Abstract
Custom-designed nucleases afford a powerful reverse genetic tool for direct gene disruption and genome modification in vivo. Among various applications of the nucleases, homologous recombination (HR)-mediated genome editing is particularly useful for inserting heterologous DNA fragments, such as GFP, into a specific genomic locus in a sequence-specific fashion. However, precise HR-mediated genome editing is still technically challenging in zebrafish. Here, we establish a GFP reporter system for measuring the frequency of HR events in live zebrafish embryos. By co-injecting a TALE nuclease and GFP reporter targeting constructs with homology arms of different size, we defined the length of homology arms that increases the recombination efficiency. In addition, we found that the configuration of the targeting construct can be a crucial parameter in determining the efficiency of HR-mediated genome engineering. Implementing these modifications improved the efficiency of zebrafish knock-in generation, with over 10% of the injected F0 animals transmitting gene-targeting events through their germline. We generated two HR-mediated insertion alleles of sox2 and gfap loci that express either superfolder GFP (sfGFP) or tandem dimeric Tomato (tdTomato) in a spatiotemporal pattern that mirrors the endogenous loci. This efficient strategy provides new opportunities not only to monitor expression of endogenous genes and proteins and follow specific cell types in vivo, but it also paves the way for other sophisticated genetic manipulations of the zebrafish genome.
Keywords: TALEN, Genome engineering, Homologous recombination, Knock-in, sox2, gfap
INTRODUCTION
Custom-designed nucleases are being used to overcome many limitations of conventional genome engineering technologies for mouse knockouts and knock-ins (Capecchi, 2005). So far, three different types of nucleases, zinc-finger (ZF) (Kim et al., 1996), transcription activator-like effector (TALE) (Boch et al., 2009; Moscou and Bogdanove, 2009) and clustered regularly interspaced short palindromic repeats/CRISPR associated (CRISPR/Cas9) (Jinek et al., 2012), have been employed for genome-editing purposes. In principle, these tools enable the induction of double-strand breaks (DSBs) in target genomic sequences. Cells can repair these DSBs through two major DNA repair systems, namely nonhomologous end joining (NHEJ) or homology-directed repair (HDR), each of which enables a different type of genomic modification. If the break is repaired by NHEJ, which is an error-prone repair mechanism, such repaired target sequences frequently harbor insertions or deletions (Bibikova et al., 2002; Cong et al., 2013; Mali et al., 2013; Miller et al., 2011). Alternatively, it is possible to edit target sequences precisely through a DSB-induced HDR mechanism by introducing into cells both nucleases and a DNA template, such as single-stranded oligonucleotides (Bedell et al., 2012; Cong et al., 2013; Radecke et al., 2010; Wang et al., 2013) or longer dsDNA donors (Cong et al., 2013; Hockemeyer et al., 2011; Urnov et al., 2005). Therefore, custom nucleases can theoretically be employed to modify genomes of any genetic model organism.
The optical transparency of zebrafish embryos and adult casper mutant fish (White et al., 2008) facilitate monitoring or visualizing a gene product or a specific cell type labeled with fluorescent proteins within an intact organism by time-lapse microscopy and at single cell or subcellular resolution. Realization of the full potential of this experimental strategy depends on the reliability and accuracy of transgenic tools used to visualize gene expression. However, the level and spatiotemporal pattern of expression of randomly integrated transgenes often diverge from those of the endogenous genes. Whereas, in some cases, engineered bacterial artificial chromosomes can recapitulate endogenous gene expression (Jessen et al., 1999; Shin et al., 2003), genome editing using nucleases offers a new method for modifying endogenous loci. The challenge is to make this method as successful as a regular transgenesis in zebrafish (Kawakami, 2005). The feasibility of homologous recombination (HR)-mediated gene replacement using TALEN in zebrafish has been recently reported (Zu et al., 2013). However, the relatively low efficiency of the reported HR-mediated genome engineering method makes it difficult for systematic generation of fluorescent protein-tagged or fluorescent protein-reporter knock-in lines in zebrafish. Therefore, there is a need to develop advanced methods to improve the efficiency of the HR-mediated genome engineering technology. Here, we carry out a systematic evaluation of a number of experimental parameters of HR-mediated genome engineering and demonstrate that the homology arm size and the configuration of the targeting vector, in particular the position of a DSB in the targeting construct, are crucial efficiency determinants. We report generation of sox2 and gfap fluorescent gene reporter lines with high germline transmission rates, demonstrating that this method can be standardized for targeting vector construction to generate knock-in zebrafish.
RESULTS
Design of a GFP reporter system for measuring the frequency of HR events in vivo
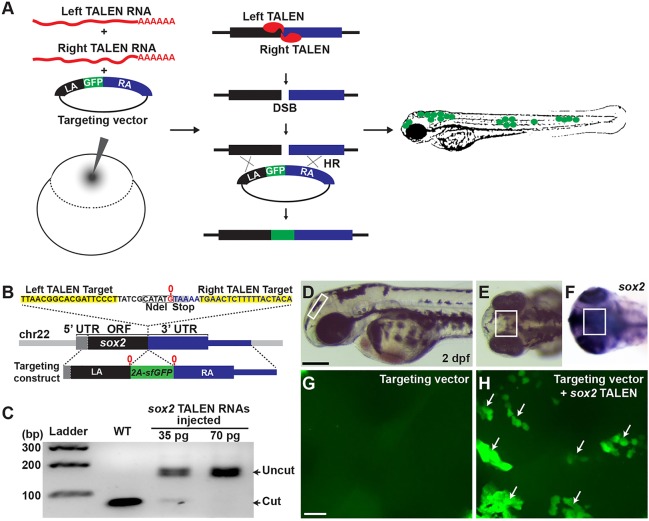
We reasoned that when DNA fragments encoding fluorescent proteins flanked by sequences from a specific gene are injected into a zebrafish zygote, they could be incorporated into the genome via TALEN-induced DSB and HDR (Fig. 1A). To test the feasibility of this approach, we chose two genes that are expressed early during zebrafish neural development, sox2 (Cunliffe and Casaccia-Bonnefil, 2006) and gfap (Marcus and Easter, 1995). We first designed a TALEN pair targeting the stop codon in the sox2 gene, a region containing an NdeI recognition site. Next, we designed a targeting reporter construct in which sequences encoding superfolder GFP (Pedelacq et al., 2006) were fused with the viral 2A peptide (Provost et al., 2007) (2A-sfGFP) and inserted in the spacer of the sox2 TALEN target site, flanked by 1168 bp left homology arm (LA) and 3716 bp right homology arm (RA) of sox2 genomic DNA fragments (Fig. 1B). Upon a precise integration of these sequences into the endogenous sox2 locus through HR, sox2-expressing cells should also express sfGFP in the cytoplasm. Therefore, we used sfGFP signal in the neural tissues that normally express sox2 at 2 dpf as an indicator of putative HR events. To test whether the designed TALENs can efficiently induce DSBs in the sox2 target sequences, we injected 35 and 70 pg of each synthetic RNA encoding a sox2 TALEN pair into one-cell stage embryos. At 1 day post fertilization (dpf), we genotyped the injected embryos by NdeI digestion of PCR products encompassing the target sequence. Whereas the PCR product from the uninjected control embryos was digested completely by NdeI, the enzyme failed to cut most of the product from the injected embryos (Fig. 1C), indicating that NdeI recognition sequence within the sox2 TALEN target sequence was effectively mutated by sox2 TALENs. To evaluate this in vivo HR reporter system, we injected the targeting construct either alone or together with the synthetic sox2 TALEN RNA pair into one-cell stage embryos. After titrating the input targeting construct (see Materials and Methods), we settled on injecting 10 pg of a targeting vector with 35 pg of each TALEN RNA. Using this dose, about 86% of injected embryos manifested normal morphology at 2 dpf (supplementary material Fig. S1). Two days after injection, the morphologically normal embryos were selected and examined for sfGFP signal in the diencephalon, where sox2 is strongly expressed (Fig. 1D-F) (Sprague et al., 2006). Whereas we did not detect sfGFP expression in embryos injected with the targeting construct alone (0%, n=10) (Fig. 1G and Fig. 2B), we frequently observed sfGFP-positive cells in the embryos co-injected with the targeting construct and TALEN RNA (80%, n=10) (Fig. 1H and Fig. 2B). This suggested that in the sfGFP-positive cells, the DNA sequences of the targeting construct encoding sfGFP were integrated into a genomic lesion induced by the sox2 TALENs.
Fig. 1.
In vivo recombination analysis. (A) Overview of HR detection system by co-injection of TALEN RNAs and a GFP-inserted targeting construct in zebrafish embryos. (B) TALEN target in the sox2 locus and a sox2-targeting construct. Black, gray and blue blocks represent ORF, 5′ UTR and 3′ UTR, respectively (sox2 has no introns). G (0 position, marked in red) just before the stop codon is duplicated in the targeting construct locus and the 2a-sfGFP fragment is inserted in between the Gs. (C) sox2 TALEN RNAs were injected without any donor DNA and its mutagenic activity was measured by PCR and restriction enzyme analysis. Each lane represents an NdeI-digested amplicon encompassing sox2 TALEN target sequence from genomic DNA isolated from five embryos. The NdeI-digested wild-type PCR products are 78 and 75 bp. (D-F) Lateral (D) and anterior-dorsal (E,F) views of 2 dpf wild-type embryo. Box marks dorsal diencephalon region. (F) Detection of sox2 transcripts by whole-mount in situ hybridization. Scale bar: 200 μm. (G,H) Confocal microscope images of the brain region that is similar to that shown in the boxed areas in D-F. (G) An embryo injected with a circular form of a sox2 reporter targeting construct. (H) An embryo co-injected with circular form of the construct and sox2 TALEN RNAs. Groups of cells expressing sfGFP in the diencephalon region are indicated by arrows. Scale bar: 20 μm.
Fig. 2.
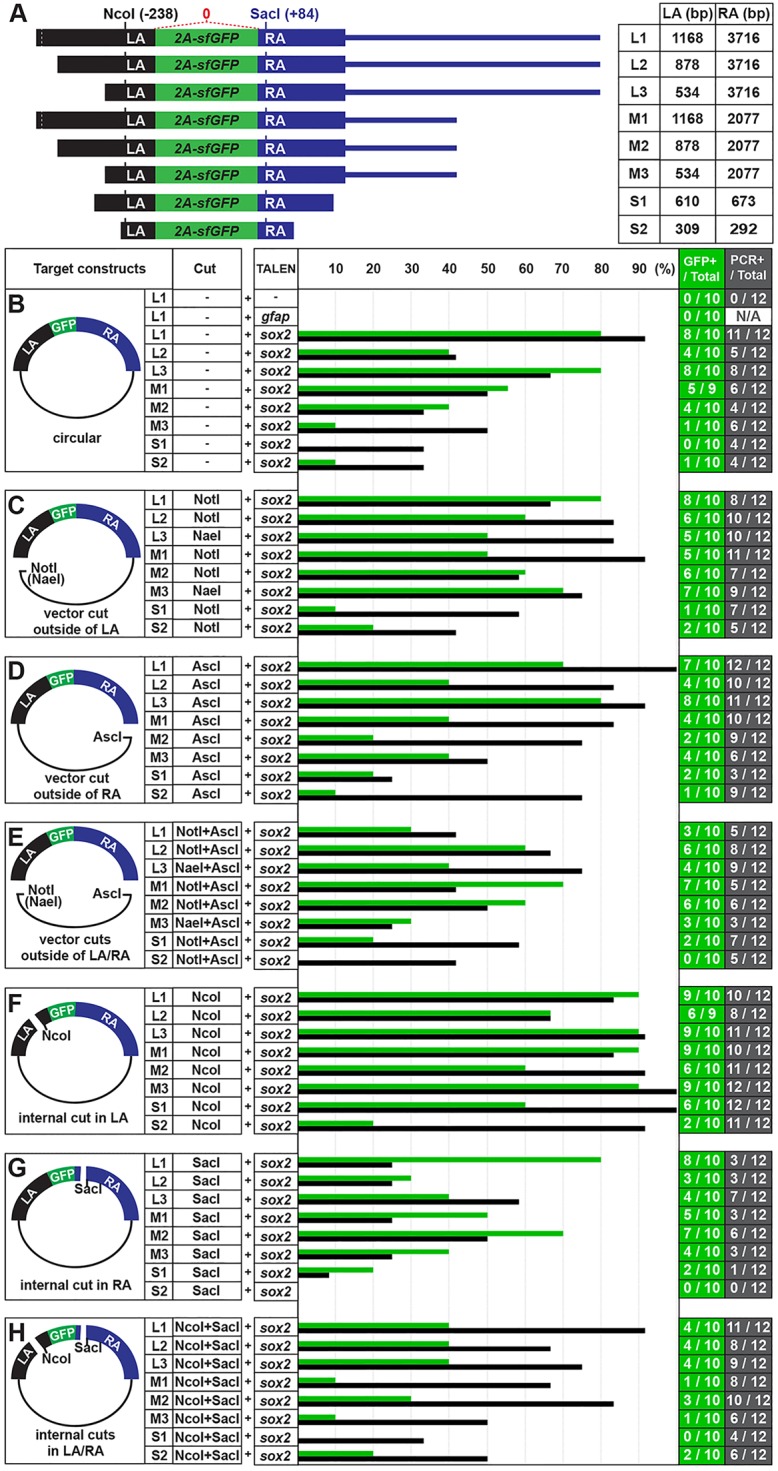
Defining the sizes of homology arms and configurations of targeting constructs for efficient recombination. (A) Eight sox2 targeting reporter constructs. Black bar marks LA and blue bar marks RA of targeting constructs. Green bar represents an sfGFP sequence linked with viral 2a peptide sequence. The NcoI site in LA is located 239 bp from and the SacI site in RA is located 85 bp from the 2a-sfGFP sequences (see Fig. 1B). (B-G) Quantification of the recombination frequency. Green bar represents the percentage of sfGFP-positive embryos determined by confocal microscopy. Black bar represents the percentage of recombination-positive embryos, as determined by PCR (see supplementary material Fig. S2). (B) Circular form of each targeting construct was injected. (C) Targeting constructs were digested with NotI (or NaeI) to cut the vector outside the LA. (D) Targeting constructs were digested with AscI to cut the vector outside the RA. (E) Targeting constructs were digested with NotI (or NaeI) and AscI to separate the targeting DNA fragment from the vector region. (F) Targeting constructs were digested with NcoI within the LA. (G) Targeting constructs were digested with SacI within the RA. (H) Targeting constructs were digested with NcoI and SacI.
To understand the relationship between the homology arm length and the frequency of recombination events, we performed co-injection experiments with eight targeting reporter constructs [designated Long1 (L1), L2, L3, Medium1 (M1), M2, M3, Short1 (S1) and S2] that differed in the length of LA and RA (Fig. 2A). We co-injected the circular form of each targeting construct with sox2 TALEN RNAs and scored the embryos for sfGFP-positive cells in the diencephalon, as described above. We observed a higher percentage of sfGFP-positive embryos in the L group, compared with the M and S groups (Fig. 2B). However, when we co-injected L1 sox2 targeting construct and TALENs targeting the stop codon of the gfap locus to test whether a random integration of the targeting construct might lead to sfGFP expression, we did not observe sfGFP-positive cells in the diencephalon of the co-injected embryos (0%, n=10) (Fig. 2B). These observations are consistent with the notion that sfGFP expression reports a HR-mediated integration of the construct into the targeted locus in somatic tissue. In addition, we interpreted these results to mean that increasing the length of the homology arm in a targeting construct can elevate the frequency of the somatic recombination events. In particular, these data suggest that a homology arm over 2 kb may be sufficient to achieve an optimal frequency of recombination events.
According to a previous report, the frequency of somatic HR events in zebrafish increases when a linear targeting DNA fragment is used rather than a circular construct (Zu et al., 2013). This raises the possibility that the structure of the targeting construct can influence HR frequency. To test this, we repeated co-injection experiments using different configurations of the targeting constructs that were linearized with NotI (or NaeI) to cut the vector outside of the LA, AscI to cut the vector outside of the RA, or both NotI (or NaeI) and AscI to separate the targeting DNA fragment from the vector region. Interestingly, we observed a higher percentage of sfGFP-positive embryos in both NotI- and NotI+AscI-digested M groups, compared with the circular M group (Fig. 2B-E). However, the percentage of sfGFP-positive embryos was either similar or lower in the AscI-digested M group or the NotI-, AscI- and NotI+AscI-digested L groups when compared with the embryos injected with the circular form of the same constructs (Fig. 2B-E).
Next, we tested whether an internal cut in the homology arm of the targeting constructs could enhance the frequency of sfGFP expression in our assays. Although we did not observe higher HR frequency when injecting the constructs with an internal cut in the RA (SacI) (Fig. 2G), or internal cuts in both LA and RA (Fig. 2H), all of the constructs with an internal cut in the LA (NcoI) produced higher proportions of GFP-positive embryos than their circular counterparts (Fig. 2F).
To verify in vivo HR assay using GFP detection, we repeated the co-injections of sox2 TALEN and the various targeting constructs, and analyzed the recombination frequency by PCR. For this, we randomly chose 12 normal-looking F0 embryos in each group, extracted the genomic DNA individually and performed PCR to amplify the recombinant genomic DNA fragment using a forward primer in the sox2 gene just outside the LA region (sF2) with an sfGFP-specific reverse primer (sR2′) (supplementary material Fig. S2A). Overall, we detected more recombination events by PCR than by using the GFP detection method. When comparing the recombination frequencies for different targeting constructs, we observed similar patterns, except in the NcoI+SacI-digested condition (Fig. 2B-H). To understand the difference, we sequenced seven of the recombinant PCR products from M1/NcoI and MI/NcoI+SacI groups. Whereas we did not detect any mutations within sox2 and sfGFP sequences in the M1/NcoI group, four out of seven PCR products contained mutations in the MI/NcoI+SacI group (data not shown). These results imply that that the PCR method can detect both HR-dependent and -independent insertions of targeting construct. Interestingly, we observed that a cut in the LA enhances the frequency of gene targeting, but generating a cut in RA did not, as assayed using GFP or a PCR detection method (Fig. 2F,G). Moreover, for targeting constructs with a cut in the LA (NaeI-digested condition), we detected highly efficient recombination frequencies (83-92%, n=12) (supplementary material Fig. S2I), further supporting the notion that a specific configuration of targeting constructs (a cut in the LA) is an important factor for efficient gene targeting. However, because in our targeting constructs the LA was generally shorter than the RA (Fig. 2), these results are also consistent with an internal cut in the short homology arm, rather than the orientation of the cut in the homology arms with respect to the insert, being an important factor for HR-mediated genome editing. To distinguish between these possibilities, we carried out additional experiments that tested several targeting constructs that had LA of 2.5 kb and RA of 1 kb, thus opposite to the configurations shown in Fig. 2. With these targeting constructs, we observed high recombination frequency for those that contained an internal cut in the short RA, but not a cut in the LA (data not shown). Therefore, these results support the view that an internal cut in the short homology arm is an important factor for HR-mediated genome editing, rather than the orientation of the cut in the homology arms with respect to the insert. Based on these results, we propose not only the length of homology arm, but also the configuration of the targeting construct can influence the frequency of HR events in somatic tissue.
Analysis of the germline transmission rates of sox2-2a-sfGFP knock-in alleles
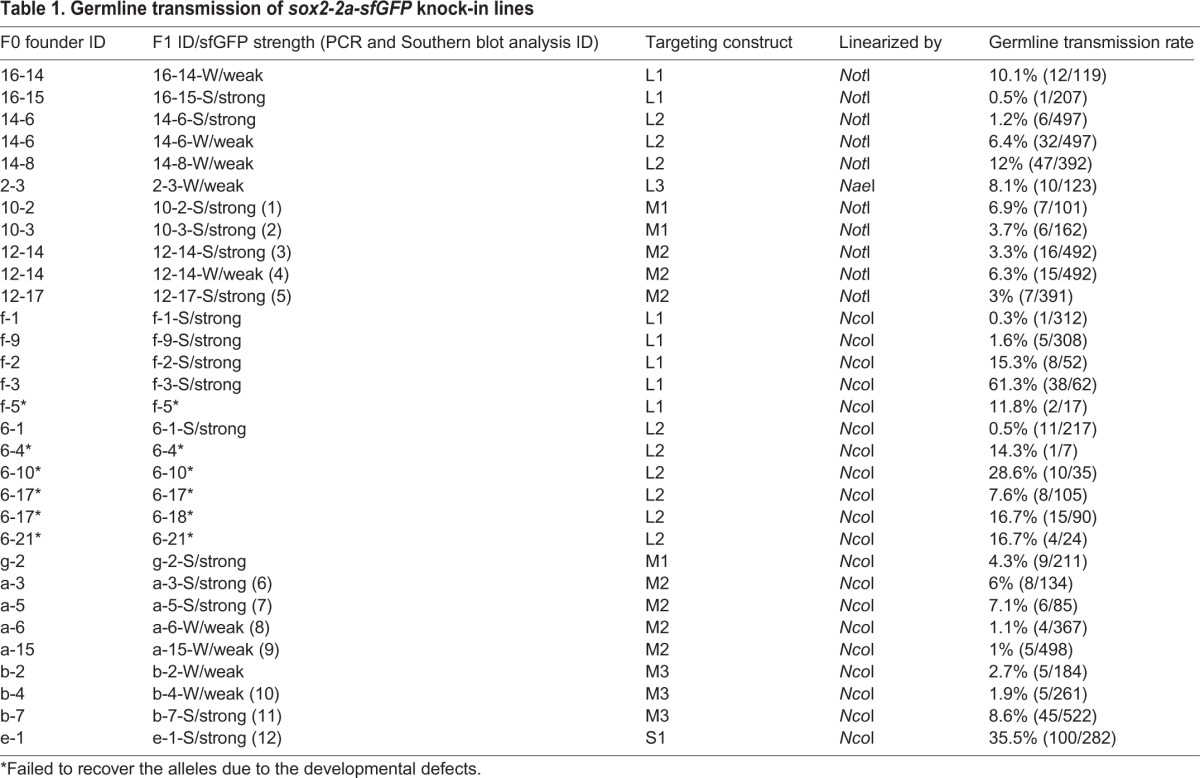
Based on our in vivo GFP reporter analysis, the constructs digested with NotI (or NaeI) or NcoI produced higher frequency of putative HR events than other conditions. Thus, we decided to determine the germline transmission rates of the seven construct types described above that were digested with NotI (or NaeI) or NcoI (Fig. 2A,C,F). To do this, we raised unselected F0-injected embryos from these 14 groups into adulthood, and outcrossed each F0 founder with a wild-type fish to obtain F1 progeny of the individual founders. Using an epifluorescence stereomicroscope, we found that 29 of 363 F0 founders produced sfGFP-positive F1 embryos (8%, Fig. 3A; Table 1). Germline mosaicism of individual F0 founders ranged from 0.3 to 61.3%, and the average was 10.5% (Table 1). In the L3/NcoI founder group, we screened only two F0 founder fish because the remaining 16 were sterile, and neither produced sfGFP-positive progeny (Fig. 3A). We also failed to recover stable lines from one F0 founder in the L1/NcoI group and from five F0 founders in the L2/NcoI group due to severe developmental defects of sfGFP-positive F1 progeny. Interestingly, we observed two different types of sfGFP-positive F1 embryos based on sfGFP signal intensity (Fig. 3B,C). All sfGFP-positive F1 progeny from 14 founders exhibited strong sfGFP expression. The progeny of another seven founders exhibited weak sfGFP expression, whereas two additional founders produced F1 embryos that showed either strong or weak sfGFP expression (Table 1). However, regardless of sfGFP expression intensity, all F1 progeny showed the same spatiotemporal sfGFP expression pattern.
Fig. 3.
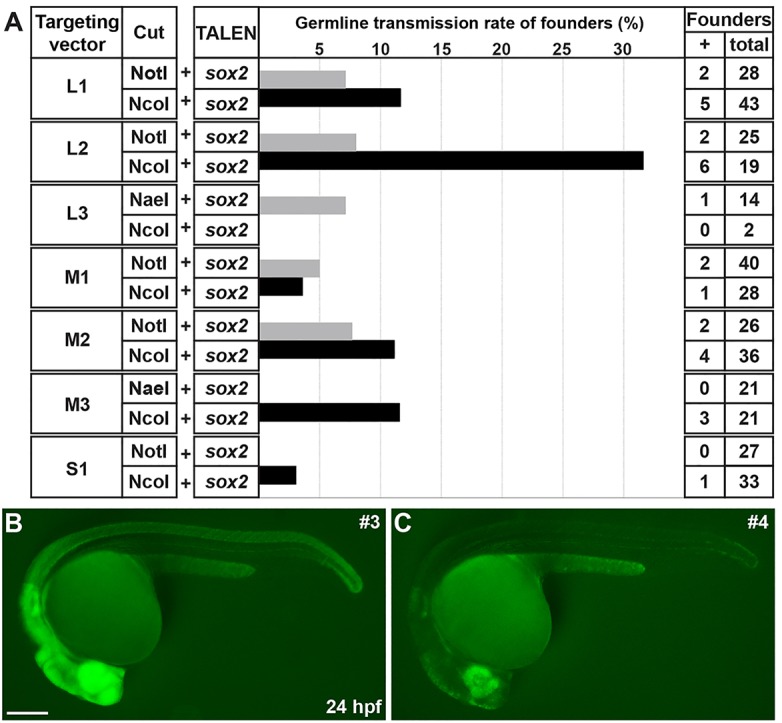
Germline transmission rates of sox2-2a-sfGFP knock-in lines. (A) The frequency of germline transmission of the founders that were co-injected with sox2 TALEN and each targeting construct, which was linearized by either NotI or NcoI. The bars represent the percentage of F0 founders that produced sfGFP-positive F1 progeny. (B,C) Comparison of sfGFP expression between strong and weak sox2-2a-sfGFP F1 embryos. Epifluorescence stereomicroscopic images of sfGFP expression. Lateral views of 1 dpf embryos. Both images were taken with the same exposure time. (B) The strong sfGFP-positive F1 embryos obtained from line 3. (C) The weak sfGFP-positive F1 embryos obtained from line 4. Scale bar: 200 μm.
Table 1.
Germline transmission of sox2-2a-sfGFP knock-in lines
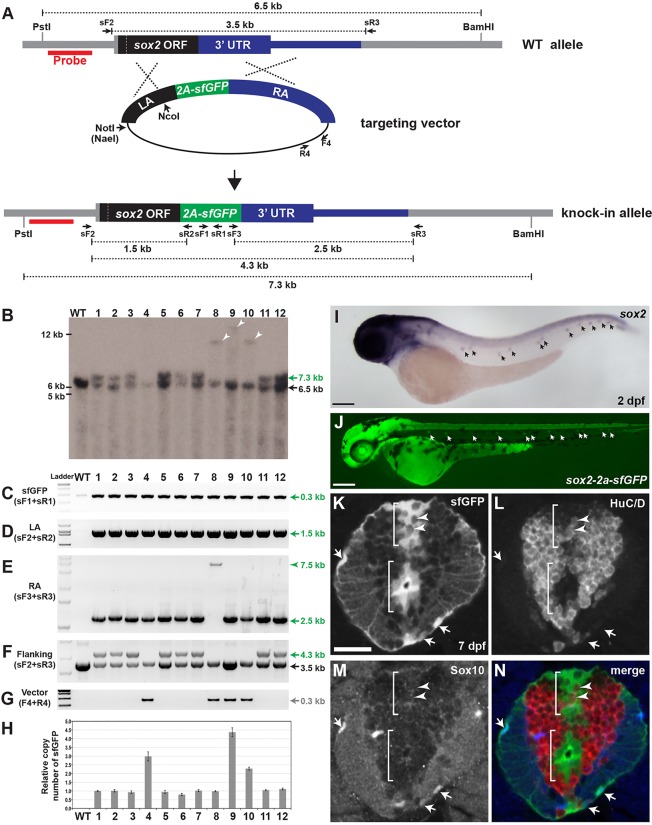
To verify that the sfGFP-positive F1 progeny are knock-in animals, we performed Southern blot analysis of genomic DNA from adult F1 fish using probe sequences from the sox2 locus that are just outside the LA homology region (685 bp) (Fig. 4A). For this experiment, we chose 12 putative knock-in F1 animals obtained from M1, M2, M3 and S1 founder groups. We confirmed that eight F1 lines had a 6.5 kb wild-type band and a 7.3 kb knock-in band in the sox2 locus (Fig. 4B). Interestingly, we also detected bigger sizes of insertion bands in genomic DNA of the four F1 lines (arrowheads in Fig. 4B; supplementary material Fig. S3).
Fig. 4.
Analysis of sox2-2a-sfGFP knock-in lines. (A) Schema of HR-mediated sfGFP insertion. Either NotI (or NaeI) or NcoI-digested targeting constructs (M1, M2, M3 and S1, see Fig. 2A) were co-injected with sox2 TALEN RNAs. Red bar indicates the probe sequences used for Southern blot. (B) Southern blot analysis. PstI/BamHI double-digested genomic DNA from individual adult fish was hybridized with the probe (red bars in A). Expected fragment size: wild type, 6.5 kb; targeted, 7.3 kb. Arrowheads indicate bands of unexpected size (lane 8, ∼10 kb; lane 9, ∼14 kb; lane 10, ∼10 kb). (C-G) PCR genotyping analysis. Primer pair locations are shown in A. WT, wild type; 1-12, sox2-2a-sfGFP F1 lines (see Table 1). (C) Detection of sfGFP (sF1+sR1 primers). (D) Amplicon extended from 5′ of the LA to within the sfGFP sequences (sF2+sR2). (E) Amplicon spanned the entire RA sequences (sF3+sR3). Arrowhead indicates a fragment of aberrant size (7.5 kb). (F) Amplicon extended from 5′ of the LA to 3′ of the RA sequences (sF2+sR3). Expected amplicon size: wild type, 3.5 kb; targeted, 4.3 kb. (G) Amplicon for vector-specific sequences (F4+R4). (H) Quantitative PCR analysis of sfGFP relative copy numbers. Data are mean±s.e.m. (I,J) Lateral views of 2 dpf embryos. (I) sox2 expression in wild type. (J) sfGFP expression in sox2-2a-sfGFPstl84. Arrows indicate neuromasts. Scale bars: 200 μm. (K-N) Transverse spinal cord section of 7 dpf sox2-2a-sfGFPstl84 larva. (K) sfGFP expression. (L) Anti-HuC/D antibody staining. (M) Anti-Sox10 antibody staining. (N) Merged image of K-M. Brackets indicate sfGFP-positive cell clusters in posterior median sulcus (top) and posterior median septum (bottom). Arrows indicate sfGFP-positive and Sox10-positive OPCs. Arrowheads indicate sfGFP-positive and HuC/D-positive neurons. Scale bar: 20 μm.
To test molecularly whether the sfGFP-positive F1 progeny are the result of HR, we performed PCR-based genotyping using several primer sets with the same genomic DNA that was used for Southern blot analysis as a template. An sfGFP-specific PCR product was detected in all the analyzed sfGFP-positive F1 animals (Fig. 4C). We were also able to amplify a fragment of the expected size from all sfGFP-positive F1 animals using a forward primer outside the LA region (sF2) with an sfGFP-specific reverse primer (sR2) (Fig. 4D), and an sfGFP-specific forward primer (sF3) with a reverse primer outside of the RA region (sR3) (with the exception of allele 8) (Fig. 4E). Using the forward primer outside the LA region (sF2) and the reverse primer outside the RA region (sF2 and sR3), we detected both the 3.5 kb PCR product for wild-type sox2 locus and the 4.3 kb PCR product for the sfGFP-knock-in sox2 locus in eight different F1 knock-in lines (Fig. 4F). We sequenced all the products, including the PCR product of unexpected size from F1 animal 8, and failed to detect any NHEJ events, indicating that sfGFP sequences were integrated into the sox2 locus via HR-mediated repair mechanism. Remarkably, with vector-specific primers (F4 and R4), we detected vector sequences incorporated into the genomic DNA of several F1 animals (4, 8, 9 and 10) that were correlated with weak sfGFP expression (Fig. 4G; Table 1). Using various PCR and sequencing analyses, we found that three F1 lines (4, 9 and 10) harbored concatemers of the targeting construct that were likely generated via double-crossover HR, whereas line 8 had a single copy of the targeting construct, suggesting it was created as a result of a single-crossover HR event (see Fig. 7E). This was confirmed by a copy number analysis of sfGFP, showing three sfGFP copies in line 4, one copy in line 8, about four copies in line 9 and two copies in line 10 (Fig. 4H).
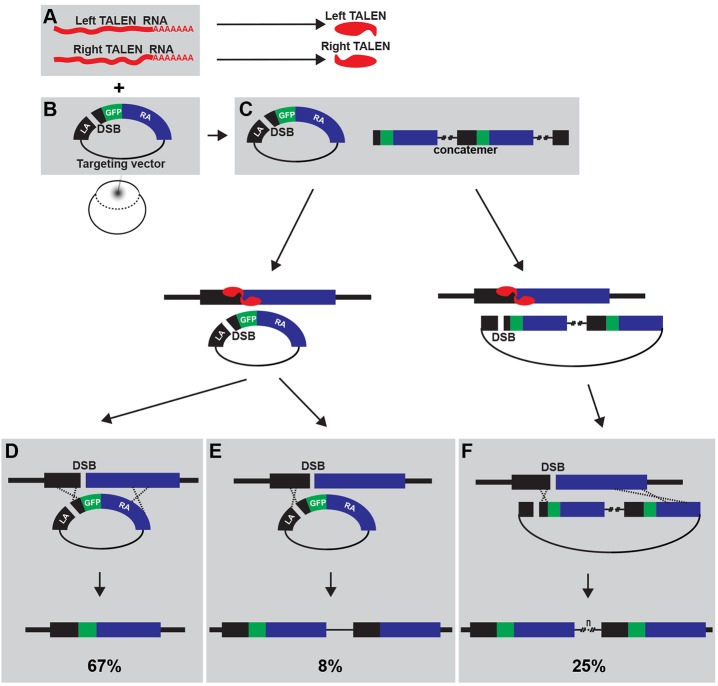
Fig. 7.
A schema of the reported strategy for efficient HR-mediated genome editing in zebrafish. Co-injection of target-specific TALEN RNAs (A) and a targeting construct that harbors DSB in the short homology arm (B) results in highly efficient HR-mediated genome editing. (C) The linearized targeting construct can exist as a single molecule as well as concatemers via a NHEJ repair mechanism in vivo. A DSB in the genome induced by TALEN is the essential first step in HR. A DSB within the short homology arm of the targeting construct is not essential, but appears to strongly enhance HR-mediated knock-in. Three outcomes were observed in our experiments (see Fig. 4B-H). (D) A single targeting construct can integrate into the genome precisely by double-crossover HR (67% of sox2-2a-sfGFP lines). (E) A single-crossover HR event can result in incorporation of the vector into the genome (8% of sox2-2a-sfGFP lines). (F) A concatemeric construct can form from the targeting vector and integrate into the genome with double-crossover HR event, resulting in incorporation of multiple variants of the targeting construct into the genome (25% of sox2-2a-sfGFP lines).
To determine whether sfGFP expression recapitulates endogenous sox2 expression, we chose the sox2-2a-sfGFP line #3 (sox2-2a-sfGFPstl84) for further analysis. Expression of both sox2 and sfGFP were detected in neural tissues including brain, spinal cord, eyes and neuromasts at 2 dpf (Fig. 4I,J). Owing to the lack of working Sox2 antibodies (see Materials and Methods), we determined the identity of sfGFP-positive cells in F1 animals indirectly, using anti-HuC/D to label neurons and anti-Sox10 antibodies to label oligodendrocyte progenitor cells (OPCs). Sox2-positive cells are expected to be neural precursors. In the spinal cord of 7 dpf sfGFP-positive larvae, we detected sfGFP-positive cells in the posterior median sulcus and septum (Fig. 4K). Although the majority of sfGFP-expressing cells appeared to be non-neuronal cells, we observed that a few sfGFP-positive cells were also labeled by anti-HuC/D antibody (Fig. 4K-N). Because it is known that Sox2 is expressed in neural precursors but is downregulated in the differentiated neurons (Lindsey et al., 2012), this suggests that sfGFP is more stable than endogenous Sox2 and persists in newly born neurons. Some Sox10-positive OPCs also expressed sfGFP (Fig. 4K-N), consistent with a previous study showing that Sox2 is detected in OPCs (Snyder et al., 2012). Moreover, we observed the majority of sfGFP-positive cells in brain regions where Sox2-expressing cells are known to reside, such as the ventricular zone of the brain, and in putative amacrine cells and Müller glia in the retina (supplementary material Fig. S4). Therefore, we conclude that sfGFP expression in the F1 sox2 knock-in animals mirrors endogenous sox2 gene expression.
Generation of a gfap-2a-tdTomato knock-in allele
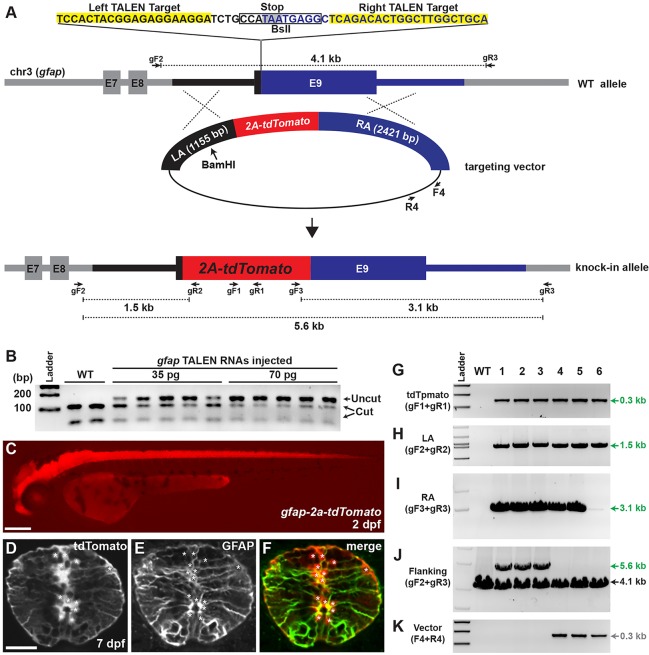
To further validate the efficiency of our knock-in strategy, we designed a TALEN pair targeting the gfap stop codon locus. For the gfap targeting construct, we used 1155 and 2421 bp of genomic DNA fragments as the LA and RA, respectively, and DNA sequences encoding 2a-tdTomato (Shaner et al., 2004) were inserted in frame with GFAP coding sequences prior to the stop codon (Fig. 5A). We confirmed the target gene disruption activity of the gfap TALEN by a PCR-based genotyping method described above for the sox2 TALEN activity test (Fig. 5B). We co-injected RNA encoding gfap TALEN pair (35 pg each) and the targeting construct (10 pg) cut with BamHI within the LA, into one-cell-stage embryos, which were raised to adulthood as F0 founders. We crossed 44 F0 founders individually with wild-type fish, and by screening with a fluorescent microscope at 2 dpf we found tdTomato-positive F1 progeny from five F0 founders (11%, Fig. 5C; Table 2). Whereas tdTomato-positive embryos from two F0 founders expressed tdTomato strongly, progeny from the other two founders expressed tdTomato weakly, and the fifth F0 founder produced both strong and weak tdTomato-expressing embryos (supplementary material Fig. S5). To confirm that GFAP was co-expressed in tdTomato-positive cells, we performed immunofluorescence using an anti-GFAP antibody (Zupanc et al., 2005). At 7 dpf, tdTomato-positive processes of neural precursors were colocalized with GFAP-positive processes in the gfap-2a-tdTomatostl85, indicating that tdTomato expression recapitulated GFAP expression (Fig. 5D-F).
Fig. 5.
Analysis of gfap-2a-tdTomato knock-in lines. (A) The strategy for generating gfap-2a-tdTomato knock-in fish. gfap TALEN target sequences and stop codon are highlighted in yellow and gray, respectively. In the targeting construct, LA and RA are marked with black and blue, respectively. Red indicates tdTomato sequences linked to viral 2a peptide sequence. The targeting construct linearized with BamHI to generate an internal cut in LA. (B) Activity test of gfap TALEN RNAs. Each lane represents a BslI-digested amplicon encompassing gfap TALEN target sequence from genomic DNA isolated from individual embryo. The BslI-digested wild-type amplicon products are 112 and 50 bp. WT, uninjected control; 35 pg and 70 pg, injection of 35 or 70 pg of gfap TALEN RNA, respectively. (C) Lateral view of 2 dpf gfap-2a-tdTomatostl85 knock-in F1 embryo. Scale bar: 200 μm. (D-F) Transverse spinal cord section of 7 dpf gfap-2a-tdTomatostl85 larva. (D) tdTomato expression. (E) Anti-GFAP antibody staining. (F) Merged image of D and E. Asterisks indicate tdTomato-positive cell bodies. Scale bar: 20 μm. (G-K) PCR genotyping analysis. Primer pair locations are shown in A. WT, wild type; 1-6, gfap-2a-tdTomato F1 lines from different founders except lines 3 and 4 (see Table 2). (G) Detection of tdTomato (gF1+gR1 primers). (H) Amplicon extended from 5′ of the LA to within the tdTomato sequences (gF2+gR2). (I) Amplicon spanned the entire RA sequences (gF3+gR3). (J) Amplicon extended from 5′ of the LA to 3′ of the RA sequences (gF2+gR3). Expected amplicon size: wild type, 4.1 kb; targeted, 5.6 kb. (K) Amplicon for vector-specific sequences (F4+R4).
Table 2.
Germline transmission of gfap-2a-tdTomato knock-in lines
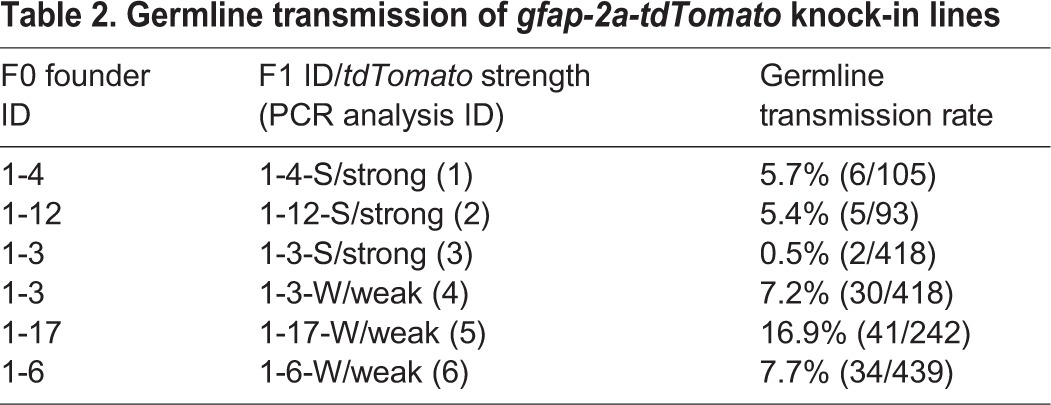
Because we observed reduced expression of the reporter gene in several sox2-2a-sfGFP knock-in lines that had vector sequences incorporated in their genome (Fig. 3C and Fig. 4G; Table 1), we genotyped the weak tdTomato-positive F1 embryos using vector-specific primers (F4 and R4) and confirmed that the weakly expressing alleles harbored vector sequences in their genomes (Fig. 5K). To test further whether tdTomato sequences were integrated via HR in the tdTomato-positive F1 fish, we performed PCR genotyping using four primer sets [tdTomato-specific forward and reverse primers (gF1+gR1); a forward primer outside of the LA region with a tdTomato-specific reverse primer (gF2+gR2); a tdTomato-specific forward primer with a reverse primer outside of the RA region (gF3+gR3); and a forward primer outside of the LA region with a reverse primer outside of the RA region (gF2+gR3) sets] (Fig. 5G-J) and sequenced the resulting amplicons from all lines except line 6. The analysis failed to detect any NHEJ events in any of the lines. These data indicate that our targeted insertion strategy was effectively working for the gfap locus with similar germline transmission rates as observed for the sox2 knock-in experiments described above. In addition, integration outcomes of the gfap targeting construct were similar to those of the sox2 knock-in lines, suggesting that the method of TALEN/HR-mediated genome editing described here is promising as a reliable method for obtaining HR-mediated zebrafish knock-ins.
Knock-in reporter mirrors the expression of endogenous target gene
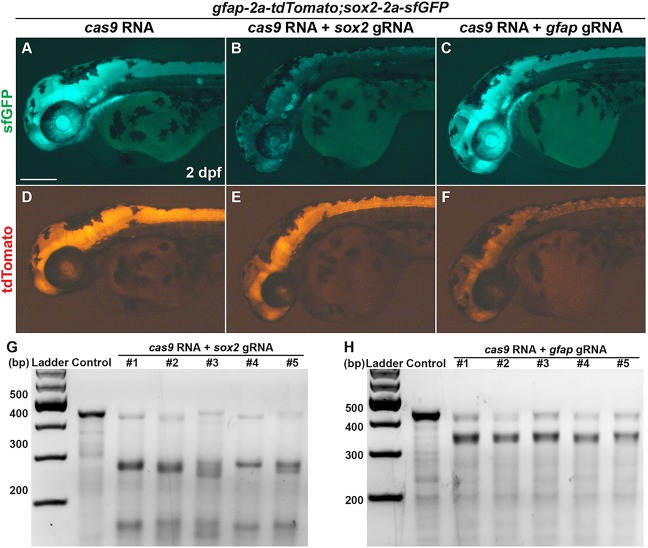
To test whether the two knock-in reporter lines do faithfully reflect the expression of the endogenous target genes, we applied CRISPR/Cas9 system to induce somatic mutations in sox2 and gfap genes in sox2-2a-sfGFPstl84;gfap-2a-tdTomatostl85 double homozygous knock-in embryos. First, we designed each guide RNA for targeting sox2 (sox2 gRNA) and gfap (gfap gRNA), and co-injected each gRNA (10 pg) along with cas9 RNA (100 pg) into one-cell stage embryos. Whereas sfGFP and tdTomato expression in cas9 RNA-injected control embryos was similar to those of uninjected embryos (Fig. 6A,D), 54% of embryos (n=48) showed a dramatic decrease of sfGFP expression in the sox2 gRNA and cas9 RNA co-injected embryos (Fig. 6B,E). Interestingly, tdTomato expression was slightly decreased in these embryos, likely as a consequence of sox2 mutations. Conversely, the expression of tdTomato was significantly decreased in the gfap gRNA and cas9 RNA co-injected embryos (76%, n=46) (Fig. 6C,F), whereas sfGFP expression was not affected. To confirm whether the gRNA target sites were mutated by CRISPR system, we performed genotyping using the T7 endonuclease I (T7EI) assay. We detected the mutations in sox2 and gfap (Fig. 6G,H) only in embryos injected with sox2 gRNA or gfap gRNA, respectively. These results provide further support for the conclusion that the expression of knock-in reporter mirrors that of endogenous target gene.
Fig. 6.
Transgene mirrors the endogenous expression of the target gene. (A-F) Lateral view of 2 dpf sox2-2a-sfGFPstl84;gfap-2a-tdTomatostl85 double homozygous knock-in embryos. All images were taken with the same exposure time. (A-C) sfGFP expression. (D-F) tdTomato expression. (A,D) Control embryo injected with 100 pg of cas9 RNA. (B,E) The embryo co-injected with 100 pg of cas9 RNA and 10 pg of gRNA targeting sox2. (C,F) The embryo co-injected with 100 pg of cas9 RNA and 10 pg of gRNA targeting gfap. Scale bar: 200 μm. (G,H) Detection of target-specific mutations by T7EI assays. Control, embryo injected with 100 pg of cas9 RNA; 1-5, individual embryos co-injected with 100 pg of cas9 RNA and 10 pg of gRNA targeting sox2 (G) or gfap (H). (G) The PCR products contain guide RNA target site of sox2. (H) The PCR products contain guide RNA target site of gfap.
DISCUSSION
Application of custom nucleases has enabled new methods for disruption and editing of specific genes; however, the low efficiency of current HR-mediated genome editing methods has limited their utility in zebrafish (Zu et al., 2013). Here, based on a systematic survey of the targeting construct parameters and measuring efficiency of HR events in live zebrafish embryos, we established an efficient method for HR-mediated genome editing in zebrafish.
We employed an in vivo HR detection assay and germline transmission analysis to determine the significance of the size of homology arms and their configuration to the efficiency of HR. We established a fluorescent reporter system, an approach similar to those employed in gene and enhancer trap studies (Balciunas et al., 2004; Kawakami et al., 2004). Owing to the limited sample size and the potential experimental variation, we repeated the experiment and determined HR frequency using a PCR method. Overall, we detected more recombination events by PCR than by GFP detection (Fig. 2). This difference between the two methods is likely due to the following three possibilities: first, experimental variation; second, it reflects the superior ability of the PCR method to detect recombination events in any cell compared with the GFP detection method, which should not detect the recombination events in cells where the endogenous sox2 gene is not transcribed; third, we found mutations in some of the recombinant PCR products, suggesting that the PCR method detected both HR-dependent and -independent insertions of the targeting construct. Indeed, a recent study demonstrated an efficient CRISPR/Cas9-mediated knock-in method via HR-independent insertion mechanism (Auer et al., 2014). However, we speculate that GFP detection assay specifically reports HR events rather than HR-independent insertions because of two lines of evidence. In F0 embryos co-injected with sox2 TALEN and S2 targeting construct, we rarely detected sfGFP expression, although the recombination events were frequently detected by PCR. This suggests that HR-independent insertions occur frequently for targeting constructs with short homology arms, but are not detected by the GFP expression assay. In addition, we did not detect sfGFP signal in embryos co-injected with gfap TALEN RNA and the sox2 targeting construct. In this second condition, sfGFP expression could occur only if HR-independent insertion placed the sfGFP in-frame with the gfap gene. Therefore, these observations strongly suggest that sfGFP expression of our assay system reflects HR rather than HR-independent insertions. However, as shown in Fig. 7, the targeting construct can be inserted into the target locus by a single HR event involving one of the homology arms. Notably, the reporter gene can be expressed in such a scenario, suggesting that sfGFP signals reflect both single and double HR events in our in vivo HR detection assay.
It was shown almost two decades ago that gene targeting can be stimulated by a genomic DSB introduced by a restriction enzyme (Smih et al., 1995). Currently, DSB in specific DNA target sequences can be generated by custom-designed nucleases enabling in vivo genome engineering. It is still unclear what additional factors should be considered to achieve highly efficient HR in vivo. Here, our zebrafish data provide evidence that the length of homology arms and configuration of targeting construct are crucial parameters in determining the efficiency of HR-mediated genome editing technology.
We found suitable sizes of homology arms (about 1 kb for one arm and 2 kb for the other arm) for a targeting construct, which ensured efficient HR and germline transmission exceeding 10%. Interestingly, inserts of 0.7 or 1.5 kb could be knocked in with similar germline transmission frequency (over 10%) when two homology arms were at least 1 and 2 kb in length. This suggests that the insert size over this range is not a crucial parameter for the efficiency of HR-mediated knock-in if the target construct contains 1 kb and 2 kb of homology arms.
The most intriguing early finding here was that a DSB in the shorter left homology arm, but not in the longer right arm, enhanced HR-mediated knock-in. In subsequent experiments, in which we inverted the positions of the short and long homology arms, we observed an enhanced HR-mediated gene targeting only when a DSB was introduced in the short RA, but not in the long LA (not shown). Thus, we propose that it is the presence of a DSB in the shorter homology arm that is crucial for HR-mediated genome editing, rather than the left-right orientation of the cut with respect to the insert. However, this hypothesis needs to be tested further in multiple targets to be considered a general mechanism and an important enhancing parameter for HR-mediated gene targeting. Our work will inform development of other HR-mediated genome editing methods. First, it will facilitate the construction of targeting vectors, because 3 kb of homologous sequences, which are necessary for effective HR, is a feasible size for PCR. Second, this size of the homology region allows one to confirm knock-in animals by fast and easy PCR-based genotyping methods. For example, using outer primers for the sequences flanking the homology arms, one can obtain an amplicon of the edited genome and confirm the precise sequences by amplicon sequencing. In addition, because most concatemeric knock-in alleles contain vector sequences, simple genotyping using vector-specific primers can easily distinguish single copy versus concatemeric alleles.
Two pioneering studies demonstrated that zebrafish genome could be edited by co-injection of a TALEN and targeting DNA, such as ssDNA and long dsDNA (Bedell et al., 2012; Zu et al., 2013). In addition to TALEN, the CRISPR/Cas9 system has proven to be an efficient customized nuclease in zebrafish (Hwang et al., 2013b). Both TALEN and CRISPR/Cas9 can efficiently induce DSBs in target sequences, suggesting that genome editing using CRISPR/Cas9 should also be possible in zebrafish. Indeed, recent studies showed that CRISPR/Cas9 system with ssDNA has the ability to edit the zebrafish genome (Hruscha et al., 2013; Hwang et al., 2013a). In addition, because several studies reported successful generation of knock-in animals using CRISPR/Cas9 and dsDNA (Dickinson et al., 2013; Gratz et al., 2013; Wang et al., 2013), zebrafish genome editing with CRISPR/Cas9 will likely be achievable in the near future. Therefore, our data should be applicable in any HR-mediated genome editing using any customized nuclease systems.
MATERIALS AND METHODS
Zebrafish husbandry and lines
Zebrafish adults and embryos were maintained according to zebrafish facility SOPs and Guide (http://zebrafish.wustl.edu/sopsandguides.htm), approved by the Animal Studies Committee at Washington University, St Louis, USA. The AB strain was used to generate sox2 and gfap knock-in lines. The sox2 and gfap knock-in lines are designated as sox2-2a-sfGFPstl84 (#3, see Fig. 4B-H,J-N; Table 1) and gfap-2a-tdTomatostl85 (#1, see Fig. 5C-K; Table 2).
TALEN design and construction
We used ZiFit (http://zifit.partners.org/ZiFiT/) (Sander et al., 2011) to design sox2 TALENs. For the assembly of RVD-containing repeats and subcloning of TALE repeats into modified TALE nuclease expression vectors, we used REAL Assembly TALEN Kit (Addgene TALEN kit 1000000017) and followed cloning steps as described previously (Sander et al., 2011). Modified TALE nuclease expression vectors were generated by sequential subcloning of NheI/EcoRI-digested TALE13 fragments from pJDS70, pJDS71, pJDS74 and pJSD78 in the TALEN Kit and EcoRI/NotI-digested EL/KK heterodimeric FokI nucleases from pCS2-EL/KK (Zhu et al., 2011) with nos1 3′ UTR fragment from GFP-nos1 3′ UTR construct (Koprunner et al., 2001) into pCS2. This strategy was used because nos1 3′ UTR can induce a rapid degradation of RNA in somatic cells whereas nos1 3′ UTR bearing TALEN RNA remains stable in germ cells (Koprunner et al., 2001).
To generate gfap TALENs, we used TALE-NT (https://tale-nt.cac.cornell.edu) (Doyle et al., 2012) for design and Golden Gate TALEN kit (Addgene TALEN kit 1000000024) for assembly of RVD repeats (Cermak et al., 2011). To generate TALE nuclease expression vectors, we modified pCS2TAL3-DD (Addgene plasmid 37275) and pCS2TAL3-RR (Addgene plasmid 37276) (Dahlem et al., 2012) by subcloning a BamHI/NotI-digested FokI nuclease fragment, which encodes either EL or KK FokI nuclease, with nos1 3′ UTR from sox2 TALEN constructs into the expression vectors.
Targeting vector construction
For sox2 targeting constructs, we initially subcloned PCR-amplified 3245 bp of sox2 genomic DNA fragment into pENTR-D/TOPO vector (designated as preM1) (Invitrogen). To insert sfGFP sequences in a sox2 stop codon (UAA), we synthesized sox2 genomic DNA-containing sfGFP sequences by the overlapping extension PCR method (Geiser et al., 2001). The PCR product was digested with NcoI and SacI, and subcloned into preM1 to make the M1 targeting construct (Fig. 2A). The remaining seven different targeting constructs were generated by a simple modification of M1 construct.
To construct gfap targeting vector, 3572 bp genomic DNA containing gfap exon 9 was amplified by PCR and subcloned into pENTR-D/TOPO plasmid. tdTomato sequences were introduced into the gfap genomic DNA-containing construct by the method described above for sox2 targeting vector. The sequences of targeting constructs are provided in supplementary material Table S1.
CRISPR/Cas9 system
We used CRISPR Design Tool (http://crispr.mit.edu) (Hsu et al., 2013) to design sox2 gRNA and gfap gRNA. The sox2 gRNA target sequence is GGAAACCGAGCTGAAGCCCC and the gfap gRNA target sequence is GGTGACCAGCCGTCACAGCA. We used pT7-gRNA (Addgene plasmid 46759) and nls-zCas9-nls (Addgene plasmid 47929) to establish a CRISPR/Cas9 system and followed the protocols as described previously (Jao et al., 2013).
Microinjection
sox2 and gfap TALEN RNAs were synthesized using SP6 mMessage mMachine Kit (Ambion). The synthetic RNAs were purified by Micro Bio-Spin P-30 Gel columns (Bio-Rad). Linearized targeting constructs were purified by QIAquick PCR-purification kit (Qiagen). For co-injection, we mixed a pair of TALEN RNAs, a targeting construct and 10× injection buffer (1 M KCl, 0.03% Phenol Red) to achieve a final concentration of 35 ng/μl for each TALEN RNA, and 10 ng/μl for targeting construct in 1× injection buffer, and injected approximately 1 nl into the cytosol of early one-cell zygotes.
In vivo recombination analysis
When we injected 35-70 pg of each TALEN RNA or 20-30 pg of targeting construct alone, the embryos developed normally. However, the majority of embryos co-injected with over 20 pg of targeting construct and 35-70 pg of TALEN RNA showed severe malformations at 1 dpf (data not shown). Hence, we carried out titration experiments to find a dose of the targeting construct that did not cause developmental defects when co-injected with TALEN RNA. We observed that the synergistic co-injection effects on embryos were minimized when the dose of the targeting construct was 10 pg or lower. Thus, in all the following experiments, one-cell stage embryos were co-injected with 10 pg of a targeting vector and 35 pg of each TALEN RNA. At 2 days after injection, 10 of the morphologically normal embryos were chosen at random, anesthetized using 0.01% ethyl 3-aminobenzoate methanesulfonic acid (Sigma-Aldrich), and mounted in 0.5% low-melting agarose (Lonza) in glass-bottomed 35 mm Petri dishes (MatTek). sfGFP signal was acquired in a 50 μm thick region of the diencephalon of individual embryos using Quorum Spinning disc Confocal/IX81 inverted microscope (Olympus) and Metamorph Acquisition software.
Targeted insertion screen
Each founder (F0) fish was outcrossed with wild-type fish to obtain F1 progeny from the individual founders. F1 progeny were screened, for sox2 knock-in lines at 1 dpf and for gfap knock-in lines at 2 dpf, using a Zeiss epifluorescence stereomicroscope. Embryos were anesthetized as described above and mounted in 1% methylcellulose (Sigma-Aldrich) for imaging using DFC365 FX camera attached to M205 FA stereomicroscope (Leica).
Whole-mount in situ RNA hybridization and immunohistochemistry
For whole-mount in situ RNA hybridization (WISH), we synthesized antisense RNA probe from a sox2 cDNA clone, which was generated by RT-PCR, using a digoxigenin RNA labeling kit (Roche). WISH was performed as described previously (Hauptmann and Gerster, 2000). After staining, the embryos were transferred into 100% glycerol (Sigma) for imaging with an AxioCam MRc camera mounted on SteREO Discovery V12 microscope (Zeiss). For immunohistochemistry, anesthetized 7 dpf larvae were fixed in 4% paraformaldehyde, embedded in 1.5% agarose and sectioned using a Leica cryostat microtome. We used rabbit anti-Sox10 [a gift from Dr Bruce Appel (University of Colorado School of Medicine, Aurora, CO, USA); 1:3000] (Park et al., 2005), mouse anti-HuC/D (Invitrogen, 16A11; 1:200) and mouse anti-GFAP (Sigma, G-A-5; 1:200) as primary antibodies, and anti-rabbit and anti-mouse IgG antibodies conjugated with Alexa Fluor 488, 568 and 647 (Invitrogen, A11001, A11011, A21237; 1:200) as secondary antibodies. Although we tried to test whether sfGFP-positive cells express Sox2 by immunohistochemistry using two different Sox2 antibodies that were previously reported to detect Sox2 in zebrafish (Germana et al., 2011; Hernandez et al., 2007), we failed to detect endogenous Sox2. The fluorescence imaging was carried out using Quorum Spinning disc Confocal/IX81-inverted microscope (Olympus) and Metamorph Acquisition software.
Genotyping, T7 endonuclease I assay, quantitative PCR and Southern blot analysis
For TALEN activity analysis and T7EI assay, genomic DNA from embryos was isolated using lysis solution [10 mM Tris-HCl (pH 8), 50 mM KCl, 0.3% Tween, 0.3% NP40 and 1 mg/ml proteinase K]. For knock-in animal genotyping, qPCR and Southern blot analysis, genomic DNA was purified using DNeasy Blood & Tissue kit (Qiagen) from frozen tissues of adult zebrafish.
Genotyping was performed using Taq DNA polymerase (NEB), LongAmp Taq DNA polymerase (NEB) and Phusion Flash high-fidelity polymerase (Thermo Scientific). For the T7EI assay, the gRNA target-containing amplicon was directly digested with T7 endonuclease I (NEB) for 2 h. For qPCR, sF1 and sR1 primers were used to amplify the sfGFP region, and a pair of primers (qF1 and qR1) amplifying a region outside the recombination site was used for normalization. PCR was performed in triplicate using SsoAdvanced SYBR Green Supermix (Bio-Rad) and CFX Connect Real time system (Bio-Rad). Relative sfGFP copy numbers were calculated using the comparative Ct method. The primers used are listed in supplementary material Table S2.
Southern blot analysis was performed using 10 μg of genomic DNA of individual wild-type and sox2 knock-in F1 fish. Briefly, the genomic DNA was digested overnight with PstI and BamHI, and precipitated with 3 M sodium acetate and 100% ethanol. The digested DNA was separated on 0.8% seakem GTG gel in 1× TBE buffer and transferred to a nylon membrane (PerkinElmer gene screen plus). The membrane was UV crosslinked and hybridized overnight with 32P random prime-labeled probe (Roche Random Prime Labeling kit). The probe sequence is provided in supplementary material Table S1.
Supplementary Material
Acknowledgements
We thank J. Keith Joung for REAL Assembly TALEN kit, Daniel F. Voytas and Adam Bogdanove for Golden gate TALEN Kit, David Jonah Grunwald for pCS2TAL3-DD/RR, and Susan R. Wente and Wenbiao Chen for CRISPR nuclease system. We also thank Ryan S. Gray, Margot Williams, Diane S. Sepich, Christina A. Gurnett, Kelly R. Monk, Michael L. Nonet and Sanjay Jain for discussion and comments on the manuscript; and the Washington University School of Medicine in St Louis Zebrafish Facility Staff for excellent animal care.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
J.S. and L.S.-K. conceived and designed the experiments. J.S. and J.C. performed the experiments and analyzed the data. J.S. and L.S.-K. wrote the paper, and all authors discussed and contributed to the final version.
Funding
This work was supported in part by grants from the National Institute of General Medicine [R01 GM55101 and GM77770 to L.S.-K.], and by the Hope Center for Neurological Disorders Transgenic Vectors Core at Washington University School of Medicine in St Louis. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.108019/-/DC1
References
- Auer, T. O., Duroure, K., De Cian, A., Concordet, J. P. and Del Bene, F. (2014). Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24, 142-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas, D., Davidson, A. E., Sivasubbu, S., Hermanson, S. B., Welle, Z. and Ekker, S. C. (2004). Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5, 62 10.1186/1471-2164-5-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell, V. M., Wang, Y., Campbell, J. M., Poshusta, T. L., Starker, C. G., Krug, R. G.II, Tan, W., Penheiter, S. G., Ma, A. C., Leung, A. Y. H.et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114-118 10.1038/nature11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, M., Golic, M., Golic, K. G. and Carroll, D. (2002). Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161, 1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., Lahaye, T., Nickstadt, A. and Bonas, U. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509-1512 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Capecchi, M. R. (2005). Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6, 507-512 10.1038/nrg1619 [DOI] [PubMed] [Google Scholar]
- Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., Baller, J. A., Somia, N. V., Bogdanove, A. J. and Voytas, D. F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P. D., Wu, X., Jiang, W., Marraffini, L. A.et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe, V. T. and Casaccia-Bonnefil, P. (2006). Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech. Dev. 123, 24-30 10.1016/j.mod.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Dahlem, T. J., Hoshijima, K., Jurynec, M. J., Gunther, D., Starker, C. G., Locke, A. S., Weis, A. M., Voytas, D. F. and Grunwald, D. J. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D. J., Ward, J. D., Reiner, D. J. and Goldstein, B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, E. L., Booher, N. J., Standage, D. S., Voytas, D. F., Brendel, V. P., Vandyk, J. K. and Bogdanove, A. J. (2012). TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117-W122 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser, M., Cebe, R., Drewello, D. and Schmitz, R. (2001). Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31, 88-90, 92. [DOI] [PubMed] [Google Scholar]
- Germana, A., Montalbano, G., Guerrera, M. C., Amato, V., Laura, R., Magnoli, D., Campo, S., Suarez-Fernandez, E., Ciriaco, E. and Vega, J. A. (2011). Developmental changes in the expression of sox2 in the zebrafish brain. Microsc. Res. Tech. 74, 347-354. [DOI] [PubMed] [Google Scholar]
- Gratz, S. J., Wildonger, J., Harrison, M. M. and O'Connor-Giles, K. M. (2013). CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly (Austin) 7, 249-255 10.4161/fly.26566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann, G. and Gerster, T. (2000). Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 137, 139-148. [DOI] [PubMed] [Google Scholar]
- Hernández, P. P., Olivari, F. A., Sarrazin, A. F., Sandoval, P. C. and Allende, M. L. (2007). Regeneration in zebrafish lateral line neuromasts: expression of the neural progenitor cell marker sox2 and proliferation-dependent and-independent mechanisms of hair cell renewal. Dev. Neurobiol. 67, 637-654 10.1002/dneu.20386 [DOI] [PubMed] [Google Scholar]
- Hockemeyer, D., Wang, H., Kiani, S., Lai, C. S., Gao, Q., Cassady, J. P., Cost, G. J., Zhang, L., Santiago, Y., Miller, J. C.et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731-734 10.1038/nbt.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha, A., Krawitz, P., Rechenberg, A., Heinrich, V., Hecht, J., Haass, C. and Schmid, B. (2013). Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982-4987 10.1242/dev.099085 [DOI] [PubMed] [Google Scholar]
- Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., Li, Y., Fine, E. J., Wu, X., Shalem, O.et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827-832 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Kaini, P., Sander, J. D., Joung, J. K., Peterson, R. T. and Yeh, J.-R. J. (2013a). Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS ONE 8, e68708 10.1371/journal.pone.0068708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., Peterson, R. T., Yeh, J.-R. J. and Joung, J. K. (2013b). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao, L.-E., Wente, S. R. and Chen, W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904-13909 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen, J. R., Willett, C. E. and Lin, S. (1999). Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nat. Genet. 23, 15-16 10.1038/12609 [DOI] [PubMed] [Google Scholar]
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K. (2005). Transposon tools and methods in zebrafish. Dev. Dyn. 234, 244-254 10.1002/dvdy.20516 [DOI] [PubMed] [Google Scholar]
- Kawakami, K., Takeda, H., Kawakami, N., Kobayashi, M., Matsuda, N. and Mishina, M. (2004). A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133-144 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Kim, Y. G., Cha, J. and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 93, 1156-1160 10.1073/pnas.93.3.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprunner, M., Thisse, C., Thisse, B. and Raz, E. (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey, B. W., Darabie, A. and Tropepe, V. (2012). The cellular composition of neurogenic periventricular zones in the adult zebrafish forebrain. J. Comp. Neurol. 520, 2275-2316 10.1002/cne.23065 [DOI] [PubMed] [Google Scholar]
- Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., Norville, J. E. and Church, G. M. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823-826 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus, R. C. and Easter, S. S.,Jr (1995). Expression of glial fibrillary acidic protein and its relation to tract formation in embryonic zebrafish (Danio rerio). J. Comp. Neurol. 359, 365-381 10.1002/cne.903590302 [DOI] [PubMed] [Google Scholar]
- Miller, J. C., Tan, S., Qiao, G., Barlow, K. A., Wang, J., Xia, D. F., Meng, X., Paschon, D. E., Leung, E., Hinkley, S. J.et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143-148 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Moscou, M. J. and Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Park, H.-C., Boyce, J., Shin, J. and Appel, B. (2005). Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J. Neurosci. 25, 6836-6844 10.1523/JNEUROSCI.0981-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. and Waldo, G. S. (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79-88 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- Provost, E., Rhee, J. and Leach, S. D. (2007). Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 45, 625-629 10.1002/dvg.20338 [DOI] [PubMed] [Google Scholar]
- Radecke, S., Radecke, F., Cathomen, T. and Schwarz, K. (2010). Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol. Ther. 18, 743-753 10.1038/mt.2009.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, J. D., Cade, L., Khayter, C., Reyon, D., Peterson, R. T., Joung, J. K. and Yeh, J.-R. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697-698 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N. G., Palmer, A. E. and Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567-1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shin, J., Park, H.-C., Topczewska, J. M., Mawdsley, D. J. and Appel, B. (2003). Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 25, 7-14 10.1023/B:MICS.0000006847.09037.3a [DOI] [PubMed] [Google Scholar]
- Smih, F., Rouet, P., Romanienko, P. J. and Jasin, M. (1995). Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 23, 5012-5019 10.1093/nar/23.24.5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, J. L., Kearns, C. A. and Appel, B. (2012). Fbxw7 regulates Notch to control specification of neural precursors for oligodendrocyte fate. Neural. Dev. 7, 15 10.1186/1749-8104-7-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, J., Bayraktaroglu, L., Clements, D., Conlin, T., Fashena, D., Frazer, K., Haendel, M., Howe, D. G., Mani, P., Ramachandran, S.et al. (2006). The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 34, D581-D585 10.1093/nar/gkj086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov, F. D., Miller, J. C., Lee, Y.-L., Beausejour, C. M., Rock, J. M., Augustus, S., Jamieson, A. C., Porteus, M. H., Gregory, P. D. and Holmes, M. C. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646-651 10.1038/nature03556 [DOI] [PubMed] [Google Scholar]
- Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F. and Jaenisch, R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910-918 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., Bourque, C., Dovey, M., Goessling, W., Burns, C. E.et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183-189 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., Smith, T., McNulty, J., Rayla, A. L., Lakshmanan, A., Siekmann, A. F., Buffardi, M., Meng, X., Shin, J., Padmanabhan, A.et al. (2011). Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development 138, 4555-4564 10.1242/dev.066779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu, Y., Tong, X., Wang, Z., Liu, D., Pan, R., Li, Z., Hu, Y., Luo, Z., Huang, P., Wu, Q.et al. (2013). TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10, 329-331 10.1038/nmeth.2374 [DOI] [PubMed] [Google Scholar]
- Zupanc, G. K. H., Hinsch, K. and Gage, F. H. (2005). Proliferation, migration, neuronal differentiation, and long-term survival of new cells in the adult zebrafish brain. J. Comp. Neurol. 488, 290-319 10.1002/cne.20571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.