Abstract
Polymorphisms of LIG4 gene may influence DNA repair ability, thus altering the genetic stability and resulting in carcinogenesis. A growing number of studies have investigated the relevance of LIG4 T9I (rs1805388) and D501D (rs1805386) polymorphisms with cancer risk, however, the results are conflicting. To obtain a comprehensive conclusion, we searched relevant literatures from PubMed, Web of Science, Ovid and Embase databases on May 15, 2014 and performed a meta-analysis. In this meta-analysis, a total of 17 articles were included. Of them, there were 15 studies with 5873 cases and 5771 controls for rs1805388 and 6 studies with 4161 cases and 4881 controls for rs1805386. Overall, our results suggested that there was no obvious relevance of LIG4 T9I polymorphism with cancer susceptibility. However, in subgroup analysis, we found the LIG4 T9I was associated with a slightly decreased cancer risk among Caucasians. As to the rs1805386, the genetic variant had no significant association with cancer risk. In conclusion, despite several limitations, this meta-analysis suggested that LIG4 T9I genetic variant is associated with a decreased risk of cancer among Caucasians, however, the rs1805386 gene polymorphism is not a risk factor of cancer.
Cancer is one of the most common causes of death in the world and results in a serious problem to global health1,2. Despite advances in treatment for cancer, the prognosis remains unsatisfied3. Thus, exploring the methods of early detection and prevention are indispensable. Currently, gene-environment interactions have been thought to be main determinant of individual risk for diseases including cancer4. Genes decide the susceptibility of individual to environment and environmental factors often damage the DNA in turn. If the host DNA repair system does not perform their functions well in repairing such destructive DNA, it might alter the stability of genome and lead to carcinogenesis. Thus, the DNA repair ability plays a critical role in maintaining the stability of human genome.
LIG4 gene, located on human chromosomes 13q33-34, encodes an ATP-dependent DNA ligase that joins single-strand breaks in a double-stranded polydeoxynucleotide in an ATP-dependent reaction5. The DNA ligase IV is the crucial enzyme to for completing the non-homologous end joining (NHEJ) by forming a complex together with X-ray repair cross complementing protein 4 (XRCC4) for a final ligation of the break in an ATP-dependent step6,7. Therefore, the loss or variant of LIG4 is supposed to contribute to genomic instability and tumorigenesis.
According to supposition mentioned above, we reviewed the related studies concerning of LIG4 variants and susceptibility of tumor. Then, we found that numerous studies investigated this issue, however, the conclusions are inconsistent. To obtain a comprehensive conclusion, we carried out a meta-analysis to systematically evaluate the relevance of LIG4 genetic variants with susceptibility of cancer.
Results
Identification of relevant studies
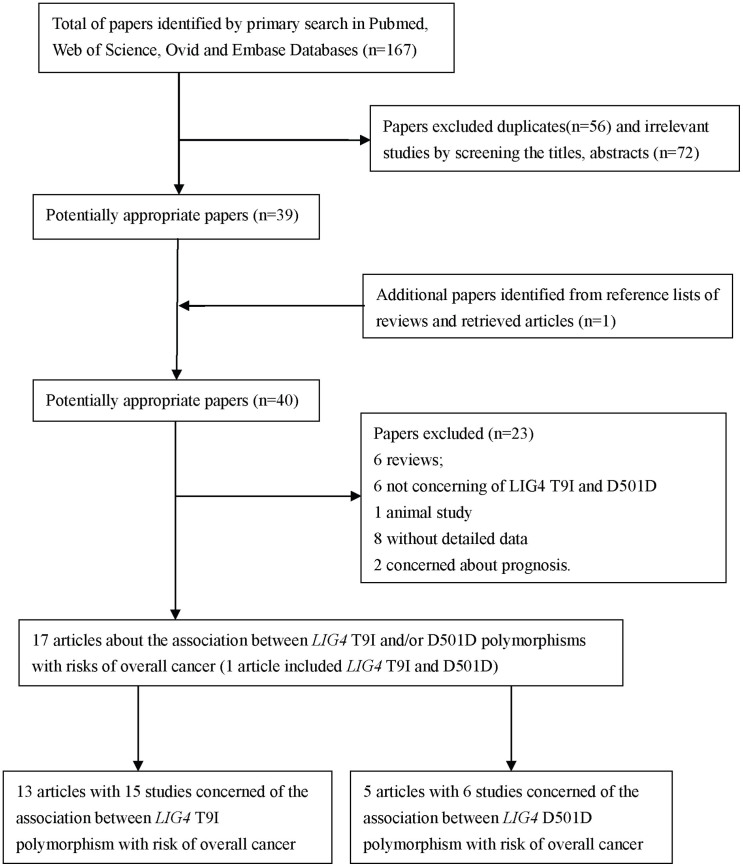
A total of 167 papers were indentified from the databases as we described above. After deleting the duplications, 111 papers were left. Then we estimated the rest articles and 72 articles were discarded because of irrelevance with this issue. And one other potential eligible paper was obtained by screening the references of reviews. Of the remains 40 papers, one article was excluded for animal study8; six papers were reviews9,10,11,12,13,14. Following this, two papers were concerned with prognosis15,16, eight articles without detailed data for further evaluation17,18,19,20,21,22,23,24. Besides, six papers estimated LIG4 polymorphism but not T9I or D501D25,26,27,28,29,30. Finally, a total of 17 studies with case-control design met the inclusion and exclusion criteria in this meta-analysis31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47. Of these 17 articles (one article included both rs1805386 and rs1805388), there were five articles with six studies for rs1805386, 13 articles with 15 studies for rs1805388. The flow diagram of searching process was shown in Figure 1.
Figure 1. Flow diagram of included/excluded studies.
Characteristics of included studies
Of these included articles, one article42 with three caner types were separated as three independent studies and three articles35,39,45 concluded rs1805386 and rs1805388. Noticeably, in the relevant articles, two articles19,24 did not find the polymorphism of rs1805386 and another article45 did supply with insufficient data for rs1805386 but for rs1805388. Thus, in the end, there were five articles with six studies for rs1805386, 13 articles with 15 studies for rs1805388 polymorphism.
Then, we established a database concerning of the information extracted from each included paper. Summaries of these studies were presented in Table 1 which included the first author's surname, publication year, ethnicity, country, number and characteristics of cases and controls, and other relevant information.
Table 1. Characteristics of studies included in this meta-analysis.
| Surname | Year | Country | Ethnicity | Cancer type | Control source | Genotyping methods | Cases/Control | D. Case | D. Control | HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1805388 | ||||||||||
| Roddam | 2002 | UK | Caucasian | acute lymphoblastic leukemia | NA | PCR | 70/220 | 0/21/49 | 13/58/149 | 0.03 |
| Roddam | 2002 | UK | Caucasian | lymphoma | NA | PCR | 362/220 | 7/108/247 | 13/58/149 | 0.03 |
| Roddam | 2002 | UK | Caucasian | multiple myeloma | NA | PCR | 269/220 | 4/61/204 | 13/58/149 | 0.03 |
| Fu | 2003 | China | Asian | breast cancer | socioeconomic status | MassARRAY | 253/376 | 16/100/137 | 28/150/198 | 0.955 |
| Garcia-Closas | 2006 | USA | Caucasian | breast cancer | Age | Taqman | 1316/1043 | 57/339/920 | 42/277/724 | 0.199 |
| Hill | 2006 | USA | Caucasian | non-Hodgkin lymphoma | Age, gender | Taqman | 1110/931 | 18/300/792 | 28/275/628 | 0.75 |
| Wu | 2006 | China | Asian | bladder cancer | Age, gender | PCR-RFLP | 606/593 | 24/168/414 | 15/194/384 | 0.099 |
| Andreae | 2007 | Germany | Caucasian | acute lymphoblastic leukemia | Age, gender | DHPLC | 107/104 | 3/19/85 | 3/33/68 | 0.673 |
| Werbrouck | 2008 | Belgium | Caucasian | head and neck cancer | Age, gender | PCR-RFLP | 152/157 | 3/30/119 | 8/44/105 | 0.242 |
| Tseng | 2009 | Taiwan | Asian | lung cancer | Age, gender | PCR | 149/152 | 20/62/67 | 12/55/85 | 0.464 |
| Li | 2009 | USA | Caucasian | pancreatic cancer | Age, gender | Taqman | 723/776 | 23/197/503 | 29/208/539 | 0.117 |
| Gomes | 2010 | Portugal | Caucasian | thyroid cancer | Age, gender | Real-Time PCR | 109/217 | 4/22/83 | 5/54/158 | 0.879 |
| Al-Hadyan | 2012 | Saudi Arabia | Asian | head and neck cancer | Age | PCR | 156/251 | 0/24/132 | 4/31/216 | 0.029 |
| Salagovic | 2012 | Slovakia | Caucasian | lymphoma | NA | PCR | 107/127 | 1/26/80 | 2/32/93 | 0.688 |
| Zhao | 2013 | China | Asian | glioma | Age, gender | PCR | 384/384 | 49/172/163 | 20/142/222 | 0.659 |
| rs1805386 | ||||||||||
| Kuschel | 2002 | Germany | Caucasian | breast cancer | NA | PCR | 1440/2016 | 20/322/914 | 50/492/1248 | 0.857 |
| Han | 2004 | USA | Caucasian | breast cancer | Age, BMI | Taqman | 977/1266 | 22/274/681 | 32/360/874 | 0.48 |
| Garcia-Closas | 2006 | USA | Caucasian | breast cancer | Age | Taqman | 1338/1057 | 55/379/904 | 34/309/714 | 0.934 |
| Jakubowska | 2010 | Germany | Caucasian | breast cancer | NA | PCR | 319/290 | 112*/207 | 94*/196 | NA |
| Jakubowska | 2010 | Germany | Caucasian | ovarian cancer | NA | PCR | 145/280 | 54*/91 | 92*/188 | NA |
| Assis | 2010 | Portugal | Caucasian | ovarian cancer | NA | Real-Time PCR | 126/198 | 7/36/83 | 5/60/133 | 0.562 |
NA: not available; D.: distribution;
*: homozygous variants + heterozygous variant (CC + TC); HWE: Hardy-Weinberg Equilibrium.
Meta-analysis results
rs1805386
We found that there was no obvious relevance of LIG4 D501D variants with overall cancer risk (homozygous: OR = 0.97, 95% CI = 0.59–1.59, P = 0.907; recessive: OR = 0.96, 95% CI = 0.88–1.06, P = 0.434; dominant: OR = 0.99, 95% CI = 0.61–1.60, P = 0.952; allele: OR = 0.95, 95% CI = 0.87–1.03, P = 0.229) (Table 2). In the subgroup analysis stratified by cancer types, no statistically significant relations were found for breast cancer and ovarian cancer (The data were not shown).
Table 2. Relevance of rs1805388 and rs1805386 with cancer risk in this Meta-analysis.
| Homozygous | Recessive | Dominant | Allele | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | No of individuals | OR (95%CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR(95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) |
| For T9I | TT vs. CC | (TT + TC) vs. CC | TT vs. (TC + CC) | T vs. C | |||||||||
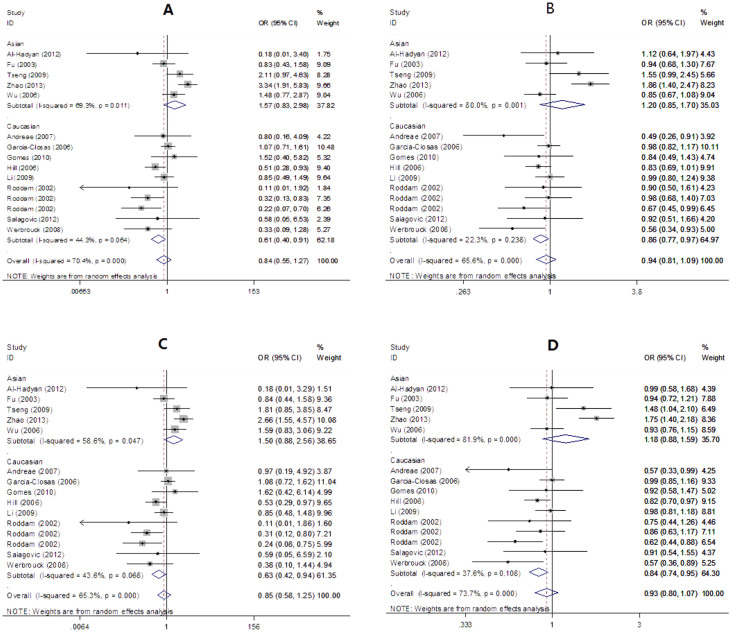
| All | 5873/5771 | 0.84 (0.55–1.27) | 0.000 | 0.704 | 0.94 (0.81–1.09) | 0.000 | 65.6 | 0.85 (0.58–1.25) | 0.000 | 65.3 | 0.93 (0.80–1.07) | 0.000 | 73.7 |
| Ethnicity | |||||||||||||
| Asian | 1941/1991 | 1.57 (0.83–2.98) | 0.011 | 0.693 | 1.20 (0.85–1.70) | 0.001 | 80.0 | 1.50 (0.88–2.56) | 0.047 | 58.6 | 1.18 (0.88–1.59) | 0.000 | 81.9 |
| Caucasian | 3932/3780 | 0.61 (0.40–0.91) | 0.064 | 0.443 | 0.86 (0.77–0.97) | 0.238 | 22.3 | 0.63 (0.42–0.94) | 0.068 | 43.6 | 0.84 (0.74–0.95) | 0.108 | 37.6 |
| For D501D | CC vs. TT | (CC + CT) vs. TT | CC vs. (CT + TT) | C vs. T | |||||||||
| All | 4161/4881 | 0.97 (0.59–1.59) | 0.044 | 63.1 | 0.96 (0.88–1.06) | 0.520 | 0.0 | 0.99 (0.61–1.60) | 0.048 | 62.1 | 0.95 (0.87–1.03) | 0.218 | 32.4 |
rs1805388
As shown in Table 2 and Figure 2, there was no relevance of LIG4 T9I variants with overall cancer risk (homozygous: OR = 0.84, 95% CI = 0.55–1.27, P = 0.401; recessive: OR = 0.94, 95% CI = 0.81–1.09, P = 0.434; dominant: OR = 0.85, 95% CI = 0.58–1.25, P = 0.410; allele: OR = 0.93, 95% CI = 0.80–1.07, P = 0.306). However, in the subgroup analysis, a statistically significant association was found among Caucasians (homozygous: OR = 0.61, 95%CI = 0.40–0.91, P = 0.016; recessive: OR = 0.86, 95% CI = 0.77–0.97, P = 0.016; dominant: OR = 0.63, 95% CI = 0.42–0.94, P = 0.023; allele: OR = 0.84, 95%CI = 0.74–0.95, P = 0.004).
Figure 2. Forest plot for overall cancer risk associated with LIG4 T9I polymorphism ((A): homozygous model; (B): recessive model; (C): dominant model; (D): allele model. subgroup analysis by ethnicity).
Heterogeneity and sensitivity analysis
Heterogeneities were observed among several studies for LIG4 D501D polymorphism and overall cancer susceptibility (homozygous: P = 0.044; dominant: P = 0.048;) and T9I (homozygous: P < 0.001; recessive: P < 0.001; dominant: P < 0.001; allele: P < 0.001). Thus, we selected the random-effect models to generate pooled ORs and corresponding 95% CIs for these models. On the other hand, no heterogeneity was observed among the other two models for D501D (recessive: P = 0.520; allele: P = 0.218) and the fixed-effect models were performed to generate ORs and 95% CIs for them. The sensitivity analysis suggested that no obvious changes were observed for the pooled ORs when single investigation was excluded respectively (data were not shown).
Publication bias
The shape of the funnel plot showed that no evidence of asymmetry was observed in the current meta-analysis by Egger's test for T9I (homozygous: P = 0.530; recessive: P = 0.919; dominant: P = 0.482; allele: P = 0.585) and D501D (homozygous: P = 0.501; recessive: P = 0.073; dominant: P = 0.499; allele: P = 0.451), which suggested that there were no significant publication bias among all these studies.
Trial sequential analysis (TSA)
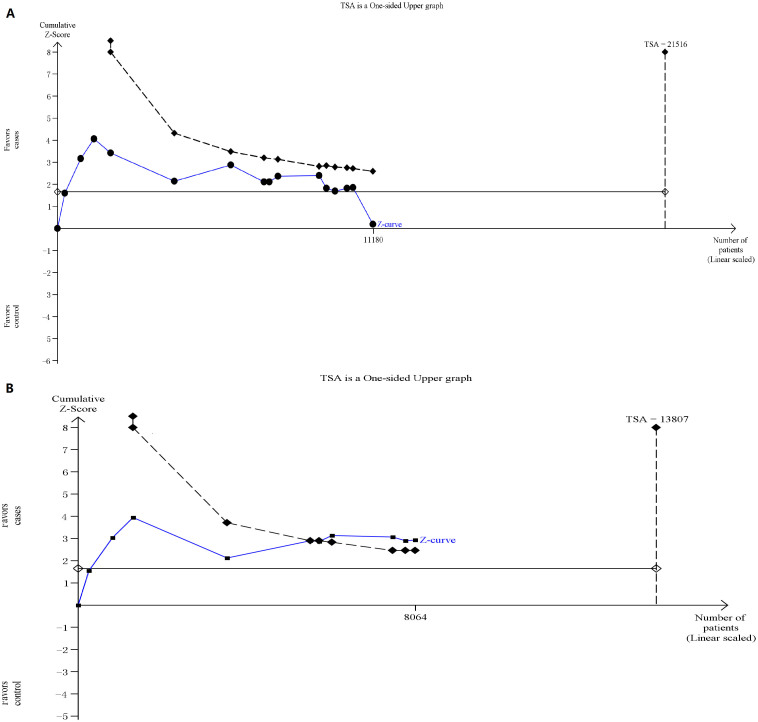
Fifteen trials (11180 subjects) were used to investigate the relevance of rs1805388 gene polymorphism with cancer susceptibility. Using the TSA (taking the data of dominant model for example), the required information size is 21516 subjects to demonstrate the issue (Figure 3A). Until now, the cumulative z-curve has not crossed the trial monitoring boundary before reaching the required information size, indicating that the cumulative evidence is insufficient and further trials are necessary. However, the cumulative z-curve crossed with TSA monitoring boundary when we performed the sub-analysis based on the ethnicity, confirming that rs1805388 is associated with a slightly decreased cancer risk among Caucasians and further relevant trials are unnecessary (Figure 3B). As for rs1805386, we chose the data of four models to perform TSA. The cumulative z-curve have crossed with TSA monitoring boundaries before the required information size is reached, indicating that the rs1805386 polymorphism is not a risk factor of cancer and no further trials are necessary (figures were not shown).
Figure 3.
(A): The required information size to demonstrate the relevance of rs1805388 gene polymorphisms with cancer susceptibility; (B): The required information size to demonstrate the relevance of rs1805388 gene polymorphisms with risk of cancer among Caucasians. The solid blue line is the cumulative Z-curve. The dashed inward-sloping line to the left represents the trial sequential monitoring boundaries.
Discussion
Genomic instability is an outstanding characteristic of cancer48. Numerous studies have demonstrated that tumor suppressor genes play a significant role in DNA double-strand break (DSB) repair and in maintenance of genomic stability; correspondingly, loss or mutation of such repair genes results in a shifty susceptibility for malignancies.
In eukaryotes, homologous recombination and NHEJ are two major pathways for DNA DSB repair and the latter is predominantly way in mammalian cells49. In the NHEJ process, LIG4, as a major functional protein, forms a complex with XRCC4 to perform the final rejoining step of NHEJ. Thus, the genetic variant of LIG4 might alter the repair capacity of DSB.
Previous investigation had demonstrated that polymorphisms in DNA repair genes might alter DNA repair capacity and thus contribute to cancer risk50. In 2002, Kuschel et al.40 found that LIG4 D501D polymorphism was associated with a decrease in breast cancer risk and Roddam et al.42 found that LIG4 T9I polymorphism may modulate predisposition to multiple myeloma. After these discoveries, a mass of investigations were performed to estimate the association between LIG4 T9I and D501D polymorphisms and the susceptibility of various cancers. To our puzzled, the results are conflicting. Thus, we performed a meta-analysis to obtain a comprehensive conclusion.
To our knowledge, this is the first time to systematically estimate the associations between LIG4 T9I and D501D polymorphisms and susceptibility of overall cancer. In this study, we found that rs1805386 and rs1805388 genetic variants had no relevance with overall cancer risk in homozygous, recessive, dominant and allele models. To further investigate the associations, we performed the subgroups analysis based on ethnicity and cancer types and found that rs1805388 variant is a decreased risk of cancers among Caucasians.
In order to make the conclusion more credible, we performed the publication bias analysis and sensitivity analysis according to Cochrane protocol. Funnel plots suggested that no obvious publication bias was observed. The sensitivity analysis indicated that the results are robust and no single study yield to obvious effects on the pooled ORs and the corresponding CIs. Besides, we performed the TSA and the results of TSA showed that the conclusions in this meta-analysis are robust.
However, several limitations in this meta-analysis should be noticed. Firstly, several studies had small sample sizes which might lessen the statistical power. Secondly, the heterogeneity was existed and thus we performed the random-effects model to obtain the wider CIs, which might weaken the reliability of conclusions. Thirdly, our results were based on unadjusted assessment of ORs, which might influence the results. Fourthly, we did not search the unpublished studies, which might miss the relevant studies. Besides, all data included in this study were from published investigations which were based on the current marker identification method of the ‘one-step-clustering'. This approach might tend to be ‘passenger signals' instead of ‘drivers' and bury the ‘real' cancer gene, which made the results less robust and accurate51,52. Based on the limitations mentioned above, the results should be considered with caution.
Overall, in spite of these limitations, this analysis reached a precise conclusion that LIG4 D501D polymorphism has no obvious relevance with cancer risk and individual with LIG4 T9I genetic variant has a decrease cancer risk among Caucasians. With the development of research methods, future studies focusing on a combinatorial use of the polygene markers and integrative network modules analysis, are necessary to make the conclusions more comprehensive.
Methods
Search strategy
We searched the PubMed, Web of Science, Ovid and Embase databases without language limitations for all related papers using the following searching strategies: 1) LIG4 or LIG 4 or ligase IV, 2) polymorphism or variant or variation or allele or genotype, and 3) cancer or carcinoma or tumor or neoplasm. And the last research was updated on May 15, 2014. All searched studies were screened and their references were retrieved to obtain other related articles. Then we downloaded the relevant papers and further screened to identify potentially eligible studies.
Inclusion/exclusion criteria
Studies included in this studies had to meet the following inclusion criteria: (1) estimating the relevance of LIG4 polymorphisms (rs1805386 and rs1805388) and cancer susceptibility; (2) case-control design; (3) sufficient data provided to assess odds ratios (ORs) and the corresponding confidence intervals (CIs); (4) when multiple publication from a particular research group reported data from overlapping samples, the study reporting the largest or latest dataset was included. Exclusion criteria: (1) review articles; (2) case reports, or case-only studies; (3) studies that estimated the risk of prognosis.
Data extraction
All data were independently reviewed and extracted from the included papers by two authors (S.X. and J.H.). Disagreements were solved by full discussion until a consensus was reached. The following characteristics were collected from each study: first author's surname, year of publication, ethnicity, country, cancer type, sample size, control source, matching contents, the Hardy-Weinberg equilibrium, genotype methods and genetic distribution of cases and controls. The subgroup analysis was performed by cancer type and ethnicity.
Statistical analysis
All the data management and analysis for this meta-analysis were performed with STATA 11.0 software (Stata corporation, College Station, TX). The strength of association between rs1805386 and rs1805388 polymorphisms and cancer susceptibility was estimated by calculating OR with the corresponding 95% CI. In order to calculate heterogeneity of studies, the Chi-Square test was used and significance was set at P value less than 0.05 level53. If the study was found to be heterogeneous (P > 0.10 for the Q-test), the fixed-effects model (the Mantel-Haenszel method) was performed to calculate the combined OR54. Otherwise, a random effect model (the DerSimonian and Laird method) was used to estimate the pooled OR55. In addition, the heterogeneity was also quantified with I2 statistics. The I2 value ranges from 0 to 100% and a larger I2 value indicating a greater heterogeneity56. The funnel plot was used to test the potential publication bias, and the funnel plot asymmetry was estimated by Egger's linear regression57. Sensitivity analyses were performed to identify the influence of the each study on the combined ORs and 95% CI.
Trial sequential analysis (TSA)
According to Cochrane Handbook for systematic reviews of interventions, meta-analyses and systematic reviews are considered to be the best available evidence if all available trials are included. However, ‘the best available evidence' might not be equal to ‘strong evidence' or ‘sufficient evidence'. It is well-known that meta-analysis may cause random errors in the series of sparse data and reduplicative testing on accumulating data. Based on these problems mentioned above, we applied the TSA to minimize the random errors and increase the robustness of conclusions58,59. In our study, we planned to calculate the required information size and estimate how many patients would be necessary to make a robust conclusion58. The required information size was based on the assumption of a plausible relative risk of 10% with low risk bias, and we adopted the risks for a type I error (α) of 5%, a type II error (β) of 20%58. Based on required information size and risk for type I and type II errors TSA monitoring boundaries were built. If a TSA monitoring boundary is crossed with Z-curve before the required information size is reached, robust evidence might have been confirmed and further trials are unnecessary. Otherwise, it is necessary to continue doing trials.
Author Contributions
All authors contributed significantly to this work. Z.-G.C. designed and revised the article. J.H. collected articles, summarized data and revised this article. S.X. collected articles, summarized data, did statistical work and drafted the manuscript. X.-F.S. collected articles, summarized data and did statistical work. H.X. and K.S. provided collected data and summarized information in part. All authors reviewed this manuscript and approved the final draft.
Acknowledgments
We thank Jie Li (School of Computer Science and Technology, Harbin Institute of Technology) for helpful explanations of network modules analysis and Multiple Survival Screening (MSS) algorithm. This work was supported in part by the grant from National Natural Science Foundation of China (Grant No. 81371162 to Z.-G.C.) and Special Financial Grant from the China Postdoctoral Science Foundation (Grant No. 2014T70836 to J.H.).
References
- Carter D. New global survey shows an increasing cancer burden. Am J Nurs 114, 17 (2014). [DOI] [PubMed] [Google Scholar]
- Popat K., McQueen K. & Feeley T. W. The global burden of cancer. Best Pract Res Clin Anaesthesiol 27, 399–408 (2013). [DOI] [PubMed] [Google Scholar]
- Siegel R., Ma J., Zou Z. & Jemal A. Cancer statistics, 2014. CA Cancer J Clin 64, 9–29 (2014). [DOI] [PubMed] [Google Scholar]
- Wilson, S., Jones, L., Coussens, C. & Hanna, K. (Eds.). Cancer and the environment: Gene–environment interaction, (National Academy Press, Washington, DC, 2002). [PubMed] [Google Scholar]
- Wei Y. F. et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol Cell Biol 15, 3206–16 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C. & van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene 26, 7731–40 (2007). [DOI] [PubMed] [Google Scholar]
- Grawunder U. et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388, 492–5 (1997). [DOI] [PubMed] [Google Scholar]
- Sharpless N. E. et al. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol Cell 8, 1187–96 (2001). [DOI] [PubMed] [Google Scholar]
- Chistiakov D. A., Voronova N. V. & Chistiakov P. A. Genetic variations in DNA repair genes, radiosensitivity to cancer and susceptibility to acute tissue reactions in radiotherapy-treated cancer patients. Acta Oncologica 47, 809–24 (2008). [DOI] [PubMed] [Google Scholar]
- Chrzanowska K. H., Gregorek H., Dembowska-Baginska B., Kalina M. A. & Digweed M. Nijmegen breakage syndrome (NBS). Orphanet J Rare Dis 7, 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce C. L. et al. Validating genetic risk associations for ovarian cancer through the international Ovarian Cancer Association Consortium. Br J Cancer 100, 412–20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah P. & Breast Canc Assoc C. Commonly studied single-nucleotide polymorphisms and breast cancer: Results from the Breast Cancer Association Consortium. J Natl Cancer I 98, 1382–96 (2006). [DOI] [PubMed] [Google Scholar]
- Tuteja N. & Tuteja R. Unraveling DNA repair in human: molecular mechanisms and consequences of repair defect. Crit Rev Biochem Mol Biol 36, 261–90 (2001). [DOI] [PubMed] [Google Scholar]
- Zhou L. P. et al. Lack of association between LIG4 gene polymorphisms and the risk of breast cancer: a HuGE review and meta-analysis. Asian Pac J Cancer Prev 13, 3417–22 (2012). [DOI] [PubMed] [Google Scholar]
- Liu Y. H. et al. Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 Genes Involved in the Double-Strand Break Repair Pathway Predict Glioblastoma Survival. J Clin Oncol 28, 2467–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbrouck J. et al. Acute normal tissue reactions in head-and-neck cancer patients treated with IMRT: influence of dose and association with genetic polymorphisms in DNA DSB repair genes. Int J Radiat Oncol Biol Phys 73, 1187–95 (2009). [DOI] [PubMed] [Google Scholar]
- Grawunder U., Zimmer D., Fugmann S., Schwarz K. & Lieber M. R. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell 2, 477–84 (1998). [DOI] [PubMed] [Google Scholar]
- Henriquez-Hernandez L. A. et al. Polymorphisms in DNA-repair genes in a cohort of prostate cancer patients from different areas in Spain: heterogeneity between populations as a confounding factor in association studies. PLoS One 8, e69735 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. M. et al. Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res 11, 4620–6 (2005). [DOI] [PubMed] [Google Scholar]
- Liu Y. H. et al. Polymorphisms of LIG4 and XRCC4 involved in the NHEJ pathway interact to modify risk of glioma. Hum Mutat 29, 381–9 (2008). [DOI] [PubMed] [Google Scholar]
- O'Driscoll M. et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell 8, 1175–85 (2001). [DOI] [PubMed] [Google Scholar]
- Pierce A. J. & Jasin M. NHEJ deficiency and disease. Mol Cell 8, 1160–1 (2001). [DOI] [PubMed] [Google Scholar]
- Reeves S. G. et al. DNA repair gene polymorphisms and risk of early onset colorectal cancer in Lynch syndrome. Cancer Epidemiol 36, 183–9 (2012). [DOI] [PubMed] [Google Scholar]
- Shin A. et al. Genotype-phenotype Relationship between DNA Repair Gene Genetic Polymorphisms and DNA Repair Capacity. Asian Pac J Cancer P 9, 501–5 (2008). [PubMed] [Google Scholar]
- Ding Z. M. et al. The LIG4 Ile658Val polymorphism does not affect the risk of cervical carcinoma. Pathol Res Pract 206, 556–9 (2010). [DOI] [PubMed] [Google Scholar]
- Romanowicz-Makowska H. et al. Genetic analysis of the polymorphisms in nonhomologous DNA end joining gene Ku70 and Ligase IV in postmenopausal breast cancer women. Prz Menopauzalny 9, 296–9 (2010). [Google Scholar]
- Sakiyama T. et al. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer 114, 730–7 (2005). [DOI] [PubMed] [Google Scholar]
- Schildkraut J. M. et al. Association between DNA damage response and repair genes and risk of invasive serous ovarian cancer. PLoS One 5, e10061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczuk A. et al. Analysis of the polymorphisms in non-homologous DNA end joining (NHEJ) gene Ku70 and Ligase IV in sporadic breast cancer in women. Pol J Pathol 61, 27–31 (2010). [PubMed] [Google Scholar]
- Yu H. P. et al. An analysis of single nucleotide polymorphisms of 125 DNA repair genes in the Texas genome-wide association study of lung cancer with a replication for the XRCC4 SNPs. DNA Repair 10, 398–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hadyan K. S., Al-Harbi N. M., Al-Qahtani S. S. & Alsbeih G. A. Involvement of single-nucleotide polymorphisms in predisposition to head and neck cancer in Saudi Arabia. Genet Test Mol Biomarkers 16, 95–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae J., Varon R., Sperling K. & Seeger K. Polymorphisms in the DNA ligase IV gene might influence the risk of acute lymphoblastic leukemia in children. Leukemia 21, 2226–7 (2007). [DOI] [PubMed] [Google Scholar]
- Assis J., Gomes M., Marques D., Pereira D. & Medeiros R. Influence of LIG4 1977 T/C polymorphism in ovarian cancer (OC) susceptibility. Ann Oncol 21, 59–59 (2010). [Google Scholar]
- Fu Y. P. et al. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res 63, 2440–6 (2003). [PubMed] [Google Scholar]
- Garcia-Closas M. et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet 119, 376–88 (2006). [DOI] [PubMed] [Google Scholar]
- Gomes B. C. et al. The role of common variants of non-homologous end-joining repair genes XRCC4, LIG4 and Ku80 in thyroid cancer risk. Oncol Rep 24, 1079–85 (2010). [PubMed] [Google Scholar]
- Han J. L., Hankinson S. E., Ranu H., De Vivo I. & Hunter D. J. Polymorphisms in DNA double-strand break repair genes and breast cancer risk in the Nurses' Health Study. Carcinogenesis 25, 189–95 (2004). [DOI] [PubMed] [Google Scholar]
- Hill D. A. et al. Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood 108, 3161–7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska A. et al. BRCA1-associated breast and ovarian cancer risks in Poland: no association with commonly studied polymorphisms. Breast Cancer Res Treat 119, 201–11 (2010). [DOI] [PubMed] [Google Scholar]
- Kuschel B. et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet 11, 1399–407 (2002). [DOI] [PubMed] [Google Scholar]
- Li D. H. et al. DNA Repair Gene Polymorphisms and Risk of Pancreatic Cancer. Clin Cancer Res 15, 740–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddam P. L. et al. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination. J Med Genet 39, 900–5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salagovic J., Klimcakova L., Ilencikova D. & Kafkova A. Association of follicular lymphoma risk with BRCA2 N372H polymorphism in Slovak population. Med Oncol 29, 1173–8 (2012). [DOI] [PubMed] [Google Scholar]
- Tseng R. C. et al. Lung Cancer Susceptibility and Prognosis Associated With Polymorphisms in the Nonhomologous End-joining Pathway Genes. Cancer 115, 2939–48 (2009). [DOI] [PubMed] [Google Scholar]
- Werbrouck J. et al. Single-nucleotide polymorphisms in DNA double-strand break repair genes: association with head and neck cancer and interaction with tobacco use and alcohol consumption. Mutat Res 656, 74–81 (2008). [DOI] [PubMed] [Google Scholar]
- Wu X. et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet 78, 464–79 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P. et al. Genetic polymorphisms of DNA double-strand break repair pathway genes and glioma susceptibility. BMC Cancer 13, 234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K. K. & Jackson S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27, 247–54 (2001). [DOI] [PubMed] [Google Scholar]
- van Gent D. C., Hoeijmakers J. H. & Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2, 196–206 (2001). [DOI] [PubMed] [Google Scholar]
- Shen M. R., Jones I. M. & Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res 58, 604–8 (1998). [PubMed] [Google Scholar]
- Wang E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett 340, 261–9 (2013). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun 1, 34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran W. The combination of estimates from different experiments. Biometrics 10, 101–29 (1954). [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–48 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–88 (1986). [DOI] [PubMed] [Google Scholar]
- Higgins J. P. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Stat Med 21, 1539–58 (2002). [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–34 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterslev J., Thorlund K., Brok J. & Gluud C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J Clin Epidemiol 61, 64–75 (2008). [DOI] [PubMed] [Google Scholar]
- Brok J., Thorlund K., Wetterslev J. & Gluud C. Apparently conclusive meta-analyses may be inconclusive--Trial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int J Epidemiol 38, 287–98 (2009). [DOI] [PubMed] [Google Scholar]