Summary
Objectives
Clostridium difficile infection (CDI) is the leading cause of infectious diarrhea in North America and Europe. The risk of CDI increases significantly in the case where antimicrobial treatment reduces the number of competing bacteria in the gut, thus leading to the increased availability of nutrients and loss of colonization resistance. The objective of this study was to determine comprehensive nutritional utilization and the chemical sensitivity profile of historic and newer C. difficile isolates and to examine the possible role of the phenotype diversity in C. difficile virulence.
Methods
Phenotype microarrays (PMs) were used to elucidate the complete nutritional and chemical sensitivity profile of six C. difficile isolates.
Results
Of the 760 nutrient sources tested, 285 compounds were utilized by at least one strain. Among the C. difficile isolates compared, R20291, a recent hypervirulent outbreak-associated strain, appears to have an expanded nutrient utilization profile when compared to all other strains.
Conclusions
The expanded nutritional utilization profile of some newer C. difficile strains could be one of the reasons for infections in patients who are not exposed to the hospital environment or not undergoing antibiotic treatment. This nutritional profile could be used to design tube feeding formulas that reduce the risk of CDI.
Keywords: Clostridium difficile, Phenotype microarray, Phenome, Phenotype, Metabolism
Introduction
Clostridium difficile is a spore-forming Gram-positive pathogen and is the leading cause of infectious diarrhea. Originally named Bacillus difficilis in the 1930s due to the difficulty in culturing it in the laboratory, it is continuing to be a difficult pathogen even today.1,2 The pathogenicity of C. difficile depends on the action of at least one of the two major exotoxins produced by the bacterium, named toxin A and toxin B.3–6 Some strains also produce a binary toxin, which belongs to the family of binary ADP-ribosylating toxins.7,8 Toxins A and B are encoded by tcdA and tcdB, which are located in a 19.6-kb region called the pathogenicity locus.6 The complications arising from Clostridium difficile infection (CDI) can range from mild self-limiting diarrhea to the fatal pseudomembranous colitis.9,10 The primary risk factors of CDI include antibiotic treatment, advanced age, severe underlying illness, prior hospitalization, tube feeding, gastrointestinal surgery, and the use of proton-pump inhibitors.11,12 However, the epidemiology of CDI has been changing in recent years. Once considered a hospital-associated infection affecting older people, CDI has now spread to younger populations. For example, children are increasingly recognized as being at risk of CDI, even without previous exposure to antibiotics or the healthcare setting.13 It is also spreading to the general community.14,15
An increase in the rate of CDI in North America and Europe is partly explained by the emergence of hypervirulent strains belonging to pulsed-field type 1 and PCR ribotype 027 (NAP1/027).16 Genome sequencing of C. difficile strains has provided many insights into the increased severity of CDI and expansion of the host range. Comparative genomic analysis of these recent hypervirulent strains against historic C. difficile strains has shown that several recent isolates have acquired new virulence determinants and therefore can produce more fulminant CDI.17–19
Although genomic analysis has provided new insights into C. difficile hypervirulence, the nutritional and chemical phenome of C. difficile strains has not been studied. The ability to utilize different nutrient sources coupled with the ability to survive the action of antibiotics, chemicals, and osmotic changes, constitute the global phenome of a bacterium. With the development of phenotype microarrays (PMs), the high-throughput determination of bacterial nutritional and chemical phenotype (phenome) is now possible.20–24 Determination of the global phenome of C. difficile strains is also important, since the severity of CDI is intricately connected to nutrient availability.25
The human gut is home to more than 100 trillion microbes.26 In terms of available nutrition, “tight economic times are the norm” for these microbes.27 Therefore, a bacterium’s survival in the gut depends on its ability to successfully compete for a seat at this crowded table and partake of the available nutrients. The risk of CDI increases significantly when antimicrobial treatment reduces the number of competing bacteria in the gut, leading to an increased availability of nutrients and loss of colonization resistance.28,29 The importance of this nutritional competition in the gut is highlighted by the fact that transplantation of fecal microflora from healthy individuals can cure recurrent CDI.30 Therefore, knowledge of the complete nutritional utilization and chemical sensitivity profile of pathogens such as C. difficile will lead to a better understanding of the conditions under which they are likely to colonize and produce infection.
In this study, we deduced the complete nutritional and chemical sensitivity profile of six C. difficile strains of varying virulence. Our results revealed that some of the recent outbreak strains have expanded metabolic potential. This expanded nutritional usage might explain the spread of community-associated CDI. Further, the metabolic phenotype results we report here could help in the design of better interventions to treat CDI patients such as those receiving tube feeding.
Materials and methods
Phenotype microarray experiments
The metabolic and chemical sensitivity profile of C. difficile strains was measured using Biolog Phenotype MicroArrays (PM). This PM technology consists of 20 PM panels. PMs 1–8 are linked to nutrient utilization (metabolism) and PMs 9–20 measure the chemical sensitivities. We analyzed the complete phenotypic profile of six divergent C. difficile strains employing all 20 PMs. Characteristics of the strains used are given in Table 1.
Table 1.
Characteristics of Clostridium difficile strains used for phenotype microarray comparisons.
| NCBI Taxon ID |
Strain name | Ribotype | Isolation year |
Isolation country |
|---|---|---|---|---|
| 272563 | C. difficile 630 | 013 | 1982 | Switzerland |
| 645462 | C. difficile CD196 | 027 | 1985 | France |
| 367459 | C. difficile QCD-32g58 | 027 | 2004 | Canada |
| 691161 | C. difficile 6534 | Unnamed | 2006 | USA |
| 499174 | C. difficile QCD-23m63 | 078 | 2007 | Canada |
| 645463 | C. difficile R20291 | 027 | 2006 | UK |
NCBI, National Center for Biotechnology Information.
All experiments were conducted in a Bactron IV anaerobic chamber (Shel Lab, OR, USA). Prior to PM experiments, all strains were grown in anaerobic brain heart infusion (BHI) broth. PM experiments were performed following standard Biolog Inc. protocols.22 Briefly, 300 μl of the bacteria grown in BHI broth was plated on Biolog Universal blood agar plates and was incubated overnight at 37 °C. A 40% transmittance cell suspension in Biolog solution IF-0a was then prepared by re-suspending the bacteria grown on Biolog Universal blood agar plates. This suspension was then diluted with Biolog mix B at a ratio of 1:16. One hundred microliters of the final cell suspension was then transferred to each well of respective PM plates. The plates were incubated at 37 °C for 48 h. Bacterial growth was then read at 750 nm using an ELISA reader. Each experiment was performed with biological triplicates.
Statistical analysis
For statistical analysis of the PM data, the means of the replicates were taken. For normalizing the data between strains, the mean of each PM well was divided by the mean of the respective negative plate controls. The value of each well was then compared to the negative control value of the respective plate using analysis of variance (ANOVA). A PM well was considered positive if its value was 40% higher than the negative control at the 5.0% significance level. The Model SEED database31 was then used to predict the genome scale metabolic phenotype of C. difficile. We compared the predicted metabolic phenotype of C. difficile and the positive PM results. This comparison showed that at a 40% growth increase cut-off from the negative control, false-positives are completely avoided. For the chemical sensitivity panels (PM 9–20), a 100% increase from the lowest value of each PM was considered positive.
Results
Nutrient utilization profile of C. difficile strains
Biolog PM metabolic panels (PMs 1–8) consist of about 200 assays of carbon source metabolism, 400 assays of nitrogen source metabolism, 100 assays of phosphorus and sulfur source metabolism, and 100 assays of biosynthetic pathways.22 Therefore, PMs enable the most comprehensive metabolic profiling of microbial cells.22 Using PMs, we compared six C. difficile strains with varying virulence (Table 1). Multiple strains were included in the comparison because genomic comparisons in the past have revealed that genome variation among C. difficile strains is massive and it can have a profound effect on the strains’ virulence properties.17,18 The nutrient utilization profile of the strains tested correlates well with this observed genome diversity.
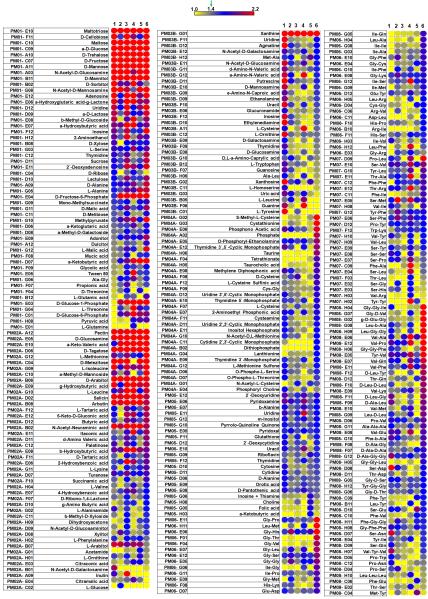
Of the 760 nutrient sources tested, 285 compounds were utilized by at least one strain (Figure 1). Simple sugars were utilized well by all strains. Several complex carbohydrates such as pectin and arbutin could also support growth similar to the levels of simple sugars. However, there were clear differences in the utilization of complex carbon (PM 1–2), nitrogen (PM 3), organic phosphorus (PM 4), sulfur (PM 4), nutritional supplements (PM 5), and peptide nitrogen sources (PM 6–8) (Supplementary Material, Supplementary file S1). Depending upon the type of nutrient in the medium, the amount of toxins produced by C. difficile varied several fold.24 Since the ability to use a nutrient by the bacterium depends on its ability to be transported into the cell, we searched TransportDB32 for C. difficile genes associated with nutrient transport. Based on TransportDB annotation, the C. difficile genome has 198 genes associated with simple sugars, complex carbohydrates, amino acids, and peptides (Supplementary Material, Supplementary file S2). Many of the nutrients had more than one gene associated with their transport, making a one-to-one assignment of nutrient and corresponding gene association difficult. However, almost all of the nutrients that had a transport-associated gene assigned were positive in our phenotype microarray measurements.
Figure 1.
Nutritional phenotypes of Clostridium difficile strains. The complete nutritional phenotype of six C. difficile strains was determined using Biolog Phenotype MicroArray panels 1–8. Compounds that were positive in at least one strain are shown. The numbering scheme given at the top of the columns represents the strains as follows: 1 = CD 630; 2 = CD 196; 3 = CD 6534; 4 = CD 23m63; 5 = CD 32g58; 6 = CD R20291. Each row represents a nutritional phenotype tested. The affinity of nutrient utilization is represented by a color range, as given in the scale bar at the top of the figure.
Among the strains compared, R20291, a recent hypervirulent outbreak-associated strain from the UK, appears to have an expanded metabolic potential when compared to all other strains. For example, this strain was found to have the highest utilization rate of multiple compounds from PMs 1–8. In particular, strain R20291 revealed a greater ability to utilize tetrathionate, taurocholate, taurine, and a large number of peptides as a nutritional source. There was no correlation between the strain type (PCR ribotype or pulsed-field type) and the metabolic utilization profile.
Chemical sensitivity profile of C. difficile strains
The Biolog PM chemical sensitivity panels (PMs 9–20) contain 100 assays of ion effects and osmolarity, 100 assays of pH effects and pH control with deaminases and decarboxylases, and 1000 assays of chemical sensitivity.22 Each chemical sensitivity assay includes four increasing doses of the test chemical. We considered a strain resistant or insensitive to a chemical when there was an at least 100% increase in growth in at least two out of these four doses (Supplementary Material, Supplementary file S3). These assays therefore test whether a strain is sensitive or insensitive to a chemical and do not give information such as minimum inhibitory concentration. As shown in Figure 2 and the Supplementary Material (Supplementary file S3), the chemical sensitivity profile of strains also showed diversity among strains compared.
Figure 2.
Chemical and pH sensitivity of Clostridium difficile strains. The chemical and pH sensitivity profile of C. difficile strains was determined using Biolog Phenotype MicroArray panels 9–14. The numbering scheme given at the top of the columns represents the strains as follows: 1 = CD 630; 2 = CD 196; 3 = CD 6534; 4 = CD 23m63; 5 = CD 32g58; and 6 = CD R20291. Each row represents a chemical tested. The scale bar at the top of the figure denotes the degree of sensitivity. The complete chemical sensitivity data of phenotype microarray panels 9–20 are given in the Supplementary Material (supplementary file S3).
Discussion
All bacteria require a source of carbon, nitrogen, phosphorus, sulfur, potassium, magnesium, calcium, iron, and vitamins for growth. For bacteria living in the human gut, these nutrients can be derived from diet, host mucosal secretions, or other resident (or dietary) microbes.27 Knowing the full metabolic requirements of a bacterium can lead to a better understanding of the conditions under which it is likely to proliferate and enable the design of interventions to prevent proliferation of the pathogen. Identifying the global phenome of more than one strain is important in the case of C. difficile, as it is a species known for large-scale genome variation. Previous studies have identified that the core genome of C. difficile is approximately 20% of the pangenome.18,33 Our PM results presented in Figure 1 are the most comprehensive test for C. difficile nutritional requirements.
PM technology normally reports cellular phenotypes colorimetrically using a tetrazolium redox dye to measure cell respiration. However measuring tetrazolium dye reduction is not feasible for C. difficile, as it is a strict anaerobe. Therefore, in our assays, we measured cell growth instead of dye reduction. This in fact makes our readings more meaningful in terms of nutrient utilization, as we measured actual cell multiplication by measuring the increase in culture turbidity.
Some of the nutritional requirements of C. difficile and those nutrients responsible for the production of C. difficile toxin are well known. For example, the presence of glucose or other rapidly metabolizable carbon sources in the bacterial growth medium strongly represses C. difficile toxin synthesis independently of strain origin.24,34 Therefore, our finding that all strains readily utilized simple sugars such as maltotriose, maltose, α-D-glucose, D-fructose, and D-mannose is in agreement with these previous findings.
The most surprising finding in our results was that strain R20291 had an expanded nutritional utilization profile when compared to all of the other strains (Figure 1). C. difficile R20291 is a hypervirulent ribotype 027 strain, isolated from a recent outbreak in Stoke Mandeville, UK.17 As our results indicate, this strain has a higher capacity to utilize almost all of the nutrient sources. This was found to be particularly pronounced in the case of peptide nitrogen sources. When a strain achieves such increased capacity to utilize a wide spectrum of nutrient sources, it is more likely to colonize a host and this might explain why this strain has been involved in severe CDI and outbreaks. Genome analysis of C. difficile strains that are historic (isolated before 1990s) and recent outbreak strains have shown that new outbreak-associated strains in the last 20 years have acquired several new genetic determinants that confer increased motility and adherence in the gut, increased resistance to bile salts, and better transmissibility manifested through sporulation.17,18 Therefore, the increased metabolic potential of R20291 could be the result of such gene acquisition.
Although it might appear trivial, the ability to use some nutrients such as taurocholic acid could have a profound effect on a strain’s dissemination and colonization. Taurocholic acid is a bile acid and C. difficile strains have varying degrees of ability to use taurocholate as a spore germinant.35 The expanded metabolic potential of this strain coupled with the ability to use compounds such as taurocholate might also explain the expansion of community-associated CDI. Although this suggestion is speculative, it is reasonable to believe that a C. difficile strain that can germinate more efficiently by responding to taurocholic acid in bile and utilize a wider number of nutrients could even colonize a host who is not exposed to antibiotics or the hospital environment.
This expanded metabolic potential is not correlated with the strain typing scheme, as other ribotype 027 strains (Table 1) did not have such a propensity. Rather it reflects the massive genome diversity among strain types and should not be taken as a general property of ribotype 027 strains. Our results, which showed that all strains compared grew well within the mild acid to alkaline range, also correlate well with those of previous studies showing that patients receiving proton pump inhibitors (PPIs) are at increased risk of CDI. Vegetative cells of C. difficile survive better in the gut when the pH of the gut fluids is elevated following the administration of PPIs.36
Overall, the chemicals presented in Figure 2 should be avoided in any interventions designed to control C. difficile, as these do not affect at least some of the C. difficile strains. However, the effects of pH and pH control by deamination have a profound effect on C. difficile growth. As shown in Figure 2, acid pH is inhibitory to C. difficile. Further, different strains have varying amino acid deamination abilities (PM10-E2-G10). Members of the genus Clostridium have the ability to use deamination as a process to use amino acids as the source of carbon. Therefore, the observed differences in growth in these conditions could have adaptive value for C. difficile strains in the gut.
Current clinical practice guidelines for CDI in adults advocate better hygiene in hospitals and among healthcare workers and the restricted use of antibiotics based on local C. difficile epidemiology, and do not recommend the use of probiotics as it is felt that sufficient evidence to support the efficacy of probiotics is lacking.37 However, the options for the treatment of elderly CDI patients and recurrent cases are limited. The diminished ability of elderly patients to mount a protective immune response coupled with loss of gut microbial diversity is a recipe for CDI.38 The nutritional affinity of C. difficile strains presented here offers a new avenue for designing better interventions for ameliorating recurrent CDI cases.
Since our results provide the most comprehensive assessment of nutritional affinity of C. difficile, adjustments to the diet can be made to reduce the amount of such nutrients reaching the gut, thereby reducing C. difficile colonization and proliferation. Such dietary interventions designed to discourage the gut invasion of pathogens have undergone clinical trials recently.39,40 The increased levels of short-chain fatty acid production by distal gut microbiota that occurs upon enrichment of dietary starch has been proposed to result in an intestinal environment unfavorable to Shigella.27,39,40 The idea of C. difficile growth control by nutritional alteration is not new and was demonstrated in animal models in the 1980s.28,41 Therefore, our results showing that C. difficile strains have a high propensity to use easily metabolizable sugars and peptides are in agreement with these previous studies.
Our results also offer an avenue for reducing CDI in patients requiring enteral tube feeding. In recent years, there has been a substantial increase in the number of endoscopically placed tubes for long-term enteral feeding of patients.42 Tube feeding has been recognized as a risk factor for CDI, as the formulation acts as a rich culture medium for C. difficile growth.43,44 The nutritional profile of C. difficile strains elucidated in this study will therefore aid in drawing up an ‘exclude list’ for such tube feeding formulations.
In summary, our results present the most comprehensive analysis of nutritional requirements and chemical sensitivities of C. difficile strains. These definitions could be used for designing better interventions for the treatment of recurrent CDI and also for formulating tube feeding formulas that could reduce the CDI risk.
Supplementary Material
Highlights of the study.
To study the effect of phenotype diversity on Clostridium difficile virulence, global phenome of historic and recent hypervirulent strains were determined using Biolog Phenotype Microarrays.
Of the 760 nutrient sources tested, 285 compounds were utilized by at least one strain.
Compared to other strains, C. difficile R20291, a recent hypervirulent outbreak associated strain, appears to have an expanded nutrient utilization profile.
The nutritional utilization profile of C. difficile strains identified in this profile study could be used to design tube feeding formulas that reduce the risk of CDI.
Acknowledgements
This work was supported in part by a subcontract from Virginia Tech (project No. 62078/A001), which has been funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and Department of Health and Human Services, under Contract No. HHSN272200900040C, and a grant from the USDA Animal Health and Disease Research Program (NYCV-478820).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Approval: Biosafety was approved (No. 16044).
Conflict of interest: No conflict of interest for all authors.
References
- 1.Hall IC, O’Toole E. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. American Journal of Diseases of Children. 1935;49:390–402. [Google Scholar]
- 2.Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359:1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 3.Jank T, Giesemann T, Aktories K. Rho-glucosylating Clostridium difficile toxins A and B: new insights into structure and function. Glycobiology. 2007;17:15R–22R. doi: 10.1093/glycob/cwm004. [DOI] [PubMed] [Google Scholar]
- 4.Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 2008;16:222–9. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan NM, Pellett S, Wilkins TD. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35:1032–40. doi: 10.1128/iai.35.3.1032-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247–63. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papatheodorou P, Hornuss D, Nolke T, Hemmasi S, Castonguay J, Picchianti M, Aktories K. Clostridium difficile binary toxin CDT induces clustering of the lipolysis-stimulated lipoprotein receptor into lipid rafts. MBio. 2013;4:e00244–13. doi: 10.1128/mBio.00244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010;23:529–49. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mylonakis E, Ryan ET, Calderwood SB. Clostridium difficile-associated diarrhea: a review. Arch Intern Med. 2001;161:525–33. doi: 10.1001/archinte.161.4.525. [DOI] [PubMed] [Google Scholar]
- 11.O’Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009;136:1913–24. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 12.O’Keefe SJ. Tube feeding, the microbiota, and Clostridium difficile infection. World J Gastroenterol. 2010;16:139–42. doi: 10.3748/wjg.v16.i2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tschudin-Sutter S, Tamma PD, Naegeli AN, Speck KA, Milstone AM, Perl TM. Distinguishing community-associated from hospital-associated Clostridium difficile infections in children: implications for public health surveillance. Clin Infect Dis. 2013;57:1665–72. doi: 10.1093/cid/cit581. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention Severe Clostridium difficile-associated disease in populations previously at low risk—four states, 2005. MMWR Morb Mortal Wkly Rep. 2005;54:1201–5. [PubMed] [Google Scholar]
- 15.Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, et al. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med. 2013;173:1359–67. doi: 10.1001/jamainternmed.2013.7056. [DOI] [PubMed] [Google Scholar]
- 16.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 17.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaria J, Ponnala L, Janvilisri T, Yan W, Mueller LA, Chang YF. Analysis of ultra low genome conservation in Clostridium difficile. PLoS One. 2010;5:e15147. doi: 10.1371/journal.pone.0015147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–13. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–55. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keymer DP, Miller MC, Schoolnik GK, Boehm AB. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Appl Environ Microbiol. 2007;73:3705–14. doi: 10.1128/AEM.02736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JW, Scaria J, Chang YF. Phenotypic and transcriptomic response of auxotrophic Mycobacterium avium subsp. paratuberculosis leuD mutant under environmental stress. PLoS One. 2012;7:e37884. doi: 10.1371/journal.pone.0037884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei XH, Bochner BR. Using phenotype microarrays to determine culture conditions that induce or repress toxin production by Clostridium difficile and other microorganisms. PLoS One. 2013;8:e56545. doi: 10.1371/journal.pone.0056545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahe S, Corthier G, Dubos F. Effect of various diets on toxin production by two strains of Clostridium difficile in gnotobiotic mice. Infect Immun. 1987;55:1801–5. doi: 10.1128/iai.55.8.1801-1805.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–47. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–4. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borriello SP. The influence of the normal flora on Clostridium difficile colonisation of the gut. Ann Med. 1990;22:61–7. doi: 10.3109/07853899009147244. [DOI] [PubMed] [Google Scholar]
- 30.Kelly CR, de Leon L, Jasutkar N. Fecal microbiota transplantation for relapsing Clostridium difficile infection in 26 patients: methodology and results. J Clin Gastroenterol. 2012;46:145–9. doi: 10.1097/MCG.0b013e318234570b. [DOI] [PubMed] [Google Scholar]
- 31.Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–82. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 32.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–9. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janvilisri T, Scaria J, Thompson AD, Nicholson A, Limbago BM, Arroyo LG, et al. Microarray identification of Clostridium difficile core components and divergent regions associated with host origin. J Bacteriol. 2009;191:3881–91. doi: 10.1128/JB.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol. 2011;79:882–99. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 35.Heeg D, Burns DA, Cartman ST, Minton NP. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One. 2012;7:e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jump RL, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother. 2007;51:2883–7. doi: 10.1128/AAC.01443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 38.Gerding DN. Clostridium difficile infection prevention: biotherapeutics, immunologics, and vaccines. Discov Med. 2012;13:75–83. [PubMed] [Google Scholar]
- 39.Alvarez-Acosta T, Leon C, Acosta-Gonzalez S, Parra-Soto H, Cluet-Rodriguez I, Rossell MR, Colina-Chourio JA. Beneficial role of green plantain [Musa paradisiaca] in the management of persistent diarrhea: a prospective randomized trial. J Am Coll Nutr. 2009;28:169–76. doi: 10.1080/07315724.2009.10719768. [DOI] [PubMed] [Google Scholar]
- 40.Rabbani GH, Ahmed S, Hossain I, Islam R, Marni F, Akhtar M, Majid N. Green banana reduces clinical severity of childhood shigellosis: a double-blind, randomized, controlled clinical trial. Pediatr Infect Dis J. 2009;28:420–5. doi: 10.1097/INF.0b013e31819510b5. [DOI] [PubMed] [Google Scholar]
- 41.Rolfe RD. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect Immun. 1984;45:185–91. doi: 10.1128/iai.45.1.185-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMahon MM, Hurley DL, Kamath PS, Mueller PS. Medical and ethical aspects of long-term enteral tube feeding. Mayo Clin Proc. 2005;80:1461–76. doi: 10.4065/80.11.1461. [DOI] [PubMed] [Google Scholar]
- 43.Iizuka M, Itou H, Konno S, Chihara J, Tobita M, Oyamada H, et al. Elemental diet modulates the growth of Clostridium difficile in the gut flora. Aliment Pharmacol Ther. 2004;20(Suppl 1):151–7. doi: 10.1111/j.1365-2036.2004.01969.x. [DOI] [PubMed] [Google Scholar]
- 44.Bliss DZ, Johnson S, Savik K, Clabots CR, Willard K, Gerding DN. Acquisition of Clostridium difficile and Clostridium difficile-associated diarrhea in hospitalized patients receiving tube feeding. Ann Intern Med. 1998;129:1012–9. doi: 10.7326/0003-4819-129-12-199812150-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.