Abstract
Autophagy is the major pathway for the delivery of cytoplasmic material to the vacuole or lysosome. Selective autophagy is mediated by cargo receptors, which link the cargo to the scaffold protein Atg11 and to Atg8 family proteins on the forming autophagosomal membrane. We show that the essential kinase Hrr25 activates the cargo receptor Atg19 by phosphorylation, which is required to link cargo to the Atg11 scaffold, allowing selective autophagy to proceed. We also find that the Atg34 cargo receptor is regulated in a similar manner, suggesting a conserved mechanism.
Keywords: Atg11, Atg19, Atg34, Cvt pathway-autophagy, Hrr25 kinase
Introduction
Macroautophagy (hereafter autophagy) is an important mechanism for the bulk degradation of cellular material. Autophagy is a sequential process beginning with the formation of a double-membrane sheet termed phagophore at the pre-autophagosomal structure (PAS). The phagophore expands to enwrap the cargo in the autophagosome, which subsequently fuses with the lysosome or vacuole where the cargo is degraded by resident hydrolases 1. This evolutionarily conserved process is integral for cellular homeostasis and the response to stress conditions such as nutrient starvation. Defects in autophagy pathways have been associated with numerous human pathologies including infectious diseases, neurodegenerative disorders, and cancer 1. Despite these fundamental functions, the regulation and coordination of cargo selection and transport to the cellular degradation site remain poorly understood.
Both non-selective “bulk” autophagy and selective autophagy of specific proteins or organelles have been described 2. One selective autophagy-related pathway in yeast is the cytoplasm-to-vacuole targeting (Cvt) pathway, which fulfills a biosynthetic function by delivering at least three resident enzymes, aminopeptidase 1 (Ape1), α-mannosidase (Ams1), and aspartyl aminopeptidase (Ape4) to the vacuole 3–5. Genetic screens in yeast have identified 38 proteins (termed Atg1 to Atg38), many of them highly conserved from yeast to mammals, that are required for different steps of autophagy 6–9. Many of these components are common to both autophagy and the Cvt pathway, although autophagy- and Cvt-specific genes exist 1.
Cargo selectivity is achieved by cargo receptors that link the cargo to the autophagic machinery. In Saccharomyces cerevisiae, four cargo receptors have been described: Atg19, Atg32, Atg34, and Atg36 acting in the Cvt pathway, mitophagy, Ams1 transport, and pexophagy, respectively 6,10–13. Cargo receptors exist also in other species, exemplified by Pichia pastoris Atg30 14 or the mammalian p62/SQSTM1 and NBR1 proteins 15,16, and the recently identified mitophagy receptor FUNDC1 17. Saccharomyces cerevisiae Atg19 binds directly to its cargo Ape1 forming the so-called Cvt complex. Subsequently, Atg19 mediates the transport of Ape1 to the vacuole by subsequent interactions with Atg11 and Atg8 18. During bulk autophagy, Atg19 and the related protein Atg34 tether their respective cargo to the isolation membrane, thereby providing some selectivity to the otherwise non-selective engulfment of cytoplasmic material during this process 13,18.
The interactions of Atg30, Atg32, and Atg36 with Atg11 and Atg8 have been reported to depend on phosphorylation 19,20. In mammals, phosphorylation of FUNDC1 has been shown to be required for binding to the Atg8 homologue LC3 17. While casein kinase 2 can regulate Atg32 function in mitophagy, the kinase(s) responsible for phosphorylation of Atg30, Atg36, and possibly other receptors such as Atg19 and Atg34 remain elusive 19,20. Using mass spectrometry, we found that Atg19 and Atg34 are highly phosphorylated at their C-terminal Atg11 binding regions in vivo. Mutational analysis showed that these phosphorylation sites are indeed required for Cvt pathway function and Ams1 transport by regulating the interaction of Atg19 and Atg34 with Atg11. We identified Hrr25 as the kinase phosphorylating Atg19, thus promoting its interaction with Atg11. These findings suggest that while receptor phosphorylation is conserved between different types of selective autophagy, the kinase responsible is pathway specific.
Results
The C-terminus of Atg19 is phosphorylated in vivo
We asked whether, similar to other cargo receptors, Atg19 function is controlled by phosphorylation. To determine Atg19 phosphorylation sites, we purified Atg19 constructs under denaturing conditions using an N-terminal His-biotin (HTB) tag 21,22. These Atg19 constructs contained additional lysine residues in the C-terminus to allow tryptic processing into fragments suitable for mass spectrometric detection (Fig 1B). The optimized proteins are functional as judged by the ability of processing prApe1 to its mature form, although some reduction in activity was observed (Supplementary Fig S1A). Mass spectrometric phosphorylation mapping revealed 26 phosphorylated residues on Atg19. Since the Atg19 C-terminus contains the interaction sites for Atg11 and Atg8 (Fig 1A), we were specifically interested in potential phosphorylation events in this region 18,23. Many of the mapped phosphorylation sites are located in the C-terminus (Fig 1B and Supplementary Fig S1B) and five of these in the Atg11 interaction region. This suggests that the interaction of Atg19 with Atg11 might be regulated by phosphorylation.
Figure 1. Atg19 phosphorylation is required for Atg11 binding and Cvt pathway function.
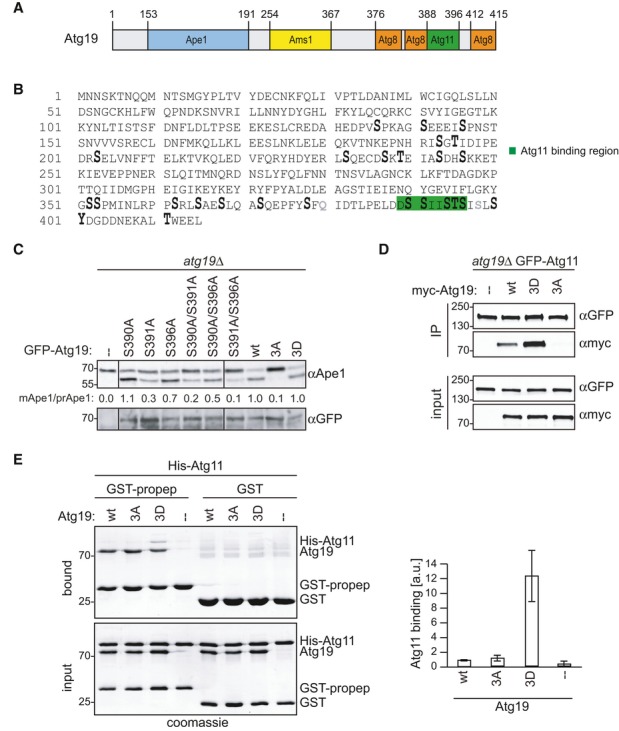
A Domain structure of Atg19.
B atg19Δ cells containing HTB-Atg19 were grown to mid-log phase. Atg19 was affinity purified and subjected to mass spectrometric phosphorylation mapping. Phosphorylation sites: enlarged; lysine substitutions: gray; Atg11 binding region: green.
C atg19Δ cells containing GFP-Atg19 wild-type or mutants as indicated were grown to mid-log phase. Processing of endogenous Ape1 was analyzed by Western blotting and quantified by calculating the ratio of cleaved versus uncleaved Ape1 normalized to the wild-type. Atg19 expression was assessed by anti-GFP Western blotting.
D atg19Δ GFP-Atg11 Atg1-TAP cells containing myc-Atg19 as indicated were grown to log phase. Atg11 was immunoprecipitated, and bound Atg19 was analyzed by Western blotting.
E GST-fused propeptide of Ape1, Atg19 as indicated, and a His-tagged fragment of Atg11 (amino acids 685–1178) were purified from Escherichia coli, mixed, and propeptide-bound proteins were isolated and analyzed by Coomassie staining. Quantification of three individual experiments is shown. Error bars represent standard deviation.
Source data are available online for this figure.
Phosphorylation in the Atg11 binding region of Atg19 is required for Cvt pathway function
To investigate whether the phosphorylation events in the Atg11 binding region are important for Atg19 function in vivo, we analyzed different non-phosphorylatable and phospho-mimicking mutants in their ability to promote the Cvt pathway. To this end, three phosphorylation sites in the Atg11 binding region, S390, S391, and S396, were mutated to non-phosphorylatable alanine residues either individually or in combination. All mutants were expressed at similar levels as the wild-type protein (Fig 1C). We assessed Cvt pathway activity by measuring Ape1 processing. The single mutations showed modest (S390A and S396A) to severe (S391A) reduction in Cvt pathway activity. All double mutations increased this defect, and no mature Ape1 was detectable in the 3A mutant. In contrast, mimicking phosphorylation on these three sites using negatively charged aspartate residues (3D) restored Cvt activity, strongly suggesting that C-terminal phosphorylation of Atg19 is essential for progression of this pathway (Fig 1C). Of four additional C-terminal phosphorylation sites analyzed, only S394 was required for Cvt function (Supplementary Fig S1C). As the 3A and 3D mutants showed a full defect and rescue, respectively, but mimicking S394 phosphorylation with an aspartate residue did not rescue Cvt activity, we excluded S394 from our mutational analysis.
Atg19 phosphorylation promotes its interaction with Atg11
Next we asked whether the Atg11-Atg19 interaction is regulated by phosphorylation on these three sites. Indeed, endogenously expressed GFP-Atg11 was able to co-precipitate myc-Atg19. This interaction was abolished for the non-phosphorylatable Atg19-3A mutant, while the phospho-mimicking Atg19-3D mutant strongly interacted with Atg11 (Fig 1D). To test whether this interaction is direct, we purified GST-Atg19 and a 6xHis-Atg11 C-terminal fragment (amino acids 685–1,178) containing the Atg19 binding region 24 from Escherichia coli. In pull-down experiments, the phospho-mimicking Atg19-3D mutant bound Atg11 approximately three times stronger than wild-type Atg19 (which remains unphosphorylated when purified from E. coli) and the Atg19-3A mutant, showing that the Atg11-Atg19 interaction is direct and promoted by phospho-mimicking mutations (Supplementary Fig S1D). To better represent the physiological situation in vitro, we mimicked Cvt cargo by decorating beads with Ape1 propeptide. Wild-type Atg19 as well as the 3A and 3D mutants bound to these GST-prApe1 propeptide containing beads to a similar extent, showing that the mutations do not affect Ape1 binding. Binding of Atg11 to propeptide-Atg19-3D complexes was approximately 12 times stronger than to complexes containing either wild-type Atg19 or Atg19-3A, further suggesting that phosphorylation promotes Atg19-Atg11 binding (Fig 1E).
Atg11 recruitment to Cvt cargo requires Atg19 phosphorylation
Since Atg19 recruits Atg11 to the prApe1 complex, we asked whether this activity is dependent on phosphorylation in vivo. To test whether the recruitment of Atg11 to Ape1 complexes depends on the phosphorylation of Atg19 in vivo, we analyzed the recruitment of GFP-Atg11 to giant Ape1 oligomers in cells expressing either wild-type Atg19, the Atg19-3A, or Atg19-3D mutants. Due to their large size, giant Ape1 oligomers cannot be fully enwrapped by the isolation membrane and are therefore not transported to the vacuole 25. Wild-type Atg19 and the phospho-mimicking Atg19-3D were able to recruit Atg11 to giant Ape1 oligomers, while the non-phosphorylatable Atg19-3A mutant failed to do so (Fig 2A). The failure of Atg19-3A to recruit Atg11 to the cargo was not due to a defect in binding to the prApe1 complex as the mutant localized indistinguishably from wild-type Atg19 (Fig 2B). Similar results were obtained when analyzing cells expressing native levels of Ape1 (Supplementary Fig S2A). As expected, Ape1 was proficient in forming oligomers in these mutants, however failed to localize to the vacuole (Supplementary Fig S2A), and no Atg8 was recruited to Ape1 complexes (Fig 2C), likely due to the lack of Atg11 binding.
Figure 2. Non-phosphorylatable Atg19 is unable to bind Atg11.
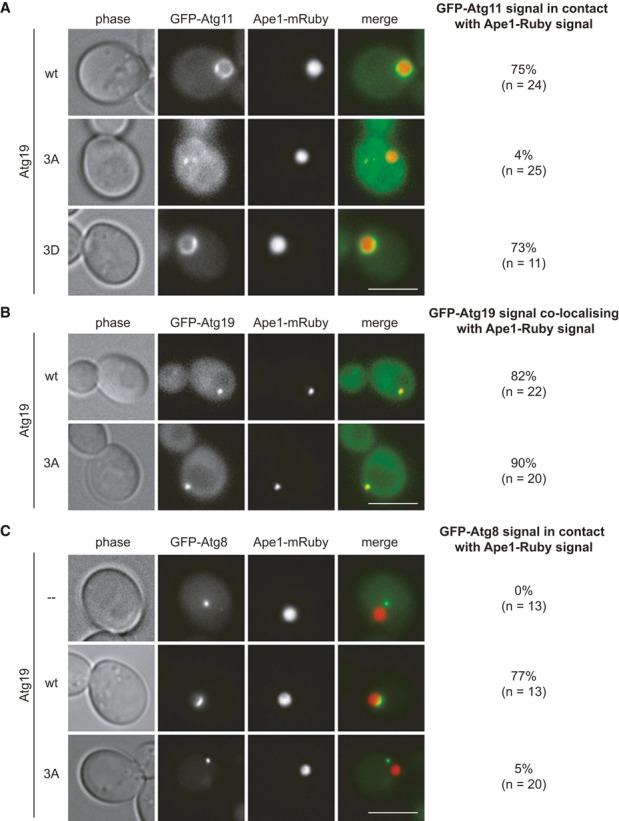
A GFP-Atg11 Ape1-mRuby atg19Δ cells containing myc-Atg19 as indicated and Cup1-Ape1 were grown to log phase. Overexpression of Ape1 was induced by addition of 250 μM copper sulfate for 3 h, and autophagy was induced by treating cells for 1 h with rapamycin. Scale bar, 5 μm.
B Ape1-mRuby atg19Δ cells containing GFP-Atg19 wild-type or GFP-Atg19-3A were analyzed in log phase. Scale bar, 5 μm.
C GFP-Atg8 Ape1-mRuby atg19Δ cells containing myc-Atg19 as indicated and Cup1-Ape1 were analyzed as in (A). Scale bar, 5 μm.
Phosphorylation on Atg34 regulates the transport of Ams1 to the vacuole
The C-terminus of Atg19 shows sequence conservation to Atg34, a cargo receptor which functions during starvation to selectively transport Ams1 to the vacuole (13, Fig 3A). Alignment of the Atg11 interaction regions of these two proteins revealed that two of the phosphorylation sites we analyzed in Atg19 are conserved (S390 and S391 in Atg19, S382 and S383 in Atg34, Fig 3A). To analyze Atg34 phosphorylation in vivo, we isolated HTB-tagged Atg34 and subjected it to mass spectrometric phosphorylation mapping. Similar to Atg19, we had to introduce an additional lysine residue to allow the generation of a suitable tryptic fragment (L390K, Fig 3B and Supplementary Fig S1E). Phosphorylation analysis of Atg34 identified 15 phosphorylated residues, among them S382 and S383 (Fig 3B and Supplementary Fig S1B).
Figure 3. Atg34 phosphorylation on homologous sites is required for Ams1 transport to the vacuole.
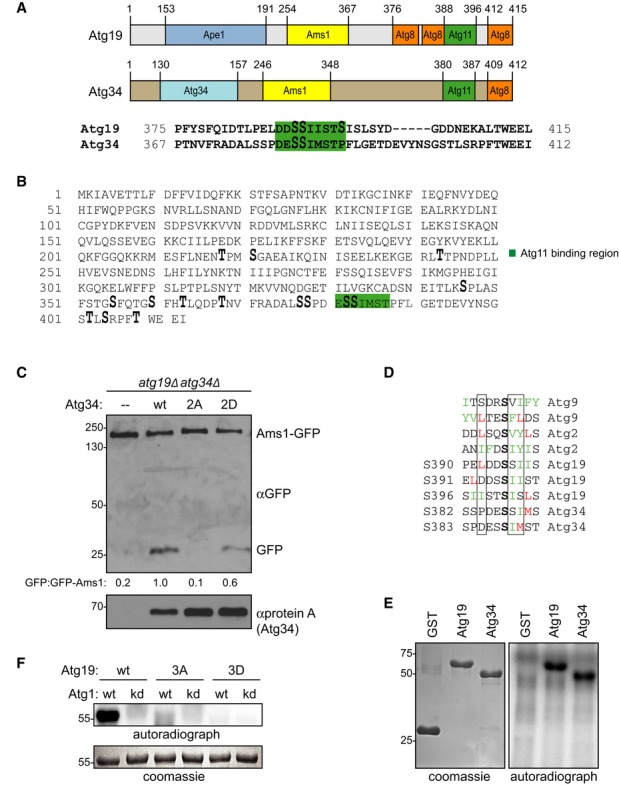
A Comparison of Atg19 and Atg34 domain structures and C-terminal amino acid sequences. Green: Atg11 binding region. Serine residues S390, S391, and S396, which are phosphorylated in Atg19, and the conserved sites in Atg34 are shown enlarged.
B atg34Δ yeast cells containing HTB-Atg34 were grown to mid-log phase and treated with rapamycin. Atg34 was affinity purified and subjected to mass spectrometric phosphorylation mapping. Phosphorylation sites: enlarged; Atg11 binding region: green; lysine substitution: gray.
C Ams1-GFP atg19Δatg34Δ cells containing protein A (Stag)-tagged Atg34 as indicated were starved for 4 h. Processing of endogenous Ams1-GFP was analyzed by Western blotting and quantified by calculating the ratio of free GFP versus uncleaved Ams1-GFP normalized to the wild-type. Expression of Atg34 proteins was assessed by anti-protein A Western blotting.
D Alignment of the Atg1 consensus sites in the Atg11 binding region of Atg19 and Atg34 with the known Atg1 phosphorylation sites of Atg9 and Atg2.
E GST, GST-Atg19, and GST-Atg34 were purified from Escherichia coli and in vitro phosphorylated with immunoprecipitated Atg1-TAP bound to IgG magnetic beads.
F In vitro phosphorylation of recombinant Atg19, Atg19-3A, and Atg19-3D using wild-type and kinase-dead Atg1-D211A as described in (E).
Source data are available online for this figure.
To assess whether these phosphorylation events function in Ams1 transport, we mutated serine residues 382 and 383 to non-phosphorylatable alanine (2A) or phospho-mimicking aspartate (2D). Since Atg19 can also promote Ams1 transport to the vacuole, we analyzed the function of the mutant Atg34 proteins in the absence of Atg19. Transport of Ams1-GFP to the vacuole was monitored by generation of free GFP due to vacuolar proteolytic cleavage 13. Atg34 wild-type and the Atg34-2D mutant were able to transport Ams1 to the vacuole. In contrast, no Ams1 was delivered to the vacuole in the Atg34-2A mutant (Fig 3C), indicating that phospho-regulation of cargo receptors at their C-terminal Atg11 binding sites is conserved.
Atg1 kinase is dispensable for Atg19-Atg11 binding in vivo
Sequence alignment of S390, S391, and S396 of Atg19 and S382 and S383 of Atg34 with known kinase consensus sequences revealed that these phosphorylation sites match the Atg1 kinase phosphorylation motif (26, Fig 3D). To determine whether Atg1 is indeed capable of phosphorylating the C-terminus of Atg19 and Atg34, we purified full-length Atg34, Atg19, the C-terminal segment of Atg19 (amino acids 365–415) and C-terminally truncated Atg19 (amino acids 1–374) from E. coli and subjected them to in vitro phosphorylation reactions using native Atg1 complexes purified from S. cerevisiae 27. Whereas Atg1 readily phosphorylated full-length Atg34 and Atg19 as well as the C-terminal fragment of Atg19, phosphorylation was strongly decreased upon C-terminal truncation of Atg19 (Fig 3E and Supplementary Fig S1F). The fact that the Atg19-3A and Atg19-3D mutants showed severely reduced phosphorylation (Fig 3F) suggested that Atg1 indeed phosphorylates these specific sites.
If Atg1 was the sole kinase promoting the Atg19-Atg11 interaction in vivo, this interaction should be lost or weakened in the absence of Atg1 activity. However, puncta formation of GFP-Atg11, reflecting its binding to the prApe1 cargo 24, was unaffected in cells expressing kinase-dead Atg1-D211A, and the Atg11-Atg19 complex was able to form, suggesting that in the absence of Atg1, other kinases can promote Atg11 recruitment (Supplementary Fig S2B, C and D). The Atg19-Atg11 interaction was still dependent on phosphorylation of serine residues 390, 391, and 396, since the Atg19-3A mutant failed to bind Atg11 also in an Atg1-D211A background (Supplementary Fig S2D). Quantitative mass spectrometry confirmed that phosphorylation at serine residues 390, 391, and 396 did not change in Atg1-D221A mutant cells compared to wild-type (Supplementary Fig S1B). Interestingly, S372 phosphorylation was dependent on Atg1 activity, but mutation of this residue had no effect on the Cvt pathway (Supplementary Fig S1C). These findings suggest that although Atg1 is able to phosphorylate S390, S391, and S396 in vitro, it is dispensable or acts redundantly with other kinases in vivo.
Hrr25 directly phosphorylates Atg19 mediating the interaction with Atg11
To identify the kinase responsible for Atg19 phosphorylation in vivo, we screened kinases with consensus motives similar to Atg1 26 and kinases reported to interact with Atg19 in large-scale studies 28–30 for a role in Atg11 localization in an Atg1 kinase-dead background. However, Atg11 localization in these mutants was indistinguishable from wild-type (Supplementary Fig S3A). Next, we screened 16 essential kinases using 34 temperature-sensitive alleles for their involvement in the Cvt pathway. Only one kinase, Hrr25, showed a defect in Ape1 maturation at the restrictive temperature, which was rescued by ectopic expression of Hrr25 (Fig 4A and Supplementary Fig S3B). hrr25-ts mutant cells already showed a severe Cvt pathway defect at 18°C, indicating that Hrr25 kinase function is strongly impaired also at the permissive temperature. This defect was partially rescued by the Atg19-3D mutant, suggesting that mimicking phosphorylation in the Atg19 C-terminus can bypass the requirement for Hrr25 kinase in the Cvt pathway (Fig 4B). Quantitative mass spectrometry confirmed that phosphorylation at serine residues 390, 391, and possibly 396 decreased in hrr25-ts mutant cells compared to wild-type (Supplementary Fig S4A). To analyze whether the Cvt defect is caused by a disruption of Atg11-Atg19 interaction, we analyzed cells for their ability to form Atg11 dots, which are formed dependent on the Atg11-Atg19 interaction 24. Whereas more than 70% of wild-type and Atg1-D211A mutant cells were proficient in Atg11 dot formation, only 5% of hrr25-ts cells formed Atg11 dots (Fig 4C and Supplementary Fig S3C), suggesting that this interaction is mediated by Hrr25. Similarly, Atg11 recruitment to native Ape1 and giant Ape1 complexes was completely impaired in the absence of Hrr25 function (Supplementary Fig S4B and D). As expected, Ape1 was proficient in forming oligomers in hrr25-ts mutants, however failed to localize to the vacuole in these cells (Supplementary Fig S4B). This defect was not caused by Atg19 mislocalization, as wild-type and hrr25-ts cells showed similar Atg19 dot formation (Supplementary Fig S4C). Also, Atg11 and Atg19 protein levels were comparable in wild-type and hrr25-ts cells (Supplementary Fig S4C and D).
Figure 4. Hrr25 kinase is responsible for Atg19 phosphorylation in the Atg11 binding region.
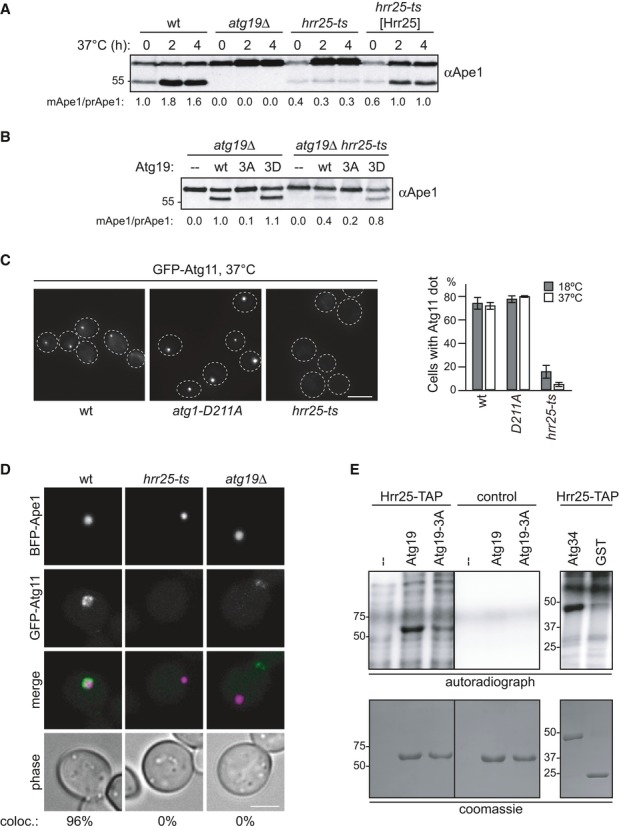
A Wild-type, hrr25-ts, and hrr25-ts cells containing HRR25 on a centromeric plasmid were grown to mid-log phase at 18°C followed by 2 and 4 h of log-phase growth at 37°C. Ape1 processing was analyzed by Western blotting.
B atg19Δ and atg19Δ hrr25-ts cells containing GFP-Atg19 as indicated were analyzed for Ape1 processing in log phase after 2 h at 37°C.
C GFP-Atg11, GFP-Atg11 hrr25-ts, and GFP-Atg11 atg1-D211A cells were analyzed for GFP-Atg11 dot formation. Quantification of at least 70 cells in three individual experiments is shown. Error bars represent standard deviation. Scale bar, 3 μm.
D GFP-Atg11, GFP-Atg11 hrr25-ts, and GFP-Atg11 atg19Δ cells containing BFP-Ape1 and Cup1-Ape1 were grown to log phase. Overexpression of Ape1 was induced by addition of 250 μM CuSO4 for 3 h. coloc.: Cells with GFP-Atg11/BFP-Ape1 co-localization. n = 50 (wt), 84 (hrr25-ts), 127 (atg19Δ). Scale bar, 3 μm.
E Hrr25 was immunoprecipitated from Hrr25-TAP cells and as a control from a non-tagged wild-type strain. In vitro phosphorylation of recombinant Atg19, Atg19-3A, Atg34, and GST using the kinase-bound beads was performed as described in Fig 3E. Both Atg19 panels are from the same gel with the same exposure.
Source data are available online for this figure.
To test whether Hrr25 is able to phosphorylate Atg19 directly, we subjected Atg19 purified from E. coli to in vitro phosphorylation by Hrr25 kinase. Atg19 was phosphorylated by Hrr25, and phosphorylation was strongly reduced for the Atg19-3A mutant (Fig 4E), showing that Hrr25 is able to directly phosphorylate Atg19 in its Atg11 binding region. Similarly, Atg34 is a target of Hrr25 in vitro (Fig 4E).
In summary, we show that phosphorylation in the Atg11 binding region of Atg19 is required for its interaction with Atg11 and transport of prApe1 to the vacuole. We identify Hrr25 as the kinase responsible for these phosphorylation events, thus promoting progression of the Cvt pathway. Likewise, phosphorylation is conserved in Atg34 and required for Ams1 transport to the vacuole. Our data suggest that phosphorylation of cargo receptors may be a general key regulatory step in selective autophagy (Fig 5).
Figure 5. Model depicting the role of cargo receptor phosphorylation in selective autophagy.
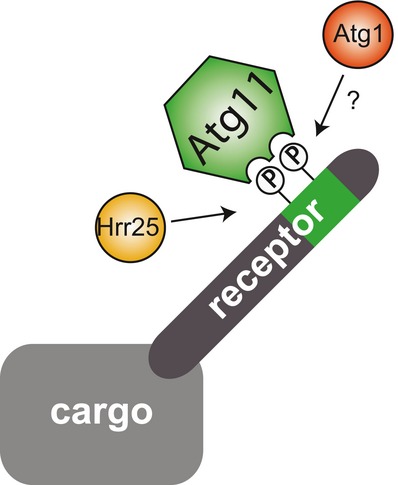
Model representing the phospho-regulation of the cargo receptor Atg19 and conceivably Atg34: Hrr25 and possibly Atg1 phosphorylate the cargo receptor in the Atg11 binding region. This promotes the interaction with Atg11.
Discussion
Our findings are in line with previous work identifying phospho-requirements on the cargo receptors Atg30, Atg32, and Atg36 19. Casein kinase 2 phosphorylates Atg32 in mitophagy, but is dispensable for the Cvt pathway 20, suggesting that different types of selective autophagy are regulated by distinct kinases.
We identified three phosphorylation sites in the Atg11 binding region of Atg19 in vivo, which are essential for its interaction with Atg11. In vitro, we found that both Atg1 and Hrr25 are able to specifically phosphorylate these serine residues. However, in Atg1-D211A mutant cells, phosphorylation of these sites persists and the Atg11-Atg19 interaction is not affected, whereas in cells lacking functional Hrr25, the interaction of Atg11 with Atg19 is abrogated under nutrient-rich conditions. This strongly suggests that Hrr25 is the major regulator of Atg19 binding to Atg11 in vivo. Atg1 kinase might serve an additional function to fine-tune the system for instance during starvation. Hrr25 kinase is a homologue of mammalian casein kinase I and has been implicated in diverse cellular processes such as ribosomal subunit biogenesis 31, chromosome segregation 32, DNA repair 33, and vesicular trafficking 34. Interestingly, Hrr25 mediates the directionality of ER-Golgi transport by phosphorylating COPII coat factors Sec23 and Sec24, and Sec23 and Sec24 mutants show defects in autophagy although autophagy function does not generally require ER-Golgi transport 35. The fact that Hrr25 phosphorylates the cargo receptor Atg19 suggests that a cross talk between COPII trafficking and the Cvt pathway may exist.
An increasing amount of data indicates that phosphorylation of cargo receptors is a universal principle to promote their association with the autophagic machinery, but it remains unclear whether cargo receptors are constitutively phosphorylated. Phosphorylation could provide a mechanism to spatially and temporally regulate the recruitment of the autophagic machinery to the cargo and distinct kinases phosphorylating only selected receptors might regulate these interactions already before cargo binding. Further studies are needed to explore the potential regulatory role of these phosphorylation events, and the kinases involved.
Materials and Methods
Yeast strains, growth conditions, protein purification, kinase assay
Yeast strains are listed in Supplementary Table S2. Yeast cell growth conditions 26, protein purification from E. coli 23, and kinase assays 27 have been described previously and are explained in more detail in Supplementary Methods.
Yeast cell extract preparation and immunoprecipitation
Protein extraction with TCA and Western blotting was carried out as described previously 21. To prepare extracts under non-denaturing conditions, cells were grown in rich or selective medium to early logarithmic phase and harvested by centrifugation. Cell extracts were prepared by freezer milling 27 (Figs 3E and F, 4E, and Supplementary Fig S1F) or by glass bead lysis (Fig 1D, and Supplementary Fig S2D). Protein A immunoprecipitations were carried out as described previously 21,27. For GFP immunoprecipitations, GFP-Trap agarose (ChromoTek) was used.
Acknowledgments
We thank Andrea Brezovich for plasmids, Natalie Romanov, and David Hollenstein for help with MS analysis, Stefan Westermann for strains and recombinant proteins, Peter Steinlein for help with microscopy and members of the Kraft and Martens laboratories for helpful discussions. Funding by the University of Vienna is gratefully acknowledged. T.B. is supported by the VIPS Program of the Austrian Federal Ministry of Science and Research and the City of Vienna. S.M. is funded by the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007–2013)/ERC Grant Agreement No. 260304 and by a grant from the Austrian Science Foundation (FWF, P 25546- B20). C.K. is supported by a “Vienna Research Groups for Young Investigators” grant from the Vienna Science and Technology Fund (WWTF, VRG10-001) and a grant from the Austrian Science Foundation (FWF, P 25522-B20).
Author contributions
TP, WR, TB, VN, DP and MS performed the experiments shown in the manuscript. CA performed initial experiments. TP, WR, TB, VN, DP, GA, SM and CK participated in the experimental design. CK wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–1412. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011;286:13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Nazarko TY, Ozeki K, Till A, Ramakrishnan G, Lotfi P, Yan M, Subramani S. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014;204:541–557. doi: 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Ku W-C, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J-C, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou Y-S, Bjørkøy G, Nunn JL, Bruun J-A, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J-C, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kurihara Y, Jin X, Goda T, Ono Y, Aihara M, Hirota Y, Saigusa T, Aoki Y, Uchiumi T, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–794. doi: 10.1038/embor.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6:1168–1178. doi: 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- Reiter W, Anrather D, Dohnal I, Pichler P, Veis J, Grøtli M, Posas F, Ammerer G. Validation of regulated protein phosphorylation events in yeast by quantitative mass spectrometry analysis of purified proteins. Proteomics. 2012;12:3030–3043. doi: 10.1002/pmic.201200185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa-Makarska J, Abert C, Romanov J, Zens B, Ibiricu I, Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane–cargo apposition during selective autophagy. Nat Cell Biol. 2014;16:425–433. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 Kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok J, Kim PM, Lam HYK, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin Z-Y, Breitkreutz B-J, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Basu U, Ray A, Majumdar R, Deng H, Maitra U. The Saccharomyces cerevisiae 60 S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem. 2008;283:9681–9691. doi: 10.1074/jbc.M710294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Hoekstra MF, Liskay RM, Ou AC, DeMaggio AJ, Burbee DG, Heffron F. HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science. 1991;253:1031–1034. doi: 10.1126/science.1887218. [DOI] [PubMed] [Google Scholar]
- Lord C, Bhandari D, Menon S, Ghassemian M, Nycz D, Hay J, Ghosh P, Ferro-Novick S. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


