Abstract
Poised (bivalent) chromatin is defined by the simultaneous presence of histone modifications associated with both gene activation and repression. This epigenetic feature was first observed at promoters of lineage-specific regulatory genes in embryonic stem cells in culture. More recent work has shown that, in vivo, mammalian germ cells maintain poised chromatin at promoters of many genes that regulate somatic development, and that they retain this state from fetal stages through meiosis and gametogenesis. We hypothesize that the poised chromatin state is essential for germ cell identity and function. We propose three roles for poised chromatin in the mammalian germ line: prevention of DNA methylation, maintenance of germ cell identity and preparation for totipotency. We discuss these roles in the context of recently proposed models for germline potency and epigenetic inheritance.
Keywords: Poised, Bivalent, Germ line, Germ cell, Chromatin, Pluripotent
Introduction
Poised, or bivalent, chromatin – chromatin domains bearing both the activation-associated histone modification H3K4me3 and the repression-associated modification H3K27me3 – was first identified at developmental gene promoters in embryonic stem cells (ESCs) (Fig. 1) (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007). In general, poised chromatin is correlated with pluripotency: pluripotent cells have high numbers of poised domains compared with more differentiated cells, and these domains tend to resolve toward a purely active or repressed state during differentiation (Mikkelsen et al., 2007; Cui et al., 2009; Hattori et al., 2013), although exceptions have been reported (Mohn et al., 2008). As a result, the poised chromatin state is thought to be functionally associated with, and important for, pluripotency (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007). Several recent studies (Hammoud et al., 2009, 2014; Mochizuki et al., 2012; Erkek et al., 2013; Lesch et al., 2013; Ng et al., 2013; Sachs et al., 2013) have collectively shown that poised chromatin is maintained at developmentally critical gene promoters at multiple stages of male and female germ cell development in the mouse (Table 1). Germ cells represent a unique in vivo cell population: although they undergo extensive cellular differentiation as unipotent cells during gametogenesis, they nevertheless contribute to a totipotent embryo at fertilization (Fig. 2A). We postulate that the maintenance of a poised chromatin state in germ cells at promoters of developmental regulatory genes spanning all somatic lineages holds the key to this apparent paradox, and represents an essential in vivo function of this epigenetic state. Here, we first review the evidence for maintenance of poised chromatin in the mammalian germ line, before discussing three mutually compatible potential roles for the poised state in germ cell biology. We then highlight the possible effects of perturbation of the germline-poised state, and outline some of the many remaining questions regarding the role and regulation of poised chromatin in the germ line.
Fig. 1.
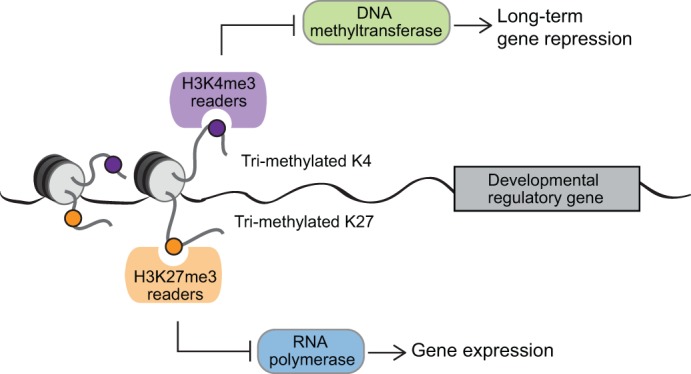
The poised chromatin state. Summary of the opposing effects of H3K4me3 and H3K27me3, and their proposed roles at poised promoters in the mammalian germ line. Protein complexes that interact with H3K4me3 inhibit the activity of DNA methyltransferases, preventing DNA methylation and the associated chromatin condensation and long-term gene repression. Protein complexes that interact with H3K27me3 prevent active transcription, thus maintaining somatic gene silencing in the germ line.
Table 1.
Published datasets examining chromatin state in the mammalian germ line
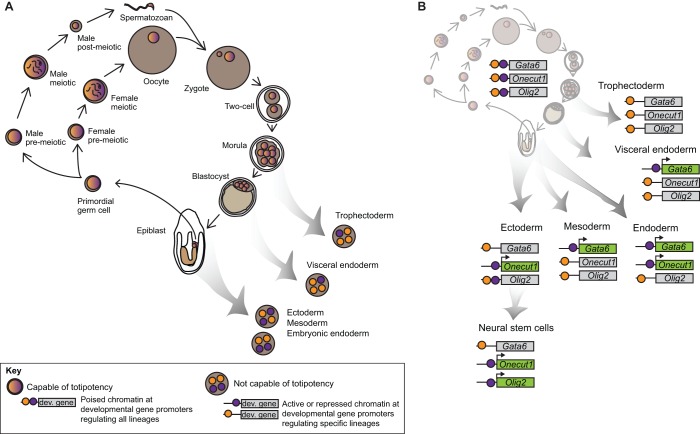
Fig. 2.
Poised chromatin in the germ line and in somatic lineages. Proposed role for poised chromatin at developmental regulators in maintaining germline identity and the ability to contribute to a totipotent zygote. (A) In developing germ cells, in the preimplantation embryo and in the early epiblast, cells maintain a poised state at a broad complement of developmental regulators, including regulators of all major somatic lineages (multicolored nuclei). Any of these cells is capable of contributing to the germ line and to any somatic or extraembryonic lineage, making them totipotent. Upon resolution of a subset of poised genes (purple and orange nuclei), cells enter a differentiating state and become restricted in their potential. (B) Three representative poised genes (Gata6, Onecut1 and Olig2) in the mouse. All of these genes are poised in cells of the germ line at multiple stages (upper left). During differentiation, these genes become restricted to particular lineages, and the poised state is resolved to either an activated (H3K4me3, purple) or repressed (H3K27me3, orange) state. Note that mesoderm and visceral endoderm have the same combination of active and repressive marks for these three genes, but would be expected to differ in chromatin state at other poised genes not shown in the figure.
Evidence for maintenance of a poised chromatin state in mammalian germ cells
In mammals, germ cells arise at around the time of gastrulation [about embryonic day (E) 6.25] from a larger pool of cells with apparently equivalent developmental potential (Tam and Zhou, 1996; Saitou et al., 2002). From that time until about E12.5, germ cells are fate restricted as gamete precursors but are thought to be ‘pluripotent-like’: they maintain expression of the pluripotency-associated regulators Oct4 (Pou5f1), Sox2 and Nanog, and they can form self-renewing pluripotent cell lines [embryonic germ (EG) lines] in culture (Rosner et al., 1990; McLaren, 2003; Yamaguchi et al., 2005; Yabuta et al., 2006). Two recent studies interrogated genome-wide H3K4me3 and H3K27me3 placement in E11.5 (Sachs et al., 2013) and E12.5 (Lesch et al., 2013) germ cells, and found that these early germ cells share an additional feature with ESCs: they retain a poised chromatin state at the promoters of a broad set of developmental regulatory genes. These poised genes include regulators of all major somatic lineages, and are not expressed in the germ line at time points for which data are available (Yamaguchi et al., 2013) (Table 2). The poised gene sets agree well across independent datasets (50-80% overlap, see Table 2). Depending on the criteria applied, estimates of the number of poised promoters range widely, from several hundred to about 4300. More surprisingly, 30-60% of poised genes retain a poised epigenetic state in both male and female germ cells following the onset of sexual differentiation and female meiosis at E13.5 (Mochizuki et al., 2012; Lesch et al., 2013; Ng et al., 2013; Sachs et al., 2013) and E14.5 (Lesch et al., 2013) (Table 2). This apparent stability during differentiation contrasts with events in in vitro stem cell populations, where promoters that are poised in pluripotent stem cells tend to resolve toward a univalent state, either active or repressed, as differentiation progresses and developmental potential becomes more restricted (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007).
Table 2.
Status of genes poised in E11.5 or E12.5 mouse germ cells at later time points
In male germ cells, retention of poised chromatin continues into postnatal stages. It is present in adult germline stem cells (Hammoud et al., 2014), which, like early fetal germ cells, can give rise to EG cell lines in culture (Kanatsu-Shinohara et al., 2004). Furthermore, differentiating adult male germ cells that have initiated or completed meiosis (pachytene spermatocytes or round spermatids, respectively) also retain a poised epigenetic state at a majority of the same genes that are poised in early fetal germ cells, thus highlighting the maintenance of the bivalent state throughout the germline cycle (Lesch et al., 2013). Retention of poised chromatin during and after meiosis, a time when extensive chromatin reorganization associated with DNA recombination occurs and pluripotency regulators such as Oct4 are turned off (Pesce et al., 1998), argues for the importance of actively maintaining this chromatin state throughout gametogenesis, even when the germ cells have lost other features of pluripotency.
These recent data now allow us to contextualize genome-wide analyses of histone modifications in mature spermatozoa in human and mouse (Hammoud et al., 2009; Brykczynska et al., 2010; Erkek et al., 2013). Although most histones are replaced by protamines during spermatogenesis, a few are retained, estimated to represent about 1% of total genome coverage in mouse (Brykczynska et al., 2010; Erkek et al., 2013) and 4-10% in human (Hammoud et al., 2009; Brykczynska et al., 2010). Overwhelmingly, retained histones are found at CpG islands near the promoters of developmental regulatory genes, including about half of the genes that are poised in fetal germ cells; many of these retained histones also carry H3K4me3 and H3K27me3 modifications (Hammoud et al., 2009; Vavouri and Lehner, 2011; Erkek et al., 2013). Together, these data indicate that the poised state is present at developmental promoters throughout germ cell development and is retained in mature gametes, at least in males. This stability suggests a fundamental role for the poised chromatin state in the germ line. We now propose three biological functions for poised chromatin in mammalian germ cells: antagonism of DNA methylation at developmental promoters, maintenance of germ cell identity and preparation for totipotency after fertilization.
Proposed role 1: prevention of DNA methylation at key developmental promoters
Early work in ESCs established a strong association between poised chromatin and DNA hypomethylation (Meissner et al., 2008). In both ESCs and germ cells, poised chromatin domains are highly correlated with CpG islands – regions with a high density of CpG dinucleotides that usually reside in the promoters of housekeeping and developmental genes and lack DNA methylation (Bernstein et al., 2006; Meissner et al., 2008). H3K4 methylation and DNA cytosine methylation are associated with opposite transcriptional states: methylated H3K4 is found in regions of open chromatin or active transcription, whereas DNA methylation promotes condensed chromatin and long-term transcriptional repression. Moreover, their deposition is mutually antagonistic: DNA methylation interferes with the recruitment of H3K4 methyltransferase complexes (Box 1), and the presence of methylated H3K4 interferes with the recruitment and activity of the DNA methyltransferases DNMT3A and DNMT3B (reviewed by Voigt et al., 2013). In stem cells, loci that lose both H3K4me3 and H3K27me3 marks after multiple passages in culture have a high likelihood of gaining DNA methylation and becoming hypermethylated (Meissner et al., 2008).
Box 1. Polycomb and Trithorax complexes
Trithorax group (TrxG) and Polycomb group (PcG) proteins are the principal methyltransferases responsible for setting up and maintaining the H3K4me3 and H3K27me3 modifications, respectively, at poised promoters. The subset of TrxG enzymes that catalyzes H3K4 trimethylation in mammals comprises SET1A, SET1B and mixed lineage leukemia (MLL) proteins 1-4. The SET1 and MLL enzymes form complexes (the SET1A/B and MLL complexes) and require additional complex components for enzymatic activity (reviewed by Shilatifard, 2012). It has recently been proposed that MLL2 is primarily responsible for H3K4 trimethylation at poised promoters (Denissov et al., 2014).
PcG proteins form two types of complexes, called Polycomb repressive complex 1 (PRC1) and PRC2, of which there are multiple variants with alternative subunits in mammals (reviewed by Simon and Kingston, 2013). PRC2 complexes catalyze trimethylation of H3K27; PRC1 complexes bind H3K27me3 and either catalyze ubiquitylation of H2A lysine 119 (H2AK119ub) or act to directly compact chromatin (Gao et al., 2012; Tavares et al., 2012), thereby reinforcing the repressive effects of PRC2.
Both TrxG and PcG complexes remain bound near sites of H3K4 and H3K27 histone methylation (Ku et al., 2008; Denissov et al., 2014). How these two antagonistic protein complexes interact in the context of poised chromatin domains in mammalian cells remains an active area of investigation (Schmitges et al., 2011; Voigt et al., 2012).
Retention of H3K4me3 at critical developmental promoters in the germ line might therefore be a means of preventing DNA methylation at these important regulatory sites. Cells of the germ line undergo two waves of de novo DNA methylation during their life cycle: once at implantation, when all cells of the embryo acquire methylation, and once following germ cell sexual differentiation, when maternal and paternal imprints are established (Smallwood and Kelsey, 2012). During these times, expression of de novo DNA methyltransferases peaks and genome-wide methylation levels increase rapidly (La Salle et al., 2004; Lucifero et al., 2007). The presence of H3K4me3 at important genomic regions during these periods might help to set these regions apart from the rest of the genome and preserve them in an unmethylated state. Protection from DNA methylation could serve two key functions: it might prevent long-term repression of important developmental genes, making them easier to activate during embryogenesis, and it might guard against the accumulation of mutations at important transcriptional regulatory regions. Methylated DNA is subject to spontaneous deamination of 5-methylcytosine to thymine, causing C→T transition mutations and leading to CpG depletion over many generations (Coulondre et al., 1978; Cohen et al., 2011). Inhibition of DNA methylation by retention of H3K4me3 at key promoter regions in the germ line would prevent these effects, and could therefore help to ensure that developmental promoters respond reliably to regulatory signals during embryogenesis on both single and multi-generational time scales. In this context, it will be important to compare the set of poised genes in germ cells with the set of methylated genes in early mouse (Smallwood et al., 2011; Smith et al., 2012) and human (Guo et al., 2014; Smith et al., 2014) embryos.
Inhibition of DNA methylation provides a possible role for H3K4me3, but not for H3K27me3, at poised promoters. However, it is significant that DNA hypomethylation and H3K4me3 deposition are typically associated with gene activation, whereas regulators of somatic development must remain silent in the germ line throughout the life cycle. This might explain the retention of H3K27me3 along with H3K4me3 at poised promoters of developmental genes: H3K27me3 might help to prevent their activation, balancing the transcriptionally permissive state associated with H3K4me3 and DNA hypomethylation (Fig. 1). By contrast, housekeeping genes associated with CpG islands are transcriptionally active in both somatic and germ cells, with corresponding promoter H3K4me3 marks; for these genes, there is no conflict between H3K4me3, DNA methylation and active transcriptional status in germ cells. A third subset of CpG island genes is DNA methylated in germ cells and ESCs. These genes are regulatory, non-housekeeping genes that lack poised chromatin. Nearly all of these genes, including Dazl, Hormad1, Sycp1, Sycp3, Taf7l, Mvh (Ddx4), Tnap, Asz1, the Rhox family and the Mage family, have known functions in the germ cells themselves, are demethylated and expressed at specific times during germ cell development, and are mis-expressed temporally and spatially when DNA methyltransferases are eliminated or depleted (Maatouk et al., 2006; Fouse et al., 2008; Meissner et al., 2008; Seisenberger et al., 2012). Intriguingly, some of these genes retain DNA methylation but also recruit RNA polymerase II and undergo active transcription during spermatogenesis, constituting an alternative, ‘atypical’ mode of transcriptional regulation at methylated DNA sites (Hammoud et al., 2014). Thus, the CpG island-associated developmental regulatory genes that acquire DNA methylation are expressed in germ cells, while those that remain poised and hypomethylated are not expressed.
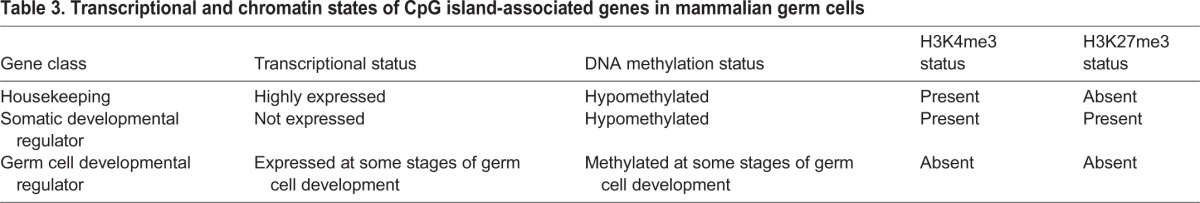
In sum, these findings suggest that functional gene classes in the germ line can be compartmentalized based on modes of epigenetic regulation, as has been previously proposed (Mochizuki et al., 2012; Erkek et al., 2013). Housekeeping genes maintain H3K4me3, are transcriptionally active and are hypomethylated. Regulators of somatic development maintain a poised, H3K4me3/H3K27me3-positive chromatin state and are transcriptionally silent but hypomethylated. Finally, regulators of germ cell development are transcriptionally regulated by mechanisms associated with DNA methylation, rather than by mechanisms associated with histone methylation (summarized in Table 3).
Table 3.
Transcriptional and chromatin states of CpG island-associated genes in mammalian germ cells
Proposed role 2: poised chromatin as mammalian germ plasm
As discussed above, poised chromatin is present not only in fetal mammalian germ cells (considered to be ‘pluripotent-like’), but also in meiotic and postmeiotic germ cells, suggesting a remarkable continuity throughout development. Both the H3K4me3 and H3K27me3 modifications are mitotically heritable (see Box 2). Continuity of a defining biochemical entity in the germ line is reminiscent of ‘germ plasm’, the specialized cytoplasmic inclusions containing specific RNAs and proteins that determine and define the germ line in many non-mammalian species. In Caenorhabditis elegans, Drosophila, Xenopus and Danio rerio, germ plasm is maternally inherited and asymmetrically segregated during early embryogenesis; information required for germ line specification is therefore passed through the egg to the germ cells and back to the egg. In these species, germ plasm is a biochemical means of specifying and maintaining the germ line across generations (Extavour and Akam, 2003; Strome and Lehmann, 2007). Like germ plasm, retention of the poised state at a broad set of genes involved in the specification of all somatic lineages might serve to set germ cells apart from surrounding somatic cell types (which may in some cases retain the poised state at a smaller, more specialized, subset of genes), effectively identifying a germ cell as a germ cell throughout the life cycle of the organism. Thus, in mammals, a specialized nuclear state rather than specialized cytoplasm might serve to specify and maintain germline identity. Interestingly, zebrafish exhibit both cytoplasmic germ plasm and poised nuclear chromatin, representing a possible evolutionary intermediate (Wu et al., 2011).
Box 2. Mitotic inheritance of H3K4me3 and H3K27me3
During mitotic division, chromosomes condense several hundredfold, and many transcription factors and chromatin regulatory factors dissociate from the chromatin (Woodcock and Ghosh, 2010). However, some H3K4me3- and H3K27me3-marked histones, as well as the histone methyltransferase complexes responsible for their deposition, remain associated with the chromosomes during mitosis (Chen et al., 2005; Terrenoire et al., 2010). In mammals, the TrxG MLL proteins (see Box 1) bind mitotic chromatin (Blobel et al., 2009). Furthermore, experiments in Xenopus and in Dictyostelium discoideum indicate a requirement for methylation of the H3K4 residue itself in mediating epigenetic memory of transcriptional activity during mitosis: when the K4 residue of histone H3 is mutated to glutamate (Ng and Gurdon, 2008) or alanine (Muramoto et al., 2010), epigenetic memory of transcriptional activity across mitotic divisions is disrupted. PcG complexes also remain associated with mitotic chromosomes, although to a lesser extent than Trithorax, and they co-localize with H3K27me3 on metaphase chromosomes in mammalian cells (Vincenz and Kerppola, 2008; Follmer et al., 2012; Fonseca et al., 2012). Although current data provide good support for the heritability of both H3K4me3 and H3K27me3 individually during cell division, the stability of the poised chromatin state during mitotic metaphase has not been thoroughly examined yet.
Indeed, August Weismann's original conception of germ plasm as a material specific to the germ cells and not the soma that is “always passed on from the germ-cell in which an organism originates in direct continuity to the germ-cells of succeeding generations”, localized it to the ‘nuclear substance’ of the germ cell, rather than to the cytoplasm (Weismann, 1893). Like both Weismann's germ plasm and the cytoplasmic substance with which the term is currently associated in various non-mammalian species, poised chromatin at a broad set of developmental genes might be continuously present in germ cells throughout the mammalian life cycle, but severely reduced and restricted to a narrow set of lineage-specific genes in somatic cell types. Loss of poised chromatin would presumably enable lineage-specific differentiation in somatic tissue, as regulators of alternative lineages lose the poised state and become repressed; conversely, retention of poised chromatin in germ cells at regulators of all somatic lineages would define them as non-somatic, and might confer on them their ability to pass nuclear material on to the next generation. Like germ plasm, poised chromatin at a broad set of developmental genes therefore links mature mammalian gametes to the early embryo, to nascent germ cells and back to mature gametes (Fig. 2A).
Poised chromatin at a set of developmental genes spanning all somatic lineages appears to be correlated with germ cell identity, but if the function of this chromatin state is analogous to that of germ plasm, then the poised state should also be functionally required for the germ cells to remain germ cells. We have already described one putative role for H3K4me3 at poised promoters (see role 1 above), although this has yet to be experimentally addressed. As a mediator of transcriptional repression, H3K27me3 also has a critical role in the germ line. Repression of somatic developmental regulators, including poised genes, is important for germ cell maintenance. In mutants such as Prdm1 (also called Blimp1) that fail to maintain repression of somatic developmental genes in the germ line, germ cells die early in their development (Ohinata et al., 2005; Kurimoto et al., 2008). Furthermore, germline-poised genes belong to a subset of ESC-poised genes that retain Polycomb repressive complex 1 (PRC1) in addition to PRC2 (see Box 1). PRC1 occupancy at PRC2-positive promoters strongly correlates with transcriptional repression. PRC1/PRC2 double-positive regions also retain H3K27me3 more efficiently upon differentiation, and their poised status is more strongly conserved between human and mouse ESCs (Ku et al., 2008). In mice, female germ cells mutant for PRC1 components exhibit impaired development and produce defective embryos (Posfai et al., 2012), and oocytes depleted for the H3K27 methyltransferase EZH2 give rise to offspring with severe growth retardation (Erhardt et al., 2003). Thus, functional evidence suggests that, like H3K4me3, H3K27me3 has an important role to play at promoters of developmental genes in germ cells. The presence of H3K4me3 and H3K27me3 together at promoters of genes regulating development of all somatic lineages defines a specialized epigenetic state that sets germ cells apart from mortal somatic lineages and, analogous to germ plasm, might be required for maintaining an essentially immortal germ cell identity.
Proposed role 3: poising for totipotency and preparing for resolution
One proposed role for the H3K4me3 mark at poised promoters in vitro is the active promotion of gene expression when differentiation begins and H3K27me3 is removed. Although still unproven, this hypothesis is supported by data demonstrating a transcription-ready state at many poised promoters. For example, 51% of poised promoters in ESCs are bound by paused polymerase, compared with 8% of non-poised promoters (data from Brookes et al., 2012), supporting the idea that they can quickly initiate transcription. Extending this model to the germ line in vivo, we speculate that pre-loading of H3K4me3 at poised promoters in the germ line might promote the rapid and efficient activation of these promoters in the early embryo as differentiation of the soma germ layers and specific cell lineages begins.
If true, this function of poised chromatin would help to resolve one of the central questions of germline biology: how do differentiated, apparently unipotent cells (the gametes) give rise to a totipotent zygote following fertilization? Retention of a transcriptionally poised chromatin state in gametes at developmental genes regulating all somatic lineages might underlie the gamete's ability to generate a zygote capable of producing each of these lineages. Others have proposed that this ability is driven by the activity of pluripotency-associated transcription factors in the germ line (Leitch and Smith, 2013); poised chromatin might be an essential complement to such factors, especially during intervals when these pluripotency factors are not expressed in germ cells. In our model, genes that were poised in the germ cells would remain poised in the embryo during the first few cell divisions, possibly due to the influence of pluripotency factors. As somatic cells in the embryo begin to differentiate, poised genes required for differentiation into a specific lineage would start to resolve toward H3K4me3- or H3K27me3-only states, depending on the identity of the differentiating tissue and the need to express or repress the corresponding genes in that tissue. At the same time, the incipient germ cell lineage would retain a complement of poised genes corresponding to all somatic lineages (Fig. 2). Thus, the broad and inclusive poised chromatin state transmitted in the germ line would enable the progressive restriction of developmental potential in somatic lineages in the embryo, promoting execution of a totipotency program. Poised chromatin might therefore mediate ‘intrinsic transgenerational inheritance’, a recently proposed concept in which a hypothetical germ cell chromatin state influences somatic development in the next generation (Gill et al., 2012).
This model can be illustrated using three representative genes: Gata6, Onecut1 and Olig2. The predicted chromatin states of these genes based on their expression patterns during embryogenesis are shown in Fig. 2B. All three have been shown to be poised in the germ line. The model predicts that Gata6 would resolve to an H3K27me3-only state in the trophectoderm, and to an H3K4me3-only state in visceral endoderm, where it is known to be expressed and to play a crucial role in the developing yolk sac (Koutsourakis et al., 1999; Rugg-Gunn et al., 2010). Gata6 is also expressed in embryonic endoderm and mesoderm, where, according to the model, it would resolve to an H3K4me3-only state, but not in ectoderm, where it would resolve to an H3K27me3-only state (Koutsourakis et al., 1999). Onecut1 plays a critical role in ectoderm and endoderm development, but is not expressed in mesoderm (Landry et al., 1997); Onecut1 would therefore resolve to H3K4me3 in endoderm and ectoderm, and to H3K27me3 in mesoderm. By contrast, Olig2 is a neural lineage-specific transcription factor that is expressed in oligodendrocyte precursors and astrocytes of the developing brain and spinal cord and is important for their differentiation (Lu et al., 2000; Zhou and Anderson, 2002; Liu and Rao, 2004). Olig2 would be expected to remain poised in early ectoderm and to resolve fully at later stages of development (e.g. in neural stem cells). In fact, Olig2 was found to resolve to an active H3K4me3-only state in neural progenitor cells differentiated from mESCs (Mikkelsen et al., 2007).
Currently available data from early mouse embryos are consistent with this model. The vast majority of our knowledge of the properties and regulation of poised genes comes from ESCs, which are derived from the inner cell mass of the blastocyst and serve as a proxy for a pluripotent, pre-differentiation state. Nearly all (88%) genes that are poised in the germ line from fetal through post-meiotic stages (Lesch et al., 2013) remain poised in ESCs (Mikkelsen et al., 2007). In vivo ChIP-seq data are difficult to obtain from the small number of cells in the preimplantation embryo, but ChIP-qPCR data are available from early mouse embryos at E5.5. At this stage, mouse embryos have separated the epiblast, which will form all tissues of the embryo proper, from the extraembryonic ectoderm and visceral endoderm, two lineages that will form extraembryonic tissues. ChIP-qPCR data from microdissected epiblast, extraembryonic ectoderm and visceral endoderm at E5.5 (Rugg-Gunn et al., 2010) indicate that germline-poised genes that are involved in differentiation of extraembryonic ectoderm (Cdx2) and visceral endoderm (Gata6, Sox17) resolve to an active or repressed state depending on the tissue in which they reside, whereas other developmental regulatory genes that are poised in the germ line (Lesch et al., 2013; Sachs et al., 2013) remain poised in the pluripotent epiblast (Rugg-Gunn et al., 2010).
Consequences of perturbing the poised state
If poised chromatin in the germ line promotes transcriptional activation in the early embryo, then disruption of the poised state in germ cells should alter gene expression dynamics in the embryo. Aberrant epigenetic information in the germ line of one generation, in the form of perturbation of poised chromatin in germ cells, could therefore be transferred to the somatic tissues of the next generation in the form of impaired or premature activation of developmental regulatory gene transcription. Consistent with this model, depletion of the PRC2 complex subunit EZH2 in oocytes results in reduced H3K27 methylation in embryos and in severe growth retardation in juvenile offspring, even when an intact embryonic Ezh2 allele is present (Erhardt et al., 2003). Additionally, maternal depletion of MLL2, the H3K4 methyltransferase hypothesized to be required for bivalent promoter methylation (Denissov et al., 2014), results in oocyte death (full knockout allele) or in impaired zygotic gene activation and early embryonic lethality (hypomorphic allele) (Andreu-Vieyra et al., 2010). Transfer of epigenetic information across generations might be direct or indirect: histones bearing H3K4me3 and H3K27me3 modifications might themselves be carried in sperm or egg and retained in the nucleus of the zygote, or they might recruit intermediate carriers, such as other chromatin proteins or non-coding RNAs, during the late stages of spermatogenesis or oogenesis. Whether direct or indirect, if information carried by poised chromatin in the germ line is inherited from gamete to zygote, it could provide a molecular basis for transgenerational epigenetic effects on offspring phenotypes in mammals (Carone et al., 2010; Ng et al., 2010).
Intriguingly, some of the same genes that are poised in germ cells are frequently DNA-hypermethylated in cancer (Ohm et al., 2007; Mack et al., 2014) and in aging cells (Rakyan et al., 2010). For example, Sfrp2 is poised in both meiotic and postmeiotic mouse spermatogenic cells, and its ortholog SFRP2 is hypermethylated in multiple human cancer types, including colon, renal and blood cancers (Ohm et al., 2007). A recent study of type A ependymomas revealed a strong DNA hypermethylation signature, along with increased levels of H3K27me3, at developmental regulatory genes in these tumors, including SFRP2 and other genes, the orthologs of which are poised in the mouse germ line (Mack et al., 2014). As described above (Proposed role 1), poised chromatin may act in part to protect the promoters of developmentally potent genes from DNA methylation in the germ line. The correlation between cancer-methylated and germline-poised genes suggests that perturbation or imbalance of the poised state in the germ line could leave promoters vulnerable to DNA methylation, perhaps altering susceptibility to cancer and aging-associated disease in adult offspring. This possibility does not exclude the initiation of aberrant DNA methylation in somatic lineages following differentiation, independent of germline influence, but might help to explain some cases of cancer-associated DNA hypermethylation. Verifying the presence of the poised chromatin state at these genes might therefore be a crucial test for germ cells derived using in vitro systems and intended for clinical use. It might also be an important quality check for sperm and oocytes isolated for use in more conventional in vitro fertilization procedures.
Conclusions
Poised chromatin is a well-defined feature of stem cells in vitro, but its meaning in vivo is only beginning to be explored. Accumulating evidence indicates that poised chromatin is present in the mammalian germ line at the promoters of genes required for differentiation of all somatic lineages across multiple stages of germ cell development. We propose that the biological role of the poised epigenetic state in the germ line is to regulate somatic developmental gene expression across generations, and that this role in the germ line might in fact be the most critical function of the poised chromatin state in vivo. The concurrent presence of H3K4me3 and H3K27me3 allows germ cells to protect crucial developmental genes from inappropriate DNA methylation that could lead to an increased risk of mutation, while simultaneously preventing the expression of somatic developmental regulators in germ cells. This poised state might help to define germ cell identity and to promote the critical transition from differentiated gamete to totipotent zygote at fertilization. Previously described examples of poised genes in differentiated somatic cells would then reflect the end stages of a long process of progressive differentiation and resolution of poised promoters, as somatic lineages diverge from the germ line. Following fertilization, a set of poised genes would resolve in somatic lineages during formation of the extraembryonic tissues and primordial germ layers, as these tissues differentiate and lose the ability to contribute to the germ line. As development continues, increasing numbers of poised genes would resolve in each lineage, until only a small subset of lineage-specific poised genes remained in multipotent progenitor cells or in terminally differentiated cell types.
A number of critical experiments remain to be performed. First, experimental evidence for prevention of DNA methylation – and consequent prevention of C→T mutation – by H3K4me3 at poised loci in the germ line will be crucial to support the proposed role of H3K4me3 as a DNA methylation inhibitor. Second, definitive evidence for a role for germline-poised chromatin during early embryonic development is lacking. Challenging, but essential, functional studies must be carried out to directly perturb the chromatin state in germ cells and connect it to phenotypic effects in early embryos. Third, it remains to be determined whether the histone modifications associated with poised chromatin remain stable at poised genes during development, or whether they recruit additional chromatin factors to act as intermediate carriers and relay information during critical transitions. Fourth, no genome-wide chromatin data exist for germ cells in the important period surrounding their specification, between E6.5 and E7.5. Others have proposed that germ cells are specified from a mesodermal intermediate at this point (Kurimoto et al., 2008; Hayashi and Surani, 2009; Aramaki et al., 2013). Demonstrating that poised chromatin is retained at developmental genes corresponding to all somatic lineages in E6.5-E7.5 germ cells would support the model that the germ line remains segregated from soma throughout this period (Leitch and Smith, 2013). Fifth, the extent to which the poised gene complements of the maternal and paternal germ lines diverge has yet to be evaluated; poising of different gene sets in the male and female germ lines might correspond to biologically significant differences in maternal and paternal influences on inheritance and development.
Finally, it is not clear how deep the germline-poised state extends into the evolutionary tree. Zebrafish sperm carry multivalent chromatin domains, including both H3K4me3 and H3K27me3, implying that poised germline chromatin is not limited to mammals (Wu et al., 2011). By contrast, there is little evidence for poised chromatin in the germ line or soma of either Drosophila or C. elegans. It is possible that a requirement for poised chromatin in the germ line is related to the timing of transcriptional initiation in developing germ cells; Drosophila and C. elegans maintain global transcriptional repression in the germ line throughout much of their development and might not require the protected, repressed epigenetic state conferred by poised chromatin on developmental genes in mammalian germ cells. Alternatively, only organisms with extensive DNA methylation at CpG sites, including zebrafish and mammals, but not Drosophila or C. elegans, might require the protective effects of poised chromatin in the germ line. Elucidation of these questions promises to illuminate fundamental aspects of the biology of inheritance, the germ line and somatic development.
Acknowledgements
We thank D. Bellott, H. Christensen, J. McCarrey, P. Nicholls, K. Romer and S. Soh for advice and critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing financial interests.
Funding
Funded by the Howard Hughes Medical Institute (HHMI) and the National Institutes of Health (NIH). Deposited in PMC for release after 12 months.
References
- Andreu-Vieyra, C. V., Chen, R., Agno, J. E., Glaser, S., Anastassiadis, K., Stewart, A. F. and Matzuk, M. M. (2010). MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 8, e1000453 10.1371/journal.pbio.1000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramaki, S., Hayashi, K., Kurimoto, K., Ohta, H., Yabuta, Y., Iwanari, H., Mochizuki, Y., Hamakubo, T., Kato, Y., Shirahige, K.et al. (2013). A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell 27, 516-529 10.1016/j.devcel.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Azuara, V., Perry, P., Sauer, S., Spivakov, M., Jørgensen, H. F., John, R. M., Gouti, M., Casanova, M., Warnes, G., Merkenschlager, M.et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532-538 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., Fry, B., Meissner, A., Wernig, M., Plath, K.et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315-326 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Blobel, G. A., Kadauke, S., Wang, E., Lau, A. W., Zuber, J., Chou, M. M. and Vakoc, C. R. (2009). A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 36, 970-983 10.1016/j.molcel.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, E., de Santiago, I., Hebenstreit, D., Morris, K. J., Carroll, T., Xie, S. Q., Stock, J. K., Heidemann, M., Eick, D., Nozaki, N.et al. (2012). Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10, 157-170 10.1016/j.stem.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska, U., Hisano, M., Erkek, S., Ramos, L., Oakeley, E. J., Roloff, T. C., Beisel, C., Schübeler, D., Stadler, M. B. and Peters, A. H. F. M. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679-687 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- Carone, B. R., Fauquier, L., Habib, N., Shea, J. M., Hart, C. E., Li, R., Bock, C., Li, C., Gu, H., Zamore, P. D.et al. (2010). Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143, 1084-1096 10.1016/j.cell.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Dundr, M., Wang, C., Leung, A., Lamond, A., Misteli, T. and Huang, S. (2005). Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168, 41-54 10.1083/jcb.200407182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, N. M., Kenigsberg, E. and Tanay, A. (2011). Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell 145, 773-786 10.1016/j.cell.2011.04.024 [DOI] [PubMed] [Google Scholar]
- Coulondre, C., Miller, J. H., Farabaugh, P. J. and Gilbert, W. (1978). Molecular basis of base substitution hotspots in Escherichia coli. Nature 274, 775-780 10.1038/274775a0 [DOI] [PubMed] [Google Scholar]
- Cui, K., Zang, C., Roh, T.-Y., Schones, D. E., Childs, R. W., Peng, W. and Zhao, K. (2009). Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4, 80-93 10.1016/j.stem.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov, S., Hofemeister, H., Marks, H., Kranz, A., Ciotta, G., Singh, S., Anastassiadis, K., Stunnenberg, H. G. and Stewart, A. F. (2014). Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141, 526-537 10.1242/dev.102681 [DOI] [PubMed] [Google Scholar]
- Erhardt, S., Su, I.-H., Schneider, R., Barton, S., Bannister, A. J., Perez-Burgos, L., Jenuwein, T., Kouzarides, T., Tarakhovsky, A. and Surani, M. A. (2003). Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130, 4235-4248 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- Erkek, S., Hisano, M., Liang, C.-Y., Gill, M., Murr, R., Dieker, J., Schübeler, D., van der Vlag, J., Stadler, M. B. and Peters, A. H. F. M. (2013). Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 20, 868-875 10.1038/nsmb.2599 [DOI] [PubMed] [Google Scholar]
- Extavour, C. G. and Akam, M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869-5884 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- Follmer, N. E., Wani, A. H. and Francis, N. J. (2012). A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet. 8, e1003135 10.1371/journal.pgen.1003135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, J. P., Steffen, P. A., Muller, S., Lu, J., Sawicka, A., Seiser, C. and Ringrose, L. (2012). In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev. 26, 857-871 10.1101/gad.184648.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse, S. D., Shen, Y., Pellegrini, M., Cole, S., Meissner, A., Van Neste, L., Jaenisch, R. and Fan, G. (2008). Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3K4/K27 trimethylation. Cell Stem Cell 2, 160-169 10.1016/j.stem.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., Zhang, J., Bonasio, R., Strino, F., Sawai, A., Parisi, F., Kluger, Y., Reinberg, D. (2012). PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344-356 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, M. E., Erkek, S. and Peters, A. H. F. M. (2012). Parental epigenetic control of embryogenesis: a balance between inheritance and reprogramming? Curr. Opin. Cell Biol. 24, 387-396 10.1016/j.ceb.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., Yan, J., Ren, X., Lin, S., Li, J.et al. (2014). The DNA methylation landscape of human early embryos. Nature 511, 606-610 10.1038/nature13544 [DOI] [PubMed] [Google Scholar]
- Hammoud, S. S., Nix, D. A., Zhang, H., Purwar, J., Carrell, D. T. and Cairns, B. R. (2009). Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473-478 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud, S. S., Low, D. H. P., Yi, C., Carrell, D. T., Guccione, E. and Cairns, B. R. (2014). Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239-253 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Hattori, N., Niwa, T., Kimura, K., Helin, K. and Ushijima, T. (2013). Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells. Nucleic Acids Res. 41, 7231-7239 10.1093/nar/gkt528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K. and Surani, M. A. (2009). Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell 4, 493-498 10.1016/j.stem.2009.05.007 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara, M., Inoue, K., Lee, J., Yoshimoto, M., Ogonuki, N., Miki, H., Baba, S., Kato, T., Kazuki, Y., Toyokuni, S.et al. (2004). Generation of pluripotent stem cells from neonatal mouse testis. Cell 119, 1001-1012 10.1016/j.cell.2004.11.011 [DOI] [PubMed] [Google Scholar]
- Koutsourakis, M., Langeveld, A., Patient, R., Beddington, R. and Grosveld, F. (1999). The transcription factor GATA6 is essential for early extraembryonic development. Development 126, 723-732. [PubMed] [Google Scholar]
- Ku, M., Koche, R. P., Rheinbay, E., Mendenhall, E. M., Endoh, M., Mikkelsen, T. S., Presser, A., Nusbaum, C., Xie, X., Chi, A. S.et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto, K., Yabuta, Y., Ohinata, Y., Shigeta, M., Yamanaka, K. and Saitou, M. (2008). Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 22, 1617-1635 10.1101/gad.1649908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle, S., Mertineit, C., Taketo, T., Moens, P. B., Bestor, T. H. and Trasler, J. M. (2004). Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 268, 403-415 10.1016/j.ydbio.2003.12.031 [DOI] [PubMed] [Google Scholar]
- Landry, C., Clotman, F., Hioki, T., Oda, H., Picard, J. J., Lemaigre, F. P. and Rousseau, G. G. (1997). HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver-enriched transcription factors. Dev. Biol. 192, 247-257 10.1006/dbio.1997.8757 [DOI] [PubMed] [Google Scholar]
- Leitch, H. G. and Smith, A. (2013). The mammalian germline as a pluripotency cycle. Development 140, 2495-2501 10.1242/dev.091603 [DOI] [PubMed] [Google Scholar]
- Lesch, B. J., Dokshin, G. A., Young, R. A., McCarrey, J. R. and Page, D. C. (2013). A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc. Natl. Acad. Sci. USA 110, 16061-16066 10.1073/pnas.1315204110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. and Rao, M. S. (2004). Olig genes are expressed in a heterogeneous population of precursor cells in the developing spinal cord. Glia 45, 67-74 10.1002/glia.10303 [DOI] [PubMed] [Google Scholar]
- Lu, Q. R., Yuk, D.-i., Alberta, J. A., Zhu, Z., Pawlitzky, I., Chan, J., McMahon, A. P., Stiles, C. D. and Rowitch, D. H. (2000). Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317-329 10.1016/S0896-6273(00)80897-1 [DOI] [PubMed] [Google Scholar]
- Lucifero, D., La Salle, S., Bourc'his, D., Martel, J., Bestor, T. H. and Trasler, J. M. (2007). Coordinate regulation of DNA methyltransferase expression during oogenesis. BMC Dev. Biol. 7, 36 10.1186/1471-213X-7-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk, D. M., Kellam, L. D., Mann, M. R. W., Lei, H., Li, E., Bartolomei, M. S. and Resnick, J. L. (2006). DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 133, 3411-3418 10.1242/dev.02500 [DOI] [PubMed] [Google Scholar]
- Mack, S. C., Witt, H., Piro, R. M., Gu, L., Zuyderduyn, S., Stütz, A. M., Wang, X., Gallo, M., Garzia, L., Zayne, K.et al. (2014). Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445-450 10.1038/nature13108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren, A. (2003). Primordial germ cells in the mouse. Dev. Biol. 262, 1-15 10.1016/S0012-1606(03)00214-8 [DOI] [PubMed] [Google Scholar]
- Meissner, A., Mikkelsen, T. S., Gu, H., Wernig, M., Hanna, J., Sivachenko, A., Zhang, X., Bernstein, B. E., Nusbaum, C., Jaffe, D. B.et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766-770 10.1038/nature07107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., Alvarez, P., Brockman, W., Kim, T.-K., Koche, R. P.et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553-560 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, K., Tachibana, M., Saitou, M., Tokitake, Y. and Matsui, Y. (2012). Implication of DNA demethylation and bivalent histone modification for selective gene regulation in mouse primordial germ cells. PLoS ONE 7, e46036 10.1371/journal.pone.0046036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn, F., Weber, M., Rebhan, M., Roloff, T. C., Richter, J., Stadler, M. B., Bibel, M. and Schübeler, D. (2008). Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755-766 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Muramoto, T., Müller, I., Thomas, G., Melvin, A. and Chubb, J. R. (2010). Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr. Biol. 20, 397-406 10.1016/j.cub.2010.01.017 [DOI] [PubMed] [Google Scholar]
- Ng, R. K. and Gurdon, J. B. (2008). Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 10, 102-109 10.1038/ncb1674 [DOI] [PubMed] [Google Scholar]
- Ng, S.-F., Lin, R. C. Y., Laybutt, D. R., Barres, R., Owens, J. A. and Morris, M. J. (2010). Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 467, 963-966 10.1038/nature09491 [DOI] [PubMed] [Google Scholar]
- Ng, J.-H., Kumar, V., Muratani, M., Kraus, P., Yeo, J.-C., Yaw, L.-P., Xue, K., Lufkin, T., Prabhakar, S. and Ng, H.-H. (2013). In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev. Cell 24, 324-333 10.1016/j.devcel.2012.12.011 [DOI] [PubMed] [Google Scholar]
- Ohinata, Y., Payer, B., O'Carroll, D., Ancelin, K., Ono, Y., Sano, M., Barton, S. C., Obukhanych, T., Nussenzweig, M., Tarakhovsky, A.et al. (2005). Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207-213 10.1038/nature03813 [DOI] [PubMed] [Google Scholar]
- Ohm, J. E., McGarvey, K. M., Yu, X., Cheng, L., Schuebel, K. E., Cope, L., Mohammad, H. P., Chen, W., Daniel, V. C., Yu, W.et al. (2007). A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237-242 10.1038/ng1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce, M., Wang, X., Wolgemuth, D. J. and Schöler, H. (1998). Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 71, 89-98 10.1016/S0925-4773(98)00002-1 [DOI] [PubMed] [Google Scholar]
- Posfai, E., Kunzmann, R., Brochard, V., Salvaing, J., Cabuy, E., Roloff, T. C., Liu, Z., Tardat, M., van Lohuizen, M., Vidal, M.et al. (2012). Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 26, 920-932 10.1101/gad.188094.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan, V. K., Down, T. A., Maslau, S., Andrew, T., Yang, T. P., Beyan, H., Whittaker, P., McCann, O. T., Finer, S., Valdes, A. M.et al. (2010). Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20, 434-439 10.1101/gr.103101.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner, M. H., Vigano, M. A., Ozato, K., Timmons, P. M., Poirie, F., Rigby, P. W. J. and Staudt, L. M. (1990). A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686-692 10.1038/345686a0 [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn, P. J., Cox, B. J., Ralston, A. and Rossant, J. (2010). Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl. Acad. Sci. USA 107, 10783-10790 10.1073/pnas.0914507107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, M., Onodera, C., Blaschke, K., Ebata, K. T., Song, J. S. and Ramalho-Santos, M. (2013). Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 3, 1777-1784 10.1016/j.celrep.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, M., Barton, S. C. and Surani, M. A. (2002). A molecular programme for the specification of germ cell fate in mice. Nature 418, 293-300 10.1038/nature00927 [DOI] [PubMed] [Google Scholar]
- Schmitges, F. W., Prusty, A. B., Faty, M., Stützer, A., Lingaraju, G. M., Aiwazian, J., Sack, R., Hess, D., Li, L., Zhou, S.et al. (2011). Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330-341 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Seisenberger, S., Andrews, S., Krueger, F., Arand, J., Walter, J., Santos, F., Popp, C., Thienpont, B., Dean, W. and Reik, W. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849-862 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard, A. (2012). The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65-95 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J. A. and Kingston, R. E. (2013). Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808-824 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood, S. A. and Kelsey, G. (2012). De novo DNA methylation: a germ cell perspective. Trends Genet. 28, 33-42 10.1016/j.tig.2011.09.004 [DOI] [PubMed] [Google Scholar]
- Smallwood, S. A., Tomizawa, S.-i., Krueger, F., Ruf, N., Carli, N., Segonds-Pichon, A., Sato, S., Hata, K., Andrews, S. R. and Kelsey, G. (2011). Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811-814 10.1038/ng.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, Z. D., Chan, M. M., Mikkelsen, T. S., Gu, H., Gnirke, A., Regev, A. and Meissner, A. (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339-344 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, Z. D., Chan, M. M., Humm, K. C., Karnik, R., Mekhoubad, S., Regev, A., Eggan, K. and Meissner, A. (2014). DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611-615 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S. and Lehmann, R. (2007). Germ versus soma decisions: lessons from flies and worms. Science 316, 392-393 10.1126/science.1140846 [DOI] [PubMed] [Google Scholar]
- Tam, P. P. L. and Zhou, S. X. (1996). The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 178, 124-132 10.1006/dbio.1996.0203 [DOI] [PubMed] [Google Scholar]
- Tavares, L., Dimitrova, E., Oxley, D., Webster, J., Poot, R., Demmers, J., Bezstarosti, K., Taylor, S., Ura, H., Koide, H.et al. (2012). RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664-678 10.1016/j.cell.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrenoire, E., McRonald, F., Halsall, J. A., Page, P., Illingworth, R. S., Taylor, A. M. R., Davison, V., O'Neill, L. P. and Turner, B. M. (2010). Immunostaining of modified histones defines high-level features of the human metaphase epigenome. Genome Biol. 11, R110 10.1186/gb-2010-11-11-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri, T. and Lehner, B. (2011). Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 7, e1002036 10.1371/journal.pgen.1002036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz, C. and Kerppola, T. K. (2008). Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc. Natl. Acad. Sci. USA 105, 16572-16577 10.1073/pnas.0805317105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, P., LeRoy, G., Drury, W. J.III, Zee, B. M., Son, J., Beck, D. B., Young, N. L., Garcia, B. A. and Reinberg, D. (2012). Asymmetrically modified nucleosomes. Cell 151, 181-193 10.1016/j.cell.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, P., Tee, W.-W. and Reinberg, D. (2013). A double take on bivalent promoters. Genes Dev. 27, 1318-1338 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann, A. (1893). The Germ-Plasm: A Theory of Heredity. New York: Charles Scribner's Sons. [Google Scholar]
- Woodcock, C. L. and Ghosh, R. P. (2010). Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2, a000596 10.1101/cshperspect.a000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.-F., Zhang, H. and Cairns, B. R. (2011). Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21, 578-589 10.1101/gr.113167.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta, Y., Kurimoto, K., Ohinata, Y., Seki, Y. and Saitou, M. (2006). Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol. Reprod. 75, 705-716 10.1095/biolreprod.106.053686 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Kimura, H., Tada, M., Nakatsuji, N. and Tada, T. (2005). Nanog expression in mouse germ cell development. Gene Expr. Patterns 5, 639-646 10.1016/j.modgep.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Hong, K., Liu, R., Inoue, A., Shen, L., Zhang, K. and Zhang, Y. (2013). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329-339 10.1038/cr.2013.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. and Anderson, D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61-73 10.1016/S0092-8674(02)00677-3 [DOI] [PubMed] [Google Scholar]