Abstract
Background
The cellular retinol binding protein I gene (CRBP) is downregulated in a subset of human breast cancers and in MMTV-Myc induced mouse mammary tumors. Functional studies suggest that CRBP downregulation contributes to breast tumor progression. What is the mechanism underlying CRBP downregulation in cancer? Here we investigated the hypothesis that CRBP is epigenetically silenced through DNA hypermethylation in human and mouse breast cancer.
Results
Bisulfite sequencing of CRBP in a panel of 6 human breast cancer cell lines demonstrated that, as a rule, CRBP hypermethylation is closely and inversely related to CRBP expression and identified one exception to this rule. Treatment with 5-azacytidine, a DNA methyltransferase inhibitor, led to CRBP reexpression, supporting the hypothesis that CRBP hypermethylation is a proximal cause of CRBP silencing. In some cells CRBP reexpression was potentiated by co-treatment with retinoic acid, an inducer of CRBP, and trichostatin A, a histone deacetylase inhibitor. Southern blot analysis of a small panel of human breast cancer specimens identified one case characterized by extensive CRBP hypermethylation, in association with undetectable CRBP mRNA and protein. Bisulfite sequencing of CRBP in MMTV-Myc and MMTV-Neu/NT mammary tumor cell lines extended the rule of CRBP hypermethylation and silencing (both seen in MMTV-Myc but not MMTV-Neu/NT cells) from human to mouse breast cancer and suggested that CRBP hypermethylation is an oncogene-specific event.
Conclusion
CRBP hypermethylation appears to be an evolutionarily conserved and principal mechanism of CRBP silencing in breast cancer. Based on the analysis of transgenic mouse mammary tumor cells, we hypothesize that CRBP silencing in human breast cancer may be associated with a specific oncogenic signature.
Background
Retinol has been implicated in the regulation of adult epithelial cell differentiation in addition to being essential for normal embryonic development, vision and reproduction [1-3]. Most of the biological effects of retinol are secondary to its metabolism to retinoic acid (RA) and RA activation of members of the RA receptor (RAR) and retinoid X receptor (RXR) family of ligand-dependent transcription factors. As such, the study of RARs and RXRs has gained widespread attention. However, a number of other genes play fundamental roles in regulating RXR-RAR function by regulating retinol and RA metabolism. Among these genes is cellular retinol-binding protein I (CRBP), which has been shown to be indispensable for life when vitamin A levels in the diet are limiting. Thus, CRBP null mice are unable to maintain adequate retinol liver stores and survive only when dietary vitamin A is abundant, a condition that is artificial and does not reflect the dietary habits of mammals in the wild [4].
In addition to being expressed in stellate cells of the liver, CRBP is expressed in many other organs and cell types including epithelia [5-7]. Several observations, including the finding that CRBP expression during mouse embryogenesis overlapped that of RARβ and RARγ, led to the suggestion that the function of CRBP is to store and release retinol where high levels of RA are developmentally required [8]. mCRBP expression is regulated by RA through a RA response element located ~1 kb upstream of the transcription start site, suggesting the existence of a positive feedback mechanism for regulating retinoid metabolism and action [9]. Glucocorticoids, cAMP, transforming growth factor β and serum factors have also been shown to impinge on CRBP expression [10-13]. The proximal hCRBP promoter region has been cloned, the transcription start site identified, and functional NF1 sites at approximately -250 and +70 demonstrated [14,15] but beyond these earlier studies there have been no functional promoter analyses.
We have recently shown that CRBP expression is downregulated in a subset of human breast cancers [7]. In follow-up studies of the potential significance of this finding, we found that the downregulation of CRBP blocks the differentiation and promotes the growth of SV40-transformed breast epithelial cells ([16] and Farias et al., manuscript submitted). The differentiation-promoting, growth-arresting effect of CRBP, witnessed in both human and mouse mammary epithelial cells, was assigned to CRBP regulation of retinol storage and RAR activation (Farias et al., manuscript submitted). These results suggested that CRBP downregulation is a contributing cause rather than a consequence of breast cancer and led us to investigate the mechanism underlying CRBP downregulation. We report here that CRBP is epigenetically silenced as a result of promoter region hypermethylation in both human and mouse breast cancer cells. Furthermore, we identify one exception to this rule and provide data that suggests that CRBP silencing is an oncogene-specific event at least in the context of mouse mammary cancer. These results extend those published by Esteller et al. while this work was in progress [17].
Results
hCRBP expression in breast cancer cells varies widely
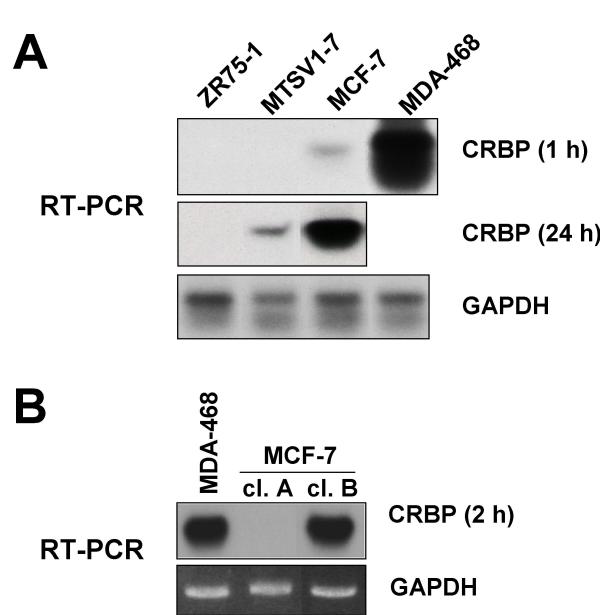
We first analyzed a panel of human breast cancer cell lines for CRBP expression using a semi-quantitative RT-PCR Southern blot protocol. CRBP expression varied from robust (MDA-MB-468), to weak (MCF-7), to very weak or undetectable (ZR75-1 and MTSV1-7; Fig. 1A). Given that CRBP downregulation occurs with an estimated frequency of 24% in human breast cancer, i.e., it is not a universal event, the identification of cell lines, such as MDA-MB-468, in which CRBP was well expressed, should not be surprising. We wondered whether the intermediate level of CRBP expression in MCF-7 cells was a consequence of clonal heterogeneity and tested this idea by random clonal selection and analysis. In support of this notion, we were able to isolate subclones that mirrored the extremes of CRBP expression, with MCF-7 clone B resembling MDA-MB-468 and MCF-7 clone A resembling ZR75-1 (Fig. 1B, compare with Fig. 1A). After densitometry and normalization to a house keeping gene, we plotted the results for all cell lines on a log scale (Figs. 2A and 3A).
Figure 1.
hCRBP expression differs widely among human breast cancer cell lines. A. RT-PCR Southern blot analysis of CRBP expression in ZR75-1, MTSV1-7, MCF-7 and MDA-MB-468 cells. B. RT-PCR Southern blot analysis of CRBP expression in MCF-7 subclones and again MDA-MB-468, to allow comparison with the cell lines studied in A. Cells were either untreated (MDA-MB-468) or treated with RA and TSA as further described in Fig. 4 (MCF-7 clone A and B cells). The data in Fig. 1 were used to generate Figs. 2A and 3A after densitometric analysis and normalization to autoradiographic exposure time (1 to 24 h) and GAPDH expression (loading control).
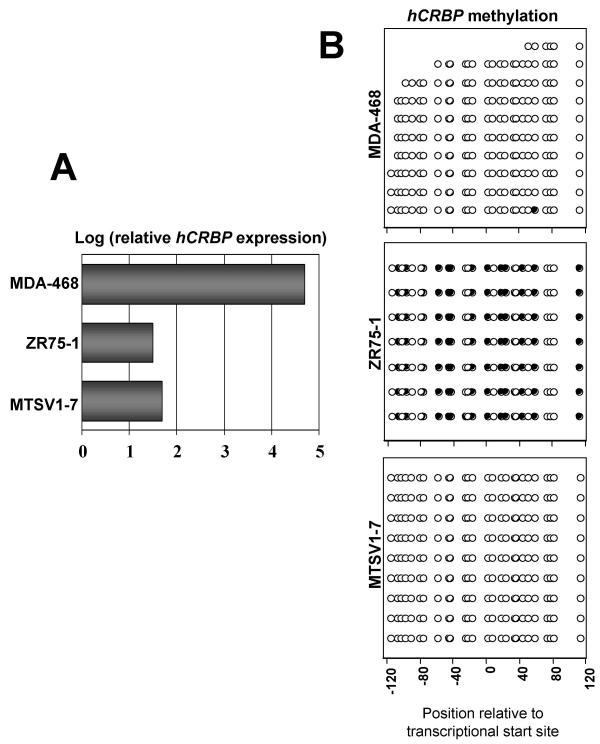
Figure 2.
hCRBP hypermethylation explains some but not all cases of hCRBP silencing. A. Relative levels of hCRBP expression determined by RT-PCR Southern analysis (see Fig. 1). The limit of detection is represented by a relative log value of 1.5 that corresponds to film background after 24 h exposure. Note that hCRBP expression in MDA-MB-468 cells is greater than that in ZR75-1 or MTSV1-7 cells by about 3 logs. B. hCRBP bisulfite sequencing results for the same cell lines as in A. Filled circles designate methylated and open circles designate unmethylated CpGs. Each row displays the methylation status of CpG sites across an individual amplicon; each "column" displays the methylation status of an individual CpG site across amplicons. The approximate position of each CpG site relative to the transcription start site is indicated in the abscissa.
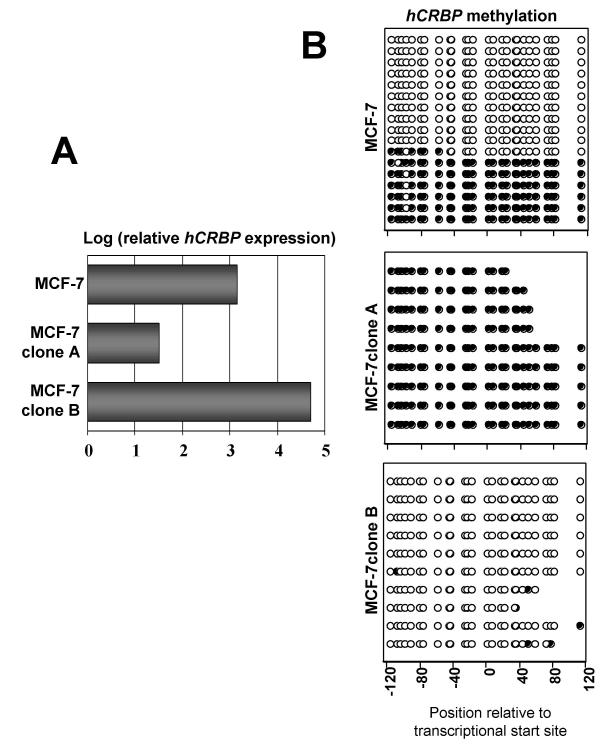
Figure 3.
hCRBP methylation explains clonal variations in hCRBP expression. A. Same as for Fig. 2A. Note that MCF-7 cells express hCRBP at a level that is intermediate to the extremes represented by MCF-7 clones A and B, which resemble in this regard ZR75-1 and MDA-MB-468 cells, respectively. B. hCRBP bisulfite sequencing results for the same cell lines as in A. Filled circles designate methylated and open circles designate unmethylated CpGs (see legend to Fig. 2B for futher details).
hCRBP methylation, with one exception, is inversely proportional to hCRBP expression
On the basis of preliminary Southern blot analyses that revealed no major hCRBP deletions or insertions in breast cancer cells (unpublished data), and taking into account the presence of a CpG island in the hCRBP regulatory region [14], we hypothesized that hCRBP downregulation might be secondary to gene hypermethylation. This hypothesis was tested by bisulfite sequencing of 26 CpG dinucleotides within the hCRBP locus (promoter and 5'-UTR region) in the cell lines whose CRBP expression we had characterized (Fig. 1). We found that all 26 sites were hypomethylated in MDA-MB-468 cells and 14 of the 26 sites were hypermethylated in ZR75-1 cells, providing the first evidence of an inverse association between CRBP methylation and expression (Fig. 2B). However, this notion was challenged by the finding that the same CRBP region was uniformly hypomethylated in MTSV1-7 cells (Fig. 2B). The bisulfite sequencing analysis of CRBP in MCF-7 parental cells and clones A and B proved very useful in that it demonstrated a very tight inverse association between CRBP methylation and expression. Thus, the intermediate level of CRBP expression in parental MCF-7 cells was associated with a mixed pattern of CRBP methylation (of 17 amplicons sequenced, 6 showed uniform hypermethylation and 10 showed uniform hypomethylation), and this mixed pattern resolved into uniform hyper or hypomethylation in clones A and B, respectively (Fig. 3B). These results suggest that, as a rule, CRBP hypermethylation is silencing, and that MTSV1-7 cells represent an exception to this rule.
Treatment with a demethylating agent rescues hCRBP expression
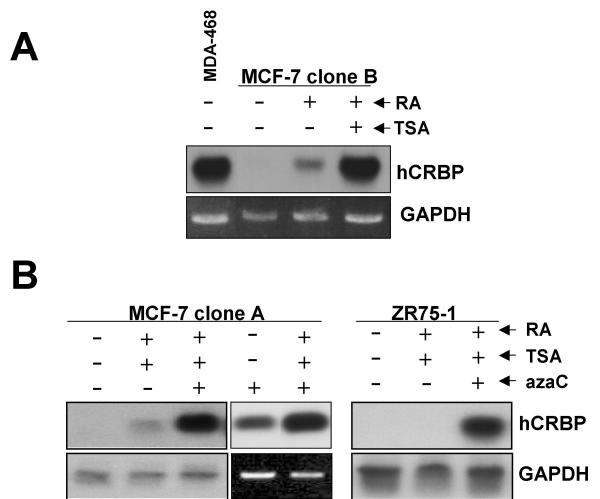
CRBP is a RA-inducible gene [9,18] and trichostatin A (TSA), a histone deacetylase inhibitor, has been shown to synergize with RAR induction of target gene transcription [19]. Cells in culture vary widely in their sensitivity to retinoids in the serum present in the culture medium; in the case of MDA-MB-468 cells (hypomethylated CRBP), there was robust CRBP expression in the absence of exogenous RA, whereas in the case of MCF-7 clone B cells (also hypomethylated CRBP) there was no CRBP expression unless exogenous RA was added and expression increased further in presence of TSA (Fig. 4A). This difference may relate to the fact that MDA-MB-468 cells are competent in RA synthesis from serum retinol whereas MCF-7 cells are not [20]. Based on the data for MCF-7 clone B cells (Fig. 4A), we predicted that treatment of MCF-7 clone A cells (hypermethylated CRBP) with 5-azacytidine (azaC), a DNA methyltransferase inhibitor, would result in some CRBP reexpression and that this effect would be potentiated by the addition of RA and TSA. We further predicted that in absence of azaC, RA and TSA would be ineffective. The data confirmed these expectations (Fig. 4B), thus supporting our hypothesis that hypermethylation is a proximal cause of CRBP silencing. In another cell line with hypermethylated CRBP (ZR75-1), reexpression could also be demonstrated to be critically dependent on azaC (Fig. 4A). Although we did not directly probe the effect of azaC on CRBP methylation, we showed earlier that the same protocol used here achieved partial demethylation of another hypermethylated gene in MCF-7 cells [21]. The work of Esteller et al. [17] suggest that our data for MCF-7 clone A is rather the exception than the rule in that azaC alone is generally sufficient to achieve CRBP reexpression when the gene is hypermethylated, suggesting that endogenous retinoids or other modulators of CRBP expression (Introduction) can drive CRBP transcription upon azaC-induced demethylation.
Figure 4.
Treatment with a demethylating agent rescues hCRBP expression in MCF-7 clone B and ZR75-1 cells. A. RT-PCR Southern blot analysis of hCRBP expression in MDA-MB-468 and MCF-7 clone B cells treated as indicated, demonstrating different sensitivity to endogenous (serum) retinoids. RA, 1 μM all-trans RA; TSA, 100 nM trichostatin A (24 h treatment). B. RT-PCR Southern analysis of hCRBP expression in MCF-7 clone A and ZR75-1 cells treated as indicated, demonstrating that a demethylating agent is required for CRBP reexpression. AzaC, 5 μM 5-azacytidine (72 h pretreatment, 96 h total); RA and TSA as above (present during last 24 h).
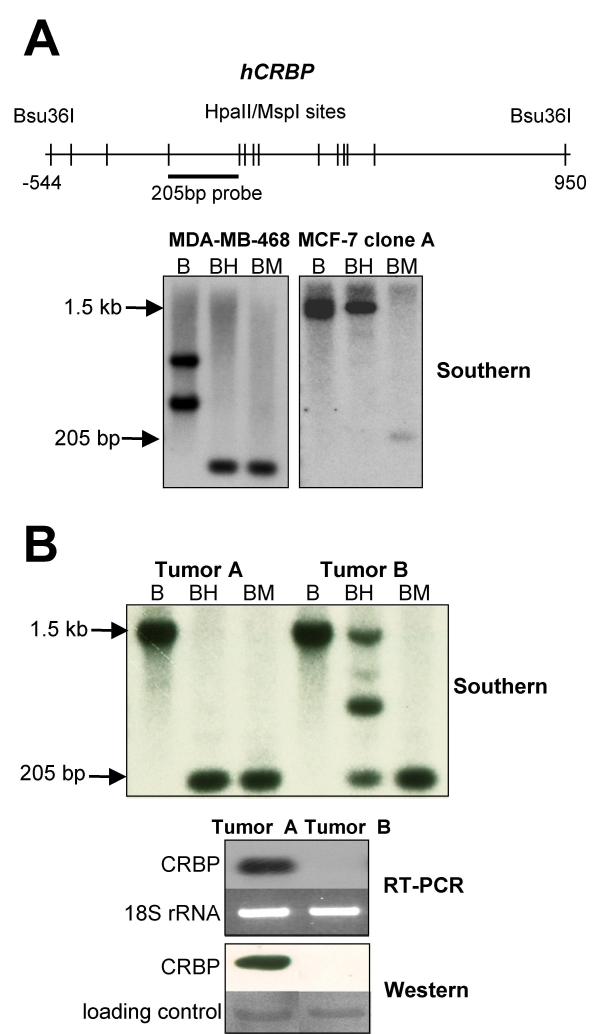
Extensive hCRBP hypermethylation in association with undetectable hCRBP in a human breast cancer specimen
Esteller et al. demonstrated by methylation-sensitive PCR (MSP) that hCRBP is hypermethylated and silenced in a subset of human breast cancers [17]. MSP is a highly sensitive technique that will detect methylation of a gene of interest even when only a small percentage of cells within the tissue have methylated alleles [22]. We wished to probe CRBP methylation using a less sensitive technique to infer whether CRBP methylation was a rare event on a per cell basis or, alternatively, was a tissue-wide event. For this purpose we used a classical Southern blot analysis approach [23] that relied on the use of methylation-sensitive isoschizomers and knowledge of the specific CpG sites that are methylated in human breast cancer cells (Figs. 2B and 3B). To test the validity of this strategy, we first applied it to MDA-MB-468 and ZR57-1 cells. Southern blotting correctly identified CRBP hypo and hypermethylation in MDA-MB-468 and ZR57-1 cells, respectively (Fig. 5A). This strategy was then used to investigate CRBP methylation in a small panel of human breast cancers (n = 10) and the results obtained for 2 cases are shown in Fig. 5B. One breast cancer specimen (tumor B) exhibited frank CRBP hypermethylation in association with low levels of CRBP mRNA, whereas tumor A did not show evidence of CRBP methylation by Southern blotting and this correlated with easily detectable CRBP mRNA levels. Based on a densitometric evaluation of the yield of the 205 bp fragment, we estimate that >50% of the cells in tumor B had at least one hypermethylated CRBP allele. When viewed in the context of the other data presented here and the work of Esteller et al. [17], this finding is of interest because it suggests that in some breast cancers there is clonal expansion of a cell population in which the CRBP gene is hypermethylated. Fig. 5B also demonstrates that there is good agreement between CRBP expression at the mRNA and protein levels. Analysis of the remaining 8 human breast cancer specimens did not identify other examples like tumor B in Fig. 5, either because of the small sample size, the low sensitivity of Southern blotting, or the low cellularity of the tumor specimen (data not shown).
Figure 5.
In some human breast cancers, hCRBP hypermethylation is widespread. A. Sketch outlining the Southern blot strategy used to evaluate hCRBP methylation in human breast cancer specimens. The strategy was tested in MDA-MB-468 and ZR75-1 cells whose hCRBP methylation status was known beforehand. B, BH and BM stand for, respectively, Bsu36I digestion alone, Bsu36I digestion followed by Hpall digestion, and Bsu36I digestion followed by MspI digestion (MspI is the methylation-insensitive isoschizomer of Hpall). In agreement with the bisulfite sequencing data, Southern blotting revealed hCRBP undermethylation in MDA-MB-468 and hypermethylation in MCF-7 clone A cells (note loss of the large Bsu36I fragment after BH digestion of MDA-MB-468 genomic DNA and retention of this fragment in the case of MCF-7 clone A). A hCRBP restriction fragment length polymorphism (an extra Bsu36I site within the 205 bp fragment) accounts for the displaced band migration pattern in MDA-MB-468 cells. B. Southern, RT-PCR and Western blotting results for 2 human breast cancer specimens. The yield of the 205 bp product was decreased by >50% after BH digestion relative to BM digestion. We infer from this that >50% of the cells in the tumor displayed hCRBP hypermethylation. The protein blot was post-stained with Ponceau S and a major band used as loading control.
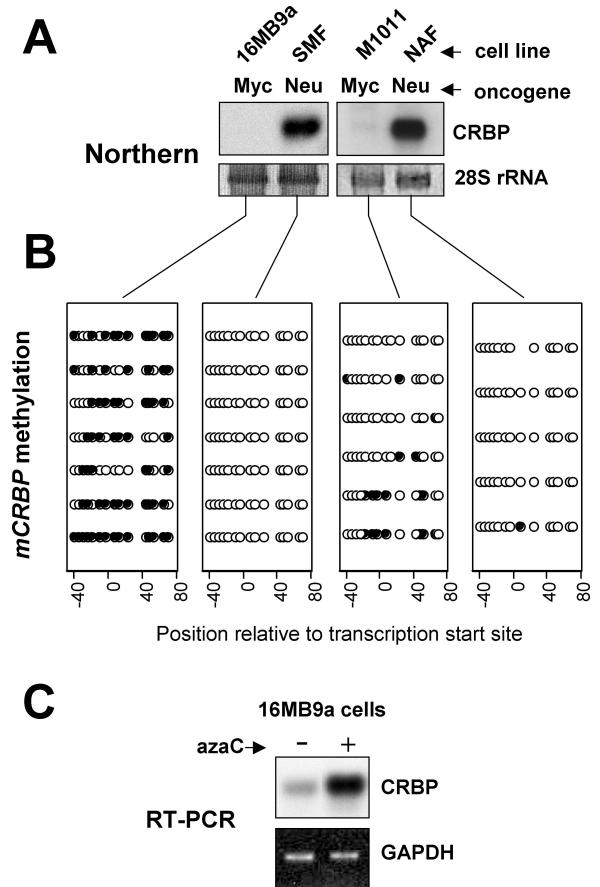
Myc but not Neu leads to mCRBP hypermethylation and silencing in mouse mammary epithelial cells
Morrison and Leder have shown that CRBP expression in transgenic mouse mammary tumors is a function of the initiating oncogene, with Myc-induced tumors lacking and Neu-induced tumors displaying CRBP expression [24]. We hypothesized that this observation might be explained by Myc but not Neu promotion of CRBP hypermethylation. To test this, we obtained two mammary tumor cell lines established from Myc and two others established from Neu-induced tumors [25] and, as a first step, confirmed that CRBP was expressed in the latter but not the former cell lines (Fig. 6A). We then analyzed CRBP methylation in the same cell lines. As predicted by our hypothesis, bisulfite sequencing of CpG dinucleotides within the mCRBP promoter and 5'-UTR region revealed extensive to moderate CRBP hypermethylation in MMTV-Myc as opposed to virtually no methylation in MMTV-Neu mammary tumor cells (Fig. 6B). As shown in Fig. 6C, treatment with azaC rescued CRBP expression in MMTV-Myc (16MB9a) cells. Thus, CRBP hypermethylation and silencing appears to represent an evolutionarily conserved event in mammary carcinogenesis. The data in Fig. 6 sets the stage for additional work investigating whether CRBP hypermethylation in human breast cancer can be associated with a specific oncogenic signature.
Figure 6.
mCRBP hypermethylation and silencing is an oncogene-specific event. A. Northern blot analysis. B. mCRBP bisulfite sequencing results for the cell lines tested in A. Each row corresponds to one sequenced amplicon. Filled circles designate methylated and open circles designate unmethylated CpGs (ee legend to Fig. 2B for futher details). C. RT-PCR demonstrating effect of azaC treatment (as in Fig. 4) on mCRBP expression in 16MB9a cells.
Discussion
Our data extend the work of Esteller et al. [17] in several respects. First, it provides an analysis of the pattern of CpG methylation across the CRBP promoter region. One interesting feature revealed by the bisulfite sequencing of CRBP in ZR75-1 and MCF-7 clone A cells, both of which express virtually no CRBP, is that 2 alternative methylation patterns are equally suppressive. Thus, in ZR75-1 cells, CRBP methylation showed intra-allelic variegation, with alternating methylated and non-methylated CpG sites (Fig. 2B). Contrast this with the uniform methylation of all CpG sites in MCF-7 clone A cells (Fig. 3B). As shown in Fig. 4, both of these patterns contributed to CRBP downregulation, suggesting that uniform methylation is not required for silencing, which is consistent with the demonstrated ability of MeCP2 to bind single methylated CpG sites [26]. CRBP methylation in MMTV-Myc mammary tumor cells defined yet another methylation pattern characterized by both intra- and inter-allelic variegation and different degrees of methylation penetrance (Fig. 6B). The conditions that lead to the establishment of these variant methylation patterns in the first place are not known.
Second, our data identified the MTSV1-7 cell line as an exception to the rule that CRBP silencing is associated with CRBP hypermethylation. The mechanism responsible for CRBP silencing in MTSV1-7 cells remains to be identified but the ease with which CRBP can be ectopically expressed in these cells from a heterologous promoter points to a defect in transcriptional initiation rather than a subsequent event [16]. In view of the association between MMTV-Myc expression and mCRBP silencing (Fig. 6), we hypothesize that c-myc overexpression may underly hCRBP silencing in MTSV1-7 cells. This possibility is supported by our finding that c-myc protein is easily detected in MTSV1-7 cells by direct Western whereas it is normally present in trace levels (Farias and Mira-y-Lopez, unpublished data). c-myc protein levels in MTSV1-7 cells may be elevated either as a result of their long-term passage in vitro [27] and/or as a result of their transduction with the SV40 small t antigen [28], which was recently shown to stabilize c-myc [29]. That possibility is further supported by the finding that immortalized mouse mammary epithelial cells lose endogenous mCRBP expression over long term in vitro passage (Farias and Mira-y-Lopez, unpublished data). On the other hand, the fact that MMTV-Myc is associated with mCRBP hypermethylation (Fig. 6) but hCRBP was not hypermethylated in MTSV1-7 cells (Fig. 2B) argues against our idea that c-myc overexpression underlies CRBP silencing in MTSV1-7 cells. However, hypermethylation has been suggested to represent a mechanism for the maintenance not the initial induction of transcriptional silencing [30]. Therefore, it is conceivable that the hCRBP locus in MTSV1-7 cells is repressed in a c-myc-dependent manner and is susceptible to but has not yet undergone hypermethylation.
Third, based on a small sample of 10 human breast cancer specimens, we succeeded in identifying one instance of widespread CRBP hypermethylation and silencing. Thus, hypermethylation was easily detected in total tumor DNA using a low sensitivity technique (Southern blotting) and this correlated with the lack of any detectable CRBP mRNA and protein in total tumor RNA and tumor lysates, respectively. Although only a single such tumor was identified, it seems reasonable to extrapolate, based on the strength of the combined evidence ([17] and this report), that a small subset of human breast cancers are characterized by pervasive CRBP methylation.
Fourth, we have found that mCRBP methylation is associated with Myc but not Neu-induced mammary tumors, suggesting that mCRBP silencing may be a requirement for MMTV-Myc tumorigenesis. We plan to test this hypothesis by monitoring CRBP expression and methylation by in situ hybridization and MSP, respectively, over the period of time during which premalignant changes accumulate in MMTV-Myc mammary tissue. We expect to find that CRBP downregulation precedes or coincides with the appearance of preneoplasia. We further expect that ectopic CRBP will prevent or delay the onset of mammary tumorigenesis. One theoretical argument against our hypothesis is that MMTV-Myc may transform a subpopulation of cells that do not express CRBP to begin with. We find this unlikely because CRBP mRNA is uniformly expressed in both layers of human breast ducts, i.e., the epithelium itself and the underlying myoepithelium [7]. Interestingly, MMTV-Myc tumors were recently identified as having both epithelial and myoepithelial components [31], suggesting that MMTV-Myc transforms a pluripotent cell population. For the reason just stated above, we hypothesize that this population expresses CRBP.
Finally, our study of CRBP methylation in mouse mammary tumor cell lines derived from Myc and Neu-induced tumors provides a hypothetical explanation for why CRBP hypermethylation is characteristic of only a subset of human breast cancers. It should be noted that CRBP is not unique in this regard, i.e., several other genes shown to be epigenetically silenced in breast cancer are silenced in a subset of, not all breast cancers [32]. The data in Fig. 6 suggests that subset-specific CRBP silencing might be explained by the nature of the initiating oncogenic event. Specifically, we propose that most human breast cancers in which c-myc is amplified and/or overexpressed might be characterized by CRBP hypermethylation and, conversely, most breast cancers with c-neu amplification and/or overexpression might be characterized by CRBP hypomethylation. Interestingly, the incidence of c-myc amplification and CRBP hypermethylation in breast cancer have both been estimated at 16% [17,33]. If further work were to validate this notion, then it would be of interest to understand how c-myc overexpression leads to CRBP silencing and how c-neu bypasses the anti-tumor function of CRBP. The finding of other defects in retinol and RA metabolism in breast cancer as well as alterations in RAR isoform expression suggest that several bypassing mechanisms may exist [34-37].
Conclusions
We conclude that: (i) CRBP hypermethylation is, as a rule, a proximal cause of CRBP silencing in a subset of human and mouse breast cancers; (ii) exceptions to this rule may exist in some human breast cancers, as indicated by our results for the MTSV1-7 cell line, in which CRBP is silenced despite being hypomethylated; (iii) the intra-allelic pattern of CRBP hypermethylation may vary and tumors may consist of mixtures of cells with hyper and hypomethylated CRBP, but in at least some cases most tumor cells (>50%) display CRBP hypermethylation; (iv) at least in the context of mouse mammary cancer, CRBP hypermethylation appears to be an oncogene-specific event. The significance of these findings should be viewed against the background of our functional analyses of CRBP, which suggest that CRBP silencing is a contributing cause rather than a consequence of breast carcinogenesis [16].
Methods
Cells and reagents
Human breast cancer cell lines, with the exception of MTSV1-7 cells [16,28], were obtained from the American Type Culture Collection (Rockville, MD) and grown as recommended. 16MB9a and M1011 (MMTV-Myc) and SMF and NAF (MMTV-Neu/NT) mammary tumor cells were cultured as described [24]. Gel purified primers were obtained from Genelink (Hawthorne, NY). RA, TSA and azaC were obtained from Sigma (St.Louis, MO). Sodium metabisulfite was obtained from BDH, England.
RNA analyses
For RT-PCR, 2 μg total RNA (Purescript kit, Gentra, Minneapolis, MN) were reverse transcribed using the Superscript Preamplification System (Gibco BRL Life Technologies, Grand Island, NY). This RNA input was chosen after preliminary experiments demonstrated a dose-dependent increase in CRBP RT-PCR product within the range of 0.5 to 4 μg total RNA. The PCR mixture consisted of 10X PCR Buffer, 1 mM MgCl2, 200 μM dNTPs, 0.4 μM each primer, 10% cDNA, and 5U of Taq (Promega, Madison, WI) in a total volume of 50 μl. hCRBP cDNA (320 bp) was amplified with sense primer TTGTGGCCAAACTGGCTCCA and antisense primer ACACTGGAGCTTGTCTCCGT and the following cycling parameters: 94 C for 5 min; 26 cycles of 94 C for 1 min, 48 C for 1 min, 72 C for 1 min; 72 C for 5 min. 18S rRNA (489 bp) was amplified using the same protocol as above except that 5% cDNA, a 3:7 mix of primers:competimers (0.4 μM total), and an annealing temperature of 57°C were used (18S rRNA primers and competimers were from Ambion, Austin, TX). GAPDH RT-PCR was performed as described [38]. The PCR products were separated on 1% agarose, stained with ethidium bromide (18S rRNA) or blotted overnight and probed with full length CRBP cDNA or a commercial GAPDH cDNA fragment (Ambion). Northern blot analyses were performed using standard methods and a full length CRBP cDNA probe as described earlier [7].
Bisulfite sequencing
Genomic DNA was isolated using the Puregene kit (Gentra Minneapolis, MN) and digested with XbaI. The bisulfite method established by Frommer et al. [39,40] was carried out on 5 μg genomic DNA digest with the following modifications. Digested DNA was denatured with 0.5 M NaOH at 75 C for 15 min, followed by deamination with freshly prepared 4.0 M, pH 5.0 sodium metabisulfite and incubated at 55 C for 8 hours in a total volume of 1.2 ml under mineral oil. The bisulfite-treated DNA was then desalted using the Wizard DNA clean-up system (Promega), desulfonated with 0.3 M NaOH at 37 C for 15 min, neutralized with 0.1 volume of 3 M NH4AC and ethanol precipitated with 2 volumes of cold ethanol. The pellet was resuspended in 50 μ1 of sterile H2O. A 780 bp hCRBP fragment was amplified using 2 rounds of fully-nested PCR, which in our experience was required to yield reproducible results. First round PCR was carried out in a 50 μl reaction mixture of 10X PCR buffer, 1.2 mM MgCl2, 160 μM dNTPs, 0.4 μM each primer, 10 μl of bisulfite-treated DNA, and 5 U of Taq (Promega). The nested PCR reactions (rounds 2 and 3) were carried out in the same way using 5% of the product obtained in the preceding round as template. The primers used were as follows: 1st round PCR, GTTGAATTTTTTAGTTTTTTTGATTTTAGT (sense) and ACACCAAAAAATTAACAAAAAAACT (antisense); 2nd round, AGGTTTTAGATAAAGGTTTGTAAGTGG (sense) and TCCCAAAATAACAAAACCCCAAAAA (antisense); 3rd round, GTTTTTTAATTTTTGAGTGGTTGT (sense) and ATCTACAACCTAAAAACTACCCTAAAA (antisense). Cycling parameters were: 94 C for 5 min; 40 (round 1) or 20 cycles (rounds 2 and 3) of 94 C for 1 min, 45 C (rounds 1 and 3) or 50 C (round 2) for 1 min, 72 C for 1 min; 72 C for 5 min. The amplified PCR product was gel purified, subcloned into the TA cloning vector (Invitrogen, Faraday, CA), transformed into TOPO kit cells (Invitrogen) using a blue-white colony X-gal selection, and individual clones submitted for sequencing. mCRBP bisulfite sequencing was performed essentially as described above for hCRBP but using semi-nested PCR and the following primers and conditions: 1st round, TGAAGGATTTTTAAGGGAAGTAAGGAG (sense) and AAATTCTCATTACTCAACATCTTCCAATAC (antisense), 40 cycles, annealing at 52 C; 2nd round, ATCCACAAACATTTTTAAAACAAACTA (sense) and same antisense primer as above, 42 cycles, annealing at 46 C.
Southern blot analysis
Human breast carcinoma specimens that had been flash frozen within 1 h of surgery were obtained through the Cooperative Human Tissue Network http://www.chtn.ims.nci.nih.gov under Institutional Review Board approval. All specimens were diagnosed as invasive breast carcinoma, with or without a carcinoma in situ component, except for one specimen which was an adenoid cystic breast carcinoma (not shown in Fig. 5). Approximately 200 mg of tissue were pulverized under liquid nitrogen and RNA and DNA isolated using Qiagen's RNA/DNA kit according to manufacturer's instructions (Qiagen, Valencia, CA). Tumor DNA (45 μg) was digested with Bsu36I, ethanol precipitated, and divided into 3 parts that were either not digested further, digested with HpaII, or digested with MsPI (restriction enzymes were from New England Biolabs, Beverly, MA). After ethanol precipitation, Southern blots were prepared and hybridized to a 205 bp hCRBP probe amplified from a cloned hCRBP genomic fragment using CCGGTCTCCTCTTCCTTTGTAGGGG (sense) and GGGACAGGGGGCTCTGCGGGG (antisense) primers.
Immunoblot analysis
A small amount of pulverized tissue was homogenized in 10 mM Tris.Cl pH 7.4, 1 mM EDTA, 7 μM beta-mercaptoethanol, the homogenate was centrifuged at 10,000 × g for 10 min, and the supernatant centrifuged further at 100,000 × g for 30 min to yield cytosol. Protein concentration was analyzed using the method of Bradford [41] with Bio-Rad reagents (Bio-Rad, Hercules, CA) and 50 μg protein were electrophoresed per lane in a 15% sodium dodecyl sulfate-polyacrylamide gel. After blotting, CRBP protein was visualized by reaction with anti-CRBP antibody followed by secondary antibody and enhanced chemiluminescence detection. The anti-CRBP antibody was raised in rabbits against a keyhole limpet hemocyanin-conjugated CRBP-specific peptide [42] and was characterized elsewhere [16].
List of abbreviations
CRBP cellular retinol binding protein type I
MMTV mouse mammary tumor virus
RA retinoic acid
RAR RA receptor
RXR retinoid x receptor
RT-PCR reverse transcription-coupled polymerase chain reaction
ER estrogen receptor
TSA trichostatin A
azaC 5-azacytidine
MSP methylation-sensitive PCR
Authors' contributions
AA established the bisulfite sequencing protocol for hCRBP, applied it to most of the human breast cancer cell lines tested, generated data toward Figs. 1 and 4, established the Southern blotting strategy used in Fig 5 and generated the data in Fig. 5A. SB performed the analysis of hCRBP methylation in MTSV1-7 cells, extended the bisulfite sequencing method to mCRBP (Fig. 6), and generated data toward Figs. 1,4 and 6C. YSK established the semi-quantitative CRBP RT-PCR method and contributed toward Fig. 1. SN generated the CRBP peptide antibody used in the experiment of Fig. 5. RML initiated and supervised all aspects of the project, helped perform some of the experiments, and wrote the manuscript.
Acknowledgments
Acknowledgements
We thank the following investigators for reagent gifts: Joyce Taylor-Papadimitriou (MTSV1-7 cells), Phil Leder (16MB9a and SMF cells), Pierre Chambon (mCRBP cDNA) and Winnie Eskild (hCRBP cDNA). Tissue samples were provided by the Cooperative Human Tissue Network which is funded by the National Cancer Institute. Other investigators may have received specimens from the same subjects. This work was supported by NCI grant CA54273 to RML, the Samuel Waxman Cancer Research Foundation, the Norman and Rosita Winston Foundation and the Chemotherapy Foundation.
Contributor Information
Alice Arapshian, Email: aa74ny@yahoo.com.
Silvina Bertran, Email: silvina.bertran@mssm.edu.
Yuvarani S Kuppumbatti, Email: ykuppumb@uci.edu.
Shigeo Nakajo, Email: nakajo@pharm.showa-u.ac.jp.
Rafael Mira-y-Lopez, Email: rafael.mira@mssm.edu.
References
- Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–54. [PubMed] [Google Scholar]
- Wolbach SB, Howe PR. Tissue changes following deprivation of fat soluble A vitamin. J Exp Med. 1925;42:753–777. doi: 10.1084/jem.42.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN, Howell JM, Pitt GAJ. Vitamin A and reproduction in rats. Proc Royal Soc. 1964;159:510–535. doi: 10.1098/rspb.1964.0017. [DOI] [PubMed] [Google Scholar]
- Ghyselinck NB, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Hakansson H, Sauvant P, et al. Cellular retinol-binding protein I is essential for vitamin A homeostasis. Embo J. 1999;18:4903–14. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D. The retinal pigment epithelium: a versatile partner in vision. J Cell Sci Suppl. 1993;17:189–95. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- Ylikoski J, Pirvola U, Eriksson U. Cellular retinol-binding protein type I is prominently and differentially expressed in the sensory epithelium of the rat cochlea and vestibular organs. J Comp Neurol. 1994;349:596–602. doi: 10.1002/cne.903490407. [DOI] [PubMed] [Google Scholar]
- Kuppumbatti YS, Bleiweiss IJ, Mandeli JP, Waxman S, Mira-y-Lopez R. Cellular retinol-binding protein expression and breast cancer. J Natl Cancer Inst. 2000;92:475–80. doi: 10.1093/jnci/92.6.475. [DOI] [PubMed] [Google Scholar]
- Ruberte E, Dolle P, Chambon P, Morriss-Kay G. Retinoic acid receptors and cellular retinoid binding proteins. II. Their differential pattern of transcription during early morphogenesis in mouse embryos. Development. 1991;111:45–60. doi: 10.1242/dev.111.1.45. [DOI] [PubMed] [Google Scholar]
- Smith WC, Nakshatri H, Leroy P, Rees J, Chambon P. A retinoic acid response element is present in the mouse cellular retinol binding protein I (mCRBPI) promoter. Embo J. 1991;10:2223–30. doi: 10.1002/j.1460-2075.1991.tb07758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskild W, Oyen O, Beebe S, Jahnsen T, Hansson V. Regulation of mRNA levels for cellular retinol binding protein in rat Sertoli cells by cyclic AMP and retinol. Biochem Biophys Res Commun. 1988;152:1504–10. doi: 10.1016/s0006-291x(88)80456-x. [DOI] [PubMed] [Google Scholar]
- Kroepelien CF, Knutsen HK, Haugen TB, Hansson V, Eskild W. Serum factors induce messenger ribonucleic acid levels for cellular retinol-binding protein in rat Sertoli cells. Endocrinology. 1993;132:968–74. doi: 10.1210/en.132.3.968. [DOI] [PubMed] [Google Scholar]
- Rush MG, Ul-Haq R, Chytil F. Opposing effects of retinoic acid and dexamethasone on cellular retinol-binding protein ribonucleic acid levels in the rat. Endocrinology. 1991;129:705–9. doi: 10.1210/endo-129-2-705. [DOI] [PubMed] [Google Scholar]
- Xu G, Bochaton-Piallat ML, Andreutti D, Low RB, Gabbiani G, Neuville P. Regulation of alpha-smooth muscle actin and CRBP-1 expression by retinoic acid and TGF-beta in cultured fibroblasts. J Cell Physiol. 2001;187:315–25. doi: 10.1002/jcp.1078. [DOI] [PubMed] [Google Scholar]
- Nilsson MH, Spurr NK, Lundvall J, Rask L, Peterson PA. Human cellular retinol-binding protein gene organization and chromosomal location. Eur J Biochem. 1988;173:35–44. doi: 10.1111/j.1432-1033.1988.tb13963.x. [DOI] [PubMed] [Google Scholar]
- Eskild W, Simard J, Hansson V, Guerin SL. Binding of a member of the NF1 family of transcription factors to two distinct cis-acting elements in the promoter and 5'-flanking region of the human cellular retinol binding protein 1 gene. Mol Endocrinol. 1994;8:732–45. doi: 10.1210/me.8.6.732. [DOI] [PubMed] [Google Scholar]
- Kuppumbatti YS, Rexer B, Nakajo S, Nakaya K, Mira-y-Lopez R. CRBP suppresses breast cancer cell survival and anchorage-independent growth. Oncogene. 2001;20:7413–9. doi: 10.1038/sj.onc.1204749. [DOI] [PubMed] [Google Scholar]
- Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, Baylin SB, Herman JG. Hypermethylation-associated Inactivation of the Cellular Retinol-Binding-Protein 1 Gene in Human Cancer. Cancer Res. 2002;62:5902–5. [PubMed] [Google Scholar]
- Fisher GJ, Reddy AP, Datta SC, Kang S, Yi JY, Chambon P, Voorhees JJ. All-trans retinoic acid induces cellular retinol-binding protein in human skin in vivo. J Invest Dermatol. 1995;105:80–6. doi: 10.1111/1523-1747.ep12313352. [DOI] [PubMed] [Google Scholar]
- Minucci S, Horn V, Bhattacharyya N, Russanova V, Ogryzko VV, Gabriele L, Howard BH, Ozato K. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1997;94:11295–300. doi: 10.1073/pnas.94.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira-y-Lopez R, Zheng WL, Kuppumbatti YS, Rexer B, Jing Y, Ong DE. Retinol conversion to retinoic acid is impaired in breast cancer cell lines relative to normal cells. J Cell Physiol. 2000;185:302–9. doi: 10.1002/1097-4652(200011)185:2<302::AID-JCP15>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Farias EF, Arapshian A, Bleiweiss IJ, Waxman S, Zelent A, Mira-y-Lopez R. Retinoic acid receptor alpha2 is a growth suppressor epigenetically silenced in MCF-7 human breast cancer cells. Cell Growth Differ. 2002;13:335–41. [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J, Roberts-Ems J, Riggs AD. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979;203:1019–21. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Morrison BW, Leder P. neu and ras initiate murine mammary tumors that share genetic markers generally absent in c-myc and int-2-initiated tumors. Oncogene. 1994;9:3417–26. [PubMed] [Google Scholar]
- Morrison AJ, Herrera RE, Heinsohn EC, Schiff R, Osborne CK. Dominant Negative N-CoR Relieves Transcriptional Inhibition of Retinoic Acid Receptor but Does Not Alter the Agonist/Antagonist Activities of the Tamoxifen-bound Estrogen Receptor. Mol Endocrinol. 2003. [DOI] [PubMed]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–81. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Wang J, Hannon GJ, Beach DH. Risky immortalization by telomerase. Nature. 2000;405:755–6. doi: 10.1038/35013171. [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Kyprianou N, Lalani EN, Staskova Z, Shearer M, Chang S, Taylor-Papadimitriou J. Efficient immortalization of luminal epithelial cells from human mammary gland by introduction of simian virus 40 large tumor antigen with a recombinant retrovirus. Proc Natl Acad Sci U S A. 1991;88:3520–4. doi: 10.1073/pnas.88.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305. doi: 10.1016/S1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–8. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8:115–27. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–95. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ruiz A, Rando RR, Bok D, Gudas LJ. Esterification of all-trans-retinol in normal human epithelial cell strains and carcinoma lines from oral cavity, skin and breast: reduced expression of lecithin :rctinol acyltransferase in carcinoma lines. Carcinogenesis. 2000;21:1925–33. doi: 10.1093/carcin/21.11.1925. [DOI] [PubMed] [Google Scholar]
- Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF-7 cells. Cancer Res. 2001;61:7065–70. [PubMed] [Google Scholar]
- Triano EA, Slusher LB, Atkins TA, Beneski JT, Gestl SA, Zolfaghari R, Polavarapu R, Frauenhoffer E, Weisz J. Class I Alcohol Dehydrogenase Is Highly Expressed in Normal Human Mammary Epithelium but not in Invasive Breast Cancer: Implications for Breast Carcinogenesis. Cancer Res. 2003;63:3092–100. [PubMed] [Google Scholar]
- Chen LI, Sommer KM, Swisshelm K. Downstream codons in the retinoic acid receptor beta -2 and beta -4 mRNAs initiate translation of a protein isoform that disrupts retinoid-activated transcription. J Biol Chem. 2002;277:35411–21. doi: 10.1074/jbc.M202717200. [DOI] [PubMed] [Google Scholar]
- Arapshian A, Kuppumbatti YS, Mira-y-Lopez R. Methylation of conserved CpG sites neighboring the beta retinoic acid response element may mediate retinoic acid receptor beta gene silencing in MCF-7 breast cancer cells. Oncogene. 2000;19:4066–70. doi: 10.1038/sj.onc.1203734. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Busch C, Sakena P, Funa K, Nordlinder H, Eriksson U. Tissue distribution of cellular retinol-binding protein and cellular retinoic acid-binding protein: use of monospecific antibodies for immunohistochemistry and cRNA for in situ localization of mRNA. Methods Enzymol. 1990;189:315–24. doi: 10.1016/0076-6879(90)89303-Y. [DOI] [PubMed] [Google Scholar]