Abstract
Autosomal recessive polycystic kidney disease (ARPKD) results from mutations in the human PKHD1 gene. Both this gene, and its mouse ortholog, Pkhd1, are primarily expressed in renal and biliary ductal structures. The mouse protein product, fibrocystin/polyductin complex (FPC), is a 445-kDa protein encoded by a 67-exon transcript that spans >500 kb of genomic DNA. In the current study, we observed multiple alternatively spliced Pkhd1 transcripts that varied in size and exon composition in embryonic mouse kidney, liver, and placenta samples, as well as among adult mouse pancreas, brain, heart, lung, testes, liver, and kidney. Using reverse transcription PCR and RNASeq, we identified 22 novel Pkhd1 kidney transcripts with unique exon junctions. Various mechanisms of alternative splicing were observed, including exon skipping, use of alternate acceptor/donor splice sites, and inclusion of novel exons. Bioinformatic analyses identified, and exon-trapping minigene experiments validated, consensus binding sites for serine/arginine-rich proteins that modulate alternative splicing. Using site-directed mutagenesis, we examined the functional importance of selected splice enhancers. In addition, we demonstrated that many of the novel transcripts were polysome bound, thus likely translated. Finally, we determined that the human PKHD1 R760H missense variant alters a splice enhancer motif that disrupts exon splicing in vitro and is predicted to truncate the protein. Taken together, these data provide evidence of the complex transcriptional regulation of Pkhd1/PKHD1 and identified motifs that regulate its splicing. Our studies indicate that Pkhd1/PKHD1 transcription is modulated, in part by intragenic factors, suggesting that aberrant PKHD1 splicing represents an unappreciated pathogenic mechanism in ARPKD.
Keywords: Autosomal recessive polycystic kidney disease, PKHD1, Alternative splicing, Exon splice enhancers
Introduction
Autosomal recessive polycystic kidney disease (ARPKD; MIM 263200) is a hereditary hepato-renal fibrocystic disorder, with an estimated incidence of 1:20,000 live births [1]. Affected children typically present in utero with enlarged, echogenic kidneys, and oligohydramnios due to poor fetal urine output. An estimated 30–50 % of affected neonates die shortly after birth as a result of severe pulmonary hypoplasia and respiratory failure. Affected perinatal survivors have varied clinical presentations. The principal causes of morbidity and mortality in these children are systemic hypertension, progressive renal insufficiency, and portal hypertension due to portal tract fibrosis [2].
Mutations in the human gene, polycystic kidney and hepatic disease 1 (PKHD1), cause all typical forms of ARPKD. PKHD1 spans more than 469 kilobases (kb) of genomic sequence and is predicted to include at least 86 exons, 71 of which are nonoverlapping, and 15 appear to have alternative splicing boundaries [3]. Mouse Pkhd1 is similar, extending over 500 kb with a minimum of 68 nonoverlapping exons, and seven alternatively spliced exons [4]. The longest open reading frame (ORF) of Pkhd1 contains 66 exons and encodes full-length fibrocystin/polyductin complex (FPC), a protein of 4,059 amino acids [4]. FPC is predicted to have a long extracellular N-terminus containing multiple Ig-like, plexin, transcription factor (IPT), and parallel beta-helix (PbH1) repeats, a single transmembrane span, and a short cytoplasmic C-terminus [4]. Human and mouse FPC share 73 % identity over their complete length, though there are segments with considerably higher (87 %) and lower levels (40 %) of identity [4].
Multiple lines of evidence suggest that PKHD1/Pkhd1 are transcriptionally complex. Efforts to assemble the complementary DNA (cDNA) of the longest PKHD1/Pkhd1 transcript identified several distinct, smaller transcripts [3]. Northern blot analysis of human tissues reveal a complex signal [3], suggesting that Pkhd1 alternative splicing may be organ/cell-type specific [4]. Interestingly, a recent study by Bakeberg et al. [5] used a knock-in strategy to generate an epitope-tagged version of the wild type Pkhd1 allele. Their northern blot analysis identified the full-length 13 kb band as well as three minor bands at 9, 7.7, and 7.5 kb. These variants are likely translated into peptides, as demonstrated by various immunoblotting studies, but the results have not been reproducible from tissue to tissue or study to study [6–8]. Thus, FPC peptide analysis has been hindered due to the lack of well-validated immunoreagents.
Alternative splicing results in distinct messenger RNA (mRNA) molecules comprised of different combinations of exons and/or introns resulting in multiple structurally and functionally distinct proteins [9]. It is estimated that 90 % of human genes are alternatively spliced [10]. Both exons and introns contain DNA binding sites called exonic/intronic splicing enhancers (ESEs/ISEs) that are recognized by splicing factors [11]. Members of the serine/arginine-rich splicing factor family (SRSF) bind to splice enhancers and facilitate spliceosome/mRNA interactions [12]. A subset of disease-causing mutations are known to affect splicing, either by directly altering canonical splice sites, or by disrupting SRSF motifs [13–15].
Given that convergent lines of evidence suggest that PKHD1/Pkhd1 undergoes extensive alternative splicing, we characterized the kidney specific transcriptional profile of Pkhd1 by cataloging its exon usage and examining key motifs that regulate splicing. Our data indicate that Pkhd1/PKHD1 transcriptional processing is modulated in part by specific SRSF intragenic motifs. Furthermore, our analyses of reported PKHD1 missense variants suggest that dysregulated PKHD1 splicing may represent an unappreciated pathogenic mechanism in ARPKD.
Methods and materials
Mice
The DBA/J2 mouse colonies were maintained at the University of Alabama at Birmingham (UAB) under protocols approved by the UAB Animal Care and Use Committee that is fully accredited by the AAALAC.
RT-PCR, subcloning, and sequencing
Total RNA from wild-type (WT), adult (2 months), and E18.5 embryonic tissues was prepared using the Qiagen RNeasy kit (Qiagen, Valencia, CA, USA). cDNA was synthesized with random hexamer primers using Invitrogen Super Script 3 cDNA synthesis kit (Life Technologies, Grand Island, NY, USA). Long-range reverse transcription PCR (RT-PCR) was performed using Roche’s Expand Long Template PCR system (Roche, Indianapolis, IN, USA) with pooled RNA (N=5) for isolation and characterization. Pkhd1 sequence specific primers positioned in exons 1 and 67 were used to amplify transcripts (Supplemental Table 1). The PCR products were resolved on agarose gels, excised, and subcloned using Invitrogen TOPO-TA 2.1 and XL PCR cloning kits (Life Technologies, Grand Island, NY, USA). The constructs were then evaluated by digestion, and representative clones were sequenced. Pkhd1 exons specific primers (Supplemental Table 5) flanking the putative novel splice sites were used for both long-range RT-PCR and for the two-round PCR as indicated. The sequence specificity of all primers was confirmed using NCBI blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (see Supplemental Tables 1 and 5).
Target enrichment and RNASeq studies
Custom enrichment was performed using a commercially available Riva chip (Prognosys Biosciences, Inc., La Jolla, CA, USA). Briefly, solution-based hybridization with Pkhd1 probes (Supplemental Table 2) selectively captured Pkhd1 cDNA for next-generation sequencing (Hudson Alpha Institute, Huntsville, AL, USA). From two separate experiments, mouse inner medullary collecting duct (mIMCD3, ATCC CRL-2123) total RNA was converted to cDNA, then fragmented and hybridized with the custom Pkhd1 probes. The targets were captured, amplified, and the cDNA was sequenced on an Illumina HiSeq2000 (136 million 50 bp pair-end reads). The RNASeq data (Supplemental Table 3) were analyzed by the Biomedical Informatics Component of the UAB Center for Clinical and Translational Science using Tophat Galaxy (https://bitbucket.org/galaxy/galaxy-central/src/tip/CITATION), and IGV [16, 17]. The Tophat analyses files are located at the following server link: https://usegalaxy.org/u/curtish-uab/h/boddu-et-al-2014-supplemental-table-3.
Bioinformatics analyses
The strength of all Pkhd1 splice sites was assessed using the Splice Site Prediction by Neural Network (http://www.fruitfly.org/seq_tools/splice.html) [18]. Analysis and comparison of the exonic splice enhancers in Pkhd1 exons was performed using the web-based algorithm ESE finder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) [19]. The ACESCAN2 Web Server was used to analyze intronic splice enhancers [20].
Minigene experiments and site-directed mutagenesis
For exon-trapping minigene analysis, constructs containing mouse Pkhd1 exons 6, 7, and either, 45 (negative control), 49, 51, or 52 were generated in the exon-trapping vector pSPL3 [21]. For evaluating the R760H missense variant, either normal or mutant PKHD1 exons 21, 22, and 23 were incorporated into the vector. The genomic DNA included at least 200 bp of intronic sequence flanking each exon. Total RNA was purified from COS cells 48 h post-transfection. cDNA was generated by reverse transcription using 4 µg of total RNA. Transcripts were amplified by PCR and analyzed by sequencing. The vector based primer sets used for PCR reaction were “a” (5-ATCTCA GTGGTATTTGTGAGC-3) and “b” (5-TCTGAGTCACCT GGACAACC-3). Site-directed mutagenesis was performed using Stratagene QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions.
Preparation of translated mRNA from polysomal fraction and RT-PCR
Translationally active mRNA was prepared from the polysomal fraction of adult WT kidney lysate using sucrose gradient centrifugation as previously described [22]. mRNA was separated, purified, and lithium chloride precipitated from the bound polysomes. cDNA was synthesized with random hexamer primers using Invitrogen Super Script 3 cDNA synthesis kit, as above. Pkhd1 exon specific primer sets were used for PCR (listed in Supplemental Tables 5 and 6). For sequencing, the amplicons were first subcloned using the Invitrogen TOPO-TA 2.1 cloning kit (Life Technologies, Grand Island, NY, USA), and sequenced.
Statistical analyses
For testing the over-representation of exon 6 skipping, we used a binominal distribution to obtain p values assuming that exon skipping started after every possible exon with equal probability in each clone category. Statistical Software R was used for these analyses.
Results
Pkhd1 transcripts vary by age and tissue
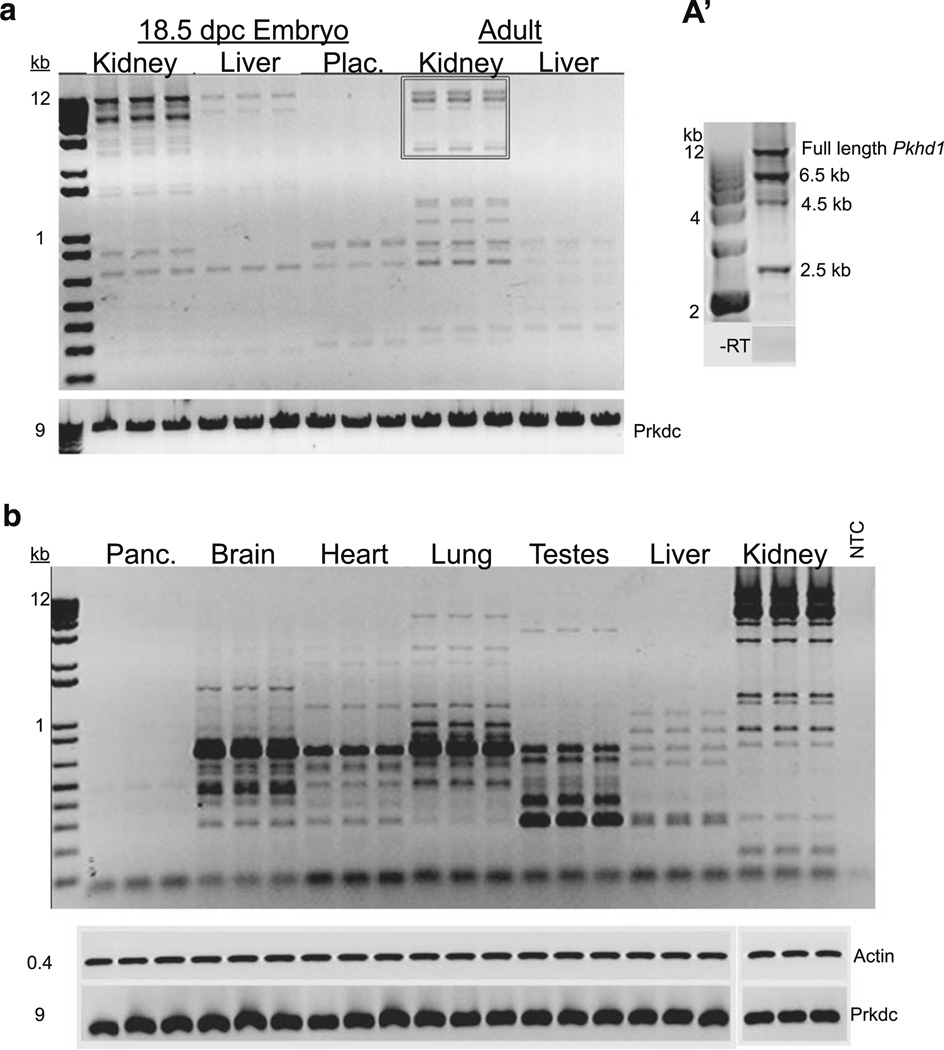
We developed and standardized a long-range RT-PCR protocol using primers within exon 1 and 67 to characterize the Pkhd1 transcriptional profile. Total RNA from wild-type (WT) adult mouse kidney and liver, as well as from 18.5-day-old embryonic kidney, liver, and placenta, were used as templates for amplification (Fig. 1a). Multiple bands were amplified in all samples, and several were excised and sequenced (see below). Each lane represents samples from five mice (n=3, 15 mice per organ), indicating the ubiquity of these transcripts. To assess developmental patterns of Pkhd1 splicing, we compared embryonic kidney, liver, and placenta to adult kidney and liver (Fig. 1a). Neither the kidney nor liver amplicon profiles from the embryonic samples corresponded to the adult samples. To examine differences among adult tissues we compared WT pancreas, brain, heart, lung, testes, liver, and kidney. Unique sets of transcripts were also observed in each of these adult tissues (Fig. 1b), suggesting tissue-specific transcription. These results suggest that Pkhd1 is differentially spliced in an age and tissue-specific fashion.
Fig. 1.
Expression of alternatively spliced Pkhd1 variants in various fetal and adult mouse tissues. a RT-PCR comparing the amplicons from kidney, liver, and placenta from WT 18.5 dpc embryos compared to adult kidney and liver. Transcripts shown in the box are those represented in a’. a’ Representative image of the predominant kidney bands (from box in A) isolated, cloned, and sequenced. b Representative RT-PCR gel image comparing amplicons from adult WT mouse pancreas, brain, heart, lung, testes, liver, and kidney using long range RT-PCR with primers positioned with exons 1 and 67 of the longest ORF of Pkhd1. RT-PCR for each tissue sample was performed in triplicate. Beta-actin serves as an input loading control and Prkdc is shown to demonstrate the integrity of large size RNA (9 kb); NTC no template control. No amplification products were seen in the no-reverse transcriptase (−RT) controls (data not shown)
Kidney-specific Pkhd1 transcripts
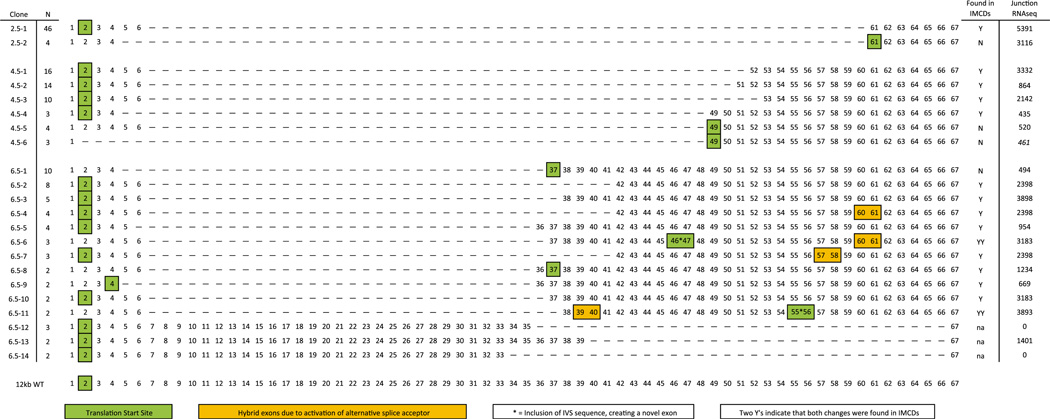
To validate these results and determine the exon composition of each amplicon, we isolated and subcloned the 2.5, 4.5, and 6.5 kb bands amplified from WT kidney cDNA (Fig. 1a’). The full-length mRNA containing 67 exons (>12 kb) was used as a reference [4]. Because each band likely represented multiple transcripts, restriction fingerprinting was performed using EcoRI, PstI, and SacI restriction endonucleases. The clones were grouped based on their band patterns (Fig. 2, “N”) and representative clones were sequenced, resulting in a catalog of 22 unique Pkhd1 transcripts (Fig. 2). A variety of alternative splicing mechanisms were observed including exon skipping, which generated 19 novel exon junctions; two novel inclusions of intronic sequences (Fig. 2 represented as asterisk); and transcripts resulting from alternative exon acceptor and donor site usage (Fig. 2, yellow boxes). The predominant skipping event occurred at exon 6. Indeed, exon 6 usage (e.g., 6–49, 6–51, 6–52, etc.) was significantly more frequent than would be predicted to occur randomly (p<1.4e−61). Further, 14/22 were predicted to use the canonical translation start site (TSS) in exon 2 (Fig. 2, green boxes) and maintained frame through the novel splice junction. NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) predicted alternative TSSs, normally downstream of the splice junction within the WT frame, for the clones whose frame was not maintained due to splicing. For example, clone 2.5–2, has a junction between exons 4–61, which causes a misalignment of the WT frame. NCBI ORF finder, however, predicts that the first ATG within exon 61 is the TSS resulting in a truncated, yet wild-type peptide.
Fig. 2.
Kidney specific Pkhd1 splice variants catalog. The plasmid library generated consisted of 22 Pkhd1 alternatively spliced amplicons. The alternative splicing events included exon skipping; two novel exon inclusions (*) of 104 bp (IVS 46) and 123 bp (IVS 55); and transcripts with truncated exons due to the utilization of cryptic acceptor sites (yellow boxes). Each of the 22 putative transcripts appeared to encode amino-acid sequences in the same frame as the WT peptide, 14 using the canonical translation start site in exon two. Predicted translation start sites indicated by green boxes. Nineteen variants were re-evaluated and amplified from WT kidney RNA using junction specific primers (clones 6.5–12, −13, and −14 not screened); 15 of these junctions were also found in mIMCD3 cDNA and are indicated by “Y.” Where multiple changes were observed in any one transcript (inclusion of IVS::asterisks and hybrid exons::yellow boxes) if both were validated in the mIMCD3 cDNA there is a second “Y”. Many of the novel junctions were also identified in our RNASeq data (Supplemental Table 1); the number of times each junction was found is indicated. N Number of times each clone was found (of total 50 clones per MW band); asterisk novel exon from an IVS; green boxes predicted translation start site; yellow boxes exon partially spliced out
We next validated our results by assessing the presence of these novel exon junctions in mIMCD3 cells, representing the nephron segment most significantly involved in the ARPKD phenotype. The initial template was generated by long-range RT-PCR, followed by a second PCR round that amplified 15/19 of our splice variants (indicated with a “Y”, Fig. 2). Taken together, these data indicate that the survey of amplicons identified by RT-PCR are alternative Pkhd1 transcripts expressed in renal epithelia.
RNASeq confirms alternative Pkhd1 transcripts
We next sought to generate a more inclusive list of Pkhd1 exon junction usage. Using high-throughput RNA sequencing (RNASeq) we evaluated Phkd1 transcripts. In initial studies, we utilized standard global RNASeq methods to evaluate exon junctions using WT kidney RNA. However, this initial dataset had too few reads to be reproducible or significant.
Therefore, we developed a target enrichment strategy using mIMCD3 RNA (see “Methods and materials”) in which cDNA corresponding to Pkhd1 was enriched before sequencing (Supplemental Table 2). We performed the analyses in duplicate. At least 275 unique exon-exon junctions were identified, each of which was identified hundreds or thousands of times (Supplemental Table 3, bold). Among these unique junctions, 20 variants from our RT-PCR catalog were confirmed by RNAseq (Fig. 2). These RNASeq data validate our focused approach and underscore the variability in Pkhd1 splicing.
Bioinformatics analysis of exon and intron splicing motifs
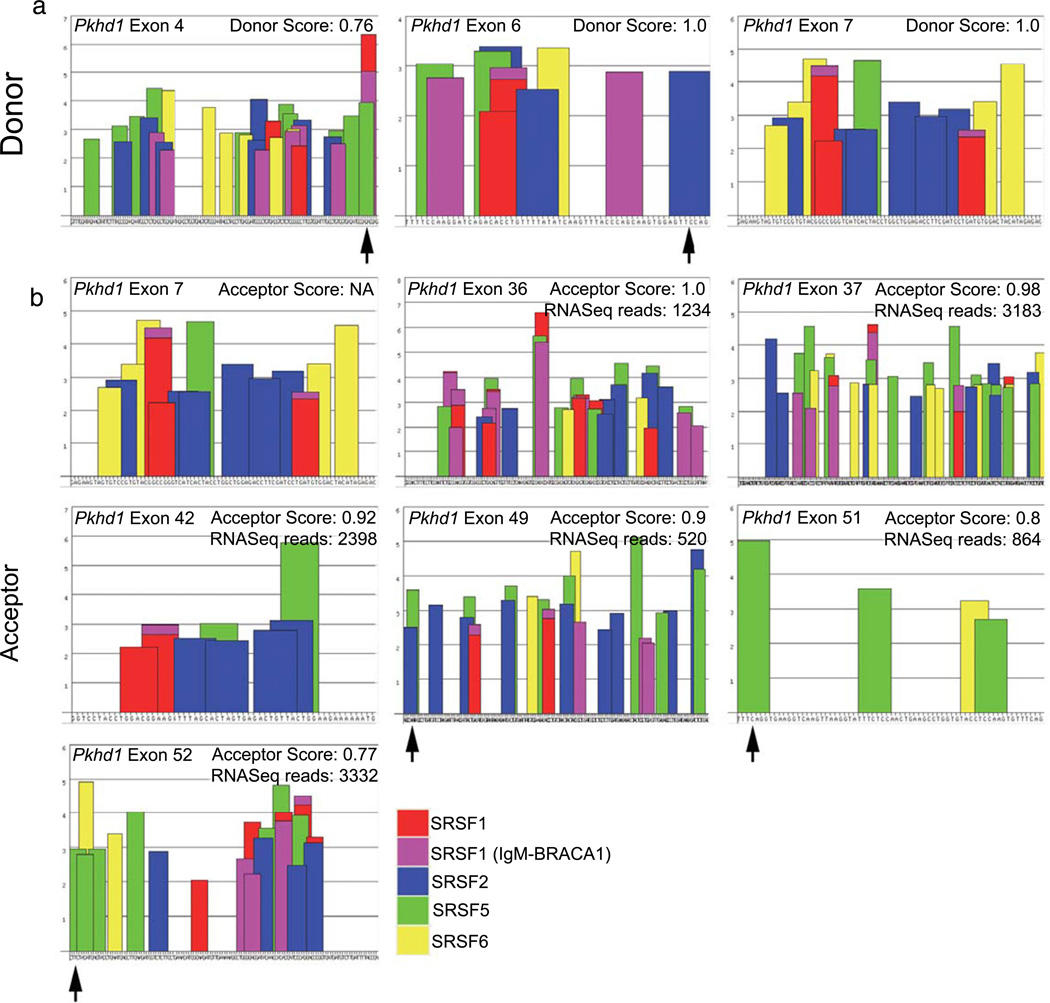
To begin exploring the mechanisms that modulate the splicing of Pkhd1, we used informatic tools. ESE Finder 3 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) identifies putative ESEs associated with common serine/arginine-rich splicing factors (Supplemental Table 4). We analyzed each of the 67 Pkhd1 exons for ESEs (data not shown). For exons that were commonly involved in alternative splicing based on our catalog (4, 6, 36, 37, and 42), we compared the donor/acceptor scores as well as the SRSF motifs to assess why certain Pkhd1 exons are more frequently involved in long range splicing events than others. In evaluating the donor motifs, we compared each exon’s splice donor site to that of exon 6, which we found to be the most common donor exon in our catalog (Fig. 3a). We identified ESEs at the 3′ end of exons 4 and 6 (Fig. 3a, arrows), but there are none within the last several nucleotides of exon 7 (Fig. 3a, no arrow), perhaps explaining why the 4 and 6 are commonly involved in long-range splicing events (Fig. 2, 17/22). We then compared the number of ESEs at the 5′ end of exons and their predicted acceptor scores. We compared exons 36, 37, and 42, which were common splice acceptors (spliced to at least three times), exons 49, 51, and 52, which had 3′ SRSF5 motifs (Fig. 3b arrows) to exon 7, a poor acceptor. Interestingly, while exons 36, 37, and 42 lacked any notable 5′ ESEs, their splice prediction scores were quite high (Fig. 3b, ≤0.92). Exons 49, 51, and 52, which had lower splice acceptor scores (≥0.9), each had ESEs at their 5′ ends (Fig. 3b, arrows). Taken together, these data suggest that for splicing to occur, an acceptor/donor site must either has a ESEs or have high predicted strength. Exons that lack both, e.g., exon 7, appear to be infrequently involved in alternative splicing.
Fig. 3.
Characterization of specific exonic signatures of Pkhd1. a Representative donor site analysis of exons 4, 6, and 7 predicts various SR motifs (bars) throughout the exon. The exon sequence is represented along the X-axis. The height of the bars represents the motif scores; the width of the bars indicates the length of the motif. SSPNN [18] analysis was used to predict the donor scores, as indicated. b Representative acceptor analysis of exons 7, 36, 37, 42, 49, 51, and 52 predicts various SRSF/SR motifs as above. The SSPNN values are indicated. The algorithm did not identify a splice acceptor in exon 7. The number of times each of the acceptor exons was spliced to exon 6, as revealed by RNASeq, is indicated. Arrows indicate the presence of a strong motif at the 3′ (donor) or 5′ (acceptor) end of a given exon
The polypyrimidine tract (PT) is another important cis-acting element that impacts splicing [23]. The PT consensus sequence (Fig. 4a, top, bold) is loosely defined as a series of >15 thymine bases immediately upstream of the intervening sequence (IVS). A strong PT is predicted to contain multiple pyrimidines and ISE motifs, whereas a PT that is interrupted by purine residues and lacks any ISEs is a weak acceptor motif [24]. Alignment of several Pkhd1 PTs illustrates that IVS6 is a poor splice acceptor motif with only eight thymine bases and several purine residues (Fig. 4a, IVS6). Conversely, IVS50 has a very strong PT with 16 thymine residues (Fig. 4a). We then assessed the intronic segments upstream of the acceptor site using ACESCAN2 [25]. The alignment reveals that IVS6 contains only three ISEs within its polypyrimidine tract, while IVS48 has 15; IVS50 has 13; and IVS51 has 13 (Fig. 4b). Together, these data suggest that several canonical exon and intron splicing motifs are contained within Pkhd1, and these motifs regulate the generation of many alternative mRNA molecules.
Fig. 4.
Characterization of the polypyrimidine signatures in Pkhd1 introns. a Alignment of the polypyrimidine tracts (PT), 24 bp upstream of the indicated exons compared to the consensus sequence (top, bold). Analysis shows that the PT in IVS6 contains more purines and fewer pyrimidines as compared to any other IVS. The tally of thymine residues is indicated in the #T column. b Analyses of the intronic splice enhancers just upstream of the acceptor site of the indicated exons were performed using ACESCAN Webserver 2
Minigene exon-trapping studies to evaluate exon and intron splicing motifs
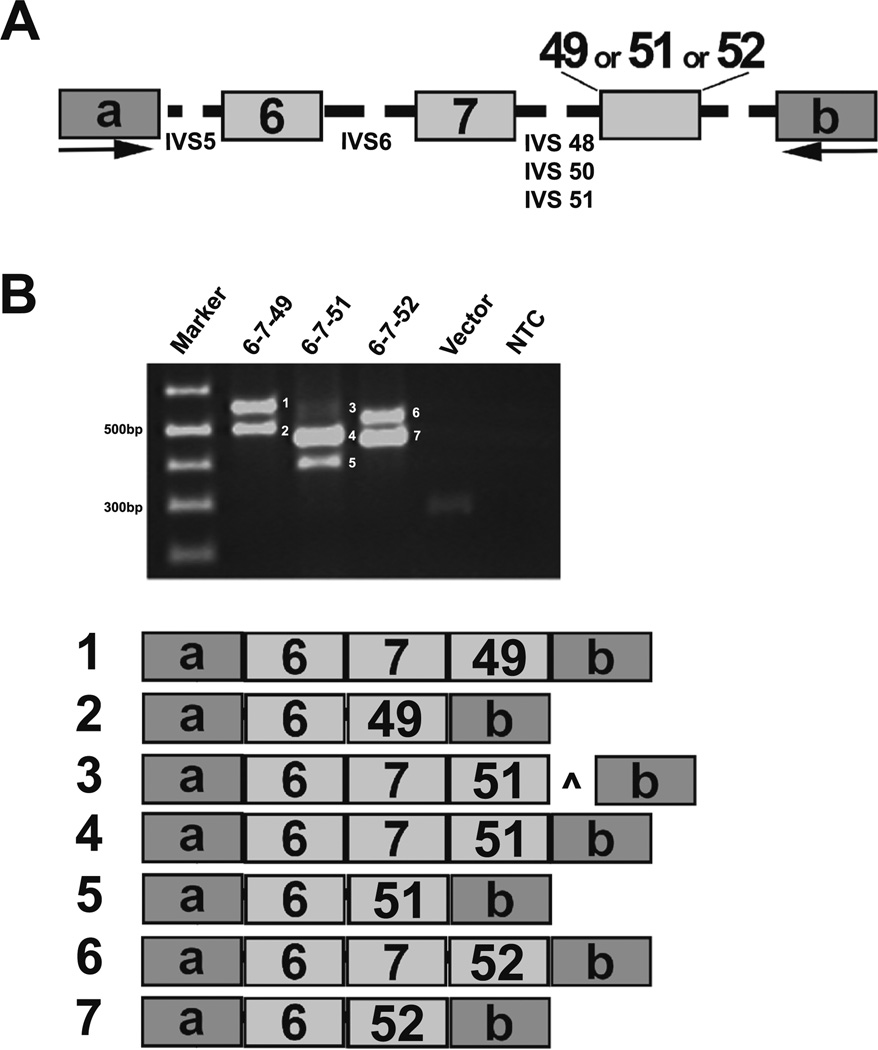
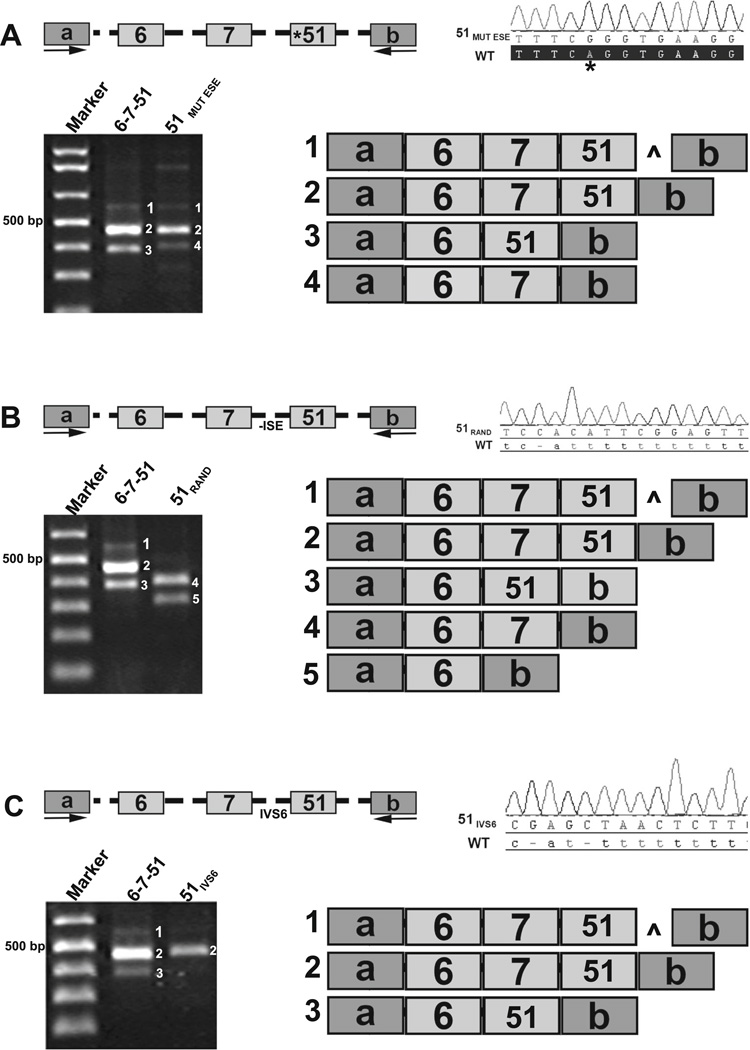
To assess the functional importance of the identified intragenic motifs in Pkhd1 splicing, we used minigene constructs in exon-trapping studies. The minigene constructs contained Pkhd1 exons 6, 7, and either 49, 51, or 52 (Fig. 5a). We first characterized the canonical splice variants with constructs containing WT exons 6–7–49, 6–7–51, and 6–7–52 (Fig. 5a, b, products 1, 4, and 6, respectively). Alternative splicing events, mostly the skipping of exon 7, were observed (Fig. 5b, products 2, 5, and 7). Surprisingly, there was a minor product that included 98 bp of IVS51 downstream of exon 51 (Fig. 5b, product 3, caret), apparently resulting from a cryptic donor site. As a negative control, we used a minigene construct containing exons 6, 7, and 45. Importantly, exon 45 is not predicted to have any SRSF5 motifs, has a weak PT, and was not observed among the alternatively spliced products in our catalog (Fig. 2). No exon splice products containing exon 45 were detected (data not shown).
Fig. 5.
Evaluation of exon and intron signatures in Pkhd1. a For exon-trapping studies, minigene constructs containing mouse Pkhd1 exons 6, 7, and 49, 51, or 52 with 200 bp of flanking intronic sequences (shown as dashed lines) were cloned into the pSPL3 vector and transfected into COS7 cells. b Representative RT-PCR amplicons demonstrating the various splicing events is shown. Each reaction showed that exon 7 was skipped frequently (products 2, 5, and 7). In addition, there was a faint product 3, which included a 98-bp of PKHD1 IVS51 indicated by caret. a, b pSPL3 vector exons
To examine the influence of SRSF5 motifs, we used site-directed mutagenesis to generate a Pkhd1 exon 51 with an ablated SRSF motif (51MUT ESE, Fig. 6a, asterisk). The WT and 51MUT ESE constructs had two splice products in common (Fig. 6a, products 1 and 2). However, splicing to generate 6–51 was not observed from the 51MUT ESE vector (Fig. 6a, product 3 absent from 51MUT ESE lane). Interestingly, the 51MUT ESE construct yielded a product without exon 51 that was not found in the WT experiment (Fig. 6a, product 4). The most common product produced from the 51MUT ESE vector was the canonical transcript, suggesting that the exon 51 SRSF5 motif may not be necessary for canonical splicing and rather may be important in alternative transcript generation.
Fig. 6.
Evaluation of mutated splicing elements in Pkhd1. a Site-directed mutagenesis of Pkhd1 exon 51 (A>G indicated by an asterisk and labeled 51MUT ESE) removed the predicted 5′ SRSF5 motif. WT exon 51 and 51MUT ESE produce two common products (products 1 and 2). The 51MUTESE construct did not skip exon 7, but did yield a product without exon 51 (compare products 3 and 4). b Site-directed mutagenesis replaced Pkhd1 WT IVS50 with a random sequence 51RAND that did not include any ISE/PT elements. Exon-trapping experiments revealed that the WT construct yielded three products (products 1, 2, and 3); whereas with the 51RAND minigene construct, none of the products contained exon 51. c Site-directed mutagenesis replaced WT Pkhd1 IVS50 with Pkhd1 IVS6 to generate a 51IVS6 minigene construct. The WT construct yielded three alternatively spliced products (products 1, 2, and 3); whereas only the canonical product was observed for the 51IVS6 construct. Caret indicates the inclusion of 98 bp from IVS51
We then evaluated the functional significance of ISEs and the PTs as detailed in Fig. 4, using site-directed mutagenesis to alter IVS50, immediately preceding exon 51. The WT IVS50 sequence, with 13 predicted ISEs (Fig. 4b) and a strong PT, was replaced with a random sequence that did not contain any ISEs and had few pyrimidine residues disrupting the acceptor site (CACATTCGGAGTTTGAGCT; 51RAND, Fig. 6b). The WT minigene construct yielded three products, each containing exon 51 (Fig. 6b products 1, 2, 3). In contrast, none of the 51RAND products contained exon 51 indicating that IVS51 was not appropriately recognized as an acceptor motif (Fig. 6b, products 4 and 5). Finally, we replaced IVS50 with the less strong IVS6 acceptor region, which is predicted to have only three ISEs and a weak PT (GAGCTAACTCTTCT TA AA A ; 5 1 IVS6, Fig. 6c). The 51IVS6 construct produced only the canonical product (Fig. 6c, product 2). These data validate our bioinformatic predictions, confirm the functionality of these specific intragenic motifs in vitro, and suggest that the PT, SRSF motifs, and additional ISE sites function primarily to enhance alternative splicing events.
Pkhd1 polysome-bound mRNAs
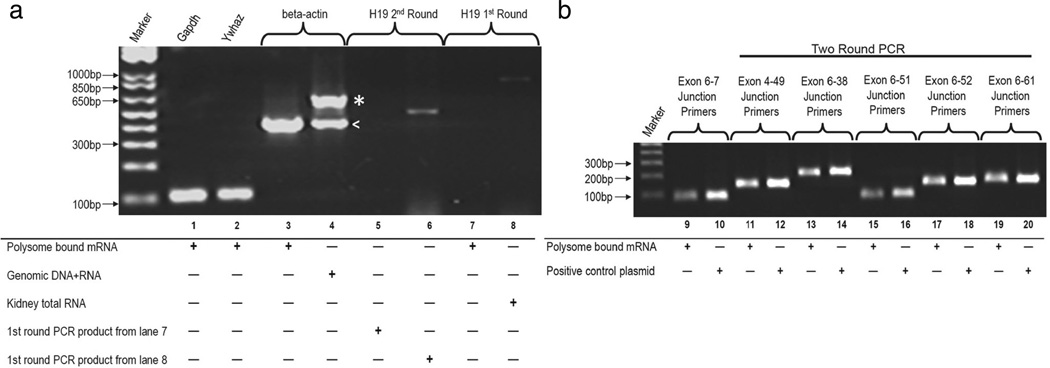
We have demonstrated that alternative Pkhd1 transcripts exist in various mouse organs and mIMCD3 cells; however, whether these transcripts are translated into proteins remains to be confirmed. Unfortunately, with no thoroughly characterized and validated antibodies, immunoblotting has been inconclusive. Thus, we used the association of Pkhd1 mRNAs with polysomes as a surrogate assay. Upon isolation of mRNA::polysome complexes from kidney tissue we evaluated those mRNAs for Pkhd1-specific isoforms (Supplemental Table 5 and 6). The integrity of the isolated polysome RNA was confirmed using primers for the Ywhaz and Gapdh genes, a beta-actin primer set was used to check for genomic DNA contamination, and primers for the h19 noncoding RNA were used as a negative control for total RNA contamination (Fig. 7a, lanes 1–8). Primer pairs flanking the Pkhd1 canonical exon junction 6–7 readily amplified the appropriate product with standard PCR (Fig. 7b lane 9). However, standard PCR did not amplify the other variants, suggesting again that the alternatively spliced transcripts are expressed at low levels. Using two-round, nested PCR on the isolated polysome RNA, we successfully amplified products corresponding to exon junctions 4–49, 6–38, 6–51, 6– 52, and 6–61 (Fig. 7b, lanes 11, 13, 15, 17, and 19). All of the amplicons were sequence verified. As an additional negative control, we used exon junction 6–45 primers as above and no amplification products were identified (data not shown). These data indicate that a subset of Pkhd1 alternative transcripts are polysome bound and are likely translated into peptides.
Fig. 7.
Polysome-bound Pkhd1 mRNAs. a The integrity of the poly-some-bound kidney mRNA was confirmed using primers for Ywhaz and Gapdh; the beta-actin primer set was used to check for genomic DNA contamination, the genomic product includes an intron and thus, is larger (indicated by asterisk, mRNA indicated by less than sign). As a negative control, two round PCR was performed on the h19 gene, a non-coding RNA. Total kidney RNA was used as a control for the h19 gene PCR reaction (lanes 5–8). b RT-PCR from isolated polysome RNA corresponding to exon junctions 6–7, 4–49, 6–38, 6–51, 6–52, and 6–61. All the amplified products were sequence confirmed, and plasmid DNA was used as a control. Primers and sequences listed in Supplemental Table 5 and 6. Where indicated, two round PCR was conducted
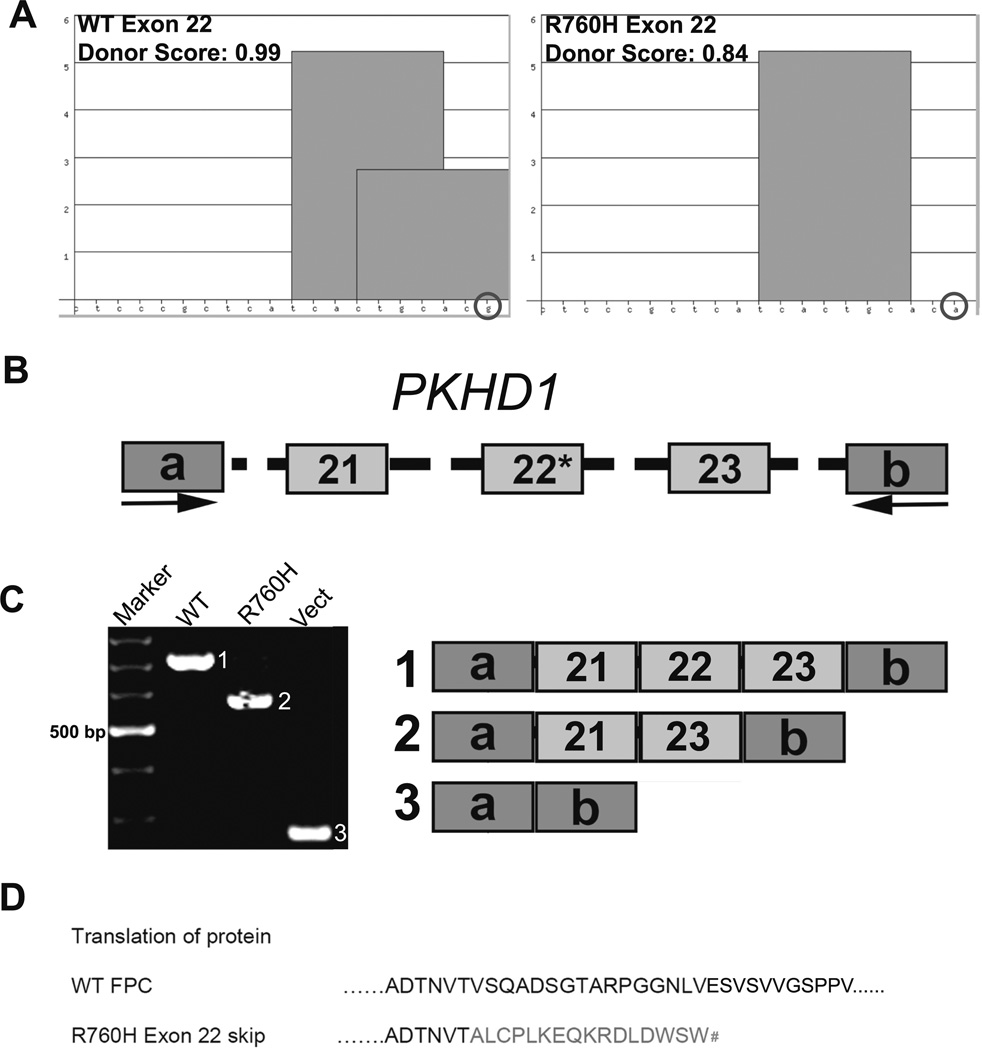
PKHD1 missense variant (R760H) shows splicing defects
Our data demonstrate that Pkhd1 is transcriptionally complex and the identified SRSF motifs, ISEs, and PTs are functionally important in alternative splicing. Therefore, we evaluated the Aachen Mutation Database for ARPKD (http://www.humgen.rwth-aachen.de) [26], as well as data from the UAB Medical Genomics Laboratory to identify patients with lesions that may have altered an SRSF splicing motif. One particular variant, R760H (c.2279G>A) with a single nucleotide change in PKHD1 exon 22, is of indeterminate pathogenicity and abolishes a 3′ SRSF5 motif in a region sparsely populated by ESEs (Fig. 8a). We again used minigene exon-trapping experiments to assess the effect of the R760H variant on PKHD1 splicing. The minigene construct with the normal sequence exclusively yielded a product of 706 bp with PKHD1 exons 21, 22, and 23 (Fig. 8c; WT/1), whereas the R760H minigene generated a single product skipping exon 22 (Fig. 8c; Mut/2). This exon skipping event is predicted to cause a pathogenic frameshift, p.V279X (Fig. 8d). These data suggest that the enhancer motifs within exon 22 of the PKHD1 gene are functional and critical for proper splicing in vitro.
Fig. 8.
PKHD1 missense variant R760H is inefficiently spliced. a ESE Finder 3 analyses identified an SRSF5 motif in the 3′ end of WT PKHD1 exon 22, while analysis of the R760H variant revealed loss of the motif. The nucleotides are indicated along the X-axis, whereas the Y-axis represents the strength of the SRSF5 motifs; SSPNN donor scores are listed. b WT or R760H minigene constructs containing human PKHD1 exons 21, 22, and 23 with 200 bp of flanking intronic sequences were cloned into pSPL3 vector (mutation indicated by R760H). c Representative RT-PCR results are shown. The WT minigene yielded only the canonical 706 bp PKHD1 product containing exons 21, 22, and 23 (WT lane, product 1). The mutant construct (asterisk) generated a product of 567 bp due to skipping of exon 22 (Mut lane, product 2). The empty vector was used as a control (Vect lane, product 3). d Translation of the wild-type and mutant peptide sequences is shown. With the skipping of exon 22, the predicted exon 21–23 junction results in a frame shift that leads to a premature stop codon (number sign). a, b Vector specific exons. All amplified products were sequence confirmed
Discussion
Several lines of evidence indicate that Pkhd1/PKHD1 is transcriptionally complex. However, it is not understood how extensively Pkhd1/PKHD1 mRNA is processed or what governs the generation of alternative transcripts. The current study was designed to address this question using RT-PCR and RNASeq. We noted age- and tissue-specific differences in mRNA profiles, and we identified several alternative Pkhd1 transcripts. We then identified multiple splice enhancer motifs predicted to modulate Pkhd1 splicing and demonstrated their importance using site-directed mutagenesis. Unfortunately, given the paucity of reliable immunoreagents to endogenous FPC, we could not validate these isoforms by immunoblotting. Thus, as a surrogate, we analyzed polysome-bound mRNAs and inferred that many of our unique transcripts are likely translated.
There are several implications of this transcriptional complexity. First, the large size and transcriptional complexity of Pkhd1 would be predicted to complicate standard gene-targeting strategies and RNA-interference-based approaches. For example, Williams et al. [27] generated a Pkhd1lacZ/lacZ mouse that carries a lacZ reporter cassette replacing Pkhd1 exons 1–3. They reported that homozygous mutant mice were viable, but progressively developed cysts. In their study, RT-PCR yielded amplicons only in the wild-type and heterozygous mice, prompting the authors to conclude that the lacZ insertion generated a null allele. In collaboration with the Igarashi lab, we performed long-range RT-PCR and identified alternative Pkhd1 products in the Pkhd1lacZ/lacZ kidneys (Supplemental Figure 1, yellow boxes, sequence confirmed). These data suggest that alternative isoforms may compensate for sequence variants and small insertions/deletions, at least in the kidney. It is important to note that, of the Pkhd1 mouse models generated to date, all have a significant liver phenotype, but the renal phenotype is either absent or remarkably less severe than in the human disease. Together, these data suggest tissue-specific tolerability of isoform loss. Notably, our data indicate that there are fewer isoforms in the liver, possibly making the liver more susceptible to loss of specific Pkhd1 transcripts. A more complete understanding of Pkhd1 transcriptional complexity will be required for proper interpretation of mouse model phenotypes and tissue specific functions.
Second, the complex transcriptome may explain the inconsistent data that have been reported regarding the detection of smaller Pkhd1 isoforms in multiple gene-targeted models and the variability in FPC detection using the available immunoreagents [6–8]. Despite data variability, several lines of investigation are beginning to support our thesis of Pkhd1 transcriptional complexity. For example, the atlas generated by the Genitourinary Development Molecular Anatomy Project (GUDMAP; http://www.gudmap.org/Menu_Index/Gene_Expression.html) [28] contains a transcript present in our catalog (clone 4.5–4; novel splice junction 4–49, 435 RNASeq reads). In addition, northern blot analysis in kidneys engineered by Bakeberg et al., to express an epitope-tagged version of the wild-type Pkhd1 allele, identified the full-length 13 kb band, as well as three minor bands at 9, 7.7, and 7.5 kb. These data are consistent with ours and support the interpretation that Pkhd1 primarily encodes the full-length transcript, with these alternative-spliced transcripts comprising a minority of the transcriptome. With our alternative approaches (e.g., two round PCR and RNASeq), we were able to isolate and characterize some of the less frequent transcripts noted by Bakeberg et al. [5] and others [29–31].
In human ARPKD, the mechanisms by which PKHD1 mutations cause clinical disease are not well understood. Review of the Aachen Mutation Database for ARPKD (http://www.humgen.rwth-aachen.de) [26] reveals that ~44 % of potentially mutant alleles involve missense changes [4, 32]. Most patients are compound heterozygotes, and the functional effect of any particular mutant allele can be difficult to discern. Nevertheless, some broad themes have emerged. Notably, patients with two truncating mutations typically have a severe phenotype leading to perinatal demise (reviewed in [33]). However, there are notable exceptions, e.g., a child who is homozygous for a large PKHD1 deletion surviving well past the neonatal period [34]. Moreover, not all missense mutations lead to a more benign outcome; indeed, a number of missense mutations result in severe phenotypes when present with a truncating mutation in trans or in the homozygous state [33]. We believe that our data indicate that the fidelity of transcriptional splicing requires the integrity of both canonical splice sites as well as ESE motifs. Our R760H analyses serve as a proof-of-concept of this point. Based on these data, we suggest that single nucleotide changes at the 5′ or 3′ ends of an exon (within ~20 bp) should be examined, not only for the impact on a specific amino acid residue but also potential effects on a conserved ESE motifs/splicing.
Many of these single nucleotide variants involve SRSF binding sites and would likely impact splicing (Supplemental Table 4). In support of this thesis, our in vitro assay of the R760H variant of exon 22 did not yield an exon 22-containing amplicon and causes an exon skipping event that leads to a frame shift mutation. Importantly, exon 22 has only two SRSF sites allowing for a straightforward experimental approach, wherein compensation by multiple overlapping sites was unlikely. Analysis of conservation of R760 reveals that only the human and chimp share this amino acid (data not shown). More importantly, however, SRSF analysis in human, chimp, mouse, dog, cat, horse, cow, and sheep reveals that each of these animals has an SRSF5 motif within the last 10 bases of the corresponding exon. Therefore, we speculate that, while the R760H amino acid substitution may have marginal pathogenic significance (as predicted by SIFT), the associated change in a highly conserved SRSF5 motif may represent the actual the pathogenic change. This experiment provides a proof of concept and supports the hypothesis that defects in PKHD1 splicing regulation may contribute to the pathogenesis of ARPKD.
In summary, our RT-PCR-based transcript library, RNASeq data, and polysome experiments provide convergent lines of evidence that Pkhd1 is transcriptionally complex, with the full-length transcript being the most highly expressed. Although expressed at lower levels, alternative transcripts need to be considered when designing experiments to dissect the role of FPC, as well as in assessing the impact of missense variants in ARPKD pathogenesis.
Supplementary Material
Key messages.
Multiple mRNA transcripts are generated for Pkhd1 in renal tissues
Pkhd1 transcription is modulated by standard splice elements and effectors
Mutations in splice motifs may alter splicing to generate nonfunctional peptides
Acknowledgments
The authors acknowledge Drs. Mary Ann Accavitti, Feng Qian, Brad Yoder, and members of the Guay-Woodford laboratory for their helpful advice. This work was supported by the NIDDK and the NIDDK intramural program (R01DK51259 and ZIA DK75042) to G.G.G., the UAB Hepato-Renal Fibrocystic Disease Core Center (P30 DK074038) to LGW, from the Foundation for Research Support of the State of São Paulo (2004/02622-0 to L.O.) and by the NIH-funded UAB Center for Clinical and Translational Science (UL1 RR025777).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00109-014-1185-7) contains supplementary material, which is available to authorized users.
The authors have no conflicts to disclose.
Contributor Information
Ravindra Boddu, Department of Cell Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA; Department of Genetics, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Chaozhe Yang, Department of Genetics, University of Alabama at Birmingham, Birmingham, AL 35294, USA; Center for Translational Science, Children’s National Medical Center, Washington, DC 20010, USA.
Amber K. O’Connor, Center for Translational Science, Children’s National Medical Center, Washington, DC 20010, USA
Robert Curtis Hendrickson, Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Braden Boone, Hudson Alpha Institute, Huntsville, AL 35806, USA.
Xiangqin Cui, Department of Biostatistics, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Miguel Garcia-Gonzalez, Complexo Hospitalario de Santiago de Compostela, Instituto de Investigación Sanitaria (IDIS), Santiago de Compostela, Spain.
Peter Igarashi, Department of Medicine, University of Texas Southwestern School of Medicine, Dallas, TX 75235, USA.
Luiz F. Onuchic, Department of Medicine, University of Sao Paulo, Sao Paulo, Brazil 01246-903
Gregory G. Germino, Division of Nephrology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA National Institute of Diabetes and Digestive and Kidney Disease, National Institutes of Health, Bethesda, MD 20892, USA.
Lisa M. Guay-Woodford, Email: LGuaywoo@childrensnational.org, Department of Genetics, University of Alabama at Birmingham, Birmingham, AL 35294, USA; Center for Translational Science, Children’s National Medical Center, Washington, DC 20010, USA; Children’s National Medical Center, 6th Floor Main Hospital, Center 6, 111 Michigan Ave NW, Washington, DC 20010, USA.
References
- 1.Zerres K, Mucher G, Becker J, Steinkamm C, Rudnik-Schoneborn S, Heikkila P, Rapola J, Salonen R, Germino GG, Onuchic L, et al. Prenatal diagnosis of autosomal recessive polycystic kidney disease (ARPKD): molecular genetics, clinical experience, and fetal morphology. Am J Med Genet. 1998;76:137–144. [PubMed] [Google Scholar]
- 2.Guay-Woodford LM, Desmond RA. Autosomal recessive polycystic kidney disease: the clinical experience in North America. Pediatrics. 2003;111:1072–1080. doi: 10.1542/peds.111.5.1072. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, Bergmann C, Senderek J, Esquivel E, Zeltner R, et al. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet. 2002;70:1305–1317. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagasawa Y, Matthiesen S, Onuchic LF, Hou X, Bergmann C, Esquivel E, Senderek J, Ren Z, Zeltner R, Furu L, et al. Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J Am Soc Nephrol. 2002;13:2246–2258. doi: 10.1097/01.asn.0000030392.19694.9d. [DOI] [PubMed] [Google Scholar]
- 5.Bakeberg JL, Tammachote R, Woollard JR, Hogan MC, Tuan HF, Li M, van Deursen JM, Wu Y, Huang BQ, Torres VE, et al. Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J Am Soc Nephrol. 2011;22:2266–2277. doi: 10.1681/ASN.2010111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Luo Y, Wilson PD, Witman GB, Zhou J. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol. 2004;15:592–602. doi: 10.1097/01.asn.0000113793.12558.1d. [DOI] [PubMed] [Google Scholar]
- 7.Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, Whelan S, Bacallao R, Torra R, LaRusso NF, Torres VE, et al. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet. 2003;12:2703–2710. doi: 10.1093/hmg/ddg274. [DOI] [PubMed] [Google Scholar]
- 8.Menezes LFC, Cai Y, Nagasawa Y, Silva AMG, Watkins ML, Da Silva AM, Somlo S, Guay-Woodford LM, Germino GG, Onuchic LF. Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int. 2004;66:1345–1355. doi: 10.1111/j.1523-1755.2004.00844.x. [DOI] [PubMed] [Google Scholar]
- 9.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterne-Weiler T, Howard J, Mort M, Cooper DN, Sanford JR. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011 doi: 10.1101/gr.118638.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA splicing and disease. Cold Spring Harbor Perspect Biol. 2011;3:a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 19.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burn TC, Connors TD, Klinger KW, Landes GM. Increased exon-trapping efficiency through modifications to the pSPL3 splicing vector. Gene. 1995;161:183–187. doi: 10.1016/0378-1119(95)00223-s. [DOI] [PubMed] [Google Scholar]
- 22.Masek T, Valasek L, Pospisek M. Polysome analysis and RNA purification from sucrose gradients. Methods Mol Biol (Clifton, NJ) 2011;703:293–309. doi: 10.1007/978-1-59745-248-9_20. [DOI] [PubMed] [Google Scholar]
- 23.Kafasla P, Mickleburgh I, Llorian M, Coelho M, Gooding C, Cherny D, Joshi A, Kotik-Kogan O, Curry S, Eperon IC, et al. Defining the roles and interactions of PTB. Biochem Soc Trans. 2012;40:815–820. doi: 10.1042/BST20120044. [DOI] [PubMed] [Google Scholar]
- 24.Coolidge CJ, Seely RJ, Patton JG. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aachen University R. Mutation Database Autosomal Recessive Polycystic Kidney Disease (ARPKD/PKHD1) Pauwelsstraβe 30, D-52074 Aachen, Germany: Department of Human Genetics, RWTH Aachen University; 2013. [Google Scholar]
- 27.Williams SS, Cobo-Stark P, James LR, Somlo S, Igarashi P. Kidney cysts, pancreatic cysts, and biliary disease in a mouse model of autosomal recessive polycystic kidney disease. Pediatric Nephrol (Berlin, Germany) 2008;23:733–741. doi: 10.1007/s00467-007-0735-4. [DOI] [PubMed] [Google Scholar]
- 28.McMahon A, Aronow B, Davidson D, Davies J, Gaido K, Grimmond S, Lessard J, Little M, Potter S, Wilder E, et al. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 29.Moser M, Matthiesen S, Kirfel J, Schorle H, Bergmann C, Senderek J, Rudnik-Schoneborn S, Zerres K, Buettner R. A mouse model for cystic biliary dysgenesis in autosomal recessive polycystic kidney disease (ARPKD) Hepatology (Baltimore, Md) 2005;41:1113–1121. doi: 10.1002/hep.20655. [DOI] [PubMed] [Google Scholar]
- 30.Gallagher AR, Esquivel EL, Briere TS, Tian X, Mitobe M, Menezes LF, Markowitz GS, Jain D, Onuchic LF, Somlo S. Biliary and pancreatic dysgenesis in mice harboring a mutation in Pkhd1. Am J Pathol. 2008;172:417–429. doi: 10.2353/ajpath.2008.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woollard JR, Punyashtiti R, Richardson S, Masyuk TV, Whelan S, Huang BQ, Lager DJ, vanDeursen J, Torres VE, Gattone VH, et al. A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int. 2007;72:328–336. doi: 10.1038/sj.ki.5002294. [DOI] [PubMed] [Google Scholar]
- 32.Sharp AM, Messiaen LM, Page G, Antignac C, Gubler MC, Onuchic LF, Somlo S, Germino GG, Guay-Woodford LM. Comprehensive genomic analysis of PKHD1 mutations in ARPKD cohorts. J Med Genet. 2005;42:336–349. doi: 10.1136/jmg.2004.024489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossetti S, Harris PC. Genotype-phenotype correlations in autosomal dominant and autosomal recessive polycystic kidney disease. J Am Soc Nephrol. 2007;18:1374–1380. doi: 10.1681/ASN.2007010125. [DOI] [PubMed] [Google Scholar]
- 34.Zvereff V, Yao S, Ramsey J, Mikhail FM, Vijzelaar R, Messiaen L. Identification of PKHD1 multiexon deletions using multiplex ligation-dependent probe amplification and quantitative polymerase chain reaction. Genetic Testing Mol Biomark. 2010;14:505–510. doi: 10.1089/gtmb.2009.0188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.