF-κB activity, cytokine effects, or other uncharacterized points of crosstalk between immune and metabolic regulation.
NF-κB transcription factors as direct metabolic regulators
In mammals, it is now clear that NF-κB activation can drive metabolic dysfunction both directly and indirectly, though the relative importance of this effect is unclear in most infections in vivo.29 In flies, some recent data hint that the Toll pathway, in addition to its well-characterized role in immune detection, may also regulate metabolic function. For example, flies lacking the essential Toll pathway component MyD88 survive longer than wild-type animals when starved; though the underlying mechanism is unclear, this suggests a link between Toll pathway signaling and metabolic homeostasis.30 This effect may be mediated by effects on peripheral insulin sensitivity. Importantly, the direct effects of Dif and dorsal in metabolism have yet to be examined.
Importance of cytokine signaling
In mammals, cytokines mediate much of the interplay between metabolism and immunity. In flies, their role is less clear. Several signals, including upd3, daw, and dpp are activated in hemocytes in response to wounding or infection.31,32 The upd3 relative upd2 can regulate metabolism via effects on insulin-like peptide secretion,33 while the dpp relative and partner gbb promotes lipid storage in the larval fat body.34 It is unclear to what extent the observed regulation of these signaling pathways in infection impacts metabolic regulation.
Other points of crosstalk
A few published studies have identified molecules with roles in both metabolic and immune function, though it is generally unclear how these physiological functions are related. The deubiquitinase CYLD is required in the fat body for normal antimicrobial responses and to regulate triglyceride storage.35 CYLD-deficient flies exhibit increased expression of the imd pathway target Diptericin in the absence of infection as well as elevated total triglyceride levels. CYLD itself physically associates with the imd pathway component KENNY, suggesting that the effects observed on immunity may be direct; it is unknown whether the metabolic effects of CYLD mutation are imd-pathway dependent. Similarly, the transcription factor ATF3 inhibits antimicrobial peptide expression and metabolic storage in larvae; it is difficult to directly compare the function of Atf3 with that of Mef2 because of the many differences in experimental approach, but the function of Atf3 appears broadly opposite to that we ascribe to Mef2.36 In this regard, it may be relevant that our computational analysis identified an association between CREB/ATF sites and antimicrobial peptides.17
Mechanism of disrupted nutrient signaling
We and others have observed that activity of the insulin-responsive kinase AKT is inhibited by infection.10,37 Though Toll pathway activation is able to inhibit AKT activity in a cell-autonomous fashion, it has been suggested that this activity is not shared by the imd pathway.4 However, we find that infections that should activate only the imd pathway also strongly reduce AKT activity, at least in adult flies (unpublished data). We also observe that infection with Gram-negative or Gram-positive bacteria reduces p70 S6 kinase activity.17 It is unclear whether p70 S6 kinase repression is secondary to AKT inhibition, and it is unclear to what extent the documented Toll pathway effect on AKT activation drives metabolic dysfunction secondary to persistent infection in vivo. In this context, the observation that many infections reduce food intake in flies as in other animals, and that this has clear functional consequences for the immune response, is intriguing.38,39
Summary
Infection-induced metabolic disruption leading to cachexia is present in nearly all animals. Our work shows that MEF2 is a key decision point mediating this event: fat body MEF2 must be switched between metabolic and immune functions and is fundamentally required for both processes. However, our knowledge of the mechanistic links between fat body metabolic and immune function—and, in a larger sense, of the ways in which immune activity in general can disrupt normal physiological homeostasis—are far from complete.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Karen Liu and Brian Stramer gave valuable comments on the manuscript.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–83. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A. 1997;94:14614–9. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ligoxygakis P. Genetics of immune recognition and response in Drosophila host defense. Adv Genet. 2013;83:71–97. doi: 10.1016/B978-0-12-407675-4.00002-X. [DOI] [PubMed] [Google Scholar]
- 7.Petersen UM, Kadalayil L, Rehorn KP, Hoshizaki DK, Reuter R, Engström Y. Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J. 1999;18:4013–22. doi: 10.1093/emboj/18.14.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M. Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell. 2004;13:19–32. doi: 10.1016/S1097-2765(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 9.Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun. 2003;71:3540–50. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr Biol. 2006;16:1977–85. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 11.Chambers MC, Song KH, Schneider DS. Listeria monocytogenes infection causes metabolic shifts in Drosophila melanogaster. PLoS One. 2012;7:e50679. doi: 10.1371/journal.pone.0050679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeya T, Broughton S, Alic N, Grandison R, Partridge L. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc Biol Sci. 2009;276:3799–807. doi: 10.1098/rspb.2009.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwenk A, Macallan DC. Tuberculosis, malnutrition and wasting. Curr Opin Clin Nutr Metab Care. 2000;3:285–91. doi: 10.1097/00075197-200007000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Tappy L, Chioléro R. Substrate utilization in sepsis and multiple organ failure. Crit Care Med. 2007;35(Suppl):S531–4. doi: 10.1097/01.CCM.0000278062.28122.A4. [DOI] [PubMed] [Google Scholar]
- 15.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–79. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark RI, Tan SW, Péan CB, Roostalu U, Vivancos V, Bronda K, Pilátová M, Fu J, Walker DW, Berdeaux R, et al. MEF2 is an in vivo immune-metabolic switch. Cell. 2013;155:435–47. doi: 10.1016/j.cell.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–41. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 19.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–36. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 20.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–93. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 21.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 22.Khiem D, Cyster JG, Schwarz JJ, Black BL. A p38 MAPK-MEF2C pathway regulates B-cell proliferation. Proc Natl Acad Sci U S A. 2008;105:17067–72. doi: 10.1073/pnas.0804868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilker PR, Kohyama M, Sandau MM, Albring JC, Nakagawa O, Schwarz JJ, Murphy KM. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol. 2008;9:603–12. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schüler A, Schwieger M, Engelmann A, Weber K, Horn S, Müller U, Arnold MA, Olson EN, Stocking C. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood. 2008;111:4532–41. doi: 10.1182/blood-2007-10-116343. [DOI] [PubMed] [Google Scholar]
- 25.Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–9. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 26.Thai MV, Guruswamy S, Cao KT, Pessin JE, Olson AL. Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. Regulation of MEF2 DNA binding activity in insulin-deficient diabetes. J Biol Chem. 1998;273:14285–92. doi: 10.1074/jbc.273.23.14285. [DOI] [PubMed] [Google Scholar]
- 27.Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–49. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, Shoelson SE. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–90. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayyaz A, Giammarinaro P, Liégeois S, Lestradet M, Ferrandon D. A negative role for MyD88 in the resistance to starvation as revealed in an intestinal infection of Drosophila melanogaster with the Gram-positive bacterium Staphylococcus xylosus. Immunobiology. 2013;218:635–44. doi: 10.1016/j.imbio.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5:441–50. doi: 10.1016/S1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 32.Clark RI, Woodcock KJ, Geissmann F, Trouillet C, Dionne MS. Multiple TGF-β superfamily signals modulate the adult Drosophila immune response. Curr Biol. 2011;21:1672–7. doi: 10.1016/j.cub.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–37. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballard SL, Jarolimova J, Wharton KA. Gbb/BMP signaling is required to maintain energy homeostasis in Drosophila. Dev Biol. 2010;337:375–85. doi: 10.1016/j.ydbio.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsichritzis T, Gaentzsch PC, Kosmidis S, Brown AE, Skoulakis EM, Ligoxygakis P, Mosialos G. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. 2007;134:2605–14. doi: 10.1242/dev.02859. [DOI] [PubMed] [Google Scholar]
- 36.Rynes J, Donohoe CD, Frommolt P, Brodesser S, Jindra M, Uhlirova M. Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol Cell Biol. 2012;32:3949–62. doi: 10.1128/MCB.00429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci U S A. 2009;106:20853–8. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown AE, Baumbach J, Cook PE, Ligoxygakis P. Short-term starvation of immune deficient Drosophila improves survival to gram-negative bacterial infections. PLoS One. 2009;4:e4490. doi: 10.1371/journal.pone.0004490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayres JS, Schneider DS. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 2009;7:e1000150. doi: 10.1371/journal.pbio.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]


