Abstract
We have carried out a global survey of age-related changes in mRNA levels in the C57BL/6NIA mouse hippocampus and found a difference in the hippocampal gene expression profile between 2-month-old young mice and 15-month-old middle-aged mice correlated with an age-related cognitive deficit in hippocampal-based explicit memory formation. Middle-aged mice displayed a mild but specific deficit in spatial memory in the Morris water maze. By using Affymetrix GeneChip microarrays, we found a distinct pattern of age-related change, consisting mostly of gene overexpression in the middle-aged mice, suggesting that the induction of negative regulators in the middle-aged hippocampus could be involved in impairment of learning. Interestingly, we report changes in transcript levels for genes that could affect synaptic plasticity. Those changes could be involved in the memory deficits we observed in the 15-month-old mice. In agreement with previous reports, we also found altered expression in genes related to inflammation, protein processing, and oxidative stress.
The hippocampal formation is one of the brain areas most affected by aging. Age-related defects in hippocampal function are found both in humans and other mammalian species. Memory deficits that are associated with aging, particularly impairment in the recall of declarative memory, are accompanied by altered synaptic plasticity in the hippocampus (Geinisman et al. 1995; Foster 1999). In normal elderly subjects, memory decline is correlated with hippocampal dysfunction (Small et al. 2002). Both in humans and rodents, the hippocampus is involved in spatial memory (Morris et al. 1982; Burgess et al. 2002), and age-related defects in spatial memory correlate with impairment of late phase hippocampal long-term potentiation (LTP; Bach et al. 1999).
There is now considerable evidence supporting the view that normal age-related memory deficits can be caused by functional changes, without the occurrence of major structural alterations such as neuronal loss (West et al. 1994; Rapp et al. 2002). It thus seems likely that functional alterations may precede structural damage in both normal aging and age-related diseases. Given that molecular mechanisms thought to underlie learning are dependent on mRNA synthesis (Dash et al. 1990; Igaz et al. 2002) and that potentially many biochemical processes affecting synaptic plasticity can be regulated at the transcriptional level, altered hippocampal function may involve specific changes in transcript levels. To test this idea, we studied changes in gene expression that may correlate with the earlier stages of age-related memory decline.
Here we report a comparison of cognitive assessment of hippocampal-dependent learning between middle-aged mice and young mice in the Morris water maze and a microarray analysis in the same animals of hippocampal gene expression.
To reduce the problems that come with using ad hoc criteria for gene selection and to have sufficient statistical power to detect small changes in expression, we applied stringent statistical methods to select genes and estimate false positive rates. Detecting small differences in expression is important for the study of heterogeneous tissues such as the hippocampus, because changes occurring in only one type of cell or limited to a certain subregion will appear as very small expression differences in samples obtained from bulk tissue (Zhao et al. 2001).
By using a conservative behavioral analysis that takes into account the confounding effect of age-related differences in swimming speed, our study reveals an incipient age-related cognitive deficit (ACD). A high proportion of the genes whose expression we found altered in the middle-aged hippocampus have been implicated in the pathogenesis of Alzheimer's disease (AD). Some of our gene expression findings reproduce earlier findings, in particular immunoinflammatory response genes and oxidative stress-related genes; however, our statistical methods place a high confidence on a much smaller number of genes than identified in a previous study of aging mouse hippocampus (Terao et al. 2002). Finally, we report a number of novel gene expression changes, many of which involve genes that could be involved in synaptic plasticity and could therefore underlie the observed ACD.
RESULTS AND DISCUSSION
Behavioral Analysis
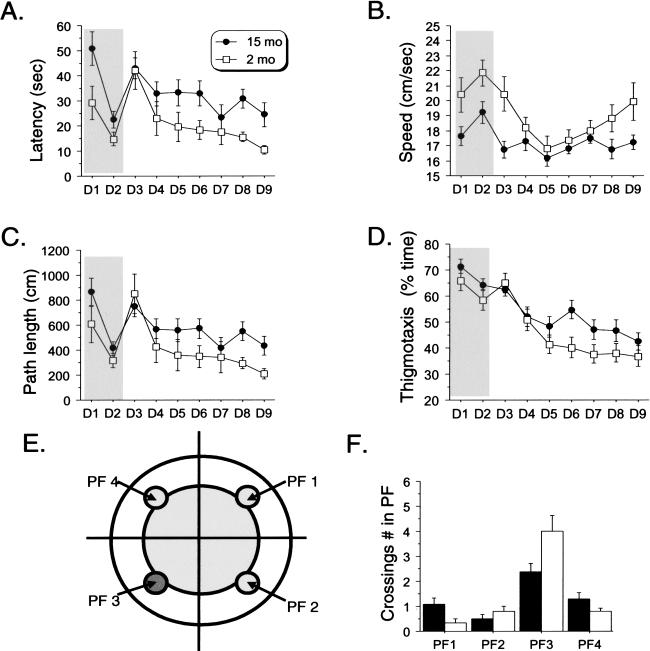
We first evaluated the effect of age on hippocampal-based declarative memory formation. We compared performance of young mice (2-month-old) and middle-aged mice (15-monthold; mean life span of C57BL/6 mice = 26 to 28 months; Jucker and Ingram 1997) in the Morris water maze. Both middle-aged and young mice learned this task, showing a decrease in their latency to reach the platform across both phases of the task. However, middle-aged mice had longer latencies throughout the experiment (Fig. 1A).
Figure 1.
Middle-aged mice display impaired spatial memory in the Morris Water maze. (A) Mean escape latency per day across days. Both middle-aged and young mice decreased their escape latency across both the visible (days 1 and 2) and hidden (days 3 through 9) versions of the task (significant day effect; all P < 0.001). Middle-aged mice showed lower latencies in both versions (visible: [F(1,21) = 5.00, P = -0.03]; Hidden: [F(1, 21) = 6.74, P = -0.02]). (B) Swimming speed. Significant effect of age [F(1, 21) = 7.39, P = -0.013], day [F(1, 8) = 7.28, P < 0.0001], and the interaction age × day [F(8, 168) = 2.12, P = 0.037]. (C) Path length. Both groups learned to localize the platform across both versions of the task (significant day effect: all P > 0.001). However, middle-aged mice showed impaired performance in the hidden version (days 4 through 9; significant Age effect: [F(1, 21) = 5.47, P = 0.03]), but not in the visible version (days 1 and 2 [F(1, 21) = 2.43, P = 0.13]). (D) Thigmotaxis. Significant age effect (days 4 through 9, [F(1, 21) = 5.22, P = 0.03]). (E) Scheme depicting platform location (dark grey circle in quadrant 3) and equivalent locations in the other quadrants. (F) Probe trial: crossings on previous exact spatial position of the platform (PF in quadrant 3). Middle-aged mice displayed lower performance (significant age effect: [F(1, 21) = 6.06, P = 0.02])
Although these data are consistent with a deficit in spatial memory in the older animals, latency may be a misleading measure of learning, as any nonspecific effect of age on swimming performance will be a confounding factor. Indeed, the middle-aged mice show overall lower swimming speeds, which were nearly constant across the entire experiment (Fig. 1B). However, there is a strong suggestion from the data that this cannot be entirely explained by motor or visual performance differences compared with young animals. Specifically, young mice had a V-shaped evolution of their speed across days of training, being faster during sessions 1 through 3 and then also during the last sessions (Fig. 1B). The decrease in swim speed during the first sessions of the spatial training (days 4 and 5) may reflect “hesitation” corresponding to a “mental” reorganization, found to be associated with the acquisition of a learned task (Teule et al. 1972; Amsel 1993; Malleret et al. 1999). This was not observed in middle-aged animals, as their swim speed remained constant across the entire experiment and equal to the swim speed of young animals during the acquisition of the spatial training. One interpretation of this finding is that middle-aged mice show a continual hesitation to solve the task. In addition, we did not find any difference in overall motivation, stress, or anxiety generated by the task in the middle-aged mice, as measured by the percentage of time spent floating (data not shown). In any case, by using only latency as a performance measure, we can make no specific conclusions as to the nature of the deficit in middle-aged mice.
As an alternative to latency, we analyzed the length of the animals' trajectories to reach the platform (path length), a more accurate measure of performance in the Morris water maze task (Gallagher and Nicolle 1993). Both groups of animals learned to find the platform, as evidenced by the decrease in path length across training sessions (Fig. 1C). However, the statistical analysis strongly supports a decrease in spatial learning performance in the middle-aged animals. Specifically, there was a significant effect of age on path length when the platform was hidden. There was no difference in path length when the platform was visible, indicating that middle-aged mice do not display any vision or motivational deficit compared with young mice. The deficit observed is thus most likely cognitive in nature.
Interestingly, this increase in path length with age was accompanied by an increase in the time spent in the peripheral zone of the pool (thigmotaxis; Fig. 1D). We must consider lower stamina as a possible explanation of augmented thigmotaxis; however, this does not seem to be the case because no increase in thigmotaxis was observed during the visible version of the task or in the beginning of the spatial training. Rather, increased thigmotaxis would indicate that middle-aged mice, perhaps because of hippocampal impairment, used a procedural (striatum-dependent) strategy to locate the platform rather than a spatial (hippocampal-dependent) one (Wolfer et al. 1998). Therefore, middle-aged mice would still be able to find the platform in a more simple way by swimming in circles, a few centimeters away from the edge of the pool (Fig. 1E, white area), without knowing the exact position of the goal (Fig. 1E, dark grey area). If this idea is correct, we would expect a poorer performance of the middle-aged animals in the probe trial that tests memory for the precise spatial location of the platform. Indeed, middle-aged mice showed significantly fewer platform field crossings than did young mice (Fig. 1F; number of crossings in PF 3: aged, 2.3 ± 1.33; young, 4.0 ± 1.87).
To summarize the behavioral analysis, we found that middle-aged mice have a mild and very specific deficit in hippocampal-dependent spatial learning assessed by several measures (path length, thigmotaxis, platform crossings in probe trial).
Transcript Profiling
We next investigated gene expression changes in the hippocampus associated with age and cognitive deficits by using oligonucleotide microarrays. Hippocampal tissue from each animal studied behaviorally was assayed independently, yielding a data set with 23 arrays (9 young and 14 aged mice).
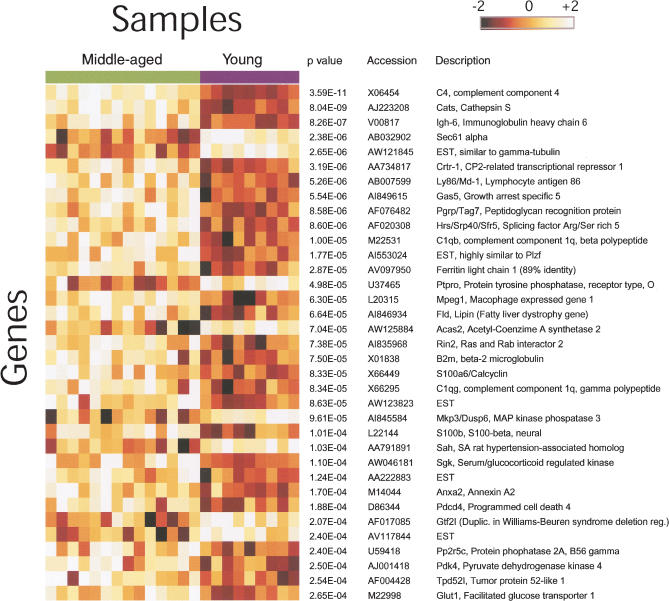
Statistical testing revealed a clear effect of age on gene expression associated with the incipient ACD (Fig. 2). By controlling the false discovery rate (FDR) at a level of 0.1 (P < 0.00026), we found 35 transcripts showing differences in mRNA level between the middle-aged and control groups. This P-value threshold is expected to yield four false positives experiment-wide (18). At a more stringent FDR (0.05, P < 0.000086, allowing for only one expected false positive experiment-wide), we identified 22 genes that show differential expression (Fig. 2, top of the list). We used real-time PCR to confirm these results for five of the most significantly changed genes, in order to show that the GeneChip analysis yields reliable results (Table 1). β-Actin, a gene with expression that showed no significant change between the two groups (P = 0.7), was chosen as a normalization reference for real-time PCR.
Figure 2.
Age-related changes in gene expression in the mouse hippocampus. Summary of the 35 genes most significantly changed in expression between young (2 months) and middle-aged (15 months) mice. The genes are listed in order of increasing P-value. Each gene is visualized as a row of colored squares, with one square for each sample. The color indicates the relative expression of the gene according to the scale bar, with lighter colors indicating higher levels of expression. Expression levels ±2 SD from the mean of the gene are depicted as white and black, respectively. Note that the majority of genes listed showed increased expression with age. Annotations for each gene and the Welch t-test P-value (uncorrected for multiple testing) are shown at the right of the figure.
Table 1.
Confirmation of Differential Gene Expression by Real-Time PCR (RT PCR)
| Gene | RT PCR δ Ct′ | RT PCR fold change | GeneChip fold change |
|---|---|---|---|
| C4 | 1.48 ± 0.28 | 2.8 ± 0.5 | 2.1 ± 0.2 |
| C1qb | 0.80 ± 0.28 | 1.7 ± 0.3 | 1.3 ± 0.1 |
| Cats | 0.83 ± 0.30 | 1.8 ± 0.4 | 1.4 ± 0.1 |
| Gas5 | 0.68 ± 0.32 | 1.6 ± 0.3 | 1.2 ± 0.03 |
| Crtr-1 | 0.94 ± 0.47 | 1.9 ± 0.6 | 1.7 ± 0.2 |
Ct values were normalized by subtracting Ctactin for each sample and then averaged within each group. The difference in mean normalized Cts between groups (δCt′) was then used to calculate fold change (2δCt′). Normalized Ct differences between middle-aged and young mice groups were statistically significant (P < 0.05).
Many of the top 35 genes have not been identified in previous studies of age-related changes in gene expression. These include CP2-related transcriptional repressor-1 (Crtr-1), Sec61α, protein phosphatase 2A B56 regulatory subunit γ isoform (Pp2A B56), mitogen-activated protein kinase phosphatase 3 (Mkp-3), serum/glucocorticoid activated kinase (Sgk), acetyl-CoA synthase 2 (AceS2), pyruvate dehydrogenase kinase 4 (Pdk 4), and growth arrest specific 5 (Gas5). Several others (e.g., cathepsin S, C4, C1q, β-2-microglobulin [β2M]) have been reported previously, adding support to the reliability of our results.
The Distinct Expression Pattern of the Hippocampus in the Middle-aged Mice and Possible Relation of the Expression Pattern to Observed Learning Deficits
Strikingly, the majority (29/35, 82%) of the genes we identified show increases (rather than decreases) in expression in middle-aged mice. This is particularly suggestive in the light of studies of molecular mechanisms of learning that show that an induction of negative regulators of synaptic plasticity can lead to deficits in memory formation. Some of the differentially expressed genes have been implicated in the molecular mechanisms of learning and memory, notably the calcium-dependent phospholipid binding protein and neurotrophic cytokine S100b. Converging lines of evidence support the implication of S100b in hippocampal-dependent forms of memory. Transgenic mice carrying multiple copies of S100B gene show impairment in hippocampal LTP and performance of hippocampal-dependent tasks (Gerlai et al. 1995; Janus et al. 1995; Roder et al. 1996) as well as altered brain aging (Whitaker-Azmitia et al. 1997). S100b-null mice display strengthened LTP and enhanced learning in the Morris water maze and contextual fear conditioning (Nishiyama et al. 2002). S100b age-related overexpression has been reported in human brain (Sheng et al. 1996), rat cortex (Linnemann and Skarsfelt 1994), senescence acceleration-prone SAMP6 mice (Griffin et al. 1998), and homozygous APPV717F transgenic mice (Sheng et al. 2000). S100b has also been implicated in Down's syndrome and in the pathogenesis of AD (Kato et al. 1991; Whitaker-Azmitia et al. 1997; Royston et al. 1999; Sheng et al. 2000; Mrak and Griffin 2001; Peskind et al. 2001; Petzold et al. 2003). Our observation that the ACD in mice is accompanied by hippocampal S100b overexpression is compatible with a possible role of S100b as a negative modulator of the molecular mechanisms of learning.
A number of genes involved in cell signaling show altered expression with age, among them, Pp2A B56. Pp2A is a ubiquitous serine/threonine phosphatase involved in many cellular, physiological, and pathological processes, including the modulation of the late phase of LTP (Woo and Nguyen 2002). It has been suggested that there is an age-dependent change in PP2A involvement in the regulation of synaptic strength (Norris et al. 1998). The function of the B56 γ-regulatory subunit in brain cells is not known. We also found underexpression of Ptpro, a receptor-type protein tyrosine phosphatase that could be involved in neurite guidance (Stepanek et al. 2001). We found that the Map kinase phosphatase 3 (Mkp-3) and serum/glucocorticoid-activated kinase (Sgk) genes show reciprocal expression patterns. This is interesting because MKP-3 is a negative regulator of ERK MAP kinase 2 (Zhou et al. 2002) and Sgk is a downstream effector of ERK MAP kinase pathway activation, and indeed, activation of ERK signaling is required for Sgk expression (Mizuno and Nishida 2001). Because we show that Mkp-3 is down-regulated in the middle-aged hippocampus, the concomitant up-regulation of Sgk may reflect increased activity in the ERK MAP kinase pathway. Because of the important role postulated for ERK2 signaling in neuronal plasticity, differentiation, and neurite outgrowth (Thomas et al. 1994; Camps et al. 1998), alterations in MAP signaling may be related to age-related cognitive decline. Our observation of down-regulation of a negative regulator or ERK1/2 may be part of a mechanism of compensation at the onset of ACD.
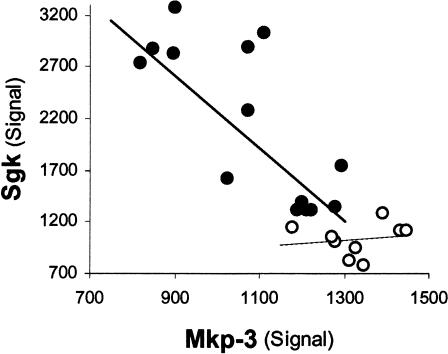
The reciprocal expression patterns described above are shown in detail in Figure 3. Mkp-3 and Sgk expression values have a correlation coefficient (rmkp3,sgk) of 0.84 across both groups. This high inverse correlation is dominated by the middle-aged mice group (rmkp3,sgk [middle-aged] = 0.78, rmkp3,sgk [young] = 0.15). The lower correlation within the young group may reflect a diminished effect of Mkp-3 upon Sgk expression in this cognitively unimpaired group.
Figure 3.
Mkp-3 and Sgk transcript levels are highly correlated in middle-aged mice in addition to showing a significant difference as compared to young mice. Sgk versus Mkp-3 hybridization signals are plotted for middle-aged (solid circles) and young (empty circles) mice. Each data point represents one chip/mouse. Sgk and Mkp-3 signals are significantly correlated within the middle-aged group (r = -0.8, P = -0.009) but not in the young animals' group (r = -0.1, P = -0.7).
Two other findings of our study may relate to impairment of synaptic function and plasticity and neurodegeneration in the middle-aged hippocampus, namely, the down-regulation of Sec61α and the overexpression of Crtr-1.
Sec61α is the main component of the transmembrane channel responsible for secretory protein translocation and membrane protein integration at the endoplasmic reticulum (ER) membrane (Knight and High 1998). It has been shown that Sec61α is localized to the endomembranes of dendritic spines in the hippocampus (Pierce et al. 2000).
CRTR-1 is highly similar (amino acid sequence identity >70%) to other members of the CP2 family of transcription factors, particularly LBP-9 and CP2/LBP-1c, which all share conserved oligomerization and DNA-binding domains. CRTR-1 is thought to act as a dominant repressor of CP2-activated promoters (Rodda et al. 2001). Interestingly, CP2/LPB-1c cooperatively interacts with C/EBP transcription factors (Bing et al. 1999). CP2/LBP-1c has been implicated as a susceptibility gene of Alzheimer's disease (Lambert et al. 2000; Lendon and Craddock 2001; Taylor et al. 2001; Luedecking-Zimmer et al. 2003). Our finding that Crtr-1 is up-regulated in middle-aged mouse hippocampus is consistent with the intriguing possibility that this transcription factor regulates other genes related to the etiology of ACD and neuropathology.
Functional Class Scoring Analysis of Gene Expression
Many of the genes that are differentially expressed in middle-aged mice are functionally related. Analysis of the results using functional class scoring (FCS, see Materials and Methods) shows that many of these co-occurrences are highly unlikely to be due to chance and, instead, are a particular feature of our data, and by implication, these gene expression changes represent a specific feature of brain aging (Table 2). The FCS analysis provides additional statistical support to the findings and can improve the sensitivity of the analysis by statistically evaluating genes in biologically meaningful groups instead of individually. It must be noted, however, that FCS relies on existing gene ontology (GO) annotations, which do not describe all possible “functional classes” of interest. FCS does not detract from the importance of the findings for individual genes but provides a statistical measure of the significance of the co-occurrence of genes with related function. The discussion in this section organizes our findings based on GO classes with high scores in our FCS analysis.
Table 2.
Top-Scoring Gene Ontology (GO) Terms Are Listed With the Corresponding P-Value and GO Identification Number and the Number of Probe Sets in the Class Represented on the Array
| GO term | Class members on array | P-Value |
|---|---|---|
| Complement activation; classical pathway (GO:0006958) Complement activation; complement component; MHC antigen; defense/immunity; antigen processing, etc. | 10 | 2.00E-05 |
| Lysosomal cysteine-type endopeptidase (GO:0004212) | 6 | 0.00054 |
| Iron homeostasis (GO:0006879) Ferric iron binding; heavy-metal ion homeostatis | 11 | 0.00264 |
| Acetyl-CoA biosynthesis (GO:0006085) Acetyl-CoA metabolism; CoA ligase | 5 | 0.0033 |
| Calcium-dependent phospholipid binding (GO:0005544) | 11 | 0.005 |
For some classes, multiple functionally related classes received high scores, and representatives of these are listed as an indented list below the main listing.
The highest scoring GO classes in our data based on FCS comprised immunoinflammatory genes and lysosomal cysteineendopeptidases. This is in agreement with other reports, including the previous microarray study of aging in the mouse hippocampus (Terao et al. 2002). Inflammatory mechanisms mostly involving activated microglia play an important role in the pathogenesis of AD (Eikelenboom and Veerhuis 1999; Akiyama et al. 2000). Some of these genes could be directly involved in the ACD. For example, β2M is expressed by hippocampal neurons, and its expression is regulated by neural activity (Corriveau et al. 1998). β2M has been implicated in the modulation of hippocampal LTP and LTD (Huh et al. 2000). Among our novel findings within these classes is the peptidoglycan recognition particle (PGRP) gene or Tag7, a cytokine shown to be induced in the brains of sleep-deprived rats (Rehman et al. 2001).
FCS analysis indicated altered expression of iron homeostasis genes. Changes in iron regulation have been linked to oxidative stress, ischemic stroke, and several neurodegenerative diseases (Jellinger et al. 1990; Beard et al. 1993; Davalos et al. 1994; Connor and Menzies 1995; Smith et al. 1997; Armengou and Davalos 2002).
As mentioned above, we found that S100b is induced in the middle-aged hippocampus. S100b belongs to the calcium-dependent phospholipid binding class, one of the high-ranking classes in our FCS analysis. Other genes in this class that were induced in the aged group are S100a6 (calcyclin) and annexin II. Calcyclin is known to interact directly with S100b (Yang et al. 1999; Deloulme et al. 2000) and annexin II (Filipek et al. 1995).
Our data also show age-related changes in expression of Acecs2 and Pdk4. These genes belong to the acetyl-CoA biosynthesis genes class, which was also flagged by FCS. Acecs2 transcription was decreased in middle-aged mice while Pdk4 was induced. Pdk4 negatively regulates the synthesis of acetyl-CoA from pyruvate (Wu et al. 2001). In addition to its role in energy production, acetyl-CoA is used to synthesize acetylcholine in cholinergic neurons. A cholinergic deficit is observed in AD (Francis et al. 1999) and is also believed to play a role in the memory decline seen in normal aging (Decker 1987; Casu et al. 2002) and in modulating LTP (Ji et al. 2001).
Interestingly, we find that many genes implicated in neurodegenerative diseases, such as Alzheimer's disease, show age-related changes in gene expression in mouse hippocampus. There is a known complex association between inflammatory processes and brain aging and degeneration. Both individual findings and FCS analysis of our data reflect that association. To what extent inflammation is related to the stage of ACD reached by 15-month-old mice requires much further investigation on the precise cellular localization and time course of the age-related changes in the brain expression profile. One possibility, compatible with our data, is that activation of glial cells in an inflammatory process involves the secretion of cytokines and neurotrophic factors that could, in turn, affect neuronal synaptic plasticity at the biochemical or transcriptional level.
General Comparison to Previous Microarray Findings
A number of the genes we identify as having changed expression in middle-aged mice have appeared in previous microarray studies of aging in mouse brain. Overall, nine of the top 35 genes we identify are found in at least one of the microarray analyses of aging mouse regions other than the hippocampus (Lee et al. 2000; Jiang et al. 2001). One other microarray study (Terao et al. 2002) examined global gene expression in the hippocampus. Inspection of the results of Terao et al. reveals six genes in common with our top 35, bringing the total overlap of our findings with previous microarray studies in mouse brain to 12 genes. One discrepancy is that Terao et al. tested and confirmed by real-time PCR age-related differential expression of H-T17 (an MHC I gene), a gene that showed no significant expression difference in our study (P > 0.1). A possible explanation for this discrepancy is that H-T17 was found by Terao et al. to be changed only at 18 months and not at 12 months. It is possible that our mice, assayed at 15 months, were too young to show this change. Finally, in a recent study, Blalock et al. (2003) combined gene expression analysis of the CA1 hippocampal subfield with behavioral assessment of cognitive function in young, middle-aged, and aged rats. No significant differences in cognitive performance were shown between young and middle-aged rats in that report, although at the level of gene expression, there are some agreements between our results, as exemplified by the complement gene C1q and the up-regulation of calcium-dependent phospholipid binding protein genes.
Many of the differences between our study and previous studies (and between those studies) may be partly attributed to the differing brain regions studied and the species, strain, and age of the animals involved. Other differences are accounted for by differences in the microarray platforms and target gene sets used, as well as in the criteria used to select genes. As an example of the latter, a major difference between the current study and that of Terao et al. is that the earlier study used only two replicates per group. As Terao et al. discussed, this limitation of their study design afforded too little power to apply the type of approach we use here incorporating multiple test correction; indeed, by using multiple test correction, they found no statistically significant evidence for alteration in gene expression (Terao et al. 2002). Similarly, Blalock et al. used a less stringent correction for multiple testing than the one we have applied, which may partly explain why they detected more genes that were affected by age.
Conclusions
We have been able to reveal a specific cognitive deficit in the Morris water maze task in middle-aged 15-month-old compared with 2-month-old C57BL/6J mice. In these mice we observed a pattern of differential gene expression mostly consisting of overexpression of genes in the middle-aged mice, including genes with possible roles in synaptic plasticity and learning. The changes in gene expression we observed correspond to the early stages of ACD, given that our study used middle-aged mice, displaying a mild ACD. In that respect, our study could contribute to the finding of targets for preventive therapeutical approaches.
MATERIALS AND METHODS
Animals and Behavioral Experiments
C57BL/6JNIA male mice, purchased from Harlan Sprague Dawley, were housed in groups and according to standard IACUC protocol. Mice received food and water ad libitum and a 12-h light/12-h dark cycle.
Training of mice in the Morris water maze task was carried out between 10:00 am and 2:00 pm in four trials a day, with each trial lasting a maximum of 120 sec with a 15-min intertrial interval. The task was performed as previously described (Malleret et al. 1999), with two training phases: 2 d with a visible platform followed by 7 d (spatial phase) with a hidden platform in the training quadrant. Probe trials (60 sec), during which the platform was removed, were performed to assess retention of the previously acquired information. Animal movements were recorded and analyzed by using a video tracking system (HVSimage).
ANOVAs were conducted for the factors age (young versus aged) and day (the average value of the four daily trials) and for the interaction between these factors.
In the probe trial, the time spent in the different quadrants (QDT), or in the exact platform zone or equivalent virtual areas in other quadrants (PF) was measured. ANOVAs were performed on these data for the factors age and area (either of the type QDT or PF).
Gene Expression Experiments
The same mice used for behavioral analysis were killed by cervical dislocation and hippocampi were dissected out and immediately frozen in liquid nitrogen. All samples were processed simultaneously to maximize the reproducibility of the results. Hippocampi were homogenized, and total RNA was extracted by using TRIzol (Invitrogen Life Technologies). RNA was precipitated with isopropanol and glycogen, washed in 75% ethanol, and resuspended in RNAse free water (Ambion). All further processing was carried out separately for each sample and according to the GeneChip Expression Analysis Technical Manual (Affymetrix). Fragmented amplified RNA was hybridized to Affymetrix Murine Genome U74Av2 arrays (12,486 probe sets). Hybridized arrays were washed and stained as described in the Affymetrix signal antibody amplification protocol. Image data were acquired with a Hewlett-Packard GeneArray scanner. Signal values were generated by using the Affymetrix Microarray Suite 5.0 software. The mean value of scaling factors across all chips within each group was 0.228 ± 0.010 for middle-aged mice and 0.227 ± 0.010 for young mice, respectively.
The full data set is available at http://microarray.cpmc.columbia.edu/pavlidis.
For real-time PCR, total hippocampal RNA from 13 middle-aged and eight young mice also used in the GeneChip experiment was treated with RNAse-free DNAse and RNasin (Promega) and used to synthesize cDNA separately for each sample and independently of the cDNA synthesis performed for GeneChips. RT PCR analysis was performed in a DNA Engine Opticon Continuous Fluorescence Detection System (MJ Research) or an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Specific primers (0.2 μM) were used for PCR with Platinum Taq DNA polymerase (Invitrogen) in the presence of SYBR Green and 3 mM MgCl2 or with Sybr Green PCR Master Mix (Applied Biosystems). As a reference, β-actin cDNA was amplified simultaneously from each sample. Primer sequences (5′ to 3′; f indicates forward; r, reverse) were as follows: act-f, AAGAGAAGCTGTGCTATGTTGC; act-r, GCTCGTTGCCAATAGTGATG; c4-f, GGTCAGACCCGCAACTTC; c4-r, CCAGGTATTTGAGTCCAGCA; c1qb-f, AACGCGAACGAGAACTATGA; c1qb-r, ACGAGATTCACACACAGGTTG; cats-f, CTGTACGGGCAATGTGAA; cats-r, CTAGCAATTCCGCAGTGA; crtr-f, ACGGAACTACGGGAGAGA; crtr-r, CATGGAAGAACATGGTAAGTAAG; gas5-f, GCACTGCAAACACAATGATTGG; and gas5-r, CACTGCATGTCCACTTGTCACA. Cycling conditions for RT PCR were as follows: six cycles with steps of 15 sec at 95°C, 10 sec at 65°C (-1°C per cycle), and 20 sec at 72°C followed by 30 cycles with steps of 15 sec at 95°C, 20 sec at 59°C, and 20 sec at 72°C, using the Opticon system; or 35 cycles with steps of 20 sec at 95°C, 20 sec at 55°C, and 30 sec at 72°C using the ABI system (gas5). Normalized Ct values for the genes tested (Ct') were calculated by subtracting Ctactin for each separate sample.
Statistical Analysis of Gene Expression
To identify differentially expressed genes, we used a statistical analysis based on the Welch t-test. The test was done independently for each gene. Each gene is assigned a P-value based on this statistic, representing the probability that the null hypothesis that the expression levels are equal between young and middle-aged mice is true. We obtained similar results by using the Student's t-test or the nonparametric Mann-Whitney U-test (data not shown). The results were corrected for multiple testing by using the false discovery rate method (Benjamini et al. 2001). Neither fold-change nor Affymetrix “absent/present” calls were considered as criteria for gene selection.
We also used FCS, a method for analyzing groups of genes based on functional annotations (Pavlidis et al. 2002). This method gives a statistical score to each of hundreds of functional gene classes, as defined by available GO annotations (Ashburner et al. 2000). FCS gives an unbiased evaluation of the results with respect to gene function. FCS is described in detail in Pavlidis et al. (2002). Briefly, for each GO class, the t-test P-values for each gene on the array that is in the class is used as the input to the algorithm. A filter was used to remove from consideration genes that are not expressed at detectable levels (30% of the probe sets on the array were removed). A raw score for the class is calculated as the mean -log(t-test P-value) for the genes in the class. This is converted into a P-value for the class by using an empirical null score distribution generated by calculating the raw scores for randomly selected sets of genes.
For real time PCR analysis, we used a one-tailed Welch t-test. Lower P-values were observed by applying the Student t-test.
Acknowledgments
We thank Dr. Etienne Sibille for his expert advice and Dr. Jie Qin for database support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.learnmem.org/cgi/doi/10.1101/lm.68204.
References
- Akiyama, H., Barger, S., Barnum, S., Bradt, B., Bauer, J., Cole, G.M., Cooper, N.R., Eikelenboom, P., Emmerling, M., Fiebich, B.L., et al. 2000. Inflammation and Alzheimer's disease. Neurobiol. Aging 21: 383-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsel, A. 1993. Hippocampal function in the rat: Cognitive mapping or vicarious trial and error? Hippocampus 3: 251-256. [DOI] [PubMed] [Google Scholar]
- Armengou, A. and Davalos, A. 2002. A review of the state of research into the role of Iron in stroke. J. Nutr. Health Aging 6: 136-137. [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology: The gene ontology consortium. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, M.E., Barad, M., Son, H., Zhuo, M., Lu, Y.F., Shih, R., Mansuy, I., Hawkins, R.D., and Kandel, E.R. 1999. Age-related defects in spatial memory are correlated with defects in the late phase of hippocampal long-term potentiation in vitro and are attenuated by drugs that enhance the cAMP signaling pathway. Proc. Natl. Acad. Sci. 96: 5280-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard, J.L., Connor, J.R., and Jones, B.C. 1993. Iron in the brain. Nutr. Rev. 51: 157-170. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N., and Golani, I. 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125: 279-284. [DOI] [PubMed] [Google Scholar]
- Bing, Z., Reddy, S.A., Ren, Y., Qin, J., and Liao, W.S. 1999. Purification and characterization of the serum amyloid a3 enhancer factor. J. Biol. Chem. 274: 24649-24656. [DOI] [PubMed] [Google Scholar]
- Blalock, E.M., Chen, K.C., Sharrow, K., Herman, J.P., Porter, N.M., Foster, T.C., and Landfield, P.W. 2003. Gene microarrays in hippocampal aging: Statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 23: 3807-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, N., Maguire, E.A., and O'Keefe, J. 2002. The human hippocampus and spatial and episodic memory. Neuron 35: 625-641. [DOI] [PubMed] [Google Scholar]
- Camps, M., Chabert, C., Muda, M., Boschert, U., Gillieron, C., and Arkinstall, S. 1998. Induction of the mitogen-activated protein kinase phosphatase mkp3 by nerve growth factor in differentiating pc12. FEBS Lett. 425: 271-276. [DOI] [PubMed] [Google Scholar]
- Casu, M.A., Wong, T.P., De Koninck, Y., Ribeiro da Silva, A., and Cuello, A.C. 2002. Aging causes a preferential loss of cholinergic innervation of characterized neocortical pyramidal neurons. Cereb. Cortex 12: 329-337. [DOI] [PubMed] [Google Scholar]
- Connor, J.R. and Menzies, S.L. 1995. Cellular management of iron in the brain. J. Neurol. Sci. 134(Suppl): 33-44. [DOI] [PubMed] [Google Scholar]
- Corriveau, R.A., Huh, G.S., and Shatz, C.J. 1998. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron 21: 505-520. [DOI] [PubMed] [Google Scholar]
- Dash, P.K., Hochner, B., and Kandel, E.R. 1990. Injection of the cAMP-responsive element into the nucleus of aplysia sensory neurons blocks long-term facilitation. Nature 345: 718-721. [DOI] [PubMed] [Google Scholar]
- Davalos, A., Fernandez Real, J.M., Ricart, W., Soler, S., Molins, A., Planas, E., and Genis, D. 1994. Iron-related damage in acute ischemic stroke. Stroke 25: 1543-1546. [DOI] [PubMed] [Google Scholar]
- Decker, M.W. 1987. The effects of aging on hippocampal and cortical projections of the forebrain cholinergic system. Brain Res. 434: 423-438. [DOI] [PubMed] [Google Scholar]
- Deloulme, J.C., Assard, N., Mbele, G.O., Mangin, C., Kuwano, R., and Baudier, J. 2000. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. J. Biol. Chem. 275: 35302-35310. [DOI] [PubMed] [Google Scholar]
- Eikelenboom, P. and Veerhuis, R. 1999. The importance of inflammatory mechanisms for the development of Alzheimer's disease. Exp. Gerontol. 34: 453-461. [DOI] [PubMed] [Google Scholar]
- Filipek, A., Wojda, U., and Lesniak, W. 1995. Interaction of calcyclin and its cyanogen bromide fragments with annexin II and glyceraldehyde 3-phosphate dehydrogenase. Int. J. Biochem. Cell Biol. 27: 1123-1131. [DOI] [PubMed] [Google Scholar]
- Foster, T.C. 1999. Involvement of hippocampal synaptic plasticity in age-related memory decline. Brain Res. Brain Res. Rev. 30: 236-249. [DOI] [PubMed] [Google Scholar]
- Francis, P.T., Palmer, A.M., Snape, M., and Wilcock, G.K. 1999. The cholinergic hypothesis of Alzheimer's disease: A review of progress. J. Neurol. Neurosurg. Psychiatry 66: 137-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, M. and Nicolle, M.M. 1993. Animal models of normal aging: Relationship between cognitive decline and markers in hippocampal circuitry. Behav. Brain Res. 57: 155-162. [DOI] [PubMed] [Google Scholar]
- Geinisman, Y., Detoledo Morrell, L., Morrell, F., and Heller, R.E. 1995. Hippocampal markers of age-related memory dysfunction: Behavioral, electrophysiological and morphological perspectives. Progr. Neurobiol. 45: 223-252. [DOI] [PubMed] [Google Scholar]
- Gerlai, R., Wojtowicz, J.M., Marks, A., and Roder, J. 1995. Overexpression of a calcium-binding protein, s100 β, in astrocytes alters synaptic plasticity and impairs spatial learning in transgenic mice. Learn. Mem. 2: 26-39. [DOI] [PubMed] [Google Scholar]
- Griffin, W.S., Sheng, J.G., and Mrak, R.E. 1998. Senescence-accelerated overexpression of s100β in brain of SAMP6 mice. Neurobiol. Aging 19: 71-76. [DOI] [PubMed] [Google Scholar]
- Huh, G.S., Boulanger, L.M., Hongping, D., Riquelme, P.A., Brotz, T.M., and Shatz, C.J. 2000. Functional requirement for class I MHC in CNS development and plasticity. Science 290: 2155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz, L.M., Vianna, M.R., Medina, J.H., and Izquierdo, I. 2002. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J. Neurosci. 22: 6781-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus, C., Janus, M., and Roder, J. 1995. Spatial exploration in transgenic mice expressing human β-s100. Neurobiol. Learn. Mem. 64: 58-67. [DOI] [PubMed] [Google Scholar]
- Jellinger, K., Paulus, W., Grundke-Iqbal, I., Riederer, P., and Youdim, M.B. 1990. Brain iron and ferritin in Parkinson's and Alzheimer's diseases. J. Neural. Transm. Park. Dis. Dement. Sect. 2: 327-340. [DOI] [PubMed] [Google Scholar]
- Ji, D., Lape, R., and Dani, J.A. 2001. Timing and location of nicotinic activity enhances or depresses hippocampal synaptic plasticity. Neuron 31: 131-141. [DOI] [PubMed] [Google Scholar]
- Jiang, C.H., Tsien, J.Z., Schultz, P.G., and Hu, Y. 2001. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc. Natl. Acad. Sci. 98: 1930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker, M. and Ingram, D.K. 1997. Murine models of brain aging and age-related neurodegenerative diseases. Behav. Brain. Res. 85: 1-26. [DOI] [PubMed] [Google Scholar]
- Kato, K., Kurobe, N., Suzuki, F., Morishita, R., Asano, T., Sato, T., and Inagaki, T. 1991. Concentrations of several proteins characteristic of nervous tissue in cerebral cortex of patients with Alzheimer's disease. J. Mol. Neurosci. 3: 95-99. [DOI] [PubMed] [Google Scholar]
- Knight, B.C. and High, S. 1998. Membrane integration of SEC61α: A core component of the endoplasmic reticulum translocation complex. Biochem. J. 331 (Pt 1): 161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, J.C., Goumidi, L., Vrieze, F.W., Frigard, B., Harris, J.M., Cummings, A., Coates, J., Pasquier, F., Cottel, D., Gaillac, M., et al. 2000. The transcriptional factor LBP-1C/CP2/LSF gene on chromosome 12 is a genetic determinant of Alzheimer's disease. Hum. Mol. Genet. 9: 2275-2280. [DOI] [PubMed] [Google Scholar]
- Lee, C.K., Weindruch, R., and Prolla, T.A. 2000. Gene-expression profile of the ageing brain in mice. Nat. Genet. 25: 294-297. [DOI] [PubMed] [Google Scholar]
- Lendon, C. and Craddock, N. 2001. Is LBP-1C/CP2/LSF a disease-modifying gene for Alzheimer's disease? Lancet 358: 1029-1030. [DOI] [PubMed] [Google Scholar]
- Linnemann, D. and Skarsfelt, T. 1994. Regional changes in expression of NCAM, GFAP, and S100 in aging rat brain. Neurobiol. Aging 15: 651-655. [DOI] [PubMed] [Google Scholar]
- Luedecking-Zimmer, E., DeKosky, S.T., Nebes, R., and Kamboh, M.I. 2003. Association of the 3′ UTR transcription factor LBP-1C/CP2/LSF polymorphism with late-onset Alzheimer's disease. Am. J. Med. Genet. 117B: 114-117. [DOI] [PubMed] [Google Scholar]
- Malleret, G., Hen, R., Guillou, J.L., Segu, L., and Buhot, M.C. 1999. 5-HT1B receptor knock-out mice exhibit increased exploratory activity and enhanced spatial memory performance in the Morris water maze. J. Neurosci. 19: 6157-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, H. and Nishida, E. 2001. The ERK MAP kinase pathway mediates induction of SGK (serum- and glucocorticoid-inducible kinase) by growth factors. Genes Cells 6: 261-268. [DOI] [PubMed] [Google Scholar]
- Morris, R.G., Garrud, P., Rawlins, J.N., and O'Keefe, J. 1982. Place navigation impaired in rats with hippocampal lesions. Nature 297: 681-683. [DOI] [PubMed] [Google Scholar]
- Mrak, R.E. and Griffin, W.S. 2001. Interleukin-1, neuroinflammation, and Alzheimer's disease. Neurobiol. Aging 22: 903-908. [DOI] [PubMed] [Google Scholar]
- Nishiyama, H., Knopfel, T., Endo, S., and Itohara, S. 2002. Glial protein s100b modulates long-term neuronal synaptic plasticity. Proc. Natl. Acad. Sci. 99: 4037-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, C.M., Halpain, S., and Foster, T.C. 1998. Alterations in the balance of protein kinase/phosphatase activities parallel reduced synaptic strength during aging. J. Neurophysiol. 80: 1567-1570. [DOI] [PubMed] [Google Scholar]
- Pavlidis, P., Lewis, D.P., and Noble, W.S. 2002. Exploring gene expression data with class scores. Pac. Symp. Biocomput. 7: 474-485. [PubMed] [Google Scholar]
- Peskind, E.R., Griffin, W.S., Akama, K.T., Raskind, M.A., and Van Eldik, L.J. 2001. Cerebrospinal fluid S100b is elevated in the earlier stages of Alzheimer's disease. Neurochem. Int. 39: 409-413. [DOI] [PubMed] [Google Scholar]
- Petzold, A., Jenkins, R., Watt, H.C., Green, A.J., Thompson, E.J., Keir, G., Fox, N.C. and Rossor, M.N. 2003. Cerebrospinal fluid S100b correlates with brain atrophy in Alzheimer's disease. Neurosci. Lett. 336: 167-170. [DOI] [PubMed] [Google Scholar]
- Pierce, J.P., van Leyen, K., and McCarthy, J.B. 2000. Translocation machinery for synthesis of integral membrane and secretory proteins in dendritic spines. Nat. Neurosci. 3: 311-313. [DOI] [PubMed] [Google Scholar]
- Rapp, P.R., Deroche, P.S., Mao, Y., and Burwell, R.D. 2002. Neuron number in the parahippocampal region is preserved in aged rats with spatial learning deficits. Cereb. Cortex 12: 1171-1179. [DOI] [PubMed] [Google Scholar]
- Rehman, A., Taishi, P., Fang, J., Majde, J.A., and Krueger, J.M. 2001. The cloning of a rat peptidoglycan recognition protein (PGRP) and its induction in brain by sleep deprivation. Cytokine 13: 8-17. [DOI] [PubMed] [Google Scholar]
- Rodda, S., Sharma, S., Scherer, M., Chapman, G., and Rathjen, P. 2001. CRTR-1, a developmentally regulated transcriptional repressor related to the CP2 family of transcription factors. J. Biol. Chem. 276: 3324-3332. [DOI] [PubMed] [Google Scholar]
- Roder, J.K., Roder, J.C., and Gerlai, R. 1996. Memory and the effect of cold shock in the water maze in s100 β transgenic mice. Physiol. Behav. 60: 611-615. [DOI] [PubMed] [Google Scholar]
- Royston, M.C., McKenzie, J.E., Gentleman, S.M., Sheng, J.G., Mann, D.M., Griffin, W.S., and Mrak, R.E. 1999. Overexpression of s100β in Down's syndrome: Correlation with patient age and with β-amyloid deposition. Neuropathol. Appl. Neurobiol. 25: 387-393. [DOI] [PubMed] [Google Scholar]
- Sheng, J.G., Mrak, R.E., Rovnaghi, C.R., Kozlowska, E., Van Eldik, L.J., and Griffin, W.S. 1996. Human brain s100 β and s100 β mRNA expression increases with age: Pathogenic implications for Alzheimer's disease. Neurobiol. Aging 17: 359-363. [DOI] [PubMed] [Google Scholar]
- Sheng, J.G., Mrak, R.E., Bales, K.R., Cordell, B., Paul, S.M., Jones, R.A., Woodward, S., Zhou, X.Q., McGinness, J.M., and Griffin, W.S. 2000. Overexpression of the neuritotrophic cytokine s1007β precedes the appearance of neuritic β-amyloid plaques in APPV717F mice. J. Neurochem. 74: 295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, S.A., Tsai, W.Y., DeLaPaz, R., Mayeux, R., and Stern, Y. 2002. Imaging hippocampal function across the human life span: Is memory decline normal or not? Ann. Neurol. 51: 290-295. [DOI] [PubMed] [Google Scholar]
- Smith, M.A., Harris, P.L., Sayre, L.M., and Perry, G. 1997. Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. Proc. Natl. Acad. Sci. 94: 9866-9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek, L., Sun, Q.L., Wang, J., Wang, C., and Bixby, J.L. 2001. Cryp-2/Cptpro is a neurite inhibitory repulsive guidance cue for retinal neurons in vitro. J. Cell Biol. 154: 867-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A.E., Yip, A., Brayne, C., Easton, D., Evans, J.G., Xuereb, J., Cairns, N., Esiri, M.M., and Rubinsztein, D.C. 2001. Genetic association of an LBP-1C/CP2/lSF gene polymorphism with late onset Alzheimer's disease. J. Med. Genet. 38: 232-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao, A., Apte-Deshpande, A., Dousman, L., Morairty, S., Eynon, B.P., Kilduff, T.S., and Freund, Y.R. 2002. Immune response gene expression increases in the aging murine hippocampus. J. Neuroimmunol. 132: 99-112. [DOI] [PubMed] [Google Scholar]
- Teule, M., Buhot, M.C., and Durhup, H. 1972. Apprentissage d'élimimination progressive chez le hamster doré. Psychologie Française 17: 175-182. [Google Scholar]
- Thomas, K.L., Laroche, S., Errington, M.L., Bliss, T.V., and Hunt, S.P. 1994. Spatial and temporal changes in signal transduction pathways during LTP. Neuron 13: 737-745. [DOI] [PubMed] [Google Scholar]
- West, M.J., Coleman, P.D., Flood, D.G., and Troncoso, J.C. 1994. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer's disease. Lancet 344: 769-772. [DOI] [PubMed] [Google Scholar]
- Whitaker-Azmitia, P.M., Wingate, M., Borella, A., Gerlai, R., Roder, J., and Azmitia, E.C. 1997. Transgenic mice overexpressing the neurotrophic factor s-100 β show neuronal cytoskeletal and behavioral signs of altered aging processes: Implications for Alzheimer's disease and Down's syndrome. Brain Res. 776: 51-60. [DOI] [PubMed] [Google Scholar]
- Wolfer, D.P., Stagljar-Bozicevic, M., Errington, M.L., and Lipp, H.P. 1998. Spatial memory and learning in transgenic mice: Fact or artifact? News Physiol. Sci. 13: 118-123. [DOI] [PubMed] [Google Scholar]
- Woo, N.H. and Nguyen, P.V. 2002. “Silent” metaplasticity of the late phase of long-term potentiation requires protein phosphatases. Learn. Mem. 9: 202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P., Peters, J.M., and Harris, R.A. 2001. Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor α. Biochem. Biophys. Res. Commun. 287: 391-396. [DOI] [PubMed] [Google Scholar]
- Yang, Q., O'Hanlon, D., Heizmann, C.W., and Marks, A. 1999. Demonstration of heterodimer formation between S100B and S100A6 in the yeast two-hybrid system and human melanoma. Exper. Cell Res. 246: 501-509. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Lein, E.S., He, A., Smith, S.C., Aston, C., and Gage, F.H. 2001. Transcriptional profiling reveals strict boundaries between hippocampal subregions. J. Comp. Neurol. 441: 187-196. [DOI] [PubMed] [Google Scholar]
- Zhou, B., Wang, Z.X., Zhao, Y., Brautigan, D.L., and Zhang, Z.Y. 2002. The specificity of extracellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. J. Biol. Chem. 277: 31818-31825. [DOI] [PubMed] [Google Scholar]