Abstract
The growing interest in cancer epigenetics is largely due to the reversible nature of epigenetic changes which tend to alter during the course of carcinogenesis. Major epigenetic changes including DNA methylation, chromatin modifications and miRNA regulation play important roles in tumorigenic process. There are several epigenetically active synthetic molecules such as DNA methyltransferase (DNMTs) and histone deacetylases (HDACs) inhibitors, which are either approved or, are under clinical trials for the treatment of various cancers. However, most of the synthetic inhibitors have shown adverse side effects, narrow in their specificity and also expensive. Hence, bioactive phytochemicals, which are widely available with lesser toxic effects, have been tested for their role in epigenetic modulatory activities in gene regulation for cancer prevention and therapy. Encouragingly, many bioactive phytochemicals potentially altered the expression of key tumor suppressor genes, tumor promoter genes and oncogenes through modulation of DNA methylation and chromatin modification in cancer. These bioactive phytochemicals either alone or in combination with other phytochemicals showed promising results against various cancers. Here, we summarize and discuss the role of some commonly investigated phytochemicals and their epigenetic targets that are of particular interest in cancer prevention and cancer therapy.
Keywords: Epigenetics, phytochemicals, DNA methylation, histone deacetylation, cancer
1. Introduction
Carcinogenesis is considered to be the outcome of deregulated genetic and epigenetic events. Epigenetic alterations are important as they link the behavior of cells to their environmental interactions and thus determine the susceptibility of a cell to transforming changes. As these changes do not involve alterations in the genome constructs, these epigenetic events occur constantly during the life of the cells. DNA methylation, histone tail modifications, chromatin remodelling and miRNA-mediated multi-gene silencing are considered to be the major epigenetic changes that are involved in maintaining cellular homeostasis and differentiation states. Multiple studies have revealed that global DNA hypomethylation events are a major characteristic of most of types of cancer and contribute to genomic instability by activating retrotransposons and other silent genomic regions [1]. On the other hand, promoter hypermethylation events occur in important tumor suppressor genes, indicating that it is inappropriate DNA methylation that is important in driving the process of carcinogenesis [2]. Histone modifications also occur, including acetylation and methylation of lysine residues, methylation of arginine residues, phosphorylation of serine or threonine residues, ubiquitination and sumoylation of lysine residues, ADP ribosylation of glutamic acid residues and isomerisation of proline residues, etc.. These modifications define the patterns of chromatin remodelling, thus determining the resultant gene expression and gene silencing patterns. The microRNAs (miRNAs), which are recently identified non-coding RNAs, also have been shown to contain gene-expression regulatory activities and are capable of functioning both to suppress and promote oncogenesis. Deregulated miRNA transcription leads to upregulation of oncogenes and silencing of tumor suppressor genes in lung, breast, head and neck as well as bone cancers [3–6].
Dietary phytochemicals or supplements are not only a rich source of minerals, vitamins and micronutrients but also contain bioactive components such as anti-oxidants, polyphenols and alkaloids, which are more than the basic nutrients. These bioactive compounds have shown great potential against many diseases including cancers through genetic and epigenetic modifications [7–10]. In this review article, we focus on the major types of epigenetic modifications, such as DNA methylation, histone modifications and miRNA-mediated gene silencing in cancer progression, and epigenetic targeting by phytochemicals in cancer prevention and therapy.
2. DNA methylation
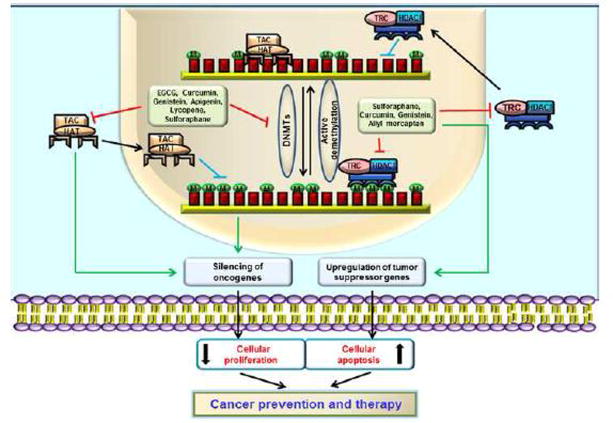
Epigenetics is defined as the study of heritable but reversible changes in gene expression that occur without alterations in the sequences of underlying DNA. Epigenetic modifications often alter gene expression and, in particular, expression of the tumor suppressor, promoter, and oncogenes that are crucial for cellular proliferation, differentiation and survival during carcinogenesis. Among epigenetic modifications, DNA methylation is the best studied modification of DNA [11]. S-adenosyl methionine (SAM) functions as a universal methyl group donor in the methyl transfer reactions catalyzed by DNA methyltransferases (DNMTs) in the eukaryotic nucleus. There are two types of DNMTs present in eukaryotes including maintenance methyltransferase (DNMT1) and the de novo methyltransferases (DNMT3A and DNMT3B). DNMT1 functions in maintaining the pre-established patterns of DNA methylation, while the de novo enzymes establish new patterns of methylation in the fully un-methylated DNA. DNA hypermethylation of tumor suppressor genes is a rather frequent event in most of the cancers both during the initiation or the progression events [2]. Gene hypermethylation also might initiate recruitment of the methylation-dependent DNA binding proteins (MBDs) to the hypermethylated DNA sites. The MBDs further help in silencing of methylated genes by recruiting repressor complexes to these regions. These proteins are commonly found occupying the hypermethylated gene promoters in multiple cancers [12]. In addition to the MBDs, a transcriptional domain in DNMT1 also recruits histone deacetylase (HDACs) and other chromatin re-modelling proteins to the target sites that can modify acetylation and methylation status of histones, thereby inhibiting transcriptional access to the chromatin [13]. An example of such aberrant methylation-mediated gene silencing was demonstrated in ultraviolet-B (UVB) radiation-induced skin tumors in the SKH-1 mouse model. This study clearly demonstrated the positive correlation between transcriptional repression of tumor suppressor p16INK4a and RASSF1A genes and the UVB-mediated hypermethylation and subsequent recruitment of MeCP2 and MBDs at gene regulatory regions in skin cancer model in vivo [14]. MeCP2 is the founding member of the MBD family of transcriptional repressors, which creates a repressive environment at the target DNA site through recruitment of HDAC-containing transcriptional repressor complexes to the methylated DNA [15]. In addition, MeCP2 is also linked with higher H3K9 methylation, which is an important heterochromatin mark [16]. Hence, DNMTs inhibitors are important in cancer therapy and some FDA-approved inhibitors of DNMTs, such as 5-azacytidine and 5-aza-2′-deoxycytidine, are already being used as therapeutic drugs against multiple cancer types [17, 18]. Many of the synthetic inhibitors have, however, been shown to cause adverse toxic effects and are narrow in their specificity. Hence, phytochemicals, which are widely available and have lesser side effects or toxicities, are being tested for their role in direct or indirect inhibition of DNMTs activity in cancer prevention and therapy. DNMTs-mediated differential effects on promoter methylation and histone acetylation on gene regulation by bioactive phytochemicals are depicted in Figure 1.
Figure 1.
Effects of bioactive phytochemicals on promoter methylation and gene regulations. Dietary phytochemicals inhibit DNMTs and induce active demethylation at CpG-rich gene promoters. The transcription activation complex (TAC), which contains HATs, in general, binds to the unmethylated gene promoter leads to silencing of oncogenes and upregulation of tumor suppressor genes. When the gene promoter is methylated by DNMTs, the TAC becomes unable to bind to the gene promoter and initiation of transcription is inhibited. The transcription repression complex (TRC) containing HDACs is recruited to the methylated DNA which in general, leads to silencing of the tumor suppressor genes. Further, the bioactive dietary supplements also directly destabilize different epigenetic remodelling complexes such as TAC and TRC to prevent their binding with respective promoter regions for gene activation/ repression. The dietary phytochemicals-mediated upregulation of tumor suppressor genes and down regulations of oncogenes are the central importance of cancer prevention and therapy. M, methyl group.
3. Histone modifications and chromatin remodelling
Eukaryotic DNA is organized in a complex structure known as chromatin, which is comprised of DNA, histones and several other DNA-binding proteins. In addition to promoting a compact structure, chromatin organization also helps in the regulation of gene expression by restricting the access of different DNA binding proteins or protein complexes to the genetic material. The processes of “opening up” of chromatin and its compaction are associated with a number of ATP-dependent multi-enzyme complexes known as chromatin remodelling complexes. The chromatin remodelling is triggered by various histone tail modifications, which determine the state of activity of chromatin. The best studied histone modification, lysine acetylation, leads to opening up of the chromatin due to the negative charge conferred by the acetyl moieties, which reduces the histone-DNA interactions. The lysine acetylation reactions are catalyzed by histone acetyltransferases (HATs), which transfer the acetyl groups from acetyl coenzyme A to the lysine moieties in the nucleosomes. HATs are classified into three families, the GCN5 N-acetyltransferase (GNAT), the MOZ/YBF2/SAS2/TIP60 (MYST) and the p300/CBP families [19, 20]. HATs also play important roles in regulating cell cycle regulatory protein expression and also can bind directly to the cell cycle regulatory apparatus [21].
Histone deacetylases (HDACs) remove the acetyl groups from lysine residues to reduce the negative charge thus leading to chromatin compaction. Four distinct classes of HDACs in humans have been identified based on their structural similarity to yeast proteins as well as their localization and acetylation activities [20]. Class I, II and IV HDACs are zinc-dependent histone deacetylases, while class III HDACs, also known as sirtuins, are NAD+-dependent HDACs. Different HDACs function in distinct ways and on different downstream targets leading to their diverse tumor suppressor and oncogenic activities. Overexpression and altered activities of HDACs are associated generally with the silencing of tumor suppressor genes, epithelial to mesenchymal transitions and metastasis [22–24]. Over-expression of HDAC1 resulted in downregulation of p53 and von Hippel-Lindau tumor suppressor genes and stimulated angiogenesis of human endothelial cells [22], while HDAC10 suppresses metastasis of cervical cancer through inhibition of matrix metalloproteinases 2 and 9 expression [25].
In general, histone acetylation is associated with gene activation and is abundant in the euchromatin whereas deacetylation is linked to gene repression and occurs in the heterochromatin. Nevertheless, gene repression or activation is not completely dependent on histone acetylation or methylation, but rather is dependent on the site and degree of methylation or acetylation on histone tails. Some of the active chromatin markers associated with gene expression are histone methylation on histone H3 at lysine 4 (H3K4), on histone H3 at lysine 36 (H3K36), on histone H3 at lysine 79 (H3K79) and on histone H4 at lysine 20 (H4K20); while the inactivation markers associated with gene repression are methylation on histone H3 at lysine 9 (H3K9) and on histone H3 at lysine 27 (H3K27) [26]. It is recognized widely that HDACs are promising targets for cancer prevention and therapy. Some of the well-studied HDAC inhibitors are trapoxin (TPX), trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA). Among them, Vorinostat or SAHA and Romidepsin (Istotax) are commercially-available FDA-approved HDAC inhibitors for treatment of cutaneous T-cell lymphoma [27]. Some other HDAC inhibitors such as Panobinostat, valproic acid and Belinostat are in different phases of clinical trials.
4. Dietary phytochemicals and their epigenetic modulatory activities
Enthusiasm for the use of dietary phytochemicals in the prevention and therapy of different diseases has increased in recent years. Possible reasons behind this interest lie in their natural origin, widespread availability, lesser side-effects and the possibility of inclusion in the routine diet. Traditionally, these phytochemicals have been utilized in the treatment of various diseases since ancient times. There now has been a tremendous increase in the knowledge concerning their mechanisms of actions and molecular targets. Interestingly, these dietary factors have shown a capability to regulate the patterns of expression of multiple genes through epigenetic modulatory mechanisms. Herein, we provide an overview in which the epigenetic mechanisms of action of dietary component in different tumor models are summarized. It should be noted, however, that the mechanisms of action of dietary components vary among various cancer types.
Polyphenols are one of the essential bioactive dietary supplements largely present in fruits, vegetables, seeds and nuts. There are approximately 8000 polyphenols present in the diet and these can be classified into ten different generalized classes according to their chemical structure [10]. The major classifications are catechins/epicatechins, stilbenes, benzoquinones, acetophenones, flavonoids, phenolic acids, proanthocyanidins, ellagitannins and anthocyanins [28]. Among these, the most commonly studied polyphenols are (−)-epigallocatechin-3-gallate (EGCG; found in green tea), resveratrol (present in grape skin) and curcumin (found in turmeric). These dietary polyphenols have been shown to have chemopreventive and therapeutic potential in preclinical models against various cancers [7, 29, 30]. Significant progress has been made in understanding the effects of these agents in altering signaling cascades and other inflammatory pathways involved in carcinogenesis. Further, these dietary polyphenols have shown to reverse the epigenetic changes occurred during the process of carcinogenesis.
4.1. Tea catechins/epicatechins
The tea plant (Camellia sinensis) is cultivated in more than 30 countries. Green, black, and oolong teas are the most common varieties of tea all of which are derived from the leaves of the tea plant defined but differ according to their manufacturing processes [31, 32]. Studies have shown that green tea possesses significant beneficial effects due to an abundance of monomeric catechins or epicatechins, which include (−)-epicatechin (EC), (−)-epicatechin-3-gallate (ECG), (−)-epigallocatechin (EGC) and (−)-epigallocatechin-3-gallate (EGCG) [32]. Of these, EGCG is the major and most active ingredient of green tea polyphenols (GTPs) and is shown to have potent anti-cancer activities both in vitro and in in vivo models [33, 34]. In particular, EGCG has been shown to inhibit cellular proliferation and to induce apoptosis in many cancer cell types through multiple mechanisms [35–37]. Several elegant studies have addressed the modes of actions of various tea polyphenols, including EGCG, in cancers. Recent studies have suggested that the anti-cancer activity of EGCG is mediated, at least in part, through its epigenetic modulatory activities, such as inhibition of DNMTs and HATs [33, 38, 39].
EGCG is involved in direct inhibition of DNMTs by forming hydrogen bonds in their active sites that hinder substrate binding [39]. EGCG also has been reported to reduce the available S-adenosyl-L-methionine (SAM), a methyl donor for DNMTs, and induce S-adenosyl-L-homocysteine (SAH), a potent inhibitor of DNMTs, in a mechanism in which EGCG mediates indirect inhibition of DNMTs [27]. EGCG-mediated indirect inhibition of SAM (but not SAH) has been demonstrated convincingly by both in vitro and in vivo studies [39, 40]. The inhibition of DNMTs can lead to reversal of the silencing of tumor suppressor genes in cancer cells, which has been found to be one of the effective treatment strategies against cancer. Fang et al. [39] showed that the treatment of EGCG in human oesophageal KYSE 510 and 150 cells leads to a concentration- and time-dependent reversal of hypermethylation and re-expression of several known tumor suppressor genes such as p16INK4a, O6-methylguanine methyltransferase (MGMT), human mutL homologue 1 (hMLH1) and retinoic acid receptor β (RARβ) [39]. Similarly, we demonstrated that the topical application of EGCG, which is protective against ultraviolet-B (UVB) radiation responses, restores UVB radiation-induced global hypomethylation in the SKH-1 hairless mouse model [41]. We also further demonstrated that treatment of cancer cells with EGCG not only reactivates tumor suppressor genes but also leads to down-regulation of human telomerase reverse transcriptase (hTERT), a catalytic subunit of telomerase, a tumor promoter and aging-related gene [42, 43]. In contrast to tumor suppressor genes, demethylation of the hTERT promoter is, in general, associated with its repression. This paradox is explained partially by the observation that the EGCG-mediated demethylation recruits gene repressor complexes to the hTERT promoter resulting in its transcriptional downregulation [43]. In addition, Wnt, an oncogenic ligand family, has been reported to be downregulated by EGCG-mediated promoter demethylation in lung cancer cells [44].
These studies suggested that the inhibition of tumor promoter genes and induction of tumor suppressor genes are of central importance in cancer prevention and therapy. In accordance, Meeran et al reported that EGCG and a pro-form of EGCG inhibit hTERT expression by inducing gene-specific demethylation and chromatin modifications in human breast cancer cells [38]. The authors demonstrated that EGCG and the pro-form of EGCG induced chromatin alterations that facilitated the binding of many hTERT repressors such as MAD1 and E2F-1 to the hTERT regulatory region, thereby contributing to their transcriptional repression [38]. In yet another study, we demonstrated that the treatment of A431 skin cancer cells with EGCG downregulates the expression and activities of DNMTs and the level of 5-methylcytosine resulting in the re-expression of key tumor suppressor genes such as p16INK4a and p21CIP1/WAF1 [33]. In addition to DNA methylation, we demonstrated that the treatment of EGCG decreased HDAC activity and increased levels of active chromatin markers, such as acetylated lysine 9 and 14 on histone H3 and acetylated lysine 5, 12 and 16 on histone H4, which also contributed to the re-expression of tumor suppressor genes in A431 skin cancer cells thereby enabling their inhibition of cellular proliferation and induction of apoptosis [33]. In another study using oral carcinoma cells, partial demethylation of RECK, a tumor suppressor gene, by EGCG was found to be well correlated with inhibition of tumor invasion, angiogenesis and metastasis [45]. Furthermore, EGCG has been demonstrated to inhibit the invasive potential of human pancreatic adenocarcinoma cells by modulation of HDAC activity [46] and EGCG has been shown to induce the degradation of DNMT3A and HDAC3 in methylation-sensitive colon cancer cells, in part, through inhibition of E3 ubiquitin ligase [47]. EGCG also was found to induce tissue inhibitor of matrix metalloproteinase-3 (TIMP-3) gene, which negatively regulates matrix metalloproteinases (MMPs) activity, through the enrichment of H3K27 trimethylation in the promoter and a concomitant increase in histone H3K9/18 acetylation in breast cancer cells [48].
Combination of HDAC inhibitors with EGCG (a known DNMT inhibitor) is another effective approach for cancer prevention and therapy. Studies conducted by Li et al. [49] showed that a combination of EGCG with an HDAC inhibitor reactivates estrogen receptor-α (ERα) expression in ERα-negative human breast cancer cells through a process associated with chromatin modifications. EGCG in combination with TSA, a HDAC inhibitor, synergistically reactivates p16INK4a expression, in part, through a reduction of promoter expression in CA46 human lymphoma cells [50]. Similarly, EGCG in combination with sodium butyrate synergistically inhibits cell cycle arrest and induces cellular apoptosis through down regulation of HDAC1, DNMT1 and inhibition of HDAC activities in colon cancer cells [51]. Not only synthetic inhibitors, but also the dietary combination of green tea polyphenol (GTPs), a DNMT inhibitor, with sulforaphane (SFN), a HDAC inhibitor, reactivate ERα expression in ERα-negative human breast cancer cells and this was found to correlate consistently with ERα promoter hypomethylation and hyperacetylation [52]. These reactivations are a promising strategy for the effective treatment of hormonal refractory breast cancer with available anti-estrogen drugs like tamoxifen. In addition to ERα reactivation, GTPs and SFN combinations also tend to enhance cisplatin-induced apoptosis and G2/M phase cell cycle arrest through upregulation of p21CIP1/WAF1, thereby enhancing the efficacy of cisplatin on both cisplatin-sensitive and cisplatin-resistant ovarian cancer cells [53]. Taken together, these data suggest that GTPs alone or in combination with dietary HDAC inhibitors can modulate epigenetic regulation of gene expression and can play a vital role in cancer chemoprevention and therapy.
4.2. Sulforaphane
Sulforaphane (SFN) is one of the major isothiocyanates present in many cruciferous vegetables and has been shown to have anti-cancer activities in several cancer models [54–57]. Although SFN has been shown to mediate anti-cancer activity through several mechanisms, including cell cycle arrest, induction of cellular apoptosis and phase-2 detoxification enzymes, there is a growing interest in their HDAC inhibitory activity [55, 56, 58, 59]. As aforementioned, HDACs are often upregulated in cancers and HDACs inhibitors play a major role in cancer prevention and therapy. SFN-mediated inhibition of DNMTs and histone methylations also have been found to play a major role in the alterations of gene expressions found in cancer prevention studies. In accordance, SFN-mediated downregulation of DNMTs also is associated positively with hTERT promoter demethylation, which is followed by binding of CTCF, a repressor protein, to the hTERT gene regulatory region in human breast cancer cells [9, 60]. Further, SFN-mediated transcriptional repression of hTERT correlates positively with its inhibition of cellular proliferation and induction of apoptosis in human breast cancer cells [60].
SFN also has been shown to inhibit DNMT1 and DNMT3b in human prostate cancer cells. Interestingly, SFN-induced demethylation at cyclin D2 promoter regions corresponded to an increase in cyclin D2 transcript levels, thereby exerting anti-proliferative effects on prostate cancer cells [61]. In addition, SFN treatment led to demethylation of the first five CpGs in the promoter region of the Nrf2 gene in TRAMP C1 cells, suggesting a modulatory role on anti-oxidative stress pathways [62]. SFN enhanced the nuclear translocation of Nrf2 and increased the mRNA and protein levels of the Nrf2 target genes in 12-O-tetradecanoylphorbol-13-acetate (TPA) in mouse skin epidermal JB6 (JB6 P+) cells [63]. Furthermore, SFN reduced the expression of DNMTs and downregulated the expression of HDAC1, HDAC2, HDAC3 and HDAC4, thereby reactivating Nrf2, a transcription factor for anti-oxidant enzymes [63]. A recent study by Wong et al. [64] evaluated genome-wide effects of SFN on promoter methylation in normal prostate epithelial cells and two prostate cancer cell lines and found that the cancer cell lines showed widespread changes in promoter methylation patterns, including both increased and decreased methylation. Although SFN has broad and complex effects on DNA methylation profiles in both normal and cancerous prostate epithelial cells the gene targets were similar within a single cell line.
4.3. Curcumin
The anti-inflammatory, anti-septic, wound-healing, anti-oxidant, anti-angiogenic and anti-cancer activities of turmeric (Curcuma longa) have been attributed to the yellow pigment curcumin, which is a diferuloylmethane polyphenolic compound. Multiple epigenetic activities of curcumin have been reported to date. Curcumin treatment has been shown to induce global hypomethylation in leukemia cells, further strengthening the notion of curcumin-mediated inhibition of DNMT activity [65]. Curcumin also has been shown to mediate inhibition of HDACs and HATs activity in multiple in vitro cancer models [65]. Significant inhibition in the expression of HDACs 1, 3 and 8, as well as of HAT p300, were found after curcumin treatment leading to repression of NF-κB and Notch 1 in Raji cells, an in vitro model of Burkitt’s lymphoma [66]. Curcumin also has been reported to alter HDAC2 expression by chemically preventing its degradation in human monocytes [67]. The α, β-unsaturated carbonyl groups in the side-chain of curcumin are considered to be structurally important for its HAT inhibitory activity [68]. Curcumin also was found to promote proteasomal degradation of p300 and related HATs in prostate cancer cells and peripheral blood lymphocytes [68]. Recently, curcumin was found to reduce the acetylation of histone H3 in the IL-6 promoter leading to its decreased expression in rheumatoid arthritis synovial fibroblasts [69]. A number of investigators have reported curcumin-mediated alterations in miRNA expression profiles. Treatment of human pancreatic cancer cells with curcumin led to upregulated expression of 11 miRNAs, with miR-22 as the most highly overexpressed miRNA, as well as the downregulation of 18 miRNAs, with miR-199a being the most significantly downregulated miRNA [70]. Curcumin also has been shown to reduce EZH2 expression and to induce the expression of several tumor suppressor miRNAs including the let-7 family members, miR-26a, miR-146a, miR-101, miR-200b/c, etc. [71]. Curcumin altered the expression of the tumor suppressor miRNA, miR-203, in a panel of bladder cancer cell lines [72]. These results suggest that curcumin is a very important epigenetically bioactive compound with multiple epigenetic modifying capabilities.
4.4. Genistein
Genistein (4′,5,7-trihydroxyisoflavone), an isoflavonoid, is found in different varieties of beans and is especially abundant in soy beans. This bioactive natural compound is the most widely studied among the flavonoid group of compounds and is well known for its anti-cancer and anti-angiogenic properties. The role of genistein as a chemopreventive phytoestrogen against carcinogenesis is well established [73]. This compound is a strong anti-oxidant and a potent tyrosine kinase inhibitor. Other important mechanisms through which genistein exert its anti-cancer effects includes prevention of mutations in DNA strands; inhibition of cancer cell proliferation and angiogenesis; and proapoptotic effects [74–76]. Genistein has been found to be capable of modulating important epigenetic events, such as DNA methylation and histone tail modifications [77–79]. The inhibitory effect on DNMTs and histone modifying activities of genistein has been established in many similar reports [80–82]. Reactivation of the tumor suppressor genes p21CIP1/WAF1, RARβ and MGMT and p16INK4a through promoter hypomethylation and active chromatin modifications after genistein treatment in breast cancer, squamous cell carcinoma and prostate cancer cells led to cell cycle arrest and cell death [77, 79]. Genistein also causes the epigenetic re-expression of another tumor suppressor gene BTG3 by altering the promoter methylation and enrichment of methyl-CpG-binding domain 2 (MeCP2) in three different renal carcinoma cell lines. In addition, genistein treatment induces the HATs activity resulting in higher enrichment of acetyl H3 and acetyl H4 as well as increased enrichment of dimethyl-H3K4 and trimethyl-H3K4 at the BTG3 promoter [83]. Genistein, in combination with other DNMT or HDACs inhibitors, has shown synergistic epigenetic reactivation of hypermethylated tumor suppressor genes [78, 84, 85]. It alas inhibits the expression of DNMT1, DNMT3a and DNMT3b and increases the enrichment of inactivation chromatin marker trimethyl-H3K9 leading to inhibition of tumor promoter hTERT, the catalytic subunit of telomerase in human breast cancer cells [78].
In addition to its epigenetic effects that have been demonstrated in vitro, an altered methylation profile of nucleosomal binding protein-1 promoter was found in genistein-treated neonatal CD-1 mice as compared to genistein untreated controls [86]. Another study demonstrated that genistein reactivates ERα expression in ERα-negative breast tumors, of importance in utilization of the available anti-estrogen treatments, in vivo in breast xenograft and spontaneous breast tumor mouse models [87]. Based on their findings, the authors further demonstrated that the ERα reactivation effect was enhanced synergistically when combined with a HDAC inhibitor in ERα-negative MDA-MB-231 breast cancer cells. In contrast, some in vivo studies have shown that genistein induced hypermethylation of cancer-related genes [88, 89]. These results correlate with a recent human trial, in which 34 healthy premenopausal women fed isoflavones, including genistein, daily through one menstrual cycle, showed hypermethylation in some of the key cancer-related genes [90].
4.5. Resveratrol
Resveratrol is abundant in grape skin, and also found in berries and peanuts, etc.. Resveratrol has been shown to possess potent anti-cancer properties. It acts on cancer cells by regulating the pathways of cell division and cell growth, apoptosis, inflammation, angiogenesis and metastasis, etc.. It has been shown to inhibit the growth of a wide variety of human cancer cells, such as breast, skin, lung, prostate and colon cancers [91–94]. Resveratrol has been shown to have a moderate inhibitory effect on DNMTs [80, 95]. Resveratrol treatment led to decreased DNMT1 and 3b expression in vitro. In a rodent model of estrogen-dependent mammary carcinoma, resveratrol treatment decreased DNMT3b expression in tumor samples but not in the normal tissues. miRNA expression was also found to be altered after resveratrol treatment in tumor vs. normal tissues in vivo [96]. Resveratrol treatment of human bladder cancer (EJ) cells was found to lead to remarkable S-phase arrest and apoptotic cell death accompanied by loss of phosphorylation of STAT-3, leading to downregulation of the STAT-3 pathway as well as decreased nuclear translocation of SIRT1 and p53 [97]. In another study, the chemopreventive effects of resveratrol were demonstrated in vivo by pre-exposing aromatic hydrocarbon receptor (AhR) agonist-treated pregnant Sprague-Dawley rats with resveratrol, which is an AhR-antagonist, which led to lesser CpG methylation of the BRCA-1 gene [95]. Collectively, these studies indicate that resveratrol possesses anti-cancerous activities and that these are mediated through multiple genetic and epigenetic modes of actions.
4.6. Other dietary phytochemicals
In addition to the various dietary phytochemicals described above, some other dietary phytochemicals have shown epigenetic modulatory activities in various cancers. For example, allyl derivatives from garlic such as allyl mercaptan, diallyl disulfide, S-allylcysteine, S-allylmercaptocysteine and allicin inhibit HDAC activities in human cancer cells [98–102]. Nian et al. [103] demonstrated competitive enzyme kinetics using a purified human HDAC8 enzyme and found that among the organosulfur compounds examined, allyl mercaptan was the most potent HDAC inhibitor. Further, the authors demonstrated that treatment of allyl mercaptan enhanced sp3 binding on the p21CIP1/WAF1 promoter, which results in p21CIP1/WAF1-mediated cell cycle arrest of cancer cells [103]. Grape seed proanthocyanidins (GSPs) are also bioactive phytochemicals that have been shown to have anti-cancer activities both in vitro and in in vivo models [104–106]. GSPs consist mostly of proanthocyanidins (>89%) with dimers, trimers, tetramers, and oligomers of monomeric catechins [105]. Treatment of A431 and SCC13 skin cancer cells with GSPs revealed inhibition of DNA methylation and histone modification leading to the activation of tumor suppressor genes [107].
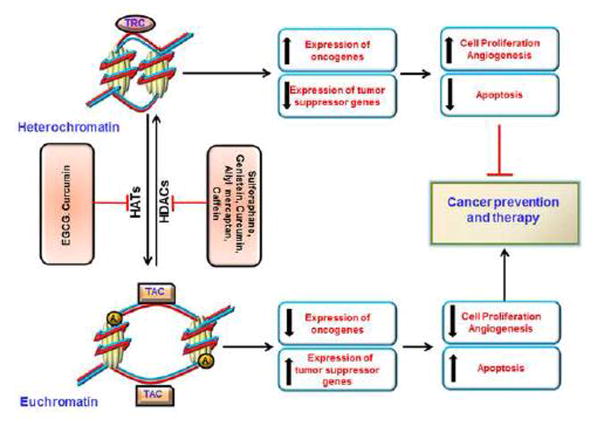
In addition to the above described epigenetic modulatory phytochemicals, there are several other compounds for which some evidence of epigenetic modulation in cancer prevention and therapy is available but require more elaborative studies to pin-point their exact epigenetic modes of action [10, 108–110]. Examples of these phytochemicals include are apigenin, caffeic acid, anacardic acid and lycopene, however, all these dietary bioactive compounds induce epigenetic modulations to variable extents and with different patterns, indicating their potential in cancer chemoprevention. In Figure 2 we summarize the epigenetic modulatory capabilities of the major bioactive dietary phytochemicals.
Figure 2.
Effect of bioactive dietary HAT and HDAC inhibitors on histone acetylation. HATs acetylate the lysine residues present in the histone tails thereby leading to formation of euchromatin state. This conformational change results in the binding of transcriptional activator complexes. The HDACs, in addition to deacetylation of these lysine residues, recruit transcriptional repressor complexes to the heterochromatin leading to gene silencing. The dietary HDAC inhibitors induce the expression of tumor suppressor genes and down regulation of oncogenes leading into suppression of carcinogenesis. A, acetyl group.
5. Advantages and present challenges to epigenetic cancer chemoprevention: future perspectives
Epigenetic therapy has emerged as a gleaming beam of hope in the field of cancer therapeutics. Epigenetic alterations are the early events during carcinogenesis, therefore understanding the course and nature of these alterations may help the researchers to set biomarker profiles for different cancers. Dietary phytochemicals have shown potential of modulating all the major epigenetic pathways such as DNA methylation, histone modifications and miRNAs. These epigenetic alterations culminate into the alterations in the activity of cellular regulatory and metabolic pathways leading to loss of carcinogenicity of transformed cells. Natural dietary phytochemicals cause lesser cytotoxicity to the normal cells and they are widely available. The dietary epigenetically active compounds have added advantage of cost-effectiveness, oral bioavailability and wide range of gene targets. In addition, they are also capable of functioning as sensitizers for chemotherapeutic drugs in the drug-resistant cancer types. The combination of chemotherapy and epigenetic therapy has shown promising results against multiple cancers.
In spite of several preclinical and clinical evidences supporting the potentiality of epigenetic therapy, there are multiple challenges which remain to be solved. First, the dietary compounds function via multiple mechanisms. This lack of specificity places a major hindrance in the druggability of these compounds. Second, since the epigenetic processes are reversible, unnecessary reversal of dietary compound-mediated epigenetic modifications might prevent effective cancer therapy. Third, although the epigenetically active compounds have shown better promises in chemoprevention, chemo-sensitization and maintenance therapy, their efficiency in monotherapy is compromised and variable among different cancers. Therefore, these compounds may not be a good choice for the first-line cancer therapy. Fourth, the early nature of epigenetic alterations requires early diagnosis of the disease to be treated by epigenetic therapy. Use of epigenetic therapy at later stages is not as effective due to accumulation of multiple genomic alterations in the tumor cells. A better understanding of the global patterns of epigenetic modifications induced by dietary compounds can establish improved insights into the chemopreventive strategies and the potential of dietary phytochemicals to inhibit carcinogenesis. Development of early diagnostic techniques might also help in the prevention and therapy of cancer by epigenetically active dietary compounds/phytochemicals.
6. Conclusion
Dietary phytochemicals are of particular interest in the field of cancer prevention and therapy. Many of these phytochemicals establish their anti-cancer activities through multiple pathways and mechanisms. Currently, there is a greater focus on their epigenetic modulatory and gene regulatory activities due to the transgenerational nature and reversibility of these mechanisms of action. These characteristics, as well as their generally low toxicity, position these bioactive natural compounds as crucial cancer chemopreventatives. This review article provides a brief insight into the mechanisms of action of some selected dietary phytochemicals and their epigenetic targets, including the inhibition of DNMTs and HDACs during the course of cancer prevention and therapy. Since cancer is a multi-stage process, it requires multi-targeted approaches for the development of an effective treatment regimen. Many of the bioactive phytochemicals possess more than one epigenetic target, and are thus capable of concomitantly upregulating tumor suppressor genes, downregulating tumor promoters and/or downregulating oncogenes in cancer. Further, exploration of combinations of bioactive phytochemicals having two different epigenetic modulatory capacities or targets is needed as such strategies could represent a major advance in the development of effective preventive and therapeutic approaches against cancer. The current body of research warrants further clinical studies that are needed to standardize the doses, routes of administration, organ specificity and bioavailability in humans. The current investigations also indicate that the use of these bioactive natural compounds regularly in the diet can be used as a preventive as well as therapeutic strategy for cancers of different organs/origins.
Highlights.
Epigenetic changes in genome make significant effects on initiation of many diseases including the risk of cancers.
Regular intake of some dietary phytochemicals appears to be the best option for the chemoprevention of cancers.
Dietary components or phytochemicals have the ability to act as inhibitors of DNA methyltransferases and histone deacetylases.
Dietary phytochemicals or ingredients can be combined with the therapeutic drugs for better treatment options.
Dietary phytochemicals show the abilities to reverse epigenetic changes responsible for the risk of cancers of many organs, and reduce the toxicities of the therapeutic drugs.
Acknowledgments
The work reported from Dr. Katiyar’s laboratory was supported by National Institutes of Health/NCI (CA140832, CA140197) and Veterans Administration Merit Review Award (1I01BX001410). The work reported from Dr. Meeran’s laboratory was supported in part by grants from the Council of Scientific and Industrial Research (CSIR), Government of India, India network grants-EpiHeD (BSC0118), UNDO (BSC0103), and BioPROS (BSC0106). It has CSIR-CDRI communication #231/2014/SMM.
Abbreviations used
- DNMT
DNA methyltransferase
- MBD
methyl binding proteins
- HDAC
histone deacetylase
- HAT
histone acetyltransferase
- EGCG
(−)-epigallocatechin-3-gallate
Footnotes
Conflict of Interest Statement
None
The content of this article does not necessarily reflect the views or policies of the funding sources.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones PA, Gonzalgo ML. Altered DNA methylation and genome instability: a new pathway to cancer? Proc Natl Acad Sci USA. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 3.Zagryazhskaya A, Zhivotovsky B. miRNAs in lung cancer: A link to aging. Ageing Res Rev. 2014 doi: 10.1016/j.arr.2014.02.009. Epub ahead of print. [DOI] [PubMed]
- 4.Tekiner TA, Basaga H. Role of microRNA deregulation in breast cancer cell chemoresistance and stemness. Curr Med Chem. 2013;20:3358–3369. doi: 10.2174/09298673113209990003. [DOI] [PubMed] [Google Scholar]
- 5.John K, Wu J, Lee BW, Farah CS. MicroRNAs in head and neck cancer. Int J Dent. 2013;2013 doi: 10.1155/2013/650218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag Res. 2014;6:15–25. doi: 10.2147/CMAR.S53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katiyar SK, Singh T, Prasad R, Sun Q, Vaid M. Epigenetic alterations in ultraviolet radiation-induced skin carcinogenesis: interaction of bioactive dietary components on epigenetic targets. Photochem Photobiol. 2012;88:1066–1074. doi: 10.1111/j.1751-1097.2011.01020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tollefsbol TO. Dietary epigenetics in cancer and aging. Cancer Treat Res. 2014;159:257–267. doi: 10.1007/978-3-642-38007-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meeran SM, Ahmed A, Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenetics. 2010;1:101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66:8342–8346. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- 13.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 14.Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis. 2011;32:597–604. doi: 10.1093/carcin/bgq282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Prokhortchouk E, Hendrich B. Methyl-CpG binding proteins and cancer: are MeCpGs more important than MBDs? Oncogene. 2002;21:5394–5399. doi: 10.1038/sj.onc.1205631. [DOI] [PubMed] [Google Scholar]
- 16.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 17.Muller A, Florek M. 5-Azacytidine/Azacitidine. Recent Results Cancer Res. 2010;184:159–170. doi: 10.1007/978-3-642-01222-8_11. [DOI] [PubMed] [Google Scholar]
- 18.Momparler RL, Cote S, Momparler LF. Epigenetic action of decitabine (5-aza-2′-deoxycytidine) is more effective against acute myeloid leukemia than cytotoxic action of cytarabine (ARA-C) Leuk Res. 2013;37:980–984. doi: 10.1016/j.leukres.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;131:822. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Dalvai M, Bystricky K. The role of histone modifications and variants in regulating gene expression in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:19–33. doi: 10.1007/s10911-010-9167-z. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Histone acetylation and the cell-cycle in cancer. Front Biosci. 2001;6:D610–629. doi: 10.2741/1wang1. [DOI] [PubMed] [Google Scholar]
- 22.Kim MS, Kwon HJ, Lee YM, Baek JH, Jang JE, Lee SW, Moon EJ, Kim HS, Lee SK, Chung HY, Kim CW, Kim KW. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Xiao W, Chen W, Luo L, Ye S, Liu Y. The epigenetic modifier trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition of lens epithelial cells. Cell Death Dis. 2013;4:e884. doi: 10.1038/cddis.2013.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SY, Jun JA, Jeong KJ, Heo HJ, Sohn JS, Lee HY, Park CG, Kang J. Histone deacetylases 1, 6 and 8 are critical for invasion in breast cancer. Oncol Rep. 2011;25:1677–1681. doi: 10.3892/or.2011.1236. [DOI] [PubMed] [Google Scholar]
- 25.Song C, Zhu S, Wu C, Kang J. Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J Biol Chem. 2013;288:28021–28033. doi: 10.1074/jbc.M113.498758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S, Khan S, Tollefsbol TO, Meeran SM. Genetics and epigenetics of lung cancer: Mechanisms and Future Perspectives. Current Cancer Therapy Reviews. 2013;9:97–110. [Google Scholar]
- 27.Shukla S, Meeran SM. Epigenetic Factors in Breast Cancer Progression. In: AA, editor. Breast Cancer Metastasis and Drug Resistance. Springer Publications; 2013. pp. 341–365. [Google Scholar]
- 28.Afaq F, Katiyar SK. Polyphenols: skin photoprotection and inhibition of photocarcinogenesis. Mini Rev Med Chem. 2011;11:1200–1215. doi: 10.2174/13895575111091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng CJ, Yen GC. Chemopreventive effects of dietary phytochemicals against cancer invasion and metastasis: phenolic acids, monophenol, polyphenol, and their derivatives. Cancer Treat Rev. 2012;38:76–87. doi: 10.1016/j.ctrv.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Khan N, Mukhtar H. Modulation of signaling pathways in prostate cancer by green tea polyphenols. Biochem Pharmacol. 2013;85:667–672. doi: 10.1016/j.bcp.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhtar H, Ahmad N. Tea polyphenols: prevention of cancer and optimizing health. Am J Clin Nutr. 2000;71:1698S–1702S. doi: 10.1093/ajcn/71.6.1698S. [DOI] [PubMed] [Google Scholar]
- 32.Katiyar S, Elmets CA, Katiyar SK. Green Tea and Skin Cancer: Photoimmunology. Angiogenesis and DNA Repair. J Nutr Biochem. 2007;18:287–296. doi: 10.1016/j.jnutbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meeran SM, Katiyar SK. Cell cycle control as a basis for cancer chemoprevention through dietary agents. Front Biosci. 2008;13:2191–2202. doi: 10.2741/2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meeran SM, Mantena SK, Elmets CA, Katiyar SK. (−)-Epigallocatechin-3-gallate prevents photocarcinogenesis in mice through interleukin-12-dependent DNA repair. Cancer Res. 2006;66:5512–5520. doi: 10.1158/0008-5472.CAN-06-0218. [DOI] [PubMed] [Google Scholar]
- 36.Baliga MS, Meleth S, Katiyar SK. Growth inhibitory and antimetastatic effect of green tea polyphenols on metastasis-specific mouse mammary carcinoma 4T1 cells in vitro and in vivo systems. Clin Cancer Res. 2005;11:1918–1927. doi: 10.1158/1078-0432.CCR-04-1976. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad N, Cheng P, Mukhtar H. Cell cycle dysregulation by green tea polyphenol epigallocatechin-3-gallate. Biochem Biophys Res Commun. 2000;275:328–334. doi: 10.1006/bbrc.2000.3297. [DOI] [PubMed] [Google Scholar]
- 38.Meeran SM, Patel SN, Chan TH, Tollefsbol TO. A novel prodrug of epigallocatechin-3-gallate: differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev Res. 2011;4:1243–1254. doi: 10.1158/1940-6207.CAPR-11-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang M, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang C. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 40.Fang M, Chen D, Yang C. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 41.Mittal A, Piyathilake C, Hara Y, Katiyar SK. Exceptionally high protection of photocarcinogenesis by topical application of (−)-epigallocatechin-3-gallate in hydrophilic cream in SKH-1 hairless mouse model: relationship to inhibition of UVB-induced global DNA hypomethylation. Neoplasia. 2003;5:555–565. doi: 10.1016/s1476-5586(03)80039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittal A, Pate MS, Wylie RC, Tollefsbol TO, Katiyar SK. EGCG down-regulates telomerase in human breast carcinoma MCF-7 cells, leading to suppression of cell viability and induction of apoptosis. Int J Oncol. 2004;24:703–710. [PubMed] [Google Scholar]
- 43.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103:509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong M, Zhi X, Jablon DM, You L. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer Res. 2009;29:2025–2030. [PubMed] [Google Scholar]
- 45.Kato K, Long NK, Makita H, Toida M, Yamashita T, Hatakeyama D, Hara A, Mori H, Shibata T. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. Br J Cancer. 2008;99:647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SO, Kim MR. (−)-Epigallocatechin 3-gallate inhibits invasion by inducing the expression of Raf kinase inhibitor protein in AsPC1 human pancreatic adenocarcinoma cells through the modulation of histone deacetylase activity. Int J Oncol. 2013;42:349–358. doi: 10.3892/ijo.2012.1686. [DOI] [PubMed] [Google Scholar]
- 47.Moseley VR, Morris J, Knackstedt RW, Wargovich MJ. Green tea polyphenol epigallocatechin 3-gallate, contributes to the degradation of DNMT3A and HDAC3 in HCT 116 human colon cancer cells. Anticancer Res. 2013;33:5325–5333. [PMC free article] [PubMed] [Google Scholar]
- 48.Deb G, Thakur VS, Limaye AM, Gupta S. Epigenetic induction of tissue inhibitor of matrix metalloproteinase-3 by green tea polyphenols in breast cancer cells. Mol Carcinog. 2014 doi: 10.1002/mc.22121. in press. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Yuan YY, Meeran SM, Tollefsbol TO. Synergistic epigenetic reactivation of estrogen receptor-alpha (ERalpha) by combined green tea polyphenol and histone deacetylase inhibitor in ERalpha-negative breast cancer cells. Mol Cancer. 2010;9:274. doi: 10.1186/1476-4598-9-274. http://www.molecular-cancer.com/content/9/1/274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu DS, Shen JZ, Yu AF, Fu HY, Zhou HR, Shen SF. Epigallocatechin-3-gallate and trichostatin A synergistically inhibit human lymphoma cell proliferation through epigenetic modification of p16INK4a. Oncol Rep. 2013;30:2969–2975. doi: 10.3892/or.2013.2734. [DOI] [PubMed] [Google Scholar]
- 51.Saldanha SN, Kala R, Tollefsbol TO. Molecular mechanisms for inhibition of colon cancer cells by combined epigenetic-modulating epigallocatechin gallate and sodium butyrate. Exp Cell Res. 2014;324:40–53. doi: 10.1016/j.yexcr.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meeran SM, Patel SN, Li Y, Shukla S, Tollefsbol TO. Bioactive dietary supplements reactivate ER expression in ER-negative breast cancer cells by active chromatin modifications. PLoS One. 2012;7:e37748. doi: 10.1371/journal.pone.0037748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Landen CN, Li Y, Alvarez RD, Tollefsbol TO. Enhancement of cisplatin-mediated apoptosis in ovarian cancer cells through potentiating G2/M arrest and p21 upregulation by combinatorial epigallocatechin gallate and sulforaphane. J Oncol. 2013;2013:872957. doi: 10.1155/2013/872957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dashwood R, Ho E. Dietary agents as histone deacetylase inhibitors: sulforaphane and structurally related isothiocyanates. Nutr Rev. 2008;66S:S36–S38. doi: 10.1111/j.1753-4887.2008.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 56.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232:227–234. [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–369. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr. 2009;139:2393–2396. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu A, Wong CP, Yu Z, Williams DE, Dashwood RH, Ho E. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin Epigenetics. 2011;3 doi: 10.1186/1868-7083-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C, Su ZY, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85:1398–1404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su ZY, Zhang C, Lee JH, Shu L, Wu TY, Khor TO, Conney AH, Lu YP, Kong AN. Requirement and epigenetics reprogramming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer Prev Res. 2014;7:319–329. doi: 10.1158/1940-6207.CAPR-13-0313-T. [DOI] [PubMed] [Google Scholar]
- 64.Wong CP, Hsu A, Buchanan A, Palomera-Sanchez Z, Beaver LM, Houseman EA, Williams DE, Dashwood RH, Ho E. Effects of sulforaphane and 3,3′-diindolylmethane on genome-wide promoter methylation in normal prostate epithelial cells and prostate cancer cells. PLoS One. 2014;9:e86787. doi: 10.1371/journal.pone.0086787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu S, Kurzrock R. Development of curcumin as an epigenetic agent. Cancer. 2010;116:4670–4676. doi: 10.1002/cncr.25414. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Shu W, Chen W, Wu Q, Liu H, Cui G. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factor kappa B and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007;101:427–433. doi: 10.1111/j.1742-7843.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- 67.Meja K, Rajendrasozhan S, Adenuga D, Biswas S, Sundar I, Spooner G, Marwick J, Chakravarty P, Fletcher D, Whittaker P, Megson I, Kirkham P, Rahman I. Curcumin restores corticosteroid function in monocytes exposed to oxidants by maintaining HDAC2. Am J Respir Cell Mol Biol. 2008;39:312–323. doi: 10.1165/rcmb.2008-0012OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, Neckers L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- 69.Wada TT, Araki Y, Sato K, Aizaki Y, Yokota K, Kim YT, Oda H, Kurokawa R, Mimura T. Aberrant histone acetylation contributes to elevated interleukin-6 production in rheumatoid arthritis synovial fibroblasts. Biochem Biophys Res Commun. 2014;444:682–686. doi: 10.1016/j.bbrc.2014.01.195. [DOI] [PubMed] [Google Scholar]
- 70.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 71.Bao B, Ali S, Banerjee S, Wang Z, Logna F, Azmi AS, Kong D, Ahmad A, Li Y, Padhye S, Sarkar FH. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335–345. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saini S, Arora S, Majid S, Shahryari V, Chen Y, Deng G, Yamamura S, Ueno K, Dahiya R. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res. 2011;4:1698–1709. doi: 10.1158/1940-6207.CAPR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banerjee S, Li Y, Wang Z, Sarkar FH. Multi-targeted therapy of cancer by genistein. Cancer Lett. 2008;269:226–242. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh AV, Franke AA, Blackburn GL, Zhou JR. Soy phytochemicals prevent orthotopic growth and metastasis of bladder cancer in mice by alterations of cancer cell proliferation and apoptosis and tumor angiogenesis. Cancer Res. 2006;66:1851–1858. doi: 10.1158/0008-5472.CAN-05-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Su SJ, Yeh TM, Chuang WJ, Ho CL, Chang KL, Cheng HL, Liu HS, Hsu PY, Chow NH. The novel targets for anti-angiogenesis of genistein on human cancer cells. Biochem Pharmacol. 2005;69:307–318. doi: 10.1016/j.bcp.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 76.Sasamura H, Takahashi A, Yuan J, Kitamura H, Masumori N, Miyao N, Itoh N, Tsukamoto T. Antiproliferative and antiangiogenic activities of genistein in human renal cell carcinoma. Urology. 2004;64:389–393. doi: 10.1016/j.urology.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 77.Li Y, Chen H, Hardy TM, Tollefsbol TO. Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS One. 2013;8:e54369. doi: 10.1371/journal.pone.0054369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Liu L, Andrews LG, Tollefsbol TO. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009;125:286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, Majid S, Igawa M, Dahiya R. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 80.Mirza S, Sharma G, Parshad R, Gupta SD, Pandya P, Ralhan R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J Breast Cancer. 2013;16:23–31. doi: 10.4048/jbc.2013.16.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Chen H. Genistein, an epigenome modifier during cancer prevention. Epigenetics. 2011;67:888–891. doi: 10.4161/epi.6.7.16315. [DOI] [PubMed] [Google Scholar]
- 82.Vanhees K, Coort S, Ruijters EJ, Godschalk RW, van Schooten FJ, Barjesteh van Waalwijk van Doorn-Khosrovani S. Epigenetics: prenatal exposure to genistein leaves a permanent signature on the hematopoietic lineage. FASEB J. 2011;25:797–807. doi: 10.1096/fj.10-172155. [DOI] [PubMed] [Google Scholar]
- 83.Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, Saini S, Tanaka Y, Dahiya AV, Khatri G, Dahiya R. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raynal NJ, Charbonneau M, Momparler LF, Momparler RL. Synergistic effect of 5-Aza-2′-deoxycytidine and genistein in combination against leukemia. Oncol Res. 2008;17:223–230. doi: 10.3727/096504008786111356. [DOI] [PubMed] [Google Scholar]
- 85.Fang M, Chen D, Sun Y, Jin Z, Christman J, Yang C. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11:7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 86.Tang WY, Newbold R, Mardilovich K, Jefferson W, Cheng RY, Medvedovic M, Ho SM. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nsbp1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology. 2008;149:5922–5931. doi: 10.1210/en.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-alpha (ERalpha) by genistein enhances hormonal therapy sensitivity in ERalpha-negative breast cancer. Mol Cancer. 2013;12 doi: 10.1186/1476-4598-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Day JK, Bauer AM, DesBordes C, Zhuang Y, Kim BE, Newton LG, Nehra V, Forsee KM, MacDonald RS, Besch-Williford C, Huang TH, Lubahn DB. Genistein alters methylation patterns in mice. J Nutr. 2002;132:2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 89.Guerrero-Bosagna CM, Sabat P, Valdovinos FS, Valladares LE, Clark SJ. Epigenetic and phenotypic changes result from a continuous pre and post natal dietary exposure to phytoestrogens in an experimental population of mice. BMC Physiol. 2008;8:17. doi: 10.1186/1472-6793-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin W, Zhang K, Clarke K, Weiland T, Sauter ER. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutr Cancer. 2014;66:270–277. doi: 10.1080/01635581.2014.868910. [DOI] [PubMed] [Google Scholar]
- 91.Mao QQ, Bai Y, Lin YW, Zheng XY, Qin J, Yang K, Xie LP. Resveratrol confers resistance against taxol via induction of cell cycle arrest in human cancer cell lines. Mol Nutr Food Res. 2010;54:1574–1584. doi: 10.1002/mnfr.200900392. [DOI] [PubMed] [Google Scholar]
- 92.Vanamala J, Reddivari L, Radhakrishnan S, Tarver C. Resveratrol suppresses IGF-1 induced human colon cancer cell proliferation and elevates apoptosis via suppression of IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer. 2010;10:238. doi: 10.1186/1471-2407-10-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu PL, Tsai JR, Charles AL, Hwang JJ, Chou SH, Ping YH, Lin FY, Chen YL, Hung CY, Chen WC, Chen YH, Chong IW. Resveratrol inhibits human lung adenocarcinoma cell metastasis by suppressing heme oxygenase 1-mediated nuclear factor-kappaB pathway and subsequently downregulating expression of matrix metalloproteinases. Mol Nutr Food Res. 2010;54:S196–S204. doi: 10.1002/mnfr.200900550. [DOI] [PubMed] [Google Scholar]
- 94.Kraft TE, Parisotto D, Schempp C, Efferth T. Fighting cancer with red wine? Molecular mechanisms of resveratrol. Crit Rev Food Sci Nutr. 2009;49:782–799. doi: 10.1080/10408390802248627. [DOI] [PubMed] [Google Scholar]
- 95.Papoutsis AJ, Selmin OI, Borg JL, Romagnolo DF. Gestational exposure to the AhR agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin induces BRCA-1 promoter hypermethylation and reduces BRCA-1 expression in mammary tissue of rat offspring: Preventive effects of resveratrol. Mol Carcinog. 2013 doi: 10.1002/mc.22095. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 96.Qin W, Zhang K, Clarke K, Weiland T, Sauter ER. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutr Cancer. 2014;66:270–277. doi: 10.1080/01635581.2014.868910. [DOI] [PubMed] [Google Scholar]
- 97.Wu ML, Li H, Yu LJ, Chen XY, Kong QY, Song X, Shu XH, Liu J. Short-term resveratrol exposure causes in vitro and in vivo growth inhibition and apoptosis of bladder cancer cells. PLoS One. 2014;9:e89806. doi: 10.1371/journal.pone.0089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lea MA, Randolph VM, Patel M. Increased acetylation of histones induced by diallyl disulfide and structurally related molecules. Int J Oncol. 1999;15:347–352. doi: 10.3892/ijo.15.2.347. [DOI] [PubMed] [Google Scholar]
- 99.Lea MA, Randolph VM, Lee JE, DesBordes C. Induction of histone acetylation in mouse erythroleukemia cells by some organosulfur compounds including allyl isothiocyanate. Int J Cancer. 2001;92:784–789. doi: 10.1002/ijc.1277. [DOI] [PubMed] [Google Scholar]
- 100.Lea MA, Rasheed M, Randolph VM, Khan F, Shareef A, DesBordes C. Induction of histone acetylation and inhibition of growth of mouse erythroleukemia cells by S-allylmercaptocysteine. Nutr Cancer. 2002;43:90–102. doi: 10.1207/S15327914NC431_11. [DOI] [PubMed] [Google Scholar]
- 101.Druesne N, Pagniez A, Mayeur C, Thomas M, Cherbuy C, Duee PH, Martel P, Chaumontet C. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25:1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- 102.Nian H, Delage B, Ho E, Dashwood RH. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50:213–221. doi: 10.1002/em.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008;29:1816–1824. doi: 10.1093/carcin/bgn165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meeran SM, Vaid M, Punathil T, Katiyar SK. Dietary grape seed proanthocyanidins inhibit 12-O-tetradecanoyl phorbol-13-acetate-caused skin tumor promotion in 7,12-dimethylbenz[a]anthracene-initiated mouse skin, which is associated with the inhibition of inflammatory responses. Carcinogenesis. 2009;30:520–528. doi: 10.1093/carcin/bgp019. [DOI] [PubMed] [Google Scholar]
- 105.Mittal A, Elmets CA, Katiyar SK. Dietary feeding of proanthocyanidins from grape seeds prevents photocarcinogenesis in SKH-1 hairless mice: relationship to decreased fat and lipid peroxidation. Carcinogenesis. 2003;24:1379–1388. doi: 10.1093/carcin/bgg095. [DOI] [PubMed] [Google Scholar]
- 106.Nandakumar V, Singh T, Katiyar SK. Multi-targeted prevention and therapy of cancer by proanthocyanidins. Cancer Lett. 2008;269:378–387. doi: 10.1016/j.canlet.2008.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vaid M, Prasad R, Singh T, Jones V, Katiyar SK. Grape seed proanthocyanidins reactivate silenced tumor suppressor genes in human skin cancer cells by targeting epigenetic regulators. Toxicol Appl Pharmacol. 2012;263:122–130. doi: 10.1016/j.taap.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee W, Zhu B. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006;27:269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 109.King-Batoon A, Leszczynska J, Klein C. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 110.Paluszczak J, Krajka-Kuzniak V, Baer-Dubowska W. The effect of dietary polyphenols on the epigenetic regulation of gene expression in MCF7 breast cancer cells. Toxicol Lett. 2010;192:119–125. doi: 10.1016/j.toxlet.2009.10.010. [DOI] [PubMed] [Google Scholar]