Abstract
Acquisition and maintenance of NK cell function is mediated by inhibitory killer-cell immunoglobulin-like receptors (KIR) through the interaction with HLA class I molecules. Recently, HLA-C expression levels were shown to be correlated with protection against multiple outcomes of HIV-1 infection; however the underlying mechanisms are poorly understood. As HLA-C is the natural ligand for the inhibitory receptors KIR2DL1 and KIR2DL2/3, we sought to determine whether HLA-C group haplotypes affect NK cell responses during primary HIV-1 infection. The phenotypes and functional capacity of NK cells derived from HIV-1(+) and HIV-1(-) individuals were assessed (N=42 and N=40, respectively). HIV-1 infection was associated with an increased frequency of KIR2DL1-3+ NK cells. Further analysis showed that KIR2DL1+ NK cells were selectively increased in individuals homozygous for HLA-C2, while HLA-C1-homozygous individuals displayed increased proportions of KIR2DL2/3+ NK cells. KIR2DL1-3+ NK cells were furthermore more polyfunctional during primary HIV-1 infection in individuals also encoding for their cognate HLA-C group haplotypes as measured by degranulation and cytokine production. These results identify a novel relationship between HLA-C and KIR2DL+ NK cell subsets and demonstrate that HLA-C-mediated licensing modulates NK cell responses to primary HIV-1 infection.
Keywords: HIV-1, Natural killer (NK) cells, killer-cell immunoglobulin-like receptors (KIR), HLA-C
Introduction
Natural killer (NK) cells represent the first line of defense against viral infections through the early production of pro-inflammatory cytokines and killing of virus-infected cells [1]. The function of NK cells is governed by multiple inhibitory and activating receptors balancing self-tolerance and effective responses in tumor surveillance and upon infections [2].
One family of receptors important for the regulation of NK cell function are the killer-cell immunoglobulin-like receptors (KIR) [3],[4]. Despite common structural features, each of the 14 described KIR in humans displays distinct functional properties with respect to ligand specificity and signal transduction, leading to the transmission of either inhibitory or activating signals upon receptor engagement. Classical and non-classical human HLA class I molecules can serve as natural ligands for several KIR [3]. In particular, for the inhibitory receptors KIR2DL1/L2/L3 and KIR3DL1, interactions with HLA class I molecules and their functional consequences have been studied intensively: While HLA-A and HLA-B molecules with a Bw4 motif can interact with KIR3DL1, HLA-C molecules serve as ligands for either KIR2DL1, KIR2DL2, and KIR2DL3 [5]– [7]. HLA-C molecules can be subdivided into two groups with distinct affinity for KIR2DL receptors based on amino acids at position 77 and 80 of the HLA class I heavy chain [5]. While KIR2DL1 exclusively binds to HLA-C group 2 molecules with high affinity, KIR2DL3 is predominantly engaged by HLA-C group 1 ligands [6],[7]. However, KIR2DL2, segregating as a distinct allele from KIR2DL3, displays specificity for both HLA-C groups [8],[9]. In addition to their contribution to self-tolerance through the interaction with their natural HLA class I ligands, expression of self-inhibitory KIR is associated with increased NK cell functionality against various cellular targets, a process termed NK cell education or licensing [10]–[12].
Specific combinations of KIR/HLA haplotypes have been shown to influence the outcome of viral infections, including HIV-1 infection [13],[14]. Genetic association studies demonstrated that the combined presence of alleles encoding for the activating receptor KIR3DS1 and HLA-Bw4 molecules with Isoleucine at position 80 was associated with delayed progression to AIDS [15]–[17]. In addition, certain alleles of the highly polymorphic inhibitory receptor KIR3DL1 were associated with high surface expression on NK cells and linked to more effective control of HIV-1 in HLA-Bw4+ individuals [18],[19]. Apart from the well-established protective effects of certain HLA-Bw4/KIR3DL1/S1 haplotypes in HIV-1 infection, recent studies draw attention to HLA/KIR interactions involving HLA-C and their respective KIR2DL ligands. The identification of KIR2DL2-associated HIV-1 sequence polymorphisms provided first evidence for KIR-associated NK cell-mediated immune pressure on HIV-1 replication [20], and subsequent studies demonstrated the ability of HIV-1-derived peptides presented by HLA-C molecules to modulate KIR2DL2 binding and KIR2DL2+ NK cell functions [21]. Furthermore, the surface expression of HLA-C molecules is strongly associated with the speed of HIV-1 disease progression [22].
Taken together, these data indicate that the interactions between KIR2DL1-3 receptors and their HLA-C ligands play a role in the control of HIV-1 infection. However, the mechanisms underlying the effects observed in HIV-1-infected individuals still remain unclear. Licensing of KIR2DL1-3-expressing NK cells in the presence of their cognate HLA ligands might play a role in the control of viral replication through inducing more rapid and efficient NK cell responses. To investigate this hypothesis, this study analyzed the frequency and response of KIR2DL1-3+ NK cells in association with the cognate HLA-C ligands in subjects with primary and early chronic HIV-1 infection and in HIV-1-negative controls.
Results
Primary HIV-1 infection is associated with HLA-C haplotype-dependent increase of KIR2DL1-3+ NK cells
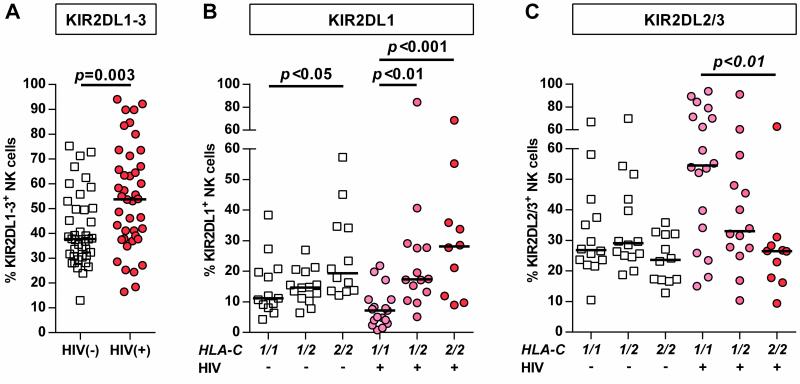
The goal of this study was to investigate the role of the receptors KIR2DL1-3 in primary HIV-1 infection and understand the influence of HLA-C group haplotypes. Initially, the frequency of NK cells expressing at least one KIR2DL1-3 receptor was measured in individuals with primary HIV-1 infection. As displayed in Figure 1 A, HIV-1 infection was associated with increased percentages of KIR2DL1-3+ NK cells (p<0.003). Subsequently, HIV-1-negative and -positive individuals were divided each into three groups defined by their HLA-C group haplotype for a more detailed analysis of KIR2DL1 and KIR2DL2/3 expression frequency. Stratification along HLA-C group haplotypes revealed significant differences in the frequency of KIR2DL1+ and KIR2DL2/3+ NK cells in HIV-1(+) individuals, while a significant difference between haplotypes in HIV-1(-) individuals was only observed for KIR2DL1 (Figure 1 B/C). Frequency of NK cells expressing KIR2DL1 was increased in HLA-C2+ HIV-1-infected individuals, while lack of HLA-C group 2 alleles was associated with the lowest percentage of KIR2DL1+ NK cells (Figure 1 B: C1/C2: p<0.01, C2/C2: p<0.001; each vs. C1/C1). Vice versa, the proportion of KIR2DL2/3+ NK cells was increased in individuals homozygous for the cognate HLA-C group 1 allotypes (Figure 1 C: C1/C1: p<0.01 vs. C2/C2,). Of note, in comparison to HIV-1(-) individuals, HIV-1(+) subjects homozygous for the respective HLA-C group allotypes displayed higher frequencies of both KIR2DL1+ and KIR2DL2/3+ NK cells (KIR2DL1: 19.4% vs. 28.1%; KIR2DL2/3: 26.9% vs. 54.6%, median) – however this did not reach statistical significance after correction for multiple comparisons. Clinical parameters such as HIV-1 viral load and CD4+ T cell count did not correlate with KIR frequencies after correction for multiple comparisons. Overall, KIR2DL1-3+ NK cells displayed an increased frequency in primary HIV-1 infection; this was mediated by KIR2DL1+ and KIR2DL2/3+ NK cells in dependence of the presence of the respective HLA-C ligands.
Figure 1. Frequency of KIR2DL1-3+ NK cells in primary HIV-1 infection stratified by HLA-C group haplotypes.
(A) Frequency of NK cells expressing at least one KIR2DL1-3 receptor in HIV-1(-) and HIV-1(+) individuals. (B) Percentage of KIR2DL1+ and (C) KIR2DL2/3+ NK cells in HIV-1(-) and HIV-1(+) individuals stratified by their HLA-C group haplotype. HIV-1(-): n=40; HIV-1(+): n=42. Haplotypes of HLA-C are indicated as follows: 1/1=C1/C1, 1/2=C1/C2, 2/2=C2/C2. Scatter plots display frequency of KIR-expressing NK cells for each individual as well as median for each group. Mann-Whitney test (A) and Kruskal-Wallis test following Dunn's post-test (B) were used to test for differences between groups.
CMV-seropositivity alone is not associated with overall increase of KIR2DL1-3+ NK cell frequency
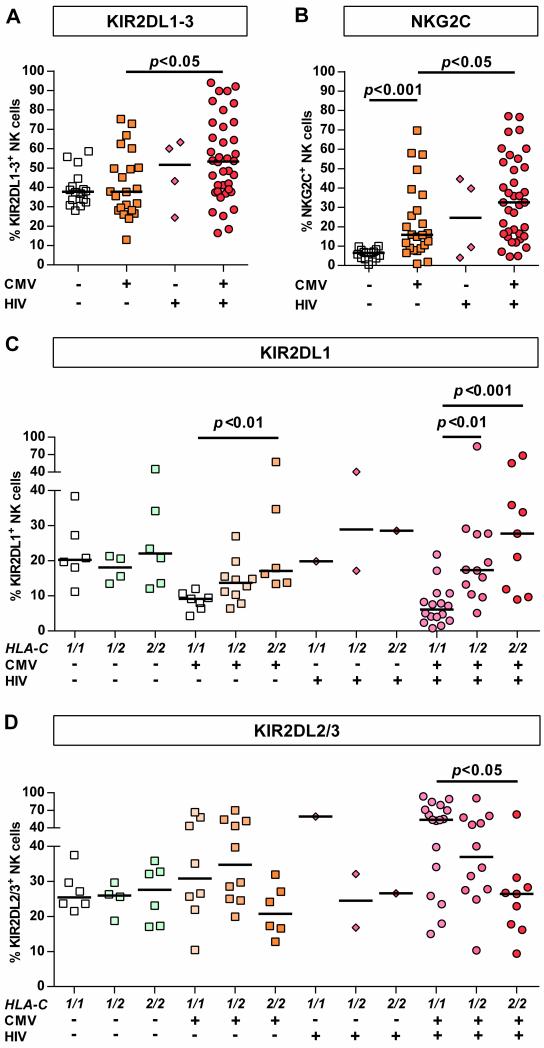
As human CMV infection is known to affect KIR expression [23], the effects of CMV-seropositivity on KIR2DL1-3 receptor expression were investigated in HIV-1(-) and HIV-1(+) subjects. Interestingly, CMV-seropositivity did not affect overall frequency of KIR2DL1-3+ NK cells in HIV-1(-) individuals whereas CMV-seropositive HIV-1-infected individuals displayed the highest percentages of KIR2DL1-3+ NK cells (Figure 2 A, p<0.05 vs. HIV-1(-)/CMV(+)). This observation was confirmed in a multiple linear regression analysis identifying HIV-1 infection as an independent covariate in contrast to CMV-seropositivity (p=0.01 vs. p=0.39). Comparison between HLA-C group haplotypes showed that CMV-seropositivity in HIV-1-negative individuals was associated with much less HLA-C-dependent skewing of KIR2DL1-3+ NK cell frequencies than in HIV-1(+) subjects (Figure 2 D/C). Due to the high prevalence of CMV-seropositivity among HIV-1-infected individuals (Table 1, p=0.002 vs. HIV-1(-), Fisher's exact test) only four CMV-seronegative individuals were enrolled, preventing statistical analysis of this subgroup. Taken together, these result show that while CMV-seropositivity is not associated with overall increase of KIR2DL1-3+ NK cell frequency, both CMV-seropositivity and HIV-1 infection affect the frequency of KIR2DL1+ and KIR2DL2/3+ NK cells in an HLA-C-dependent fashion. HIV-1 infection significantly increased this effect, when compared to frequencies in CMV-seropositive HIV-1(-) individuals.
Figure 2. KIR2DL1-3+ and NKG2C+ NK cell frequencies in HIV-1(+) and HIV-1(-) individuals stratified by CMV serostatus.
Frequency of (A) KIR2DL1-3+ and (B) NKG2C+ NK cells in HIV-1(-) and HIV-1(+) CMV-seronegative/positive individuals. Proportion of (C) KIR2DL1+ and (D) KIR2DL2/3+ NK cells in HIV-1(-) and HIV-1(+) individuals stratified by their HLA-C group haplotype and CMV serostatus. HIV-1(-): n=40; HIV-1(+): n=42. Haplotypes of HLA-C are indicated as follows: 1/1=C1/C1, 1/2=C1/C2, 2/2=C2/C2. Scatter plots display frequency of receptor-expressing NK cells for each individual as well as median for each group. Kruskal-Wallis test following Dunn's post-test was used to test for differences between groups.
Table 1.
Clinical and demographic data of HIV-1(+) and HIV-1(−) individuals.
| All samples | C1/C1 haplotype | C1/C2 haplotype | C2/C2 haplotype | |
|---|---|---|---|---|
| HIV-1(+) subjects (primary infection) | ||||
| Number | 42 | 18 | 14 | 10 |
| Age in years, median (IQR) | 39 (31 - 44) | 39 (26 - 44) | 42 (36 - 45) | 31 (29 - 43) |
| Male, number (%) | 40 (95.2) | 18 (100) | 13 (92.9) | 9 (90.0) |
| CMV-seropositivity (%) | 38 (90.5) | 17 (94.4) | 12 (85.7) | 9 (90.0) |
| Subjects on HAART (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| HIV-1 viral load in copies/ml, median (IQR)# | 116,500 (22,550-600,000) | 99,550 (18,820-813,000) | 341,355 (22,550-573,750) | 150,400 (8,989-635250) |
| CD4+ T cells in cells/μl, median (IQR)# | 489 (400 - 631) | 498 (403 - 756) | 499 (370 - 602) | 468 (318 - 763) |
| % CD4+ T cells, median (IQR)# | 26 (20 - 32) | 29 (24 - 33) | 27 (17 - 33) | 21 (21 - 26) |
| Follow up of clinical data of selected HIV-1(+) subjects (early chronic infection) | ||||
| Number | 36 | 16 | 12 | 8 |
| Subjects on HAART (%) | 26 (72.2) | 10 (62.5) | 11 (91.7) | 5 (62.5) |
| HIV-1 viral load in copies/ml, median (IQR) | 49 (49-3,340) | 49 (49-4,690) | 49 (49-79) | 287 (49-29,405) |
| CD4+ T cells in cells/μl, median (IQR) | 701 (495 - 829) | 755 (573 - 938) | 639 (503 - 914) | 665 (460 - 791) |
| % CD4+ T cells, median (IQR) | 38 (29 - 43) | 37 (27 - 44) | 41 (27 - 48) | 35 (29 - 38) |
| HIV-1(−) subjects | ||||
| Numbers | 40 | 14 | 14 | |
| Age in years, median (IQR) | 32 (25 - 44) | 34 (26 - 45) | 35 (23 - 50) | 28 (24 - 37) |
| Male, number (%) | 29 (72.5) | 9 (64.3) | 11 (78.6) | 9 (75) |
| CMV-seropositivity (%) | 24 (60) | 8 (57.1) | 10 (71.4) | 6 (50) |
IQR: Interquartile range
Differences between groups: Viral load: p=0.93, CD4 abs.: p=0.76, CD4 %: p=0.37; Kruskal-Wallis test.
KIR2DL1-3+ expression is accompanied by NKG2C co-expression but not limited to NKG2C+ NK cells
Altered expression of the activating receptor NKG2C and its inhibitory counterpart NKG2A have been reported in HIV-1-infected subjects co-infected with CMV [24]. We therefore investigated whether NKG2A/NKG2C expression in HIV-1 infection was also associated with changes in expression of KIR2DL1 and KIR2DL2/3 on NK cells, as it had been previously described in chronic hepatitis C patients [25]. As shown in Figure 2 B increased percentage of NKG2C+ NK cells was observed in CMV-seropositive individuals (p<0.001 vs. CMV(-)/HIV-1(-)) that was even more enhanced in subjects with primary HIV-1 infection (p<0.05 vs. CMV(+)/HIV-1(-)). In contrast, frequency of NKG2A+ NK cells did not differ between the analyzed groups (Supporting Information Fig. 2 A). Moreover, correlation analysis between frequency of KIR2DL1-3+ and NKG2C+ NK cells revealed a strong positive association (p<0.0001) in HIV-1(+) individuals, while frequency of NKG2A+ NK cells was inversely correlated (p<0.0001) (Supporting Information Fig. 2 B). Of note, no association was observed in HIV-1(-) individuals (data not shown). Next, distinct NK cell subpopulations were defined based on the presence or absence of NKG2A and NKG2C on the NK cell surface. Assessment of KIR2DL1-3+ NK cell frequencies in these subsets revealed that HLA-C-dependent increase of KIR2DL1-3+ NK cell frequency was not only limited to the NKG2C+/NKG2A(−) NK cell subset but was also observed in the NKG2C(−) NK cell compartment and regarding KIR2DL1 in the NKG2A+ NK cell subset as well (Supporting Information Fig. 2 C/D). Overall, these data suggest that the increased frequency of KIR2DL1-3+ NK cells in primary HIV-1 infection is generally not restricted to a particular NK cell subset.
Effects of KIR co-expression and gene content on HLA-C-dependent enrichment of KIR2DL1-3+ NK cells
Individual NK cells are able to express multiple KIR in a stochastic manner generating a unique set of NK cell phenotypes with various KIR expression patterns in a given individual that is able to influence NK cell function. Therefore, the co-expression of inhibitory KIR and their potential effect on HLA-C-associated enrichment of KIR2DL1-3+ NK cells in HIV-1 infection were explored. In addition to KIR2DL1 and KIR2DL2/3, KIR3DL1 was included in the analysis to correct for potential HLA-Bw4-driven expansion of KIR3DL1+ NK cells. As illustrated in Supporting Information Fig. 3 A/B, frequencies of KIR2DL1 and KIR2DL2/3 single-positive NK cells increase with the number of the respective HLA-C group allele ligands expressed, indicating a HLA-C gene-dose effect that was not affected by the co-expression of other included inhibitory KIR. Correlation analyses between KIR2DL1+ and KIR2DL2/3+ NK cell frequencies revealed are strong inverse correlation in a given individual with primary HIV-1 infection, which was further enhancement in the absence of the KIR2DL2 gene (Supporting Information Fig. 3 C/D: KIR2DL2+: r=-0.51; p=0.018; KIR2DL2(−): r=-0.9; p<0.0001). In contrast, no such strong negative correlation was observed in HIV-1(-) individuals (KIR2DL2+: r=-0.29; p=0.4; KIR2DL2(−): r=-0.51; p=0.09). In addition, as all enrolled KIR2DL2(−) individuals lacked KIR2DS2, a potential skewing of our results because of cross-reactivity of the anti-KIR2DL2/3 antibody clone DX27 with KIR2DS2 was considered negligible.
HLA-C-dependent enrichment of KIR2DL3+ NK cells in vitro and after seroconversion
Finally, analysis of pre- and post HIV-1 infection samples available from two enrolled subjects showed an increased frequency of NK cells expressing self-inhibitory KIR after HIV-1 seroconversion (Supporting Information Fig. 4). Similar observations were made in an in vitro co-culture of NK cells with autologous HIV-1-infected CD4+ T cells from HIV-1(-) donors (Supporting Information Fig. 5). Taken together, these data suggest an HIV-1-associated redistribution of NK cells expressing self-inhibitory KIR2DL1-3 receptors during primary HIV-1 infection.
Self-inhibitory KIR2DL1-3+ NK cells in primary HIV-1 infection contribute predominantly to NK cell polyfunctionality
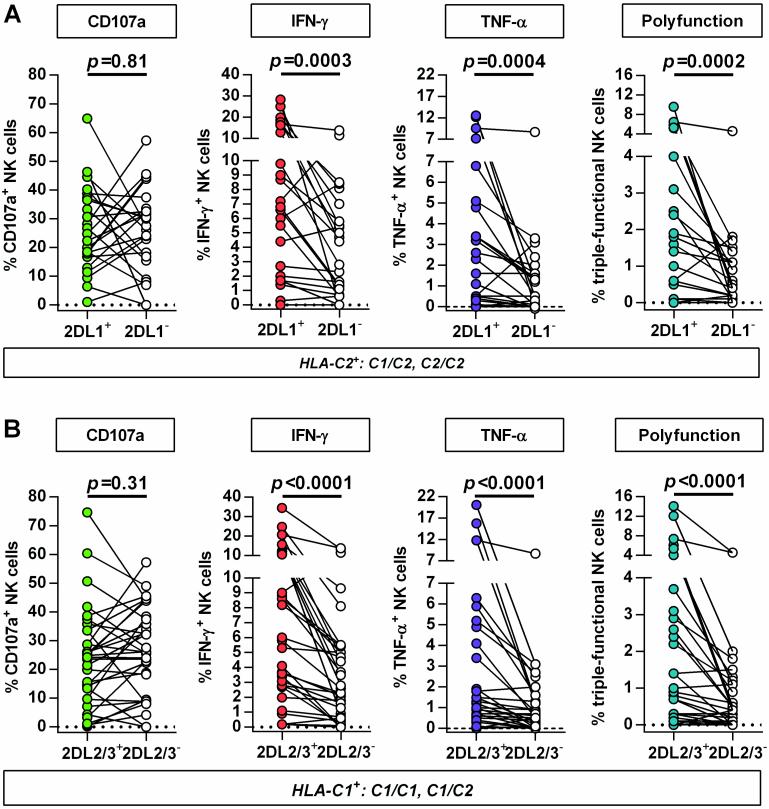
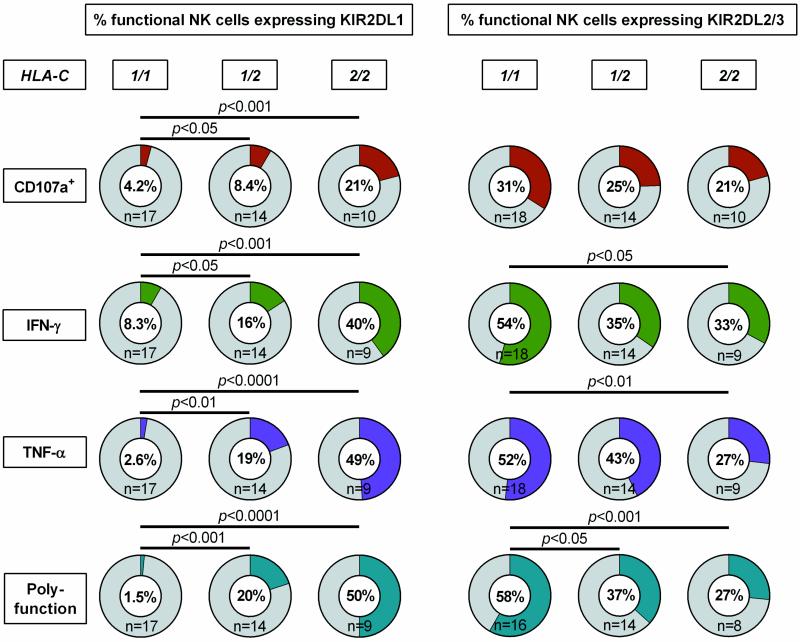
To investigate the impact of primary HIV-1 infection on HLA-C-modulated KIR2DL1-3+ NK cell function, CD107a expression and cytokine production (IFN-γ, TNF-α) of NK cells was assessed after co-incubation with MHC class I-devoid 721.221 cells. As illustrated in Figure 3 responses of KIR2DL1 and KIR2DL2/3 single-positive NK cells were compared with KIR2DL1-3-negative NK cell subsets in individuals expressing the cognate HLA-C ligands. For both KIR2DL1- and KIR2DL2/3-expressing NK cells, no differences in the level of degranulation were observed between the KIR positive and negative subsets (KIR2DL1: p=0.81; KIR2DL2/3: p=0.31). In contrast, frequencies of IFN-γ-, TNF-α-producing or polyfunctional cells were significantly increased in NK cells expressing the respective self-inhibitory KIR2DL1-3 receptor (KIR2DL1: IFN-γ p=0.0003, TNF-α p=0.0004, polyfunction p=0.0002; KIR2DL2/3: IFN-γ p<0.0001, TNF-α p<0.0001, polyfunction p<0.0001; each vs. KIR(−)). Of note, NK cell responses did not differ between HIV-1(-) subjects and individuals with primary HIV-1 infection (data not shown). In addition, KIR2DL1+ and KIR2DL2/3+ NK cells represented the majority of IFN-y-, TNF-α-producing and polyfunctional NK cells in individuals homozygous for the respective HLA-C group alleles (Figure 4). Taken together, NK cells from individuals with primary HIV-1 infection maintain a licensed phenotype and are capable of responding to MHC class I-devoid target cells similar to NK cells derived from HIV-1-negative donors. Moreover, KIR2DL1+ and KIR2DL2/3+ NK cells represent a dominant fraction of the cytokine-producing and polyfunctional NK cell population in individuals homozygous for the respective HLA-C ligand.
Figure 3. Comparison of NK cell responses between KIR2DL1-3+ and KIR2DL1-3(−) NK cell subsets in primary HIV-1 infection.
Functional assessment of KIR2DL1-3-expressing NK cell subsets in HIV-1(+) individuals carrying at least one copy of the cognate HLA-C group allele (KIR2DL1+/HLA-C2+; KIR2DL2/3+/HLA-C1+): (A) Paired comparison between KIR2DL1 single-positive NK cells and KIR-negative NK cells (n=24); (B) paired comparison between KIR2DL2/3 single-positive NK cells and KIR-negative NK cells (n=32). Single-positive KIR NK cells are defined as expressing either KIR2DL1, or KIR2DL2/3 but not KIR3DL1. KIR-negative NK cells are defined by the absence of KIR2DL1, KIR2DL2/3 and KIR3DL1 on the cell surface. Scatter plots display frequency of CD107a+, IFN-γ and TNF-α producing NK cells as well as percentage of polyfunctional NK cells (CD107a+/IFN-γ+/TNF-α+) for each individual. Wilcoxon matched-pairs signed rank test for paired values was used to test for differences between groups.
Figure 4. Frequency of functional NK cells expressing KIR2DL1-3+ stratified by type of response and HLA-C group haplotypes.
Pie charts display median percentage of functional NK cells expressing KIR2DL1 or KIR2DL2/3 stratified by HLA-C group haplotypes of the respective HIV-1(+) individuals. Functional NK cell subsets are defined either as CD107a+ NK cells, IFN-γ or TNF-α+ producing NK cells or polyfunctional NK cells (CD107a+/IFN-γ+/TNF-α+). Haplotypes of HLA-C are indicated as follows: 1/1=C1/C1, 1/2=C1/C2, 2/2=C2/C2. Kruskal-Wallis test following Dunn's post-test was used to test for differences between groups.
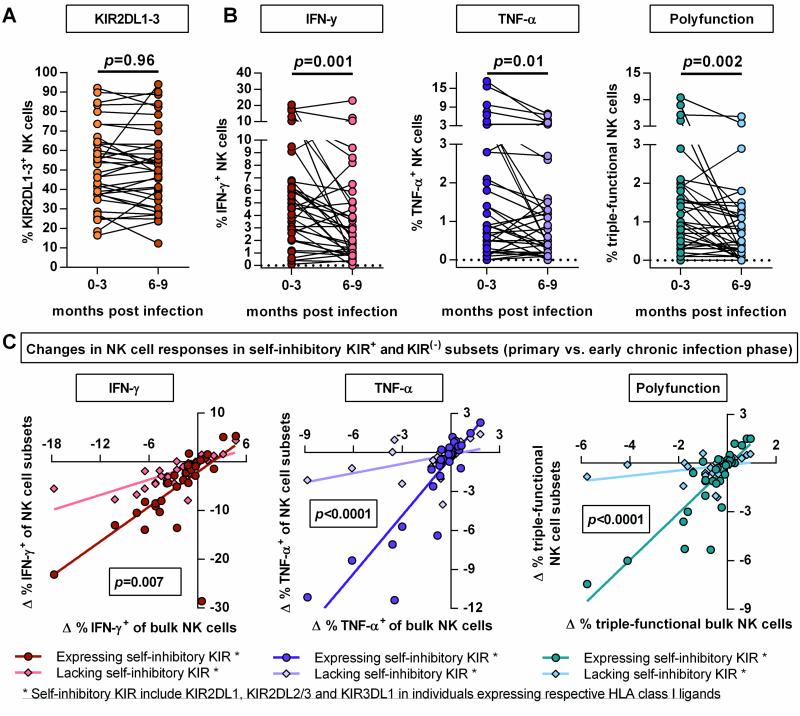
Transition to early chronic HIV-1 infection is associated with reduced cytokine-related responses of self-inhibitory KIR2DL1-3+ NK cells
Finally, the effects of persistent HIV-1 infection on the KIR2DL1-3+ NK cell repertoire and NK cell function were studied by assessing the phenotype and function of NK cells collected in the early chronic infection phase (6-9 months post infection). As shown in Figure 5 A, the overall percentage of KIR2DL1-3+ NK cells as well as the individual KIR2DL1+ and KIR2DL2/3+ NK cell subsets (data not shown) did not change between primary and early chronic phase of HIV-1 infection. This observation was independent of the initiation of antiretroviral therapy or residual viremia (viremic: p=0.98, aviremic: p=0.85) and no significant correlation between NK cell function and clinical parameters such as HIV-1 viral load or CD4+ T cell count was observed after correction for multiple comparisons. With regards to potential changes in NK cell function, the capability of NK cells to degranulate was not altered in response to 721.221 cells (data not shown). In contrast, the percentage of IFN-γ-, TNF-α-producing and polyfunctional NK cells was reduced in early chronic infection as compared to NK cells derived from an earlier stage of infection (Figure 5 B: IFN-γ p=0.001, TNF-α p=0.01, polyfunction p=0.002). Further paired comparison between the responses of NK cells expressing self-inhibitory KIR (KIR2DL1-3/3DL1) and cells lacking self-inhibitory KIR revealed that the loss of cytokine production and polyfunctionality in the early chronic infection phase was more pronounced in the NK cell subpopulation expressing self-inhibitory KIR (Figure 5 C; IFN-γ: p=0.007, TNF-α p<0.0001, polyfunction: p<0.0001). Overall, early chronic HIV-1 infection was associated with reduced cytokine-related NK cell responses, in particular in the self-inhibitory KIR-expressing NK cell compartment, despite stable KIR2DL1-3+ NK cell frequencies.
Figure 5. Paired comparison of NK cell phenotype and function between primary and early chronic HIV-1 infection.
Changes in frequency of KIR2DL1-3+ NK cells (A), percentages of IFN-γ, TNF-α producing and polyfunctional (CD107a+/IFN-γ+/TNF-α+) NK cells (B). Scatter plots display frequency of above mentioned NK cell subpopulations for each individual (n=36). (C) Correlation plots displaying the differential between NK cell responses in primary and in early chronic infection for bulk as well as self-inhibitory KIR+ and KIR(−) NK cell subsets. * Self-inhibitory KIR+ NK cells are defined as follows: NK cells expressing KIR2DL1-3 and/or KIR3DL1 in individuals carrying the cognate HLA-C group allele or HLA-Bw4 allele. * Self-inhibitory KIR(−) NK cells are characterized by the lack of the mentioned self-inhibitory KIR in the respective individuals. Wilcoxon matched-pairs signed rank test for paired values was used to test for differences between groups. Differential was calculated as follows: Δ % response = [% response (early chronic infection) - % response (primary infection)]. Linear regression analysis was used to test for differences between the slopes of the NK cell subsets.
Discussion
The present study provides evidence for an HLA-dependent increase in frequency of self-inhibitory KIR+ NK cells in HIV-1 infection. A comprehensive analysis including additional factors such as CMV-seropositivity revealed that the increased percentage of KIR2DL1-3+ NK cells is predominantly comprised by KIR2DL1+ and KIR2DL2/3+ NK cells in the presence of their respective HLA-C ligands and is strongly associated with co-expression of NKG2C. Furthermore, we demonstrate that KIR2DL1-3+ NK cells maintain a licensed phenotype and contribute largely to NK cell polyfunctionality in primary infection but subsequently lose some of the capability to respond to MHC class I-devoid target cells in the course of infection. Overall, this study indicates that redistribution of KIR+ NK cell subsets during primary HIV-1 infection is more widespread than previously described and provides evidence for HLA-C-mediated licensing of NK cells that express the cognate KIR receptors KIR2DL1-3 in HIV-1 pathogenesis.
Epidemiological studies indicated that the combined presence of certain KIR/HLA haplotypes can affect the outcome of HIV-1 infection [15],[19]. While the underlying mechanisms still remain elusive, accumulating evidence suggest that alterations of the KIR repertoire and effector function of KIR+ NK cells might play an important role. Based on the comparison of pre- and post-infection samples, Carr and colleagues recently reported an increased frequency of KIR+ NK cells following HIV-1 seroconversion [26]. Although no stratification into distinct KIR sub-populations or HLA class I haplotypes was performed, the study provided strong evidence for a general enrichment of KIR+ NK cells in the peripheral blood upon acute HIV-1 infection. Our and previously reported results [27] indicate that the increased proportion of KIR+ NK cells in primary HIV-1 infection is driven by distinct KIR+ sub-populations and potentially HLA-restricted. Our data further suggest that enrichment of self-inhibitory KIR+ NK cells might not exclusively occur in individuals encoding for protective KIR/HLA combinations, but rather reflect a general mechanism in response to acute viral infections. These data do however not exclude the possibility that rather than an expansion an HLA-dependent contraction of other NK cell sub-populations serve as an underlying mechanism. Increased frequency of KIR+ NK cells was also accompanied with the expansion of NKG2C+ NK cells in patients with acute Hanta virus infection and in individuals previously undergoing CMV infection [23],[28]. HIV-1 driven increase of KIR2DL1-3+ NK cells was closely linked to NKG2C+ co-expression and reactivation of CMV infection in the course of HIV-1 infection cannot be ruled out as an underlying mechanism due the high prevalence of CMV-seropositivity among HIV-1-infected individuals and therefore limited access to CMV-seronegative subjects [29],[30]. However, we observed that enrichment of self-inhibitory KIR2DL1-3+ NK cells was not limited to the NKG2C+ subset but also occurred in NKG2C- NK cell subsets, indicating that HIV-1 infection provides a strong trigger for HLA-C-dependent enrichment of KIR2DL1-3+ NK cells that goes beyond the so far described factors.
Our results indicate that both CMV and HIV-1 infection serve as extrinsic factors driving enrichment of NK cells expressing self-inhibitory KIR+ in an HLA-C-dependent manner. However, the high variance in the KIR2DL1 and KIR2DL2/3+ NK cell frequencies suggests that other genetic and/or environmental parameters might play a role in shaping the KIR repertoire in a given individual. As an intrinsic factor, KIR copy numbers have been identified to be positively associated with KIR expression and it has been shown that copy number variation of KIR3DL1/S1 influences HIV-1 control [31]–[33]. Additionally, Schönberg et al. have shown that the HLA-C-dependent skewing of the KIR2DL1-3 repertoire was lost in the presence of KIR2DL2 [34]. In fact, comparison of KIR2DL2+ and KIR2DL2(−) individuals in our dataset revealed an enhanced redistribution of between KIR2DL1 and KIR2DL2/3 frequency in HIV-1-infected KIR2DL2(−) individuals. The promiscuous ability of KIR2DL2 to interact with HLA-C group 1 and group 2 molecules might serve as an underlying mechanism.
Finally, it is noteworthy to review the design and results of the present study in the context of recent studies investigating specificities of commercially available KIR antibodies. Czaja and others reported cross-reactivity of KIR antibodies with activating KIR isoforms including clones recognizing KIR2DL2/3 (DX27, cross-reactivity with KIR2DS2) and KIR2DL1 (143221, cross-reactivity with KIR2DS5) [35]. In addition, Beziat et al. described the development of a flow cytometric staining procedure and optimized workflow to accommodate atypical expression patterns derived from the cross-reactivity of allelic variants KIR2DL3*005 and KIR2DL3*015 with the KIR2DL1/S1 antibody EB6 [36],[37]. In our study identification of these allelic variants was not possible due the different KIR-antibody panel used. However, analysis of KIR2DL1+ and KIR2DL2/3+ NK cell frequencies from individuals either carrying or lacking KIR2DL2 or KIR2DS1/S2/S5 showed similar HLA-C dependent expression patterns, thus indicating that the overall increase of KIR2DL1-3+ NK cells in HIV-1 infection was not attributed to cross-reactivity of the antibodies used to KIR2DS2 or KIR2DS1/S2/S5-expressing NK cells.
Our results showed that increasing frequencies of self KIR2DL1-3+ NK cells were linked to the number of the respective HLA-C alleles suggesting a gene-dose effect that potentially contributes to increased surface expression of the cognate HLA-C ligands. Considering other confounding factors, larger studies will be required to assess the effect of HLA-C expression levels on KIR expression frequencies in HIV-1 infection [38]. It has been shown that the KIR-associated NK cell repertoire of neonates has not been biased towards a preferential expansion of self-inhibitory KIRs in the presence of the cognate HLA class I molecules [39]. Therefore, the results displayed in this and previous studies indicate that a strong environmental trigger, such as HIV-1 infection, is needed to drive sustained alterations of the KIR-expressing NK cell repertoire. However, the extent and nature of these changes seem to be genetically determined by the underlying KIR and HLA gene locus. As such, it is possible that a strong enrichment of KIR2DL1-3+ NK cells could indeed contribute to the control of HIV-1 infection, however difficult to identify due to the abundance of the KIR2DL1 and KIR2DL2/3 genes in the human population [40].
Apart from the phenotypic changes of the NK cell repertoire in the course of primary HIV-1 infection, the effector functions of licensed NK cells are likely diminished by the sustained chronic infection and general immune activation. Thus, emergence of dysfunctional NK cells in the course of HIV-1 infection could be potentially linked to an impaired licensing process or exhaustion of licensed NK cells. Indeed, expansion of CD56(−)/CD16+ NK cells with poor cytotoxic capacity has been associated with increased expression of inhibitory KIR [41],[42]. In our study KIR2DL1-3+ NK cells display a licensed phenotype and are capable to respond to MHC class I-deficient target cells to the same degree as NK cells derived from healthy donors. However, we demonstrate for the first time a subsequent loss of function of licensed NK cells in the transition to chronic infection. Previous reports have studied NK cell licensing in HIV-1 infection, predominantly analyzing individuals with the protective KIR3DL1+/HLA-Bw4+ haplotypes. In most of these, NK cells derived from slow progressors or viremic controllers display a licensed phenotype in comparison to NK cells obtained from HLA-Bw6+ individuals [43]–[47]. However, functional impairment of licensed NK cells including antibody-dependent cell-mediated cytotoxicity [48] and decreased frequency of polyfunctional NK cells in subjects with chronic HIV-1 infection have been reported [45]. Whether changes of the KIR repertoire in the course of HIV-1 infection are linked to impaired NK cell function needs to be further investigated.
Taken together, the comprehensive phenotypic and functional analyses of NK cells performed in this study suggest a complex interplay between intrinsic and extrinsic factors shaping the NK cell repertoire in the context of HLA-C. Our results indicate that HIV-1-driven enrichment of NK cells expressing self-inhibitory KIR2DL1-3 receptors is framed by the underlying HLA and KIR genotypes. In contrast to HLA-Bw4/KIR3DL1 haplotypes, other potentially protective interactions involving inhibitory KIR and HLA-C molecules have not been identified. However, it has been shown that NK cells can exert immunological pressure through the inhibitory KIR2DL2 receptor and that these are able to recognize HIV-1 peptides presented by HLA-C molecules [20],[21]. The present study demonstrates that HLA-C-mediated licensing of KIR2DL1-3+ NK cells contributes crucially to NK cell polyfunctionality in individuals with primary HIV-1 infection, providing a potential mechanism for the described protective effect of highly expressed HLA-C alleles.
Materials and Methods
Study subjects
This study included 42 HIV-1-infected and 40 HIV-1-negative individuals. Samples were obtained within the first 3 months of HIV-1 infection for individuals with primary infection. Patients were untreated at the time of the first sample collection. For a subset of individuals, follow-up samples were available at least 6 months (26 weeks (26, 34), median (IQR)) after first sample collection (early chronic infection). Clinical and demographic data of subjects are displayed in Table 1. HLA class I and KIR genotypes are shown in the Supporting Information Table 1/2.
Ethics statement
All subjects were recruited either at Massachusetts General Hospital, Fenway Health Center, or Brigham and Women's Hospital. All subjects gave written informed consent for participation in this study. The study was approved by the MGH Institutional Review Board (2003P001991, 2010P002121, 2012P000826).
Human blood samples and cell lines
Cryopreserved PBMC and plasma were used to assess phenotype and function of primary NK cells. PBMC were thawed and subsequently cultured overnight in RPMI complete media [RPMI 1640 (Sigma), 100 IU/ml Penicillin, 100 μg/ml Streptomycin, 2 mM L-glutamine (Corning Cellgro), and 10% (v/v) FBS (Sigma)] supplemented with 0.1 ng/ml IL-15 (R&D Systems). In addition, HLA class-I-deficient 721.221 cells were cultured in RPMI complete media and used as target cells for functional studies. Long- and short-term culturing was conducted at 37°C with 5% (v/v) CO2.
Human CMV serology
For the qualitative determination of the CMV serostatus CMV IgG Human ELISA kit (abcam) was used according to the manufacturer's protocol.
Antibodies and multi-parameter flow cytometry
A variety of antibodies were used for flow cytometric identification and phenotypic analysis of NK cells: anti-human CD3, CD14, CD19, CD16, CD56 and Blue Viability Dye; anti-human KIR2DL1, KIR2DL2/3, KIR2DL3, KIR3DL1, NKG2A and NKG2C. Anti-human CD107a, IFN-γ and TNF-α were used to assess quality and quantity of NK cell responses. A comprehensive list of the above mentioned antibodies is provided in Supporting Information Tab. 3. The gating strategy for identification and phenotype of NK cells is illustrated in Supporting Information Fig. 1.
Assessment of NK cell function
Quantitative and qualitative measurement of NK cell responses was performed by co-incubation of PBMC with either MHC class I-deficient 721.221 cells which have previously used to study NK cell function in the context of NK cell licensing [11],[12]. PBMC were co-cultured at an effector to target ratio of 5:1 or left alone in RPMI complete media supplemented with 0.1 ng/ml IL-15 in the presence of 2.5 μg/ml anti-human CD107a for 5 hours. After 1 hour incubation, 1.5 μl/mL of GolgiStop (BD) and 2.5 μg/ml of Brefeldin A (Sigma) were added. Cells were then stained for viability and surface marker expression. Intracellular cytokine staining was performed using anti-human IFN-γ and TNF-α and FIX & PERM (Invitrogen) according to the manufacturer's protocol. Following washing and additional fixation with 2% (w/v) paraformaldehyde (Affymetrix) cells were analyzed by flow cytometry.
Data acquisition and statistical analysis
Acquisition of flow cytometric data was performed on a BD LSR Fortessa (BD Biosciences) and analyzed using FlowJo software v7.6.5 (Tree Star,Inc). Single-positive KIR-expressing NK cells were defined by Boolean gating. Specific NK cell responses were calculated by the following equation: % specific resp. = 100*[% resp. (721.221) – % resp. (media)]/[100 - % resp. (media)]. Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software,Inc.) and SPSS 22 (IBM). Comparison between two groups was performed using Mann–Whitney test or Wilcoxon matched-pairs signed rank test for paired values. For comparison between >2 groups Kruskal-Wallis test with Dunn's post-test to correct for multiple comparisons was used. Spearman rank and multiple linear regression analyses were used to test for correlation between parameters. p values below <0.05 were considered significant.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the support and assistance of the Ragon Institute Imaging Core for the flow cytometry experiments. The authors gratefully acknowledge the scientific advice by Mary Carrington, Maureen Martin and Ying Qi. The authors thank Angelique Hölzemer and Wilfredo Garcia Beltran for helpful discussions.
This study was supported by the National Institutes of Health (NIH) (R01-AI067031-08, P01-AI104715), the Doris Duke Charitable Foundation (#208682), and the Ragon Institute of MGH, MIT and Harvard. Sagar A. Vaidya was supported by the NIH (T32-AI07387-23, P30-AI060354-10).
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. DOI: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 2.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. DOI: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moretta L, Moretta A. Killer immunoglobulin-like receptors. Curr. Opin. Immunol. 2004;16:626–633. doi: 10.1016/j.coi.2004.07.010. DOI: 10.1016/j.coi.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 5.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, Conte R, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by ‘group 2’ or ‘group 1’ NK clones. J. Exp. Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. DOI: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, Barbaresi M, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54:221–229. doi: 10.1007/s00251-002-0463-7. DOI: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 9.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 10.Elliott JM, Yokoyama WM. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 2011;32:364–372. doi: 10.1016/j.it.2011.06.001. DOI: 10.1016/j.it.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charoudeh HN, Schmied L, Gonzalez A, Terszowski G, Czaja K, Schmitter K, Infanti L, et al. Quantity of HLA-C surface expression and licensing of KIR2DL+ natural killer cells. Immunogenetics. 2012;64:739–745. doi: 10.1007/s00251-012-0633-1. DOI: 10.1007/s00251-012-0633-1. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Sunwoo JB, Yang L, Choi T, Song Y-J, French AR, Vlahiotis A, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. 2008;105:3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashirova AA, Thomas R, Carrington M. HLA/KIR Restraint of HIV: Surviving the Fittest. Annu. Rev. Immunol. 2011;29:295–317. doi: 10.1146/annurev-immunol-031210-101332. DOI: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamil KM, Khakoo SI. KIR/HLA Interactions and Pathogen Immunity. J. Biomed. Biotechnol. 2011;2011:1–9. doi: 10.1155/2011/298348. DOI: 10.1155/2011/298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–34. doi: 10.1038/ng934. DOI: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 16.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, et al. Differential natural killer cell mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. DOI: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, Nixon DF, Barbour JD. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol. 2008;82:4785–92. doi: 10.1128/JVI.02449-07. DOI: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song R, Lisovsky I, Lebouché B, Routy J-P, Bruneau J, Bernard NF. HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets. PLoS Pathog. 2014;10:e1003867. doi: 10.1371/journal.ppat.1003867. DOI: 10.1371/journal.ppat.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–40. doi: 10.1038/ng2035. DOI: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. DOI: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fadda L, Körner C, Kumar S, van Teijlingen NH, Piechocka-Trocha A, Carrington M, Altfeld M. HLA-Cw*0102-restricted HIV-1 p24 epitope variants can modulate the binding of the inhibitory KIR2DL2 receptor and primary NK cell function. PLoS Pathog. 2012;8:e1002805. doi: 10.1371/journal.ppat.1002805. DOI: 10.1371/journal.ppat.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, et al. Influence of HLA-C Expression Level on HIV Control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. DOI: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beziat V, Liu LL, Malmberg J-A, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121:2678–2688. doi: 10.1182/blood-2012-10-459545. DOI: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, Moretta A, et al. Chronic HIV-1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co-infection. AIDS Lond. Engl. 2010;24:27–34. doi: 10.1097/QAD.0b013e3283328d1f. DOI: 10.1097/QAD.0b013e3283328d1f. [DOI] [PubMed] [Google Scholar]
- 25.Béziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, Hervier B, et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 2012;42:447–457. doi: 10.1002/eji.201141826. DOI: 10.1002/eji.201141826. [DOI] [PubMed] [Google Scholar]
- 26.Naranbhai V, Altfeld M, Karim SSA, Ndung'u T, Karim QA, Carr WH. Changes in Natural Killer Cell Activation and Function during Primary HIV-1 Infection Ansari AA, ed. PLoS ONE. 2013;8:e53251. doi: 10.1371/journal.pone.0053251. DOI: 10.1371/journal.pone.0053251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, Miller JS, et al. HLA Class I Subtype-Dependent Expansion of KIR3DS1+ and KIR3DL1+ NK Cells during Acute Human Immunodeficiency Virus Type 1 Infection. J. Virol. 2009;83:6798–6805. doi: 10.1128/JVI.00256-09. DOI: 10.1128/JVI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Björkström NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michaëlsson J, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 2011;208:13–21. doi: 10.1084/jem.20100762. DOI: 10.1084/jem.20100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross MG, Burns DM, Grundy JE, Griffiths PD. Infection with human immunodeficiency virus (HIV) and cytomegalovirus in a London health district 1980-4. Genitourin. Med. 1987. 63:28–31. doi: 10.1136/sti.63.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, Snider JJ, Gill MJ. Cytomegalovirus Disease in HIV Infection: Twenty Years of a Regional Population's Experience. Clin. Infect. Dis. 2006;42:1808–1809. doi: 10.1086/504435. DOI: 10.1086/504435. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic Control of Variegated KIR Gene Expression: Polymorphisms of the Bi-Directional KIR3DL1 Promoter Are Associated with Distinct Frequencies of Gene Expression Roopenian DC. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. DOI: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, Trowsdale J, et al. Influence of KIR gene copy number on natural killer cell education. Blood. 2013;121:4703–4707. doi: 10.1182/blood-2012-10-461442. DOI: 10.1182/blood-2012-10-461442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, Ge D, et al. Copy Number Variation of KIR Genes Influences HIV-1 Control Emerman M. PLoS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. DOI: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M. Analyses of HLA-C-specific KIR repertoires in donors with group A and B haplotypes suggest a ligand-instructed model of NK cell receptor acquisition. Blood. 2010;117:98–107. doi: 10.1182/blood-2010-03-273656. DOI: 10.1182/blood-2010-03-273656. [DOI] [PubMed] [Google Scholar]
- 35.Czaja K, Borer A-S, Schmied L, Terszowski G, Stern M, Gonzalez A. A comprehensive analysis of the binding of anti-KIR antibodies to activating KIRs. Genes Immun. 2014;15:33–37. doi: 10.1038/gene.2013.58. DOI: 10.1038/gene.2013.58. [DOI] [PubMed] [Google Scholar]
- 36.Falco M, Romeo E, Marcenaro S, Martini S, Vitale M, Bottino C, Mingari MC, et al. Combined Genotypic and Phenotypic Killer Cell Ig-Like Receptor Analyses Reveal KIR2DL3 Alleles Displaying Unexpected Monoclonal Antibody Reactivity: Identification of the Amino Acid Residues Critical for Staining. J. Immunol. 2010;185:433–441. doi: 10.4049/jimmunol.0903632. DOI: 10.4049/jimmunol.0903632. [DOI] [PubMed] [Google Scholar]
- 37.Béziat V, Traherne J, Malmberg J-A, Ivarsson MA, Björkström NK, Retière C, Ljunggren H-G, et al. Tracing dynamic expansion of human NK-cell subsets by high-resolution analysis of KIR repertoires and cellular differentiation. Eur. J. Immunol. 2014:n/a–n/a. doi: 10.1002/eji.201444464. DOI: 10.1002/eji.201444464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas R, Apps R, Qi Y, Gao X, Male V, O'hUigin C, O'Connor G, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. DOI: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schonberg K, Fischer JC, Kogler G, Uhrberg M. Neonatal NK-cell repertoires are functionally, but not structurally, biased toward recognition of self HLA class I. Blood. 2011;117:5152–5156. doi: 10.1182/blood-2011-02-334441. DOI: 10.1182/blood-2011-02-334441. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–D919. doi: 10.1093/nar/gkq1128. DOI: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, O'Shea MA, et al. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U A. 2005;102:2886–91. doi: 10.1073/pnas.0409872102. DOI: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Björkström NK, Ljunggren H-G, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. 2010;31:401–406. doi: 10.1016/j.it.2010.08.003. DOI: 10.1016/j.it.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Eller MA, Koehler RN, Kijak GH, Eller LA, Guwatudde D, Marovich MA, Michael NL, et al. Human Immunodeficiency Virus Type 1 Infection Is Associated with Increased NK Cell Polyfunctionality and Higher Levels of KIR3DL1+ NK Cells in Ugandans Carrying the HLA-B Bw4 Motif. J. Virol. 2011;85:4802–4811. doi: 10.1128/JVI.00111-11. DOI: 10.1128/JVI.00111-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, Bernard NF. HIV Protective KIR3DL1 and HLA-B Genotypes Influence NK Cell Function Following Stimulation with HLA-Devoid Cells. J. Immunol. 2010;184:2057–2064. doi: 10.4049/jimmunol.0902621. DOI: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- 45.Kamya P, Tallon B, Melendez-Pena C, Parsons MS, Migueles SA, Connors M, Miconiatis S, et al. Inhibitory Killer Immunoglobulin-like Receptors to self HLA-B and HLA-C ligands contribute differentially to Natural Killer cell functional potential in HIV infected slow progressors. Clin. Immunol. Orlando Fla. 2012;143:246–255. doi: 10.1016/j.clim.2012.01.001. DOI: 10.1016/j.clim.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Kamya P, Boulet S, Tsoukas CM, Routy J-P, Thomas R, Côté P, Boulassel M-R, et al. Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J. Virol. 2011;85:5949–5960. doi: 10.1128/JVI.02652-10. DOI: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomescu C, Duh F-M, Hoh R, Viviani A, Harvill K, Martin MP, Carrington M, et al. Impact of protective killer inhibitory receptor/human leukocyte antigen genotypes on natural killer cell and T-cell function in HIV-1-infected controllers. AIDS Lond. Engl. 2012;26:1869–1878. doi: 10.1097/QAD.0b013e32835861b0. DOI: 10.1097/QAD.0b013e32835861b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsons MS, Wren L, Isitman G, Navis M, Stratov I, Bernard NF, Kent SJ. HIV Infection Abrogates the Functional Advantage of Natural Killer Cells Educated through KIR3DL1/HLA-Bw4 Interactions To Mediate Anti-HIV Antibody-Dependent Cellular Cytotoxicity. J. Virol. 2012;86:4488–4495. doi: 10.1128/JVI.06112-11. DOI: 10.1128/JVI.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.