Abstract
The thymus and parathyroid glands arise from a shared endodermal primordium in the third pharyngeal pouch (3rd pp). Thymus fate is specified in the ventral 3rd pp between E9.5 and E11, whereas parathyroid fate is specified in the dorsal domain. The molecular mechanisms that specify fate and regulate thymus and parathyroid development are not fully delineated. Previous reports suggested that Tbx1 is required for thymus organogenesis because loss of Tbx1 in individuals with DiGeorge syndrome and in experimental Tbx1 deletion mutants is associated with thymus aplasia or hypoplasia. However, the thymus phenotype is likely to be secondary to defects in pharyngeal pouch formation. Furthermore, the absence of Tbx1 expression in the thymus-fated domain of the wild-type 3rd pp suggested that Tbx1 is instead a negative regulator of thymus organogenesis. To test this hypothesis, we generated a novel mouse strain in which expression of a conditional Tbx1 allele was ectopically activated in the thymus-fated domain of the 3rd pp. Ectopic Tbx1 expression severely repressed expression of Foxn1, a transcription factor that marks the thymus-fated domain and is required for differentiation and proliferation of thymic epithelial cell (TEC) progenitors. By contrast, ectopic Tbx1 did not alter the expression pattern of Gcm2, a transcription factor restricted to the parathyroid-fated domain and required for parathyroid development. Ectopic Tbx1 expression impaired TEC proliferation and arrested TEC differentiation at an early progenitor stage. The results support the hypothesis that Tbx1 negatively regulates TEC growth and differentiation, and that extinction of Tbx1 expression in 3rd pp endoderm is a prerequisite for thymus organogenesis.
Keywords: TBX1, FOXN1, Thymus, Mouse
INTRODUCTION
T-cell development takes place in the thymus, which provides a unique and indispensable stromal microenvironment composed of thymic epithelial cells (TECs), fibroblasts, endothelial cells, macrophages and dendritic cells. TECs are the predominant cell type in the thymic microenvironment and perform a number of nonredundant functions that are essential for proper thymocyte growth and maturation (reviewed by Boehm, 2008; Rodewald, 2008; Manley et al., 2011; Thompson and Zuniga-Pflucker, 2011; Anderson and Takahama, 2012). Cortical TECs (cTECs) and medullary TECs (mTECs) present self-peptides that mediate the positive and negative selection processes required to produce T cells that express a diverse array of antigen-specific receptors, yet are self-tolerant. TECs also produce cytokines that promote growth and survival of thymocytes, chemokines that direct thymocyte migration and Notch ligand that activates signaling pathways required for thymocyte differentiation and T-cell lineage commitment. Given the various crucial roles played by TECs in T-cell development and the loss of TECs that occurs during age-related thymus involution, there is considerable interest in elucidating the molecular mechanisms that commit primitive endodermal progenitors to a TEC fate and regulate thymus organogenesis.
The thymus and parathyroid glands arise from shared primordia that develop bilaterally from third pharyngeal pouch (3rd pp) endoderm. Correct patterning and cell specification within the shared pouch are essential for development of both organs. The ventral posterior region of the 3rd pp is specified to a thymus fate, whereas the dorsal anterior region is specified to a parathyroid fate. Although the thymus and parathyroid domains are specified by E9.5-10.5, they cannot be morphologically distinguished (Gordon et al., 2001, 2004; Patel et al., 2006). By E11.5, the thymus- and parathyroid-fated domains can be recognized by Foxn1 and Gcm2 expression respectively. However, although these transcription factors regulate differentiation, they do not specify thymus versus parathyroid fate (Bleul et al., 2006; Liu et al., 2007; Nowell et al., 2011). A Hox-Pax-Eya-Six cascade is implicated upstream of thymus specification, but has not been directly linked to the establishment of thymus fate (Manley and Condie, 2010). Additional transcription factors with restricted expression patterns in the 3rd pp have been identified, but their role in thymus fate is not yet established (Wei and Condie, 2011). Thus, the transcription factors and molecular pathways that specify thymus fate and regulate thymus organogenesis are not yet defined.
The T-box transcription factor TBX1 is widely considered to play an indispensable role in thymus development. During ontogeny, Tbx1 is expressed throughout the pharyngeal endoderm and arch mesenchyme, but is not expressed by neural crest-derived mesenchyme (Vitelli et al., 2002). Deletion or mutation of Tbx1 is a common denominator in the human chromosome 22q11.2 deletion syndrome (22q11DS) that results in DiGeorge and velocardiofacial syndromes (Jerome and Papaioannou, 2001; Merscher et al., 2001; Yagi et al., 2003; Baldini, 2005). In addition to cardiovascular defects and craniofacial abnormalities, these patients typically have thymus aplasia or hypoplasia (Hollander et al., 2006). Furthermore, thymus aplasia is a characteristic feature of Tbx1 homozygous deletion mutants, a finding consistent with the prevalent view that TBX1 potentiates thymus development (Jerome and Papaioannou, 2001; Baldini, 2005; Hollander et al., 2006). However, this interpretation is complicated by the fact that Tbx1 is required for segmentation of the pharyngeal apparatus; in its absence, the pharyngeal pouches do not develop (Chapman et al., 1996; Jerome and Papaioannou, 2001; Lindsay et al., 2001; Vitelli et al., 2002; Arnold et al., 2006). Therefore, the athymia observed in the absence of Tbx1 is a secondary consequence of defective pouch formation; as a result, the potential role of Tbx1 in thymus organogenesis cannot be determined from analysis of Tbx1-null embryos. Moreover, by E10.5 Tbx1 expression in the 3rd pp is restricted to the parathyroid domain and excluded from the thymus domain (Vitelli et al., 2002; Manley et al., 2004; Dooley et al., 2007), suggesting that Tbx1 expression may be necessary for parathyroid formation, but may antagonize rather than promote thymus development. A previous study reported the occurrence of thymus hypoplasia resulting from Tbx1Cre-mediated activation of a conditional Tbx1 transgene during embryogenesis; however, the mechanism responsible for this phenotype was not determined (Vitelli et al., 2009).
To test the hypothesis that Tbx1 expression in 3rd pp endoderm suppresses thymus fate and/or TEC differentiation, we generated a novel mouse strain in which a stop-floxed Tbx1 allele was knocked into the ubiquitously expressed Rosa26 locus. We used Foxn1Cre to activate Tbx1 expression in the thymus-fated domain of the 3rd pp. Here, we show that ectopic Tbx1 expression in the ventral 3rd pp suppresses Foxn1 expression, inhibits TEC proliferation and blocks TEC differentiation at an early progenitor stage, but does not reverse thymus fate. The results support the hypothesis that Tbx1 negatively regulates TEC growth and differentiation, and that extinction of Tbx1 expression in 3rd pp endoderm is a prerequisite for thymus organogenesis.
RESULTS
Foxn1Cre activates ectopic expression of the R26iTbx1 allele in the ventral domain of the 3rd pp
Tbx1 is expressed throughout 3rd pp endoderm at E9.5, but by E10.5 is restricted to the dorsal, parathyroid-fated domain (Vitelli et al., 2002; Manley et al., 2004; Zhang et al., 2005; Liu et al., 2007). Foxn1-expressing cells are present in the dorsal 3rd pp and extend into pharyngeal endoderm in the absence of SHH, a positive regulator of Tbx1 expression (Garg et al., 2001; Yamagishi et al., 2003; Moore-Scott and Manley, 2005). Taken together, these data suggest that TBX1 antagonizes thymus development. Therefore, we used a gain-of-function approach to test this hypothesis. Tbx1 cDNA and an IRES-GFP tag were inserted into a modified Rosa26 targeting vector containing a floxed stop cassette to generate a knock-in strain hereafter referred to as R26iTbx1 (supplementary material Fig. S1). This allele activates expression of both Tbx1 and EGFP after Cre-mediated deletion of the stop cassette.
We used Foxn1Cre to activate ectopic Tbx1 expression in the ventral thymus-fated domain of the 3rd pp beginning at ∼E11.25 (Gordon et al., 2007). Thymus fate is specified in 3rd pp endoderm as early as E9.5-10.5 prior to expression of Foxn1, which is essential for TEC differentiation and proliferation (reviewed by Manley and Condie, 2010; Gordon and Manley, 2011). Therefore, Foxn1Cre activates expression of the R26iTbx1 allele in ventral 3rd pp endoderm after the cells commit to a thymus fate and are beginning to differentiate. Immunohistochemical (IHC) analysis of GFP expression in the Foxn1Cre/+;R26iTbx1/+ 3rd pp verified the predicted ectopic expression of R26iTbx1 in the ventral, but not in the dorsal, domain and confirmed the absence of a GFP signal in Foxn1Cre/+;R26+/+ controls (supplementary material Fig. S2). Consistent with previous reports, 3rd pp and thymus development were normal in Foxn1Cre/+;R26+/+ controls (supplementary material Fig. S2; see Fig. 3) (Gordon et al., 2007; Chen et al., 2009). Continued expression of the R26iTbx1 allele in fetal thymi throughout ontogeny was confirmed by FACS analysis of GFP expression, which also verified restriction of the GFP signal to EpCAM+ CD45− TECs (supplementary material Fig. S2).
Fig. 3.
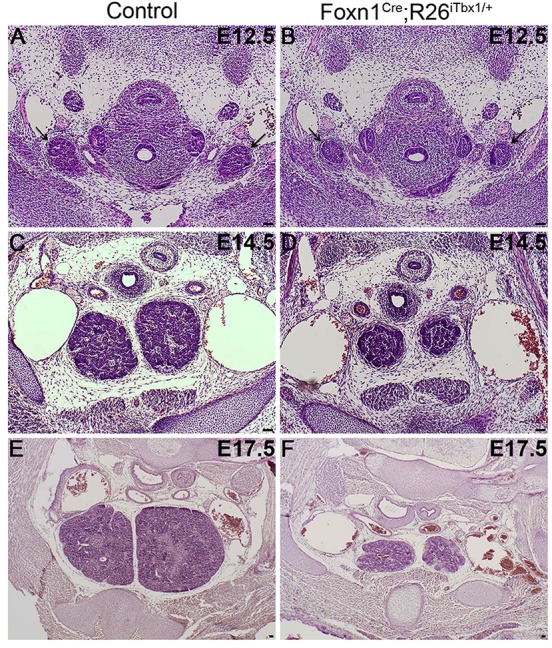
Thymic lobes are hypoplastic throughout ontogeny in Foxn1Cre;R26iTbx1/+ embryos. (A-F) Hematoxylin and Eosin-stained transverse sections show that Foxn1Cre;R26iTbx1/+ thymic lobes (arrows) are smaller than controls at each developmental stage. (A,B) E12.5; (C,D) E14.5; (E,F) E17.5. n=5 control and n=5 Foxn1Cre;R26iTbx1/+ 3rd pps. Scale bars: 50 μm.
Ectopic TBX1 expression suppresses FOXN1 but does not alter GCM2 expression
We asked whether ectopic expression of Tbx1 in the ventral domain altered patterning of the 3rd pp at E11.5. IHC staining of serial sections from Foxn1Cre;R26+/+ controls confirmed that endogenous TBX1 is restricted to the dorsal parathyroid-fated domain (Fig. 1A), whereas FOXN1 is restricted to the ventral thymus-fated domain (Fig. 1A,C). FOXN1 and TBX1 co-expressing cells were not found in the control 3rd pp. In contrast to the restricted expression of TBX1 in the control 3rd pp at this stage, we found an increased number of TBX1-positive cells located in both the ventral and dorsal regions of the Foxn1Cre;R26iTbx/+ 3rd pp (Fig. 1B,E). The presence of ectopic TBX1 protein in the ventral 3rd pp is consistent with the GFP staining pattern observed in the Foxn1Cre;R26iTbx/+ 3rd pp (supplementary material Fig. S1). Ectopic TBX1 in the ventral domain resulted in a profound and rapid reduction in the frequency of 3rd pp cells containing detectable levels of FOXN1 (Fig. 1B,D,E). The few cells in the ventral 3rd pp that did express detectable levels of FOXN1 generally co-expressed TBX1 (Fig. 1B).
Fig. 1.
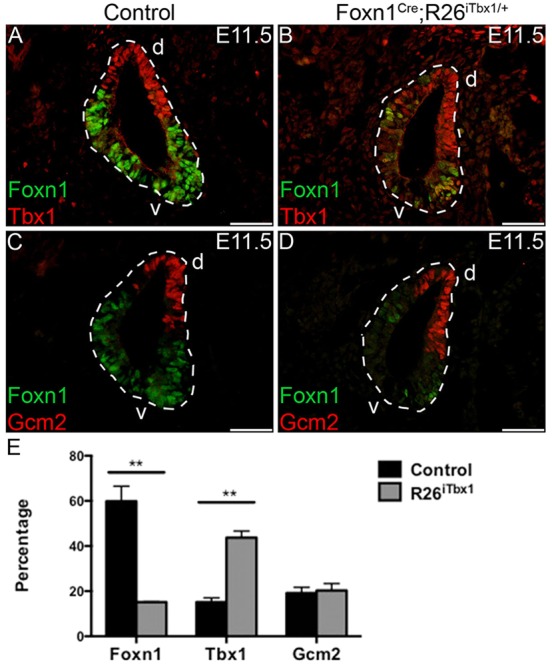
Ectopic TBX1 in the ventral 3rd pp reduces the number of FOXN1-positive cells. (A-D) Representative immunohistochemical stains of sagittal sections from the 3rd pp of Foxn1Cre;R26+/+ control and Foxn1Cre;R26iTbx1/+ E11.5 embryos. (A,B) TBX1 (red) is restricted to the dorsal (d) parathyroid-fated domain of the control, but is present throughout the mutant 3rd pp. There is a marked reduction in FOXN1-positive cells (green) in the mutant 3rd pp. A few cells in the mutant 3rd pp are FOXN1 positive and co-stain for TBX1 (yellow). (C) GCM2 (red) is present in parathyroid-fated cells in the dorsal domain of the control 3rd pp. (D) Ectopic TBX1 does not affect localization or frequency of GCM2-positive cells in the mutant 3rd pp. Scale bars: 50 μm. (E) The percentage±s.d. of 3rd pp cells that are positive for FOXN1, TBX1 or GCM2 (n=4 control and n=4 Foxn1Cre;R26iTbx1/+ 3rd pps). **P<0.02. v, ventral.
Gcm2 was previously proposed to be a downstream target of Tbx1 (Ivins et al., 2005; Liu et al., 2007). However, ectopic Tbx1 expression had no discernible effect on the number or localization of GCM2-positive cells (Fig. 1D,E). GCM2 was localized to the dorsal anterior region of the control 3rd pp, and this restricted expression pattern was maintained in the Foxn1Cre;R26iTbx/+ 3rd pp. Therefore, these data demonstrate that ectopic expression of Tbx1 is not sufficient to induce Gcm2 expression in the ventral domain of the 3rd pp, suggesting that additional regulators are involved in controlling Gcm2 expression.
Ectopic TBX1 expression in the E11.5 3rd pp does not reverse thymus fate
Owing to the severe reduction in FOXN1-positive cells and failure to expand GCM2, the majority of cells in the ventral domain of the Foxn1Cre;R26iTbx/+ 3rd pp express neither FOXN1 nor GCM2 (Fig. 1B,D,E). To determine whether these cells are no longer committed to a thymus fate, we examined expression of additional factors that are expressed by thymus-specified cells. We previously reported that at E11.5, thymus-, but not parathyroid-fated, cells in the 3rd pp express IL7, a cytokine that is essential for thymocyte differentiation and survival (Zamisch et al., 2005; Repass et al., 2009). Moreover, IL7 is expressed by immature TECs in the thymic rudiment of FOXN1-deficient nude (Foxn1nu/nu) mice (Zamisch et al., 2005). Therefore, IL7 is a Foxn1 independent marker of thymus-fated cells. Quantitative RT-PCR analysis demonstrated comparable levels of Il7 mRNA in 3rd pp cells of Foxn1Cre;R26iTbx/+ and control embryos (Fig. 2A).
Fig 2.
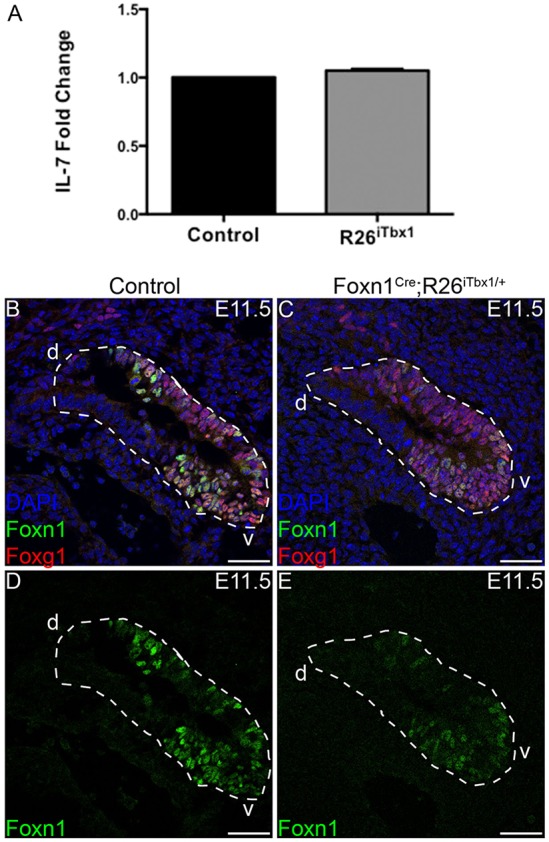
Ectopic TBX1 does not affect expression of IL7 or FOXG1 in the ventral 3rd pp. (A) Real-time quantitative PCR analysis shows equivalent Il7 mRNA levels in the 3rd pp dissected from E11.5 control or Foxn1Cre;R26iTbx1/+ embryos (n=6 control and n=6 Foxn1Cre;R26iTbx1/+). (B) Representative immunohistochemical stain showing a sagittal section of E11.5 3rd pp. FOXG1 (red) and FOXN1 (green) are co-expressed in the ventral (v) domain; DAPI (blue). (C) Representative immunohistochemical stain of a sagittal section of the mutant 3rd pp showing abundant FOXG1 in the ventral domain despite the severe reduction in FOXN1. (D,E) Single-color images of FOXN1 staining. n=4 control and n=4 Foxn1Cre;R26iTbx1/+ 3rd pps). Scale bars: 50 μm. d, dorsal.
FOXG1 is a transcription factor that is initially expressed in the ventral domain of the 3rd pp by E10.5, preceding Foxn1 (Wei and Condie, 2011). Similar to Il7, Foxg1 is expressed in the Foxn1nu/nu thymic rudiment (Q. Wei and B. Condie, unpublished). IHC analysis of serial sections in the control 3rd pp showed that FOXG1 overlaps with FOXN1 in the ventral domain and is excluded from the dorsal GCM2 domain (Fig. 2B,D). Interestingly, FOXG1 is abundantly expressed by the FOXN1-negative cells in the Foxn1Cre;R26iTbx/+ ventral 3rd pp domain (Fig. 2C). The robust expression of IL7 and FOXG1 in the Foxn1Cre;R26iTbx/+ ventral 3rd pp demonstrates that ectopic expression of Tbx1 does not repress or reverse thymus fate, but instead is selectively acting through repression of Foxn1.
Ectopic TBX1 expression reduces proliferation of GCM2-negative cells in the ventral 3rd pp
Despite the severely reduced levels of Foxn1 expression in the 3rd pp of Foxn1Cre;R26iTbx/+ embryos, thymic primordia formed, detached from the pharynx and migrated to a normal position above the heart in a timely manner (Fig. 3) (Gordon and Manley, 2011). These data are consistent with the conclusion that cells in the ventral 3rd pp remain specified to a thymus fate after ectopic Tbx1 expression. However, Foxn1Cre;R26iTbx/+ fetal thymi are severely hypoplastic throughout ontogeny and remain so in the postnatal period (Fig. 3; data not shown).The hypoplastic thymus phenotype is not surprising given that FOXN1 is essential for TEC proliferation (Itoi et al., 2001; Su et al., 2003; Chen et al., 2009; Nowell et al., 2011).
To determine the cellular mechanism(s) by which ectopic TBX1 expression results in thymus hypoplasia, we analyzed the frequency of proliferating and apoptotic cells in the ventral domain of the 3rd pp. Owing to the paucity of FOXN1-positive cells caused by ectopic TBX1, we determined the percentage of BrdU-incorporating cells in GCM2-positive compared with GCM2-negative 3rd pp domains. Ectopic Tbx1 expression decreased the frequency of proliferating cells in the GCM2-negative domain (Fig. 4A-C). By contrast, we found no difference in the frequency of proliferating GCM2-positive cells. To determine whether ectopic Tbx1 expression enhanced apoptosis in the 3rd pp, we stained sections for cleaved caspase 3. No difference was observed in the frequency of apoptotic 3rd pp cells in Foxn1Cre;R26iTbx/+ compared with control embryos (data not shown). These data demonstrate that TEC proliferation, but not survival, is compromised by ectopic expression of Tbx1 in the thymus-fated domain.
Fig. 4.
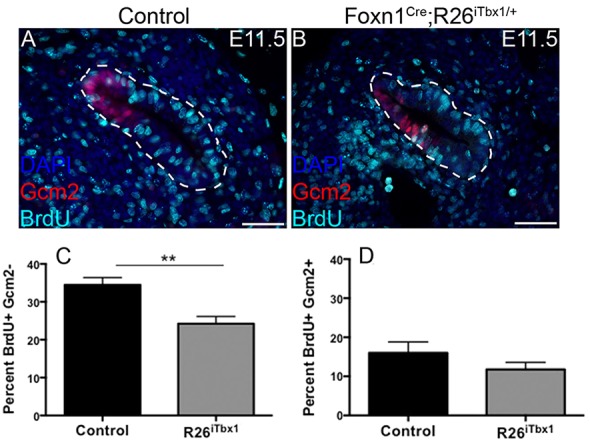
Proliferation is reduced in the Foxn1Cre;R26iTbx1/+ thymus-fated 3rd pp domain. (A,B) BrdU (10 mg/ml) was injected into pregnant females 1.5 h prior to obtaining E11.5 embryos. Immunohistochemical staining for BrdU (teal) identifies proliferating cells in the GCM2-positive (red), parathyroid-fated domain and the GCM2-negative thymus-fated domain of the 3rd pp from control (A) and Foxn1Cre;R26iTbx1/+ (B) embryos. Scale bars: 50 μm. (C,D) Bar graphs showing the frequency of BrdU-labeled cells that are GCM2 negative or GCM2 positive. Data show percentage±s.d. for eight control and eight Foxn1Cre;R26iTbx1/+ 3rd pps. **P<0.005.
TEC differentiation is impaired by ectopic TBX1 expression
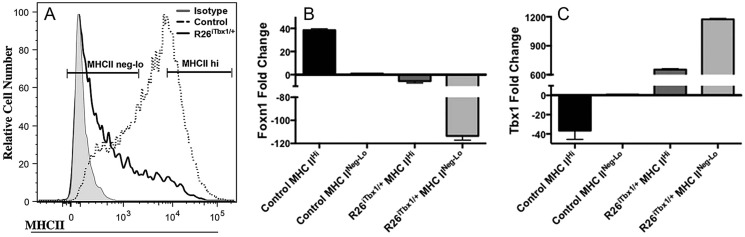
Given that FOXN1 is required for TEC differentiation (Blackburn et al., 1996; Bleul et al., 2006; Chen et al., 2009; Corbeaux et al., 2010; Nowell et al., 2011), we predicted that this process would be impaired in Foxn1Cre;R26iTbx/+ thymi. To test this hypothesis, we first assessed expression of MHC Class II (MHCII), an early differentiation marker that is expressed on wild-type TECs beginning at E12.5 in a Foxn1-dependent manner (Chen et al., 2009; Shakib et al., 2009; Xiao and Manley, 2010; Nowell et al., 2011). Whereas most TECs in E15.5 control thymi expressed high levels of MHCII, there were five to ten times fewer MHCIIhi TECs in Foxn1Cre;R26iTbx/+ mutants, indicating an early block in TEC differentiation (Fig. 5A). We isolated TECs by cell sorting into MHCIIhi and MHCIIneg-lo populations for qRT-PCR analysis using the sort gates shown in Fig. 5A. As previously reported (Chen et al., 2009), MHC IIhi TECs from controls expressed higher levels of Foxn1 transcripts than MHCIIneg-lo TECs (Fig. 5B). Neither TEC subset isolated from control thymi expressed significant levels of Tbx1 (Fig. 5C). By contrast, both MHCIIhi and MHCIIneg-lo TECs from Foxn1Cre;R26iTbx/+ thymi expressed very low levels of Foxn1 and high levels of Tbx1 mRNA (Fig. 5B,C). These data are consistent with the notion that TBX1 suppression of Foxn1 causes decreased TEC differentiation.
Fig. 5.
Ectopic TBX1 blocks TEC differentiation and suppresses Foxn1 expression. (A) Flow cytometric analysis of MHC class II expression on EpCAM+ CD45− control and Foxn1Cre;R26iTbx1/+ TECs. The majority of control TECs are MHCIIhi, whereas most Foxn1Cre;R26iTbx1/+ TECs are MHCIIneg-lo. (B,C) Representative bar graphs of qRT-PCR analysis showing Foxn1 (B) or Tbx1 (C) expression in E15.5 control and Foxn1Cre;R26iTbx1/+ TECs. MHCIIhi and MHCIIneg-lo were obtained by FACS sorting using the indicated gates. Data are mean±s.d for six control and six Foxn1Cre/+;R26iTbx/+ experiments.
FOXN1-positive TECs express cTEC and mTEC markers, and segregate from TBX1-positive TECs
By E15.5, a small number of FOXN1-positive TECs were present in the Foxn1Cre;R26iTbx/+ thymi; these TECs had low TBX1 levels, presumably owing to downregulation of or failure to activate R26iTbx. The small number of FOXN1-positive TECs in the E15.5 mutant thymus localize predominantly to the central region of each lobe (Fig. 6B), whereas FOXN1-expressing TECs are distributed throughout control thymi (Fig. 6A). The majority of TECs in E15.5 Foxn1Cre;R26iTbx/+ thymic lobes continued to express high levels of TBX1 and low to undetectable levels of FOXN1. These cells were concentrated towards the outer region of each lobe (Fig. 6B). A similar pattern of TBX1 and FOXN1 expression was observed at E17.5 (supplementary material Fig. S3).
Fig. 6.
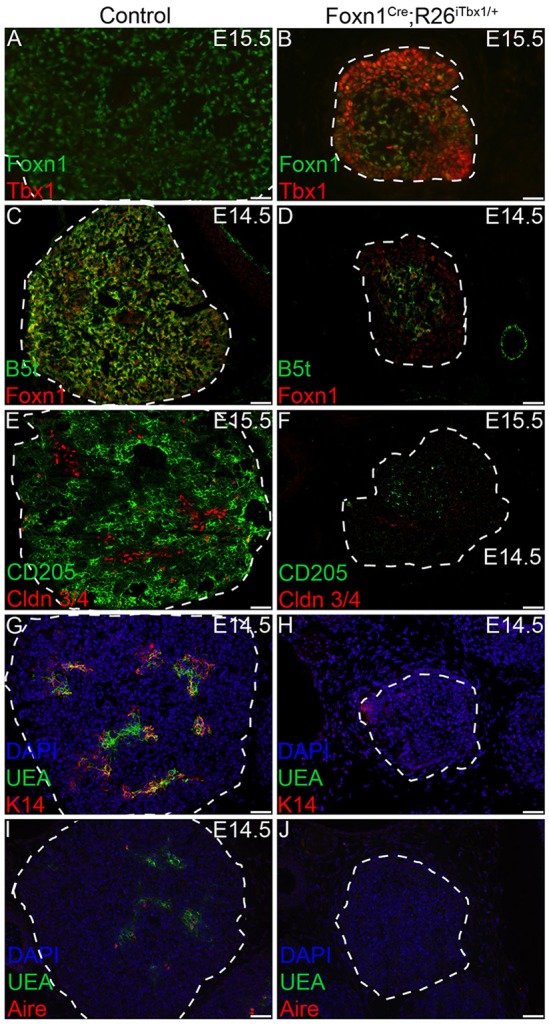
A small number of FOXN1-positive TECs are present in the center of Foxn1Cre;R26iTbx1/+ fetal thymic lobes. (A-J) Representative immunohistochemical stains of transverse sections from control and Foxn1Cre;R26iTbx1/+ E14.5 or E15.5 thymic lobes show distinct patterns of TEC localization. (A) FOXN1-positive (green) TECs are dispersed throughout the control thymus and TBX1-positive cells are not found. (B) The small number of FOXN1-positive TECs (green) in Foxn1Cre;R26iTbx1/+ mutants localize to the center of each lobe and generally segregate away from the abundant TBX1-positive TECs (red) that are concentrated in the outer region of the each lobe. (C-F) Only a few of the mutant TECs are positive for β5t and for CD205, which mark the cTEC lineage (green); these cells localize primarily in the center of each lobe where FOXN1-positive cells are found (see B). Scale bars: 50 µm.
To determine whether TECs in Foxn1Cre;R26iTbx/+ thymi were committed to either cTEC or mTEC lineages, we analyzed additional lineage-specific TEC markers. The thymoproteosome subunit β5t, as well as the surface receptor CD205, are expressed in a FOXN1-dependent manner at early stages of cTEC differentiation and are therefore expressed poorly or not at all in the Foxn1 null (nude) thymus (Nowell et al., 2008; Shakib et al., 2009; Ripen et al., 2011). As expected, the majority of cTECs in the control thymus were positive for both CD205 and β5t (Fig. 6C,E). By contrast, only a subset of TECs in Foxn1Cre;R26iTbx/+ thymi expressed β5t or CD205. TECs expressing these markers were primarily found in the center of each lobe, where FOXN1-positive cells were also located (Fig. 6D,F).
K14 expression and binding of the lectin Ulex europeaus agglutinin 1 (UEA1) are markers that distinguish mTEC subsets (Klug et al., 1998). Both K14-positive and UEA1-binding mTECs were present in the developing medullary regions of control thymi (Fig. 6G,I). However, only exceedingly rare cells expressed either of these mTEC markers in the Foxn1Cre;R26iTbx/+ thymic lobes (Fig. 6H,J). Furthermore, the mutant TECs failed to express the nuclear protein AIRE (an autoimmune regulator), which regulates transcription of tissue restricted antigens and is a marker for mTEC maturation (Fig. 6J) (Gillard et al., 2007; Org et al., 2008; Gardner et al., 2009). Claudin (CLDN) 3 and CLDN4 are expressed on TEC precursors that are committed to the mTEC lineage (Hamazaki et al., 2007). There was a paucity of CLDN3/4-positive TECs in Foxn1Cre;R26iTbx/+ thymi (Fig. 6F). Interestingly, the few CLDN3/4 positive precursors were positioned towards the outer region of the thymic lobes where TECs expressing higher levels of TBX1 and lower levels of FOXN1 are found. Taken together, these data suggest that ectopic TBX1 compromises differentiation of both cTEC and mTEC lineages.
Ectopic TBX1 results in an accumulation of early TEC progenitors
PLET1 is a cell-surface protein expressed by founder cells in the 3rd pp that give rise to both cTEC and mTEC lineages (Bennett et al., 2002; Gill et al., 2002; Bleul et al., 2006; Rossi et al., 2006; Depreter et al., 2008). A relatively high frequency of TECs in both Foxn1Cre;R26iTbx/+ and control thymic rudiments express PLET1 at E12.5 (Fig. 7A,B), consistent with previously published data (Nowell et al., 2011). The frequency of PLET1-positive TECs decreases during ontogeny as TECs undergo differentiation and proliferation (Nowell et al., 2011). Thus, PLET1 expression is confined to a rare subset of TECs in control thymi by E14.5 (Fig. 7C). In striking contrast, a higher frequency of PLET1-positive cells persists at E14.5 in the Foxn1Cre;R26iTbx/+ thymi (Fig. 7D). Furthermore, the PLET1-positive cells in the mutant thymi are found in the outer region of the lobes, similar to the localization pattern observed for TBX1-expressing cells (Fig. 7D). These data are consistent with ectopic TBX1 expression blocking TEC differentiation at an early progenitor stage. Flow cytometric analysis confirmed an increased frequency of PLET1-positive cells in E15.5 Foxn1Cre;R26iTbx/+ compared with control thymi (Fig. 7E). However, the absolute number of PLET1-positive cells was comparable in Foxn1Cre;R26iTbx/+ and control thymi (Fig. 7F,G). The maintenance of the same number of PLET1-positive TECs in the hypoplastic Foxn1Cre;R26iTbx/+ thymi at late stages of ontogeny is more consistent with TBX1 blocking differentiation of existing progenitors, rather than reverting differentiated TECs to a progenitor state.
Fig. 7.
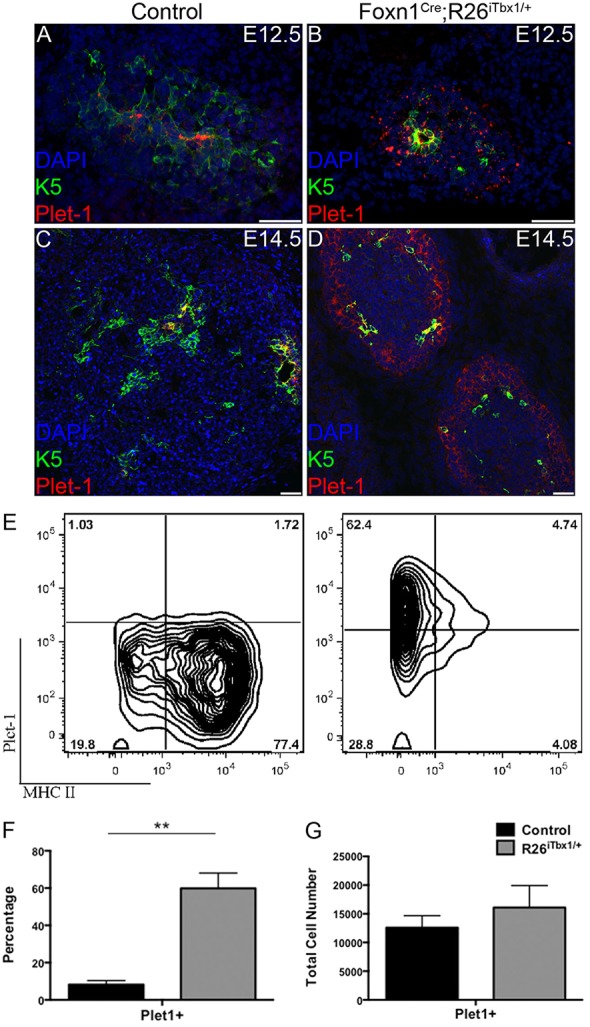
Foxn1Cre;R26iTbx1/+ fetal thymi contain a high frequency of TEC progenitors. (A-D) Representative immunohistochemical stains of transverse sections from control and Foxn1Cre;R26iTbx1/+ E12.5 or E14.5 thymic lobes showing PLET1- (red) and K5- (green) positive cells. Note the high frequency of PLET1-positive TEC progenitors in the outer region of Foxn1Cre;R26iTbx1/+ thymic lobes (D). Scale bars: 50 μm. (E) FACS plots showing MHC II and PLET1 expression on EpCAM+ CD45− control and Foxn1Cre;R26iTbx1/+ TECs. Numbers in each quadrant show percentage of cells. (F,G) The frequency (F) or number (G) of PLET1-positive cells in E14.5 control and Foxn1Cre;R26iTbx1/+ thymic lobes. Data are mean±s.d. for four control and four Foxn1Cre;R26iTbx1/+ thymic lobes. **P<0.001.
Thymocyte development and localization is altered in the Foxn1Cre;R26iTbx/+ thymic microenvironment
TECs are required for thymocyte proliferation, survival and differentiation (Ritter and Boyd, 1993; van Ewijk et al., 1994; Nitta et al., 2011; Anderson and Takahama, 2012). Given the profound arrest in TEC development resulting from ectopic Tbx1 expression, we expected to find defects in thymocyte development. Total thymocyte cellularity was decreased by ∼5-fold in in E15.5 Foxn1Cre;R26iTbx/+ compared with control thymi (Fig. 8A). Flow cytometric analyses were performed to assess thymocyte differentiation. At this developmental stage, thymocytes have not yet differentiated beyond the immature CD4 and CD8 double-negative (DN) precursor subset (Fig. 8B). Expression of KIT and CD25 distinguishes DN subsets at sequential maturation stages, referred to as DN1-DN4. Flow cytometric analysis revealed a partial block at the DN3 to DN4 transition in Foxn1Cre;R26iTbx/+ thymi (Fig. 8C). The reduction in thymocyte cellularity, together with the partial differentiation arrest at the DN3 stage, suggest that the aberrant TEC microenvironment in Foxn1Cre;R26iTbx/+ thymi fails to provide adequate growth and differentiation signals to support normal thymocyte development. IHC analysis of E14.5 thymi revealed that thymocytes, identified by expression of the Ikaros (IKZF1) transcription factor (Georgopoulos et al., 1992), were distributed throughout the control thymic lobes (Fig. 8D). By contrast, the scarce thymocytes present in Foxn1Cre;R26iTbx/+ fetal thymus lobes were concentrated in the central region in juxtaposition to TECs expressing higher levels of FOXN1 (compare Fig. 6B with Fig. 8E). Similar colocalization of thymocytes and TECs expressing low levels of FOXN1 was observed in the center of E17.5 mutant thymic lobes (supplementary material Fig. S3). This spatial relationship emphasizes the importance of TEC-thymocyte crosstalk in T-cell development.
Fig. 8.
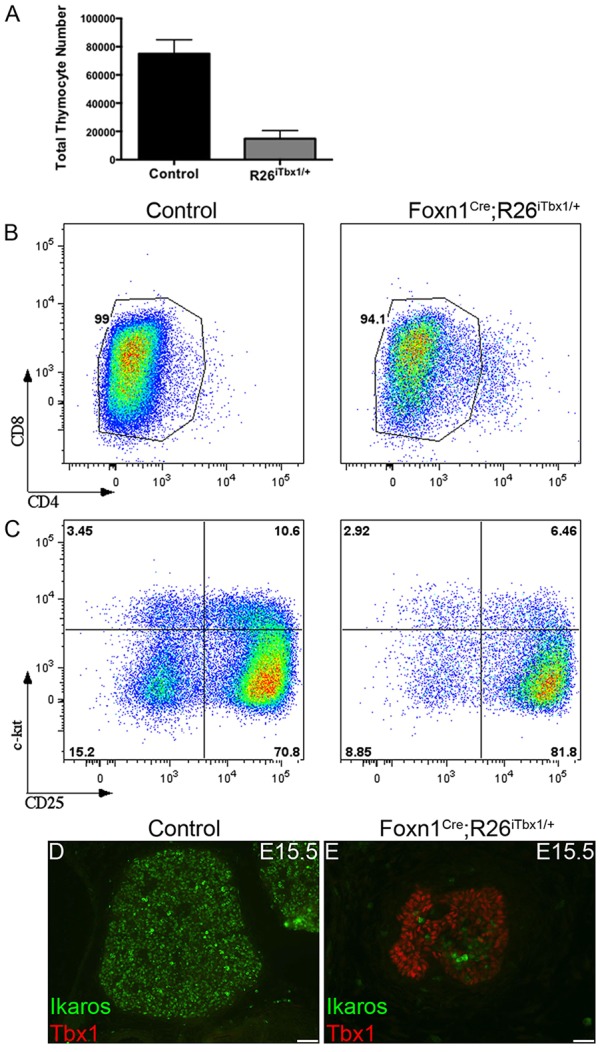
Thymocyte localization, cellularity and development are aberrant in Foxn1Cre;R26iTbx1/+ fetal thymi. (A) The number±s.d. of thymocytes recovered from control (n=2) and Foxn1Cre;R26iTbx1/+ (n=3). (B) FACS plots showing that most E15.5 thymocytes are CD4 or CD8 double-negative (DN) cells. (C) FACS plots showing the distribution of DN subsets based on KIT and CD25 expression. There is a partial block at the DN3 (KIT−CD25+) to DN4 (KIT−CD25+) developmental transition in Foxn1Cre;R26iTbx1/+ thymocytes. (D,E) Representative immunohistochemical stains of transverse sections from control and Foxn1Cre;R26iTbx1/+ E15.5 thymic lobes show that thymocytes identified as Ikaros positive (green) are scattered through control thymic lobes, but are restricted to the central region of mutant lobes where FOXN1-positive cells are found (see Fig. 6B). Scale bars: 50 μm.
DISCUSSION
These results clearly demonstrate that ectopic expression of Tbx1 in the thymus-fated domain of the 3rd pp suppresses Foxn1 expression and inhibits TEC proliferation and differentiation. These data directly contradict the prevailing model in which TBX1 is required for thymus development. Our data, together with the downregulation of Tbx1 that occurs in the ventral domain of the wild-type 3rd pp (Vitelli et al., 2002; Manley et al., 2004; Liu et al., 2007), strongly support the conclusion that TBX1 is a negative regulator of thymus development.
Ectopic expression of TBX1 in ventral 3rd pp endoderm suppresses FOXN1
All of the defects in TEC maturation and proliferation observed in Foxn1Cre;R26iTbx/+ thymi are consistent with the suppression of Foxn1 expression, which is known to promote these processes. Downregulation of Foxn1 occurred as early as E11.5, almost immediately upon ectopic expression of Tbx1. The inverse pattern of FOXN1 and TBX1 protein levels in E15.5 Foxn1Cre;R26iTbx/+ TECs is further evidence that TBX1 negatively regulates Foxn1 expression. TBX1 may shut down Foxn1 expression by an indirect, rather than direct, mechanism, as there is not an obvious TBX-binding site in the Foxn1 promoter (BLAT; UCSC Genome Bioinformatics). These findings are consistent with other work showing that ectopic activation of SHH signaling in 3rd pp endoderm both induces Tbx1 expression and represses Foxn1 (V.E.B. and N.R.M., unpublished).
The few FOXN1 positive TECs present in these mutants appear to be the progeny of 3rd pp progenitors that failed to effectively express ectopic TBX1, as these cells have very low or undetectable levels of TBX1. This failure could be due to inefficient or late deletion of the stop codon in the R26iTbx1 allele, or due to secondary shutdown of the Rosa26 locus. These FOXN1-positive cells tend to aggregate in the center of the lobe, forming a microenvironment that supports differentiation of a small number of thymocytes, with variable efficiency.
Ectopic TBX1 expression does not reverse thymus fate
By E11.5, most cells in the wild-type 3rd pp have acquired either a thymus or parathyroid fate, and express either Foxn1 or Gcm2 (Gordon et al., 2001). By contrast, the reduction of FOXN1-positive cells in the Foxn1Cre;R26iTbx/+ 3rd pp, coupled with the lack of a compensatory expansion of Gcm2 expression, results in a sizeable domain containing cells that fail to express either marker. However, as Foxn1 is not required to establish thymus fate, these cells could still retain thymus fate specification. This appears to be the case, as these ventral cells do express Il7 and FOXG1, both of which are expressed by thymus-fated cells independently of Foxn1-mediated regulation (Zamisch et al., 2005; Wei and Condie, 2011) (B. G. Condie, personal communication). These data also demonstrate that ectopic Tbx1 cannot reverse thymus fate after it is established, as Foxn1Cre is expressed in 3rd pp cells that are already committed to a thymus fate. The downregulation of Tbx1 in the ventral domain during pouch outgrowth at E9-9.5, prior to expression of thymus-specific markers, raises the possibility that this downregulation is essential to establish thymus fate. Activation of R26iTbx1 in 3rd pp endoderm at an earlier developmental stage will be required to test whether TBX1 can block initial thymus fate commitment.
Ectopic TBX1 expression arrests TEC differentiation at an early progenitor stage
TEC differentiation is arrested at a PLET1-positive stage in Foxn1-null (nude) mutants (Blackburn et al., 1996; Depreter et al., 2008). Plet1-expressing TECs have been shown to contain a TEC progenitor activity in transplant experiments at both early and later stages of fetal thymus development (Bennett et al., 2002; Gill et al., 2002). These data strongly suggest that PLET1 likely marks a very early stage of committed thymus progenitor cells (reviewed by Manley and Condie, 2010). The loss of FOXN1 in Foxn1+/Cre;R26+/iTbx1 fetal thymi was associated with a marked increase in the frequency of PLET1-positive TECs. However, the number of PLET1-positive TECs was comparable in mutant and control fetal thymi. These data indicate that ectopic TBX1 results in arrested differentiation of PLET1-positive progenitors, rather than induction of their proliferation or reversal of a differentiated phenotype to a progenitor-like state.
Ectopic TBX1 expression uncouples FOXN1 and MHC Class II expression
Previous studies have suggested that Foxn1 directly or (more likely) indirectly regulates expression of MHC Class II genes. MHC Class II levels are severely downregulated on TECs from Foxn1Δ/Δ mutants (Tennant et al., 1982; Chen et al., 2009). Furthermore, a direct relationship was observed between the frequency of MHC Class II-positive TECs and Foxn1 levels in an allelic series of strains expressing progressively lower levels of Foxn1 (Nowell et al., 2011). Therefore, we were surprised to find that the minor fraction of MHCIIhi Foxn1Cre;R26iTbx/+ TECs expressed exceedingly low levels of Foxn1 mRNA. The mechanism responsible for this finding is not clear, but this result does indicate that FOXN1 levels alone are not sufficient to determine the levels of MHC Class II genes. It is possible that ectopic Tbx1 in Foxn1Cre;R26iTbx/+ mutants disrupts the normal regulatory pathway linking Foxn1 and MHC Class II genes. Regardless of the mechanism, Foxn1 and MHC Class II gene expression are uncoupled by ectopic Tbx1 expression in TECs.
Ectopic TBX1 expression does not induce GCM2 expression
GCM2 is a transcription factor that marks the parathyroid-fated domain of the 3rd pp and is required for parathyroid survival and differentiation (Gunther et al., 2000; Gordon et al., 2001; Liu et al., 2007). Tbx1 and Gcm2 expression domains overlap in the dorsal 3rd pp, and previous reports have suggested that Gcm2 may be a downstream target of Tbx1, based on microarray data in Tbx1-null mutants (Ivins et al., 2005) or on the persistence of Tbx1 expression in Gcm2-null mutants (Liu et al., 2007). However, our data show that ectopic expression of TBX1 was not sufficient to induce an expansion of Gcm2 expression into the ventral 3rd pp. This result is consistent with the finding that expression of a constitutively active allele of Smo throughout the 3rd pp expanded the Tbx1 expression domain ventrally, but did not expand Gcm2 expression (V.E.B. and N.R.M., unpublished). Thus, activation of additional positive regulators and/or suppression of negative regulators must be involved in regulating parathyroid fate specification and/or Gcm2 expression during 3rd pp patterning.
MATERIALS AND METHODS
Mice
We generated a conditional Tbx1 knock-in strain by targeting a Tbx1 inducible expression cassette to the endogenous Rosa26 locus to generate a R26iTbx1 allele. In this Tbx1 inducible cassette, a full-length mouse Tbx1 cDNA and an IRES-GFP were inserted downstream of a floxed stop cassette (vector provided by Dr A. McMahon). Expression of the R26iTbx1 allele was activated in TECs by crossing R26iTbx1 females with Foxn1Cre males (Gordon et al., 2007). R26iTbx1/+ and R26+/+ littermate controls were maintained on a mixed C57Bl/6 genetic background at the MD Anderson Cancer Center Science Park in accordance with the guidelines set forth by the Association for the Accreditation of Laboratory Animal Care. Timed pregnancies were set up in the evening and monitored daily thereafter. The day of vaginal plug was considered embryonic day (E) 0.5. Morphological cues were used to determine embryonic stage. Yolk sac DNA was genotyped using Rosa26 forward 5′-TTGCAATACCTTTCTGGGAGTT-3′, Rosa26 reverse 5′-AACCCCAGATGACTCCTATCCT-3′ and β-galactosidase reverse 5′-GACAGTATCGGCCTCAGGAAG-3′. All experiments were performed in accordance with the MD Anderson Animal Care and Use Committee.
Histochemistry and immunohistochemistry
Embryos harvested for frozen sections were fixed in 4% PFA for 15 min (E11.5) or 30 min E13.5-15.5, washed in PBS, dehydrated in sucrose and embedded in OCT. Serial sections (7 μm) were air dried and washed in PBS. Tissue was incubated overnight at 4°C in 120 μl containing primary antibody, 5% donkey serum and 0.05% Triton X-100 in PBS. Slides were washed in PBS and incubated with 120 μl of secondary antibody and 0.05% Triton X-100 in PBS for 1 h at room temperature, in the dark. Slides were washed in PBS, incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 3 min at room temperature, and washed in PBS. Slides were mounted with ProLong Gold anti-fade reagent (Life Technologies) and coverslipped for imaging.
Embryos collected for paraffin embedding were fixed for 1 h in 4% PFA, washed in PBS, dehydrated in an ethanol gradient, permeabilized using xylenes and embedded in paraffin. Serial sections were cut at 7 μm on a Leica ThermoShandon Finesse 325 microtome. Tissue was washed in xylene and rehydrated through a methanol gradient to tap water. Immmunostaining for paraffin-embedded tissue required antigen retrieval. Paraffin-embedded tissue was incubated in AR buffer [10 mM sodium citrate (pH 6), 0.05% Tween 20] at 95°C for 30 min. Slides were allowed to cool to room temperature. Paraffin-embedded tissue was incubated in primary and secondary antibodies as described above. Paraffin-embedded tissues collected for Hematoxylin and Eosin (H&E) staining were processed according to standard techniques. Primary antibodies used were goat anti-Foxn1 (1:200, Santa Cruz, G-20), rabbit anti-Tbx1 (1:100, Abcam, ab18530), rabbit anti-Gcm2 (1:200, Abcam, ab64723), rabbit anti-Foxg1 (1:50, Abcam, ab18259), rat anti-BrdU (1:10, Serotec, OBT0030S), goat anti-β5t (1:200, MBL, PD021), rat anti-Ly75 (1:100, Abcam, ab51820), K14 (1:400, Covance, PRB-155P-100), biotinylated UEA1 (1:200, Vector, B-1065), rabbit anti-Aire-1 (1:50, Santa Cruz, M-300), rabbit anti-claudin 3 and anti-claudin 4 (1:200, Invitrogen, 341700 and 364800), K5 (1:400, Covance, PRB-160P-100), PLET1 (kindly provided by Dr Clare Blackburn, University of Edinburgh), and Ikaros (1:100, Santa Cruz, M-20). All secondary antibodies were conjugated with DyLight 488 (705-546-147), Dylight 594 (711-586-152), Cy3 (711-166-152) or Cy5 (712-176-153) (Jackson ImmunoResearch, 1:400).
BrdU (300 μl of 10 mg/ml) was injected by the intraperitoneal (IP) route into pregnant females and embryos were harvested after a 90 min chase. Cells from serial sections were counted based on expression of FOXN1, GCM2, TBX1 and/or BrdU using Cell Profiler (Broad Institute) or Imaris (Bitplane).
Flow cytometry
For TEC analysis, E15.5 thymi were digested with 0.125% collagenase (Sigma) and 0.1% DNase (Roche) as previously described (Gray et al., 2002). For analysis of thymocyte subsets, E15.5 thymi were pressed through a 70 μm strainer (Fisher). Cells were stained with fluorochrome-conjugated antibodies in FACS buffer [PBS (pH 7.2), 0.005 M EDTA, 2% FBS] for 20 min on ice and washed with FACS buffer. Propidium iodide (Invitrogen) was added (0.5 μg/ml) to each sample prior to analysis to exclude dead cells. Anti-CD326 (clone G8.8) conjugated to PE/Cy7; anti-I-A/I-E (clone M5/114.15.2) and anti-CD25 (clone PC61) conjugated to Pacific Blue; anti-CD326 (clone G8.8) conjugated to allophycocyanin; and anti-CD117 (clone 2 B8) conjugated to allophycocyanin-Cy7 were purchased from BioLegend. Anti-CD44 (clone IM7) conjugated to allophycocyanin; anti-CD8α (clone 53-6.7) conjugated to PE-Cy7; and anti-CD45 (clone 30-F11) conjugated to PerCp-Cy5.5 were purchased from eBioscience. Anti-CD4 (clone RM4-5) conjugated to Qdot 605 was purchased from Invitrogen. To exclude erythrocytes, granulocytes, dendritic cells, macrophages and NK cells from the thymocyte analyses, the following antibodies, all conjugated to PE-Cy5 were purchased form eBioscience: TER-119, CD11c (clone N418, 0.17 μg/μl), CD11b (clone M1/70, 0.17 μg/μl), NK-1.1 (clone PK136, 0.17 μg/μl) and Ly-6G (clone RB6-8C5, 0.17 μg/μl). Cells were analyzed or sorted on a FACS Aria II (BD Biosciences). Data were analyzed using FlowJo software (Tree Star).
RNA isolation, cDNA synthesis and real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
FACS-sorted R26iTbx1 and control TECs from a given litter were pooled and lysed in Trizol (Ambion). RNA was isolated using chloroform and linear acrylamide (Ambion) and RNA was resuspended in DEPC water until ready for cDNA synthesis. First-strand cDNA was synthesized using a SuperScript III First-Strand cDNA synthesis kit (Invitrogen). For quantitative reverse transcriptase PCR (qRT-PCR) analysis of Foxn1, Tbx1 and α-tubulin (endogenous control), we used TaqMan Gene Expression Master Mix (Applied Biosystems) and TaqMan primers (Applied Biosystems) in a LightCycler 480 (Roche). In each experiment, comparators were set to a value of 1. All experiments were repeated three times and analyzed using the ΔΔCt method.
Statistics
Prism 6.0 (GraphPad version 6.0b) and Microsoft Excel were used for statistical analyses. We used a Student's t-test to evaluate differences between controls and mutants regarding the number of cells expressing FOXN1, GCM2, TBX1 and/or BrdU. P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank Julie Gordon for helpful discussions and sharing her expertise on third pharyngeal pouch dissection and Pam Whitney for flow cytometric analysis. We are grateful to Andrew McMahon for providing the Rosa26 targeting vector.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
K.A.G.R. and K.T.C. designed and performed the experiments and analyzed the data under the supervision of E.R.R.; V.E.B. confirmed results and contributed images to Figs 2 and 6; Z.L. and M.L. assisted in making the construct and generating and testing the knock-in mice; E.R.R. and N.R.M. conceived the project; K.A.G.R., E.R.R. and N.R.M. wrote and edited the manuscript.
Funding
This work was supported by the National Institutes of Health (NIH) [R01 HD056315 to E.R.R. and N.R.M., and R56AI107096 to E.R.R.]. K.T.C. was supported by the NIH [T32CA009480]. K.A.G.R. was supported by fellowships from the Schissler Foundation and the American Legion Auxiliary. This research was also supported in part by the NIH through MD Anderson's Cancer Center Support [CA016672]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.111641/-/DC1
References
- Anderson G., Takahama Y. (2012). Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 33, 256-263 10.1016/j.it.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Arnold J. S., Werling U., Braunstein E. M., Liao J., Nowotschin S., Edelmann W., Hebert J. M., Morrow B. E. (2006). Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development 133, 977-987 10.1242/dev.02264 [DOI] [PubMed] [Google Scholar]
- Baldini A. (2005). Dissecting contiguous gene defects: TBX1. Curr. Opin. Genet. Dev. 15, 279-284 10.1016/j.gde.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Bennett A. R., Farley A., Blair N. F., Gordon J., Sharp L., Blackburn C. C. (2002). Identification and characterization of thymic epithelial progenitor cells. Immunity 16, 803-814 10.1016/S1074-7613(02)00321-7 [DOI] [PubMed] [Google Scholar]
- Blackburn C. C., Augustine C. L., Li R., Harvey R. P., Malin M. A., Boyd R. L., Miller J. F., Morahan G. (1996). The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors. Proc. Natl. Acad. Sci. USA 93, 5742-5746 10.1073/pnas.93.12.5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul C. C., Corbeaux T., Reuter A., Fisch P., Mönting J. S., Boehm T. (2006). Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 441, 992-996 10.1038/nature04850 [DOI] [PubMed] [Google Scholar]
- Boehm T. (2008). Thymus development and function. Curr. Opin. Immunol. 20, 178-184 10.1016/j.coi.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Chapman D. L., Garvey N., Hancock S., Alexiou M., Agulnik S. I., Gibson-Brown J. J., Cebra-Thomas J., Bollag R. J., Silver L. M., Papaioannou V. E. (1996). Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev. Dyn. 206, 379-390 [DOI] [PubMed] [Google Scholar]
- Chen L., Xiao S., Manley N. R. (2009). Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood 113, 567-574 10.1182/blood-2008-05-156265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeaux T., Hess I., Swann J. B., Kanzler B., Haas-Assenbaum A., Boehm T. (2010). Thymopoiesis in mice depends on a Foxn1-positive thymic epithelial cell lineage. Proc. Natl. Acad. Sci. USA 107, 16613-16618 10.1073/pnas.1004623107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depreter M. G. L., Blair N. F., Gaskell T. L., Nowell C. S., Davern K., Pagliocca A., Stenhouse F. H., Farley A. M., Fraser A., Vrana J., et al. (2008). Identification of Plet-1 as a specific marker of early thymic epithelial progenitor cells. Proc. Natl. Acad. Sci. USA 105, 961-966 10.1073/pnas.0711170105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley J., Erickson M., Larochelle W. J., Gillard G. O., Farr A. G. (2007). FGFR2IIIb signaling regulates thymic epithelial differentiation. Dev. Dyn. 236, 3459-3471 10.1002/dvdy.21364 [DOI] [PubMed] [Google Scholar]
- Gardner J. M., Fletcher A. L., Anderson M. S., Turley S. J. (2009). AIRE in the thymus and beyond. Curr. Opin. Immunol. 21, 582-589 10.1016/j.coi.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V., Yamagishi C., Hu T., Kathiriya I. S., Yamagishi H., Srivastava D. (2001). Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev. Biol. 235, 62-73 10.1006/dbio.2001.0283 [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., Moore D. D., Derfier B. (1992). Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258, 808-812 10.1126/science.1439790 [DOI] [PubMed] [Google Scholar]
- Gill J., Malin M., Holländer G. A., Boyd R. (2002). Generation of a complete thymic microenvironment by MTS24(+) thymic epithelial cells. Nat. Immunol. 3, 635-642 10.1038/ni812 [DOI] [PubMed] [Google Scholar]
- Gillard G. O., Dooley J., Erickson M., Peltonen L., Farr A. G. (2007). Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J. Immunol. 178, 3007-3015 10.4049/jimmunol.178.5.3007 [DOI] [PubMed] [Google Scholar]
- Gordon J., Manley N. R. (2011). Mechanisms of thymus organogenesis and morphogenesis. Development 138, 3865-3878 10.1242/dev.059998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J., Bennett A. R., Blackburn C. C., Manley N. R. (2001). Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech. Dev. 103, 141-143 10.1016/S0925-4773(01)00333-1 [DOI] [PubMed] [Google Scholar]
- Gordon J., Wilson V. A., Blair N. F., Sheridan J., Farley A., Wilson L., Manley N. R., Blackburn C. C. (2004). Functional evidence for a single endodermal origin for the thymic epithelium. Nat. Immunol. 5, 546-553 10.1038/ni1064 [DOI] [PubMed] [Google Scholar]
- Gordon J., Xiao S., Hughes B., III, Su D.-M., Navarre S. P., Condie B. G., Manley N. R. (2007). Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7, 69 10.1186/1471-213X-7-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. H. D., Chidgey A. P., Boyd R. L. (2002). Analysis of thymic stromal cell populations using flow cytometry. J. Immunol. Methods 260, 15-28 10.1016/S0022-1759(01)00493-8 [DOI] [PubMed] [Google Scholar]
- Günther T., Chen Z.-F., Kim J., Priemel M., Rueger J. M., Amling M., Moseley J. M., Martin T. J., Anderson D. J., Karsenty G. (2000). Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406, 199-203 10.1038/35018111 [DOI] [PubMed] [Google Scholar]
- Hamazaki Y., Fujita H., Kobayashi T., Choi Y., Scott H. S., Matsumoto M., Minato N. (2007). Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat. Immunol. 8, 304-311 10.1038/ni1438 [DOI] [PubMed] [Google Scholar]
- Holländer G., Gill J., Zuklys S., Iwanami N., Liu C., Takahama Y. (2006). Cellular and molecular events during early thymus development. Immunol. Rev. 209, 28-46 10.1111/j.0105-2896.2006.00357.x [DOI] [PubMed] [Google Scholar]
- Itoi M., Kawamoto H., Katsura Y., Amagai T. (2001). Two distinct steps of immigration of hematopoietic progenitors into the early thymus anlage. Int. Immunol. 13, 1203-1211 10.1093/intimm/13.9.1203 [DOI] [PubMed] [Google Scholar]
- Ivins S., Lammerts van Beuren K., Roberts C., James C., Lindsay E., Baldini A., Ataliotis P., Scambler P. J. (2005). Microarray analysis detects differentially expressed genes in the pharyngeal region of mice lacking Tbx1. Dev. Biol. 285, 554-569 10.1016/j.ydbio.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Jerome L. A., Papaioannou V. E. (2001). DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 27, 286-291 10.1038/85845 [DOI] [PubMed] [Google Scholar]
- Klug D. B., Carter C., Crouch E., Roop D., Conti C. J., Richie E. R. (1998). Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc. Natl. Acad. Sci. USA 95, 11822-11827 10.1073/pnas.95.20.11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay E. A., Vitelli F., Su H., Morishima M., Huynh T., Pramparo T., Jurecic V., Ogunrinu G., Sutherland H. F., Scambler P. J., et al. (2001). Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410, 97-101 10.1038/35065105 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yu S., Manley N. R. (2007). Gcm2 is required for the differentiation and survival of parathyroid precursor cells in the parathyroid/thymus primordia. Dev. Biol. 305, 333-346 10.1016/j.ydbio.2007.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N. R., Condie B. G. (2010). Transcriptional regulation of thymus organogenesis and thymic epithelial cell differentiation. Prog. Mol. Biol. Transl. Sci. 92, 103-120 10.1016/S1877-1173(10)92005-X [DOI] [PubMed] [Google Scholar]
- Manley N. R., Selleri L., Brendolan A., Gordon J., Cleary M. L. (2004). Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev. Biol. 276, 301-312 10.1016/j.ydbio.2004.08.030 [DOI] [PubMed] [Google Scholar]
- Manley N. R., Richie E. R., Blackburn C. C., Condie B. G., Sage J. (2011). Structure and function of the thymic microenvironment. Front. Biosci. 17, 2461-2477 10.2741/3866 [DOI] [PubMed] [Google Scholar]
- Merscher S., Funke B., Epstein J. A., Heyer J., Puech A., Lu M. M., Xavier R. J., Demay M. B., Russell R. G., Factor S., et al. (2001). TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104, 619-629 10.1016/S0092-8674(01)00247-1 [DOI] [PubMed] [Google Scholar]
- Moore-Scott B. A., Manley N. R. (2005). Differential expression of Sonic hedgehog along the anterior-posterior axis regulates patterning of pharyngeal pouch endoderm and pharyngeal endoderm-derived organs. Dev. Biol. 278, 323-335 10.1016/j.ydbio.2004.10.027 [DOI] [PubMed] [Google Scholar]
- Nitta T., Ohigashi I., Nakagawa Y., Takahama Y. (2011). Cytokine crosstalk for thymic medulla formation. Curr. Opin. Immunol. 23, 190-197 10.1016/j.coi.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Nowell C. S., Richie E., Manley N. R., Blackburn C. C. (2008). Thymus and parathyroid organogenesis. In Principles of Tissue Engineering (ed. Lanza R., Langer R., Vacanti J.), pp. 647-662 Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- Nowell C. S., Bredenkamp N., Tetélin S., Jin X., Tischner C., Vaidya H., Sheridan J. M., Stenhouse F. H., Heussen R., Smith A. J. H., et al. (2011). Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 7, e1002348 10.1371/journal.pgen.1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org T., Chignola F., Hetényi C., Gaetani M., Rebane A., Liiv I., Maran U., Mollica L., Bottomley M. J., Musco G., et al. (2008). The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370-376 10.1038/embor.2008.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. R., Gordon J., Mahbub F., Blackburn C. C., Manley N. R. (2006). Bmp4 and Noggin expression during early thymus and parathyroid organogenesis. Gene. Expr. Patterns 6, 794-799 10.1016/j.modgep.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Repass J. F., Laurent M. N., Carter C., Reizis B., Bedford M. T., Cardenas K., Narang P., Coles M., Richie E. R. (2009). IL7-hCD25 and IL7-Cre BAC transgenic mouse lines: new tools for analysis of IL-7 expressing cells. Genesis 47, 281-287 10.1002/dvg.20497 [DOI] [PubMed] [Google Scholar]
- Ripen A. M., Nitta T., Murata S., Tanaka K., Takahama Y. (2011). Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit beta5t. Eur. J. Immunol. 41, 1278-1287 10.1002/eji.201041375 [DOI] [PubMed] [Google Scholar]
- Ritter M. A., Boyd R. L. (1993). Development in the thymus: it takes two to tango. Immunol. Today 14, 462-469 10.1016/0167-5699(93)90250-O [DOI] [PubMed] [Google Scholar]
- Rodewald H.-R. (2008). Thymus organogenesis. Annu. Rev. Immunol. 26, 355-388 10.1146/annurev.immunol.26.021607.090408 [DOI] [PubMed] [Google Scholar]
- Rossi S. W., Jenkinson W. E., Anderson G., Jenkinson E. J. (2006). Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature 441, 988-991 10.1038/nature04813 [DOI] [PubMed] [Google Scholar]
- Shakib S., Desanti G. E., Jenkinson W. E., Parnell S. M., Jenkinson E. J., Anderson G. (2009). Checkpoints in the development of thymic cortical epithelial cells. J. Immunol. 182, 130-137 10.4049/jimmunol.182.1.130 [DOI] [PubMed] [Google Scholar]
- Su D.-M., Navarre S., Oh W.-J., Condie B. G., Manley N. R. (2003). A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat. Immunol. 4, 1128-1135 10.1038/ni983 [DOI] [PubMed] [Google Scholar]
- Tennant R. W., Otten J. A., Myer F. E., Rascati R. J. (1982). Induction of retrovirus gene expression in mouse cells by some chemical mutagens. Cancer Res. 42, 3050-3055. [PubMed] [Google Scholar]
- Thompson P. K., Zúñiga-Pflücker J. C. (2011). On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin. Immunol. 23, 350-359 10.1016/j.smim.2011.08.007 [DOI] [PubMed] [Google Scholar]
- van Ewijk W., Shores E. W., Singer A. (1994). Crosstalk in the mouse thymus. Immunol. Today 15, 214-217 10.1016/0167-5699(94)90246-1 [DOI] [PubMed] [Google Scholar]
- Vitelli F., Morishima M., Taddei I., Lindsay E. A., Baldini A. (2002). Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum. Mol. Genet. 11, 915-922 10.1093/hmg/11.8.915 [DOI] [PubMed] [Google Scholar]
- Vitelli F., Huynh T., Baldini A. (2009). Gain of function of Tbx1 affects pharyngeal and heart development in the mouse. Genesis 47, 188-195 10.1002/dvg.20476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Condie B. G. (2011). A focused in situ hybridization screen identifies candidate transcriptional regulators of thymic epithelial cell development and function. PLoS ONE 6, e26795 10.1371/journal.pone.0026795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Manley N. R. (2010). Impaired thymic selection and abnormal antigen-specific T cell responses in Foxn1(Delta/Delta) mutant mice. PLoS ONE 5, e15396 10.1371/journal.pone.0015396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H., Furutani Y., Hamada H., Sasaki T., Asakawa S., Minoshima S., Ichida F., Joo K., Kimura M., Imamura S.-I., et al. (2003). Role of TBX1 in human del22q11.2 syndrome. Lancet 362, 1366-1373 10.1016/S0140-6736(03)14632-6 [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Maeda J., Hu T., McAnally J., Conway S. J., Kume T., Meyers E. N., Yamagishi C., Srivastava D. (2003). Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 17, 269-281 10.1101/gad.1048903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamisch M., Moore-Scott B., Su D.-M., Lucas P. J., Manley N., Richie E. R. (2005). Ontogeny and regulation of IL-7-expressing thymic epithelial cells. J. Immunol. 174, 60-67 10.4049/jimmunol.174.1.60 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Cerrato F., Xu H., Vitelli F., Morishima M., Vincentz J., Furuta Y., Ma L., Martin J. F., Baldini A., et al. (2005). Tbx1 expression in pharyngeal epithelia is necessary for pharyngeal arch artery development. Development 132, 5307-5315 10.1242/dev.02086 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.