Abstract
Background
Transmitted drug resistance (TDR) is increasing in some areas of Africa. Detection of TDR may predict virologic failure of first-line non-nucleoside reverse-transcriptase inhibitor (NNRTI)-based antiretroviral therapy (ART). We evaluated the utility of a relatively inexpensive oligonucleotide ligation assay (OLA) to detect clinically relevant TDR at time of ART initiation.
Methods
Pre-ART plasmas from ART-naive Kenyans initiating an NNRTI-based fixed-dose combination ART in a randomized adherence trial conducted in 2006 were retrospectively analyzed by OLA for mutations conferring resistance to NNRTI (K103N, Y181C, and G190A) and lamivudine (M184V). Post-ART plasmas were analyzed for virologic failure (≥1,000 copies/mL) at 6 month intervals over 18-month follow-up. Pre-ART plasmas of those with virologic failure were evaluated for drug resistance by consensus and 454-pyrosequencing.
Results
Among 386 participants, TDR was detected by OLA in 3.89% [95% Confidence Interval (CI), 2.19-6.33], and was associated with a 10-fold higher rate of virologic failure [Hazard Ratio (HR), 10.39; 95% CI, 3.23-32.41; p<0.001) compared to those without TDR. OLA detected 24 TDR mutations (K103N, n=13; Y181C, n=5; G190A, n=3; M184V, n=3) in 15 subjects (NNRTI, n=15; 3TC, n=3). Among 51 participants who developed virologic failure, consensus sequencing did not detect additional TDR mutations conferring high-level resistance, and pyrosequencing only detected additional mutations at frequencies <2%. Mutant frequencies <2% at ART initiation were significantly less likely to be found at the time of virologic failure compared to frequencies ≥2% (22% vs. 63%; p<0.001).
Conclusions
Detection of TDR by a point mutation assay may prevent use of sub-optimal ART.
Keywords: transmitted drug resistance, oligonucleotide ligation assay, HIV, Kenya, antiretroviral therapy
Introduction
Access to antiretroviral therapy (ART) has substantially increased in many areas of sub-Saharan Africa over the last decade,1,2 yet patients are failing ART and HIV resistance to some antiretroviral medications is increasing.3,4 As a result, ART-naïve individuals are becoming infected with transmitted drug resistant virus (TDR) and failing first-line ART based on non-nucleoside reverse transcriptase inhibitors (NNRTI).5 Public health officials in Africa will need to decide when resistance testing prior to ART initiation is warranted to prevent early treatment failure.6
HIV resistance testing by consensus sequencing or newer ultra-deep sequencing methods are costly and require equipment and technical capacity beyond what is accessible at many African HIV clinics. An oligonucleotide ligation assay (OLA), which is relatively inexpensive compared to conventional sequencing, has been shown to detect HIV resistance selected by single-dose nevirapine that is associated with virologic failure.7 It is not known whether the implementation of OLA prior to ART initiation might decrease virologic failure in Africa.
We designed a retrospective study to describe the prevalence of TDR in Nairobi, Kenya in 2006 and to determine whether OLA-detected TDR was associated with virologic failure. In addition, we examined whether low-frequency mutations or mutations at codons not evaluated by OLA contributed to virologic failure.
Methods
Setting
A retrospective analysis of TDR using data and specimens collected from participants in a randomized controlled trial on adherence in Nairobi, Kenya was conducted. The design and methods of the adherence study are described elsewhere.8 In brief, 400 ART-naïve individuals eligible to start ART at the Coptic Hope Center for Infectious Diseases were enrolled in 2006 and equally randomized to: adherence counseling; alarm device; combined adherence counseling and alarm device; or a control group that received neither intervention.8,9 First-line ART regimen was initiated with a fixed-dose combination of nevirapine (NVP), lamivudine (3TC) and stavudine (d4T). The trial demonstrated that adherence counseling significantly increased adherence and reduced virologic failure while alarm device had no effect.8
Study design
Sociodemographic information, adherence data, and blood samples from the adherence trial were used in this analysis. After initiating ART, participants had monthly pill counts and ART refills. Plasma HIV RNA levels (Gen-Probe, San Diego, CA, USA) and CD4 counts (FACScan, Becton Dickinson, Franklin Lanes, NJ, USA) were determined at baseline prior to ART initiation, and at 6, 12, and 18 months of ART. Virologic failure was defined as plasma HIV RNA copies ≥1,000 copies/mL at least 4 months after ART initiation.10
To measure TDR, plasma collected at baseline from all participants was evaluated for NNRTI and 3TC drug resistance mutations by OLA. Participants who experienced virologic failure were evaluated for additional mutations at baseline by consensus sequencing and 454- pyrosequencing of their pre-ART plasma specimen. Plasma samples at the time of virologic failure were analyzed by OLA and consensus sequencing.
Nucleic acid extraction and PCR amplification of HIV pol
RNA was extracted from 0.5-1mL of pre-ART plasma (NucliSENS miniMAG, bioMérieux, Durham, NC, USA). After reverse transcription (Superscript III, Invitrogen, Grand Island, NY, USA) with random hexamers, HIV pol spanning codons 1-239 of RT was amplified (MyTaq DNA Polymerase, Bioline USA Inc, Taunton, MA, USA) by nested polymerase chain reaction (PCR) (1st round primers; forward: GARAGACAGGCTAATTTTTTAGGGA, and reverse: AAYTTCTGTATATCATTGACAGTCCA; 2nd round primers forward: CAAATCACTCTTTGGCARCGACC and reverse: CAYTTGTCAGGATGGAGTTCATA).
Oligonucleotide ligation assay (OLA)
Amplicons from baseline and virologic failure were tested by an OLA for point mutations conferring resistance to NNRTI (K103N, Y181C and G190A) and 3TC (M184V).11,12 The OLA used probes optimized for subtypes A, D and C prevalent in Kenya. Standards of plasmids mixed to contain 0%, 2%, 5%, 10%, 25%, 50%, 75% or 100% mutant in a wild-type background were included in each assay plate, and comparisons of the optical densities of each specimen to those of the standards was used to quantify the proportion of mutant in the HIV population.13
Dideoxy-nucleotide consensus sequencing and parallel 454-pyrosequencing
Amplicons from individuals with virologic failure underwent dideoxy-nucleotide consensus sequencing to identify mutations both at baseline and at the first time point when virologic failure was detected (GenBank sequence accession numbers: KF544089-KF544288, KJ395348 and KJ395349).14,15 Massive parallel 454-pyrosequencing was performed on baseline plasma samples of individuals with virologic failure and/or OLA-detected mutations. After reverse transcription of plasma HIV RNA as described above, the cDNA was quantified by HIV LTR real-time PCR.16 Approximately 1,000 templates of amplifiable HIV were submitted to a median of three nested PCR (range, 1 to 20 reactions) with subsequent pooling of amplicon. First round PCR used the primers described above and FastStart High Fidelity PCR System (Roche Diagnostics, Mannheim, Germany). Subsequently, two regions of HIV reverse transcriptase were amplified in separate second round PCR with 454-barcoded-fusion primers (454 Life Sciences, Roche Diagnostics Corporation, Branford, CT, USA), with 14 discrete multiplex identifiers (MID) or barcodes:
Amplicon 1 (RT codons 21-134)
Forward: CGTATCGCCTCCCTCGCGCCATCAG-MIDGTTAAACAGTGGCCATTGACAGA
Reverse: CTATGCGCCTTGCCAGCCCGCTCAG-MIDACTAGGTATGGTGAATGCAGTATA
Amplicon 2 (RT codons 150-242)
Forward: CGTATCGCCTCCCTCGCGCCATCAG-MIDCACAGGGATGGAAAGGATCAC
Reverse: CTATGCGCCTTGCCAGCCCGCTCAG-MIDCTGGACTGTCCATYTGTCAGGATG
Amplicons were visualized on agarose gels, purified using a High Pure PCR Product Purification Kit (Roche Applied Science, Mannheim, Germany), and quantified using a Quant-iT PicoGreen dsDNA Reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA). Pools containing barcoded amplicons from 14 subjects, diluted to 1x107 molecules/μL, underwent emulsion PCR and 454-sequencing using a 2-region gasket and the GS FLX Titanium System per manufacturer's instructions (454 Life Sciences, Roche Diagnostics Corporation, Branford, CT, USA).
The output was processed through a pipeline aligning sequences to a subtype-specific consensus reference.17 Nucleotide frequencies at each position were calculated in forward, reverse and overall reads to determine the frequency of mutants at codons conferring resistance to NRTI and NNRTI as defined in the Stanford Database.14
Errors introduced by PCR and 454-sequencing were estimated by including amplicon of an HIV-1 subtype A plasmid in each 454-sequencing plate. A control for errors in reverse transcription was not included. The limit of mutant quantification was estimated based on the number of HIV templates from each specimen submitted to the assay and the error rate observed across 454-sequencing of the plasmid. Testing of a median of 1,000 (range 275-1,300) HIV templates per specimen by pyrosequencing and an average substitution error rate of 0.06 +/− 0.01% SD observed in the plasmid control set the limit of mutant detection at >0.1%.
Statistical methods
Wilcoxon rank sum and Fisher's exact tests were used to compare continuous and categorical characteristics between those with and without TDR. Among those who initiated ART, time to virologic failure was estimated using Kaplan-Meier (KM) analysis and groups with and without TDR were compared using a log-rank test. Participants were censored if they died or were lost to follow-up before the 18-month study endpoint.
A discrete time Cox proportional hazards regression model assessed differences in rates of virologic failure. A preliminary model included TDR and randomization assignment. Adjusted models also included baseline characteristics associated with TDR and virologic failure, as well as age, employment status, baseline viral load, randomization assignment, and adherence. Adherence was examined as a time-varying covariate as average adherence in the prior 6 months.
Mutant frequency (<2% versus ≥2%) detected by pyrosequencing at baseline was assessed for subsequent detection at viral failure by consensus sequencing using generalized estimating equations (GEE) models with binomially-distributed errors, log link, and exchangeable correlation matrix with robust errors. Among the subjects with virologic failure, linear regression was used to assess time to viral failure between no (0%), “very low” (>0% to <2% frequency), and “low-high” (≥2%) levels of mutations by pyrosequencing. Stata SE v12 (Statacorp, College Station, TX, USA) was used in all analyses; significance tests were two-sided, with alpha=0.05.
Ethical review
The study protocol was reviewed and approved by the institutional review boards at the University of Washington (Seattle, WA, USA), Seattle Children's Hospital (Seattle, WA, USA), and Kenyatta National Hospital (Nairobi, Kenya).
Results
Study population
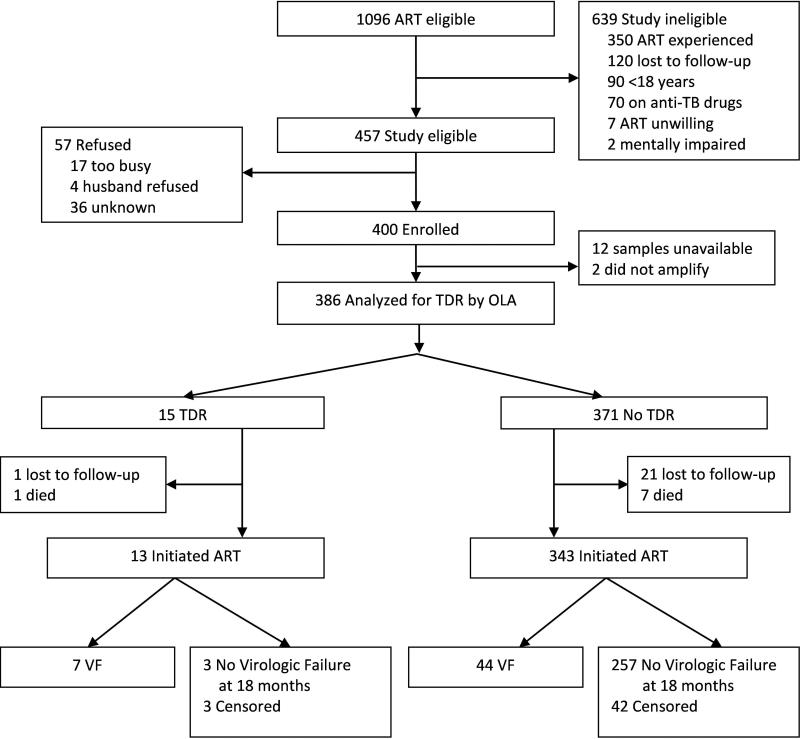
Of 1,096 adults eligible to initiate ART at the Hope Center in 2006, 437 qualified for the study and 400 were enrolled (Figure 1). Of the 400 enrollees, 386 had baseline pre-ART plasma samples tested by OLA and 356 participants initiated ART and were assessed for virologic failure. Prior to initiating ART, 22 participants were lost to follow-up and 8 died. A total of 307 participants (86%) completed 18 months of study follow-up. Between ART initiation and the 18-month endpoint, 21 participants were lost to follow-up and 28 died.
Figure 1.
Trial Profile
Transmitted drug resistance and virologic failure
Among the 386 participants evaluated at baseline, 15 had one or more mutations detected by OLA for a TDR prevalence of 3.89% [95% Confidence Interval (CI), 2.19-6.33]. The median age among those with TDR was 37 years [Interquartile Range (IQR), 32-43], 60% were female, 47% were married, and the median CD4 count was 100 cells/mm3 (IQR, 23-155) (Table 1). Compared to those without TDR, those with TDR were significantly more likely to have had more lifetime sexual partners (median, 7 vs. 4; p=0.004), never attended school (13% vs. 2%; p=0.044), and been casual laborers (33% vs. 13%; p=0.045). CD4 count, plasma HIV-1 RNA, and randomization assignment did not differ between those with and without TDR.
Table 1.
Baseline comparison between those with and without transmitted drug resistance (TDR) detected by oligonucleotide ligation assay (OLA) in pre-ART blood plasma.
| Characteristic | TDR (N=15) | No TDR (N=371) | |
|---|---|---|---|
| n (%) or Median (IQR) | n (%) or Median (IQR) | p-value | |
| Age (years) | 37 (32-43) | 36 (30-42) | 0.45 |
| Female | 9 (60) | 246 (66) | 0.59 |
| Married or attached | 7 (47) | 192 (52) | 0.80 |
| Education (years) | 11 (8-12) | 12 (8-13) | 0.19 |
| Education | 0.030 (overall) | ||
| None | 2 (13) | 7 (2) | 0.04 |
| Primary | 4 (27) | 124 (34) | 0.78 |
| Secondary | 8 (53) | 144 (39) | 0.29 |
| College | 1 (7) | 93 (25) | 0.13 |
| Employment | 0.05 (overall) | ||
| Salaried job | 1 (7) | 117 (32) | 0.045 |
| Self-employed | 4 (27) | 82 (22) | 0.75 |
| Casual laborer | 5 (33) | 49 (13) | 0.045 |
| Unemployed | 5 (33) | 121 (33) | 1.00 |
| Rooms in house | 2 (1-4) | 2 (1-4) | 0.76 |
| Rent | 2500 (570-7000) | 2000 (1000-4500) | 0.87 |
| People in house | 4 (1-6) | 4 (2-5) | 0.80 |
| Flush toilet | 8 (53) | 173 (47) | 0.79 |
| Cost of travel to clinic ≥US$0.70 | 8 (53) | 231 (62) | 0.59 |
| Distance to clinic (km) | 9.5 (6.0-13.9) | 10.3 (6.2-14.9) | 0.64 |
| Age at first sex (years) | 17 (16-20) | 18 (16-20) | 0.76 |
| Lifetime sexual partners | 7 (4-10) | 4 (2-6) | 0.004 |
| Money or favors for sex | 3 (20) | 35 (9) | 0.18 |
| Plasma HIV log10 copies/ml | 5.85 (5.12-6.20) | 5.67 (5.24-6.07) | 0.70 |
| CD4 count, cells/ml | 100 (23-155) | 116 (61-183) | 0.29 |
| Randomization assignment | 0.18 (overall) | ||
| Counseling | 1 (7) | 94 (25) | 0.13 |
| Alarm | 7 (47) | 91 (25) | 0.07 |
| Counseling and alarm | 3 (20) | 93 (25) | 1.00 |
| Control | 4 (27) | 93 (25) | 1.00 |
Median (IQR) is presented unless otherwise specified
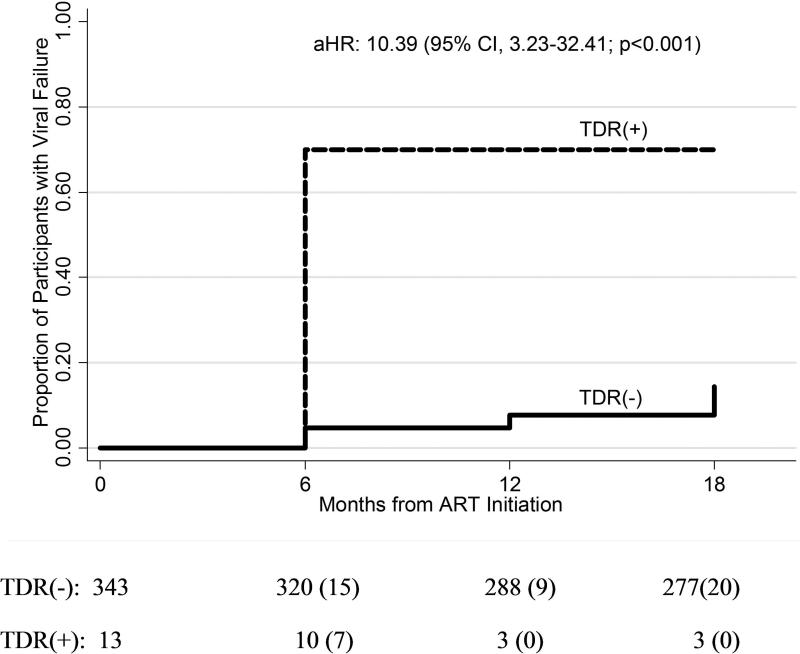
Among the 356 participants who initiated ART, 13 (3.65%) had TDR detected by OLA and 51 (14.3%) experienced virologic failure. Overall, the cumulative probability of virologic failure at 18 months was 0.16 (95% CI, 0.12-0.21). The cumulative probability of virologic failure among those with TDR was significantly higher (0.70; 95% CI, 0.42-0.93) compared to those without TDR (0.14; 95% CI, 0.11-0.19) (Figure 2). Characteristics associated with TDR in this subset were similar to those in the overall cohort. Those with TDR experienced virologic failure at a rate 7.99 times (95% CI, 2.95-21.64; p<0.001) higher than those who did not have TDR. In a model adjusting for age, employment status, baseline viral load, adherence, and randomization assignment, the risk of virologic failure associated with TDR was 10.39 (95% CI, 3.23-32.41; p<0.001).
Figure 2.
Kaplan-Meier curve of time to virologic failure between those with presumed transmitted drug resistance (TDR) detected in codons M184V, K103N, Y181C, and/or G190A by oligonucleotide ligation assay (OLA) (dashed line) and those without TDR by OLA (solid line).
Note: The number of participants at risk at each time point is listed and the number experiencing virologic failure is in parentheses.
Detection of baseline mutations by OLA
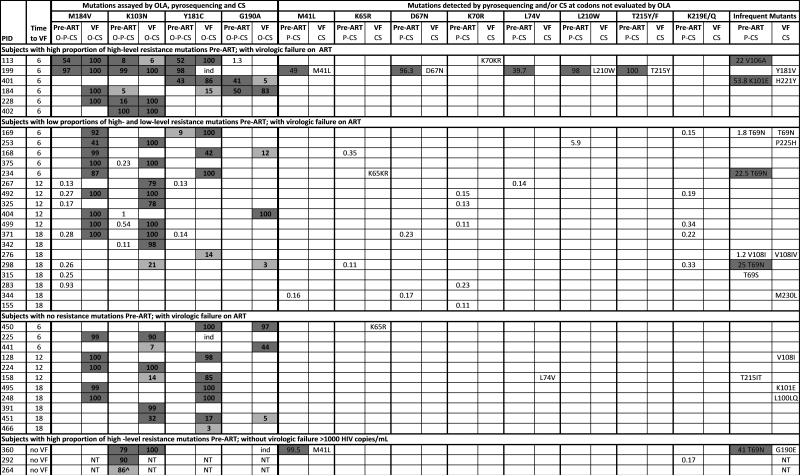
OLA, sensitive to a mutant frequency of 2%, detected 24 mutations in 15 participants at baseline. These 15 had mutations associated with resistance to NNRTI and 3 also had mutations associated with resistance to 3TC (Figure 3). The median mutant frequency in each subject's viral population was 67% (range, 2%-100%).
Figure 3.
Drug resistance mutations detected by OLA, pyrosequencing and consensus sequencing at baseline, and by OLA and consensus sequencing at virologic failure.
Note: Pre-ART specimens were tested by OLA (O), pyrosequencing (P), and consensus sequencing (CS); virologic failure (VF) specimens were tested by OLA and CS only. Numbers indicate the % mutant virus quantified by OLA (bold font, limit of detection = 2%) or pyrosequencing (regular font, limit of detection 0.1%). Dark grey indicates mutation detected by all methods evaluated. Light grey indicates mutation not detected by consensus sequencing, except as indicated for subject #264. Numbers with no shading indicate mutations detected by pyrosequencing only. NT = not tested (due to undetectable HIV viral load). ind = indeterminate result by OLA due to the presence of different mutation. ^ = mutation detected by OLA and consensus sequencing but not by pyrosequencing due to sequence polymorphisms in the region of the pyrosequencing PCR primers.
Of the 15 participants who had mutations detected by OLA at baseline, virologic outcome could not be ascertained for 5 participants; 3 died and 2 were lost before viral load was measured. Of these 5 participants, 3 participants had a single mutation (K103N), 1 participant had 2 mutations (K013N and M184V), and 1 participant had 3 mutations (K103N, Y181C, and G190A). Among the 10 with virologic outcome, 7 experienced virologic failure: 3 of these participants had single mutations (K103N, n=2; Y181C, n=1); 2 participants had 2 mutations (K103N and G190A, n=1; Y181C and G190A, n=1); and 2 participants had 3 mutations (K103N, M184V, and Y181C). The median mutant viral load in the pre-ART specimen of those who failed was 536,167 copies/mL (range, 12,976 – 2,128,896 copies/mL). Three participants with mutations detected by OLA at baseline did not experience virologic failure after 18 months on ART and each had only a single mutation (K103N); their mutant viral loads were 112,316, 642,240, and 585,469 copies/mL.18
Among the 7 participants who had mutations detected by OLA at baseline and experienced virologic failure, all failed by month 6 (the first time point evaluated after ART initiation) and 6 of the 7 had adherence ≥95%.
Detection of baseline mutations among virologic failures by consensus and pyrosequencing
Of the 51 participants who experienced virologic failure, consensus sequencing detected 23 mutations in 10 participants and pyrosequencing detected 56 mutations in 24 participants at baseline (Figure 3). Consensus sequencing confirmed baseline mutations in 6 of the 7 participants identified by OLA, and detected baseline mutations at sites associated with “potential low-level resistance” to didanosine and abacavir or “low-level resistance” to zidovudine and stavudine in 4 additional participants at codons not evaluated by OLA (T215T/I, n=1; T69S, n=1; T69T/N, n=2) (hivdb.stanford.edu).
In addition to the 7 participants who had mutations identified by OLA, pyrosequencing detected 17 additional participants with either very low (<2%) frequency mutations at the codons tested by OLA (M184V, K103N, Y181C, G190A), or mutations (frequencies 0.1%-100%) in other codons conferring drug-resistance. Eleven individuals had mutations at the OLA codons, all at frequencies <2%. The remaining 6 individuals had mutations at non-OLA codons, all at frequencies <2%, except for 3 individuals: one with L210W detected at a frequency of 5.9%, and two with T69N detected; one at a frequency of 22.5% and the second at 25.0% (Figure 3).
Detection of baseline mutations at virologic failure
Of the 13 mutations detected in 7 participants by OLA at baseline, OLA detected 11 (85%) and consensus sequencing detected 10 (77%) at the time of virologic failure. Two mutations were detected by OLA at baseline but not at failure. In one participant, K103N at a frequency of 5% co-existed with G190A; only the latter was detected at failure along with Y181C and M184V. In another, Y181C was detected at baseline with additional mutations, and at failure it had shifted to Y181V (Figure 3). Of the mutations classified as “potential low-level resistance” or “low-level resistance” in the Stanford HIV Drug Resistance Database (hivdb.stanford.edu), none were detected by consensus sequencing at virologic failure.
Mutant variants detected in the pre-ART plasma at a frequency of <2% by pyrosequencing were significantly less likely than those >2% frequency to be found by consensus sequencing at the time of virologic failure. Of the 32 mutations detected by pyrosequencing at baseline at frequency <2%, 7 (22%) were detected by consensus sequencing at the time of virologic failure. Of the 24 mutations at frequency ≥2%, significantly more, 15 (63%), were found at virologic failure (63% vs. 22%; p<0.001). This was also true when restricted to codons evaluated by OLA (K103N, Y181C, G190A, and M184V). Among the 13 mutations detected in “OLA codons” by pyrosequencing (and OLA) at frequency ≥2%, 10 (77%) were detected by consensus sequencing at virologic failure compared to 5/14 (36%) at baseline frequencies <2% (77% vs. 36%; p=0.049).
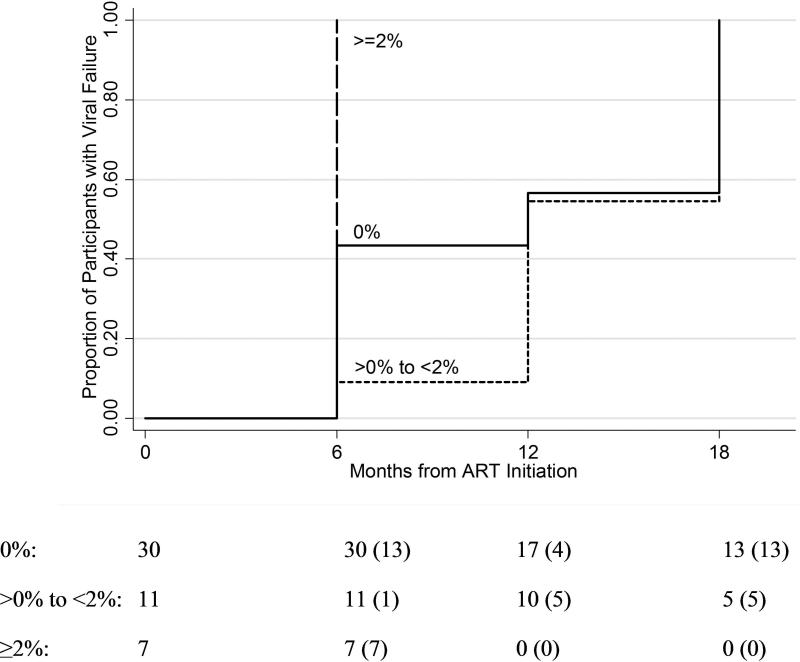
Baseline mutant frequency and time to virologic failure
When examining OLA codons (K103N, Y181C, G190A, and M184V), the mean time to virologic failure was significantly earlier among participants with baseline mutation frequencies ≥2% by pyrosequencing compared to those with minority mutations (>0% to <2%) (p=0.002) or no mutations (0%) (p=0.008) (Figure 4). In an analysis adjusting for age, employment status, baseline viral load, adherence, and randomization assignment, participants with mutant frequencies ≥2% experienced virologic failure 9.6 months earlier compared to those with mutational frequencies between 0-2% (p=0.001) and 7.0 months earlier compared to those with no mutations (p=0.004). There was no significant difference in time to virologic failure between participants with resistance mutations present at frequencies <2% compared to those with no mutations detected (p=0.194).
Figure 4.
Kaplan-Meier curve of time to virologic failure in participants grouped by whether or not M184V, K103N, Y181C, and/or G190A was detected in baseline (pre-ART) viral population, and if detected, the proportion of the mutant population: 0% (solid line), >0% to <2% (short dashed line), and ≥2% (long dash line).
Note: The number of participants at risk at each time point is listed and the number experiencing virologic failure is in parentheses.
Discussion
In this analysis of 386 ART-naïve adults initiating ART in Nairobi, Kenya in 2006, we found a TDR prevalence of 3.9%. TDR was determined using OLA that detected mutations conferring high-level resistance to NNRTI and 3TC and was significantly associated with virologic failure in this study. Those with TDR detected by OLA at one or multiple codons (M184V, K103N, Y181C, and G190A) were 10 times more likely to experience virologic failure than those without TDR. This evidence suggests that OLA performed prior to ART initiation may guide antiretroviral prescribing practices to reduce rates of virologic failure.
The detection of TDR by this study in Nairobi, Kenya is consistent with increased prevalence of antiretroviral resistance found in many resource-limited settings since the widespread rollout of ART.3 To our knowledge, this study is the largest single site cohort study of TDR detected among ART-naïve patients in East Africa. While the prevalence of 3.9% is less than 13.2% found among 68 Kenyans in Mombasa in 2009-10,19 7.5% among 53 in Nairobi in 2005,20 and 4.5% among 200 in Nairobi in 2008-9,21 it is greater than 1.1% found among 182 Kenyans in Kilifi in 2008-10 and 3.1% among 76 across Kilifi, Nairobi, and Mtwapa in 2006-9.22,23 In relation to these figures, the study's TDR prevalence is low given the clinic's urban setting where ART coverage is relatively high. This may be due to enrollment of only ART-naïve adults, examination of antiretroviral resistance at only 4 codons, and testing of specimens from 2006. It is expected that TDR prevalence will continue to increase paralleling expansion of ART and evolving recommendations for earlier initiation of treatment.24
The association between the detection of TDR and virologic failure is not new and has been noted in both resource-rich and constrained settings.25-27 However, detection of TDR by OLA, a point mutation resistance test, is novel. Inexpensive antiretroviral resistance testing is needed in resource-limited settings, where most HIV patients cannot afford resistance testing by consensus sequencing. OLA may be as much as 10-20 times less expensive than consensus sequencing, and has been successfully implemented by laboratories in resource-limited countries.12,28
Consensus sequencing and pyrosequencing at baseline did not appear to contribute more information than OLA. Among those who experienced virologic failure, consensus sequencing did not detect additional subjects with pre-ART mutations associated with “high-level resistance” beyond those identified by OLA testing (hivdb.stanford.edu). In addition, the baseline mutants detected by consensus sequencing were not selected at virologic failure, and suggests that these mutations may not have been clinically relevant in these subjects.
These data suggest that the codons tested by OLA were appropriate for this setting and time period. Baseline mutations detected by pyrosequencing but not by OLA were primarily at low frequencies (<2%) in participants’ viral populations, and were significantly less likely to be selected at the time of virologic failure compared to those at a frequency ≥2%, which were detectable by OLA. Given this finding as well as the shorter time to virologic failure among those with mutant frequencies ≥2%, it appears that baseline mutations at low frequencies <2% did not significantly contribute to virologic failure.
Weaknesses in this study include the limited number of samples tested by consensus sequencing and pyrosequencing due to cost constraints. Although consensus sequencing was not performed on all samples, detection of TDR by consensus sequencing probably would have been just as predictive of virologic failure as OLA. Although the prevalence of TDR was reported, the study is likely to have underestimated the actual prevalence, as a comprehensive survey of all drug resistance codons was not performed since our primary focus was to evaluate the utility of the OLA to predict virologic failure. Based on consensus sequencing and pyrosequencing, OLA did not appear to miss the detection of mutations associated with virologic failure. However, additional codons will be needed to screen for TDR by OLA when antiretroviral regimens change within a community. Setting the limit of mutant detection by pyrosequencing at >0.1% may be considered too low. While the 0.1% estimate accounts for errors introduced by PCR and pyrosequencing, it does not account for those introduced by reverse transcription. We chose to report the detection of mutations detected above the error rate of our plasmid controls to allow the reader to compare detection of mutations around the cutoff of OLA. Samples collected at 6-month intervals may not have captured the exact time of virologic failure, and among participants with poor adherence to antiretrovirals, the absence of selective pressure may have allowed resistance mutations to be outcompeted by wild type viruses with greater replication capacity. Additionally, our testing of pre-ART plasma may not have accurately measured the rate of TDR in this cohort. It is possible that HIV infection was founded by drug-resistant variants in some participants, but in the time interval between acute infection and collection of the pre-ART specimen these mutants could have reverted to wild type variants.
The increasing rate of TDR observed in Africa could threaten efficacy of first-line ART. Cost-effective strategies to contain drug-resistant HIV are needed for resource-poor countries. This study suggests that pre-ART testing for HIV drug resistance with a point mutation assay, which is less expensive than conventional sequencing, may be a viable strategy to help preserve the efficacy of first-line ART. Among those with >95% adherence, detection of TDR by OLA at ART initiation and subsequent regimen modification would have averted 6/13 (46%) virologic failures at 6 months follow-up (data not shown). Further development of simple, inexpensive assays to test for multiple drug-resistance mutations appears warranted, as do feasibility and cost-effectiveness studies of these assays in low-resource communities.
Acknowledgments
M Chung designed and implemented the study, supervised the on-site data management, interpreted the data, and wrote the paper. L Frenkel led the laboratory analysis, designed the study, interpreted the data, and wrote the paper. I Beck conducted the laboratory analysis, interpreted the data, and helped write the paper. S Dross conducted the laboratory analysis, interpreted the data, and helped write the paper. K Tapia analyzed the data and helped interpret the results. J Kiarie helped implement the study, oversee data collection, and helped write the paper. B Richardson analyzed the data and interpreted the results. J Overbaugh helped conduct and analyze the laboratory data and write the paper. S Sakr helped implement and design the study. G John-Stewart helped interpret the data and write the paper.
We thank the patients, research personnel, clinic and laboratory staff, and data management teams in Nairobi and Seattle for their efforts as well as the Coptic Hope Center for Infectious Diseases, and Dr. Joseph Fitzgibbon of the National Institutes of Health for his intellectual contributions.
Source of Funding: This work was supported by grants and an American Recovery and Reinvestment Act supplement from the National Institutes of Health [R01-AI058723 to LMF and K23-AI065222 to MHC]. Support of the study was provided by the University of Washington Center for AIDS Research [P30AI027757]. The Coptic Hope Center for Infectious Diseases is supported by the President's Emergency Plan for AIDS Relief through a cooperative agreement [U62/CCU024512] from the Centers for Disease Control and Prevention.
Footnotes
This data was presented at the 18th Conference on Retroviruses and Opportunistic Infections (CROI 2011) held February 27 – March 2, 2011 in Boston, MA, USA (Oral Abstract #41).
Conflicts of Interest
None of the authors has a conflict of interest in this study.
References
- 1.UNAIDS . Global report: UNAIDS report on the global AIDS epidemic 2012. UNAIDS; Geneva: 2012. [Google Scholar]
- 2.Jahn A, Floyd S, Crampin AC, et al. Population-level effect of HIV on adult mortality and early evidence of reversal after introduction of antiretroviral therapy in Malawi. Lancet. 2008 May 10;371(9624):1603–1611. doi: 10.1016/S0140-6736(08)60693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RK, Jordan MR, Sultan BJ, et al. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012 Oct 6;380(9849):1250–1258. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther. Oct 10. 2012 doi: 10.3851/IMP2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth RE, Wensing AM, Tempelman HA, Moraba R, Schuurman R, Hoepelman AI. Rapid accumulation of nonnucleoside reverse transcriptase inhibitor-associated resistance: evidence of transmitted resistance in rural South Africa. AIDS. 2008 Oct 18;22(16):2210–2212. doi: 10.1097/QAD.0b013e328313bf87. [DOI] [PubMed] [Google Scholar]
- 6.Jordan MR, Bennett DE, Wainberg MA, et al. Update on World Health Organization HIV drug resistance prevention and assessment strategy: 2004-2011. Clin Infect Dis. 2012 May;54(Suppl 4):S245–249. doi: 10.1093/cid/cis206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jourdain G, Wagner TA, Ngo-Giang-Huong N, et al. Association between detection of HIV-1 DNA resistance mutations by a sensitive assay at initiation of antiretroviral therapy and virologic failure. Clin Infect Dis. 2010 May 15;50(10):1397–1404. doi: 10.1086/652148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung MH, Richardson BA, Tapia K, et al. A randomized controlled trial comparing the effects of counseling and alarm device on HAART adherence and virologic outcomes. PLoS Med. 2011 Mar;8(3):e1000422. doi: 10.1371/journal.pmed.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung MH, Drake AL, Richardson BA, et al. Impact of prior HAART use on clinical outcomes in a large Kenyan HIV treatment program. Curr HIV Res. 2009 Jul;7(4):441–446. doi: 10.2174/157016209788680552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery S, Bodrug S, Richardson BA, et al. Evaluation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya. J Clin Microbiol. 2000;38(7):2688–2695. doi: 10.1128/jcm.38.7.2688-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck IA, Mahalanabis M, Pepper G, et al. Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J Clin Microbiol. 2002 Apr;40(4):1413–1419. doi: 10.1128/JCM.40.4.1413-1419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Micek MA, Blanco AJ, Beck IA, et al. Nevirapine resistance by timing of HIV type 1 infection in infants treated with single-dose nevirapine. Clin Infect Dis. 2010 May 15;50(10):1405–1414. doi: 10.1086/652151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck IA, Deng W, Payant R, et al. Validation of an Oligonucleotide Ligation Assay for Quantification of Human Immunodeficiency Virus type-1 (HIV) Drug-Resistant Mutants Using Massively Parallel Sequencing. J Clin Microbiol. 2014 Apr 16; doi: 10.1128/JCM.00306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006 Jun 1;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck IA, Crowell C, Kittoe R, et al. Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J Acquir Immune Defic Syndr. 2008 Aug 1;48(4):418–427. doi: 10.1097/QAI.0b013e31817ed7d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arvold ND, Ngo-Giang-Huong N, McIntosh K, et al. Maternal HIV-1 DNA load and mother-to-child transmission. AIDS Patient Care STDS. 2007 Sep;21(9):638–643. doi: 10.1089/apc.2006.0169. [DOI] [PubMed] [Google Scholar]
- 17.Deng W, Maust BS, Westfall DH, et al. Indel and Carryforward Correction (ICC): a new analysis approach for processing 454 pyrosequencing data. Bioinformatics. 2013 Oct 1;29(19):2402–2409. doi: 10.1093/bioinformatics/btt434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman DD, Zhou Y, Margot NA, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS. 2011 Jan 28;25(3):325–333. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- 19.Sigaloff KC, Mandaliya K, Hamers RL, et al. Short communication: High prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2012 Sep;28(9):1033–1037. doi: 10.1089/AID.2011.0348. [DOI] [PubMed] [Google Scholar]
- 20.Lihana RW, Khamadi SA, Lubano K, et al. HIV type 1 subtype diversity and drug resistance among HIV type 1-infected Kenyan patients initiating antiretroviral therapy. AIDS Res Hum Retroviruses. 2009 Dec;25(12):1211–1217. doi: 10.1089/aid.2009.0007. [DOI] [PubMed] [Google Scholar]
- 21.Hamers RL, Wallis CL, Kityo C, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: a multicentre observational study. Lancet Infect Dis. 2011 Oct;11(10):750–759. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 22.Price MA, Wallis CL, Lakhi S, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses. 2011 Jan;27(1):5–12. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan AS, Mwaringa SM, Obonyo CA, et al. Low Prevalence of Transmitted HIV Type 1 Drug Resistance Among Antiretroviral-Naive Adults in a Rural HIV Clinic in Kenya. AIDS Res Hum Retroviruses. 2013 Jan;29(1):129–135. doi: 10.1089/aid.2012.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization . Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. World Health Organization Press; Geneva: Jun, 2013. 2013. [PubMed] [Google Scholar]
- 25.Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002 Aug 8;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 26.Wittkop L, Gunthard HF, de Wolf F, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011 May;11(5):363–371. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- 27.Hamers RL, Schuurman R, Sigaloff KC, et al. Effect of pretreatment HIV-1 drug resistance on immunological, virological, and drug-resistance outcomes of first-line antiretroviral treatment in sub-Saharan Africa: a multicentre cohort study. Lancet Infect Dis. 2012 Apr;12(4):307–317. doi: 10.1016/S1473-3099(11)70255-9. [DOI] [PubMed] [Google Scholar]
- 28.Wagner TA, Kress CM, Beck I, et al. Detection of HIV-1 drug resistance in women following administration of a single dose of nevirapine: comparison of plasma RNA to cellular DNA by consensus sequencing and by oligonucleotide ligation assay. J Clin Microbiol. 2010 May;48(5):1555–1561. doi: 10.1128/JCM.02062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]