Abstract
Chloromethane (CH3Cl) is produced on earth by a variety of abiotic and biological processes. It is the most important halogenated trace gas in the atmosphere, where it contributes to ozone destruction. Current estimates of the global CH3Cl budget are uncertain and suggest that microorganisms might play a more important role in degrading atmospheric CH3Cl than previously thought. Its degradation by bacteria has been demonstrated in marine, terrestrial, and phyllospheric environments. Improving our knowledge of these degradation processes and their magnitude is thus highly relevant for a better understanding of the global budget of CH3Cl. The cmu pathway, for chloromethane utilisation, is the only microbial pathway for CH3Cl degradation elucidated so far, and was characterized in detail in aerobic methylotrophic Alphaproteobacteria. Here, we reveal the potential of using a two-pronged approach involving a combination of comparative genomics and isotopic fractionation during CH3Cl degradation to newly address the question of the diversity of chloromethane-degrading bacteria in the environment. Analysis of available bacterial genome sequences reveals that several bacteria not yet known to degrade CH3Cl contain part or all of the complement of cmu genes required for CH3Cl degradation. These organisms, unlike bacteria shown to grow with CH3Cl using the cmu pathway, are obligate anaerobes. On the other hand, analysis of the complete genome of the chloromethane-degrading bacterium Leisingera methylohalidivorans MB2 showed that this bacterium does not contain cmu genes. Isotope fractionation experiments with L. methylohalidivorans MB2 suggest that the unknown pathway used by this bacterium for growth with CH3Cl can be differentiated from the cmu pathway. This result opens the prospect that contributions from bacteria with the cmu and Leisingera-type pathways to the atmospheric CH3Cl budget may be teased apart in the future.
Keywords: bacteria, chloromethane, comparative genomics, isotope fractionation, diversity
INTRODUCTION
Halocarbons such as chloromethane (CH3Cl) and bromomethane are known for their ozone depletion potential (Harper, 2000). CH3Cl, the most abundant volatile halocarbon in the atmosphere (∼600 ppt), is responsible for approximately 15% of halogen-dependent ozone destruction in the stratosphere (Harper, 2000; Montzka and Reimann, 2011). The largest sources of CH3Cl emissions to the atmosphere include terrestrial vegetation (Hamilton et al., 2003; Yoshida et al., 2004; Keppler et al., 2005) and in particular the phyllosphere (i.e., aboveground parts of vegetation, Saito and Yokouchi, 2008), biomass burning, and the oceans (Montzka and Reimann, 2011). Conversely, the dominant sink for CH3Cl is via reaction with hydroxyl radicals in the troposphere and represents 84% of the total, estimated at 4.1 Tg Cl yr-1 (Yoshida et al., 2004). However, certain methylotrophic bacteria capable of using CH3Cl as their sole source of carbon and energy for growth may also participate in this process, but the magnitude of their contribution remains to be characterized. Chloromethane-degrading bacteria are quite widespread, with representatives affiliated to the genera Aminobacter, Hyphomicrobium, Leisingera, Methylobacterium, Roseovarius (Alpha-Proteobacteria), Pseudomonas (Gamma-Proteobacteria) and Acetobacterium (Actinobacteria), isolated from diverse environments such as soils (Doronina et al., 1996; Miller et al., 1997; Coulter et al., 1999; McAnulla et al., 2001), activated sludge (Hartmans et al., 1986; Traunecker et al., 1991; Freedman et al., 2004), freshwaters (McAnulla et al., 2001), and seawater (Schäfer et al., 2005).
The only pathway for CH3Cl degradation known so far is corrinoid- and tetrahydrofolate-dependent, and was characterized in detail for the aerobic facultative methylotrophic strain Methylobacterium extorquens CM4 (Vannelli et al., 1999). This pathway, termed cmu (abbreviation for chloromethane utilization), involves a set of genes that were subsequently detected in several other chloromethane-degrading strains (reviewed in Schäfer et al., 2007; also see Nadalig et al., 2011). The first step of the cmu pathway involves the methyltransferase/corrinoid-binding CmuA protein, which transfers the CH3Cl methyl group to a corrinoid cofactor, and CmuB, another methyltransferase which catalyzes the transfer of the methyl group from the methylated corrinoid to tetrahydrofolate (H4F). Methyl-H4F is then oxidized to methylene-H4F and further to CO2 via formate to conserve energy, or exploited for biomass production. However, other yet to be characterized metabolic pathways may be involved in the degradation of CH3Cl in the environment. For example, Leisingera methylohalidivorans MB2 grows with methyl halides but was reported not to contain close homologs of cmu genes (Schäfer et al., 2007).
Evidence for a given metabolic pathway may be obtained through the use of stable isotope techniques, and this has been used to distinguish different sources and sinks for CH3Cl (Harper et al., 2001, 2003; Czapiewski et al., 2002; Keppler et al., 2004, 2005; Saito and Yokouchi, 2008; Greule et al., 2012; Redeker and Kalin, 2012). Degradation of CH3Cl by cell suspensions of strains with the cmu pathway is also associated with specific carbon fractionation (Miller et al., 2001) but also with hydrogen isotope fractionation (Nadalig et al., 2013). Thus, isotopic approaches combined with genomic approaches may prove decisive in constraining the bacterial contribution to the global CH3Cl budget.
In the present study, we review available bacterial genome sequences for the presence of cmu genes, thereby uncovering several bacteria that have not been described to degrade CH3Cl. In parallel and as a proof of concept for the potential of isotope methods to characterize yet unknown pathways for CH3Cl degradation, we determined hydrogen and carbon isotopic fractionation patterns of CH3Cl during growth of the chloromethane-degrading strain L. methylohalidivorans MB2 lacking cmu genes, as compared to that observed for cmu pathway strains M. extorquens CM4 and Hyphomicrobium sp. MC1.
MATERIALS AND METHODS
BIOINFORMATIC ANALYSIS
Comparative genome analysis was performed with the software tools available on the Microscope platform at Genoscope (Vallenet et al., 2009), using the assembled sequences of M. extorquens CM4 (GenBank accession numbers CP001298, CP001299, CP001300), Hyphomicrobium sp. MC1 (FQ859181), Desulfomonile tiedjei (CP003360, CP003361), Thermosediminibacter oceani (CP002131), Thermincola potens (CP002028), and L. methylohalidivorans MB2 (CP006773, CP006774, CP006775), and the draft sequences for Desulfotomaculum alcoholivorax (GenBank AUMW00000000; 66 contigs), Desulfurispora thermophila (GenBank AQWN00000000; 19 contigs; Table 1).
Table 1.
Characteristics of the bacterial strains discussed in this study.
| Strains | Methylobacterium extorquens CM4 | Hyphomicrobium sp. MC1 | Desulfotomaculum alcoholivorax DSM 16058 | Desulfurispora thermophila DSM 16022 | Desulfomonile tiedjei DSM 6799 | Thermincola potens JR | Thermosedimi-nibacter oceani DSM 16646 | Leisingera methylohalidivorans MB2 DSM 14336 |
|---|---|---|---|---|---|---|---|---|
| Affiliation | Alphaproteobacteria | Alphaproteobacteria | Deltaproteobacteria | Deltaproteobacteria | Deltaproteobacteria | Clostridia | Clostridia | Alphaproteobacteria |
| Origin | Industrial soil | Sewage sludge | Fluidized-bed reactor | Fluidized-bed reactor | Sewage sludge | Anodic biofilm | Deep-sea sediment | Sea water |
| Genome size (MB) | 6.18 | 4.75 | 3.47 | 2.82 | 6.53 | 3.16 | 2.28 | 4.65 |
| GC (%) | 68 | 59 | 47 | 55 | 50 | 46 | 47 | 62 |
| Total CDS | 6035 | 4955 | 3588 | 2815 | 5664 | 3343 | 2460 | 4608 |
| Plasmids | 2 | 0 | Unknown | Unknown | 1 | 0 | 0 | 2 |
| Sequence status | Assembled, finished | Assembled, finished | 66 contigs Permanent draft | 19 contigs Permanent draft | Assembled, finished | Assembled, finished | Assembled, finished | Assembled, finished |
| Genbank accession number |
CP001298 CP001299 CP001300 | FQ859181 | AUMW00000000 | AQWN00000000 | CP003360 CP003361 | CP002028 | CP002131 | CP006773 CP006774 CP006775 |
| Reference | Marx et al. (2012) | Vuilleumier et al. (2011) | Kaksonen et al. (2008) | Kaksonen et al. (2007) | DeWeerd et al. (1990) | Byrne-Bailey et al. (2010) | Pitluck et al. (2010) | Buddruhs et al. (2013) |
BACTERIAL STRAINS AND GROWTH CONDITIONS
Strains M. extorquens CM4 and Hyphomicrobium sp. MC1 were laboratory stocks and cultivated in a mineral medium for methylotrophic bacteria (M3; Roselli et al., 2013) containing (L-1 of distilled water) KH2PO4 (6.8 g), (NH4)2SO4 (0.2 g), NaOH (5 M) (5.85 mL), yielding a final pH of 7.2. After autoclaving, 1 mL L-1 medium each of calcium nitrate solution (25 g L-1) and of trace elements solution containing (mg L-1) FeSO4 7H2O (100), MnSO4 H2O (100), ZnSO4 (29.5), Co(NO3)2 6H2O (25), CuCl2 H2O (25), Na2MoO4 2H2O (25), NH4VO3 (14.4), NiSO4 6H2O (10), H3BO3 (10), and 0.5 mL L-1of H2SO4 (95%) were added. Strain L. methylohalidivorans MB2 (DSM 14336) was obtained from DSMZ (Braunschweig, Germany) and cultivated in a mineral medium (MAMS) containing (L-1 of distilled water) NaCl (16 g), (NH4)2SO4 (1 g), MgSO4 7H2O (1 g), CaCl2 2H2O (0.2 g), KH2PO4 (0.36 g), and K2HPO4 (2.34 g) as described (Schaefer et al., 2002). After autoclaving, 1 mL L-1 medium of trace elements solution was added. Strains CM4, MC1, and MB2 were grown with CH3Cl gas [10 mL (Fluka), effectively yielding approximately 10 mM final concentration], in 300 mL Erlenmeyer vials fitted with sealed mininert valve caps (Sigma) and containing 50 mL of medium. Cultures were incubated at 30°C on a rotary shaker (100 rpm). Abiotic controls (no bacteria added) were prepared and incubated in the same way. Growth was followed by absorbance measurement at 600 nm.
The headspace of cultures was sampled regularly (0.1 mL) for determination of CH3Cl concentration by gas chromatography, and 1 mL headspace samples were also taken at each point and conserved in 12 ml Exetainers® (Labco Limited, Lampeter, UK) for subsequent isotopic measurements. Concentration of chloride was measured in supernatants of cultures using the spectrophotometric method of Jörg and Bertau (2004), except for L. methylohalidivorans MB2 because of the high chloride content of MAMS medium.
ANALYSIS OF CONCENTRATIONS AND STABLE ISOTOPE VALUES OF CHLOROMETHANE
Concentration and stable carbon and hydrogen isotope values for CH3Cl were performed by gas chromatography coupled with flame ionization detector (GC-FID) and isotope ratio mass spectrometry (IRMS), respectively, as described previously (Nadalig et al., 2013), except that helium flow entering the gas chromatograph in isotopic analysis was increased to 1.8 ml min-1.
The conventional “delta” notation, which expresses the isotopic composition of a material relative to that of a standard on a per mil (‰) deviation basis, was used. Values of δ2H (‰) are relative to that for V-SMOW (Vienna Standard Mean Ocean Water), and values of δ13C (‰) are relative to that for V-PDB (Vienna Pee Dee Belemnite). Carbon and hydrogen isotope fractionations associated with CH3Cl degradation by L. methylohalidivorans MB2, M. extorquens CM4 and H. sp. MC1 were determined from the slopes (bC and bH) of the linear regression of isotope variation (13C and δ2H) of CH3Cl on the logarithm of the remaining CH3Cl concentration (ln f):
Fractionation factors αC and αH were calculated as α = 1,000/(b+1,000), and also expressed as isotope enrichment factors (εC and εH), calculated as ε = (α-1)103. Errors represent 95% confidence intervals calculated on the least-squares regression.
RESULTS
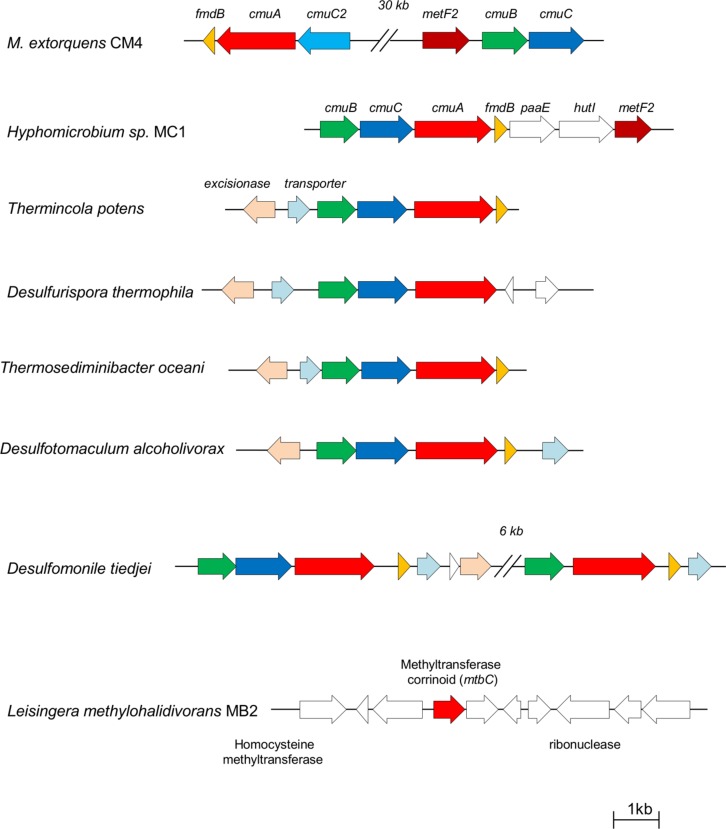
Several chloromethane-degrading bacteria with the cmu pathway have been characterized (Schäfer et al., 2007), and a complete and assembled genome sequence is available for two of them, i.e., M. extorquens CM4 (Marx et al., 2012) and Hyphomicrobium sp. MC1 (Vuilleumier et al., 2011). Two types of organization of cmu genes were identified (Nadalig et al., 2011). The usual gene organization involves a putative cmuBCA operon and was found in all experimentally characterized chloromethane-degrading bacteria with the cmu pathway except the reference chloromethane-degrading strain M. extorquens CM4, which harbors cmu genes in two clusters (Figure 1). The chloromethane-degrading strain L. methylohalidivorans MB2, in contrast, was known to lack cmu genes (Schäfer et al., 2007), so the recent report of its assembled genome sequence (Buddruhs et al., 2013) was of particular interest.
FIGURE 1.
Comparison of cmu gene organization in sequenced genomes of chloromethane-degrading bacteria. Arrows represent protein-coding genes, and homologous genes are shown in the same color. Gene clusters are drawn to scale.
COMPARATIVE GENOMICS
An exhaustive survey of the presence of cmu genes in available sequenced bacterial genomes was carried out, yielding several novel insights (Table 2). First, all strains with cmu homologs contained all three genes cmuA, cmuB, and cmuC essential for growth with CH3Cl using the cmu pathway. Second, these three genes were detected as a cmuBCA gene cluster (Figure 1) in the genomes of five bacterial strains that had not been reported to possess cmu genes or CH3Cl degradation activity. Strikingly and in contrast to all strains growing with CH3Cl using the cmu pathway described so far, all these strains are anaerobes. Three of them are Gram-negative bacteria from the class Deltaproteobacteria, i.e., Desulfotomaculum alcoholivorax (Kaksonen et al., 2008), Desulfurispora thermophila (Kaksonen et al., 2007) and Desulfomonile tiedjei (DeWeerd et al., 1990), and two belong to the class Clostridia, i.e., the Gram-positive Thermincola potens (Byrne-Bailey et al., 2010) and the Gram-negative Thermosediminibacter oceani (Pitluck et al., 2010). Notably, Desulfomonile tiedjei has a second cmu cluster containing only cmuB and cmuA (Figure 1) 6 kb away from a cmuBCA gene cluster. Levels of identity with homologs of the CM4 strain at the protein level are high, and range between 64 and 84%, 60 and 64%, and 34 and 39% for cmuA, cmuB and cmuC gene products, respectively (Table 2). Pairwise identity comparisons of the proteins encoded by cmu genes show that homologs of strains Desulfotomaculum alcoholivorax, Desulfurispora thermophila, Thermincola potens and Thermosediminibacter oceani are most related to each other, with identities at the protein level between 84–93%, 82–92%, and 66–87% for cmuA, cmuB, and cmuC gene products, respectively, and that CM4 homologs represent outliers for all three genes. It is interesting to note that the gene products of two copies of cmuA and cmuB of Desulfomonile tiedjei (78 and 77% protein identity, respectively) are not each others’ closest homologs. In addition, no evidence for substantial identity at the DNA level was detected between the two cmu gene clusters of this strain (data not shown). Further, CmuA encoded by the cmuBCA cluster of Desulfomonile tiedjei is closer to homologs from other strains (>80% identity at the protein level) than that encoded by the partial cmu cluster cmuBA (<75% identity).
Table 2.
Key CDS related to the cmu pathway in investigated genomesa.
| Protein | Methylobacterium extorquens CM4b | Hyphomicrobium sp. MC1 |
Desulfotomaculum alcoholivorax DSM 16058 |
Desulfurispora thermophila DSM 16022 |
Desulfomonile tiedjei DSM 6799 |
Thermincola potens JR |
Thermosediminibacter oceani DSM 16646 |
Leisingera methylohalidivorans MB2 DSM 14336 |
|---|---|---|---|---|---|---|---|---|
| CmuA | Mchl_5697 | HYPMCv2_2273 (84%) |
DESALv160093 (69%) |
AQWNv1_40006 (67%) |
Desti_5447 (67%) Desti_5437 (64%) |
TherJR_0143 (68%) |
Toce_1533 (68%) |
Leime_2531c (32%) |
| CmuB | Mchl_5727 | HYPMCv2_2275 (64%) |
DESALv160091 (62%) | AQWNv1_40005 (62%) |
Desti_5449 (60%) Desti_5438 (60%) |
TherJR_0145 (61%) |
Toce_1535 (62%) |
n.d.d |
| CmuC CmuC2 |
Mchl_5728 Mchl_5698 |
HYPMCv2_2274 (39/35%)e |
DESALv160092 (36/38%) |
AQWNv1_40004 (35/34%) |
Desti_5448 (37/36%) |
TherJR_0144 (37/37%) |
Toce_1534 (35/35%) |
n.d. |
| FmdB | Mchl_5696 | HYPMCv2_2271 (45%) |
DESALv160094 (38%) |
n.d. | Desti_5446 (42%) Desti_5436 (43%) |
TherJR_0142 (42%) |
Toce_1532 (37%) |
n.d. |
| FolD | Mchl_5700 | HYPMCv2_2266 (47%) |
DESALv110219 (40%) |
AQWNv1_30228 (42%) |
Desti_2379 (46%) |
TherJR_1709 (41%) TherJR_1706 (39%) |
Toce_0805 (40%) |
Leime_3180 (52%) Leime_4077 (52%) |
| HutI | Mchl_5694 | HYPMCv2_2269 (53%) |
DESALv150151 (33%) |
AQWNv1_70146 (34%) |
n.d. | n.d. | Toce_1473 (33%) |
Leime_0109 (39%) |
| MetF2 | Mchl_5726 | HYPMCv2_2268 (31%) |
n.d. | n.d. | n.d. | n.d. | n.d. | Leime_2796 (35%) |
| MetF | Mchl_1881 | HYPMCv2_2119 (67%) |
n.d. | n.d. | n.d. | n.d. | n.d. | Leime_1763 (46%) |
| PurU | Mchl_5699 | HYPMCv2_2267 (62%) |
DESALv110045 (34%) |
AQWNv1_20249 (32%) |
n.d. | TherJR_0829 (33%) |
Toce_1499 (33%) |
Leime_2536 (36%) |
| Serine pathway (10 reactions) |
Complete | Complete | 4 | 4 | 5 | 3 | 5 | 9 |
| Ethylmalonyl-CoA pathway (14 reactions) |
Complete | Complete | 10 | 9 | 10 | 2 | 6 | Complete |
| H4F pathway (3 reactions) |
Complete | Complete | Complete | Complete | Complete | Complete | Complete | Complete |
| H4MPT pathway (7 reactions) |
Complete | Complete | 0 | 0 | 0 | 0 | 0 | 0 |
aSequence identity (>30%) at the protein level to M. extorquens CM4 CDS in brackets. All CDS homologs of CM4 genes are chromosomally encoded except hutI in L. methylohalidivorans MB2 which is plasmid-encoded.
bAll CM4 CDS are plasmid-encoded except metF.
cOver the full length (211 aa) of the homolog.
dn.d., not detected.
eSequence identity at the protein level to CmuC/CmuC2 of M. extorquens CM4 in brackets.
Analysis of the L. methylohalidivorans MB2 genome (Buddruhs et al., 2013) confirmed the original report of Schäfer et al. (2007) that this CH3Cl strain did not contain bona fide cmu genes. As mentioned in the genome report, the closest homolog to cmuA is a gene coding a short (232 residues) corrinoid methyltransferase protein (MtbC) with only 32% identity to the C-terminal domain of CmuA (Table 2). However, no full-length homologs to cmuB and cmuC were detected in the genome sequence (Table 2). Taken together, these data confirm that the metabolic pathway used by L. methylohalidivorans MB2 to grow with CH3Cl is different to that of other known chloromethane-degrading strains with the cmu pathway.
The presence of downstream genes in the cmu pathway in strains containing cmuABC genes was also evaluated (Table 2). Genes potentially involved in the tetrahydrofolate (H4F) dependent pathway for oxidation of methyl-H4F to formate via methylene-H4F are present in all genomes (Table 2), but close homologs of metF encoding methylene-H4F reductase were not detected except in strain MC1. Notably, only Alphaproteobacterial strains CM4, MC1, and L. methylohalidivorans MB2 possess the genes involved in the serine and ethylmalonyl-CoA pathways involved in growth of strains CM4 and MC1 with C1 compounds. Moreover, the tetrahydromethanopterin (H4MPT) pathway crucial for growth of Methylobacterium with methanol (Marx et al., 2005), but thought to be dispensable for growth with CH3Cl (Studer et al., 2002), is only present in M. extorquens CM4 and Hyphomicrobium sp. MC1 which also grow with methanol, but absent in L. methylohalidivorans MB2, which is unable to grow with methanol, as well as in all other strains containing cmu genes investigated here. Finally, a search for genes common to chloromethane-degrading strains (including or excluding L. methylohalidivorans MB2) failed to reveal genes other than essential housekeeping genes (data not shown). This suggests that identification of the genes involved in CH3Cl degradation or in adaptation to CH3Cl metabolism is not possible by comparative genomics analysis alone.
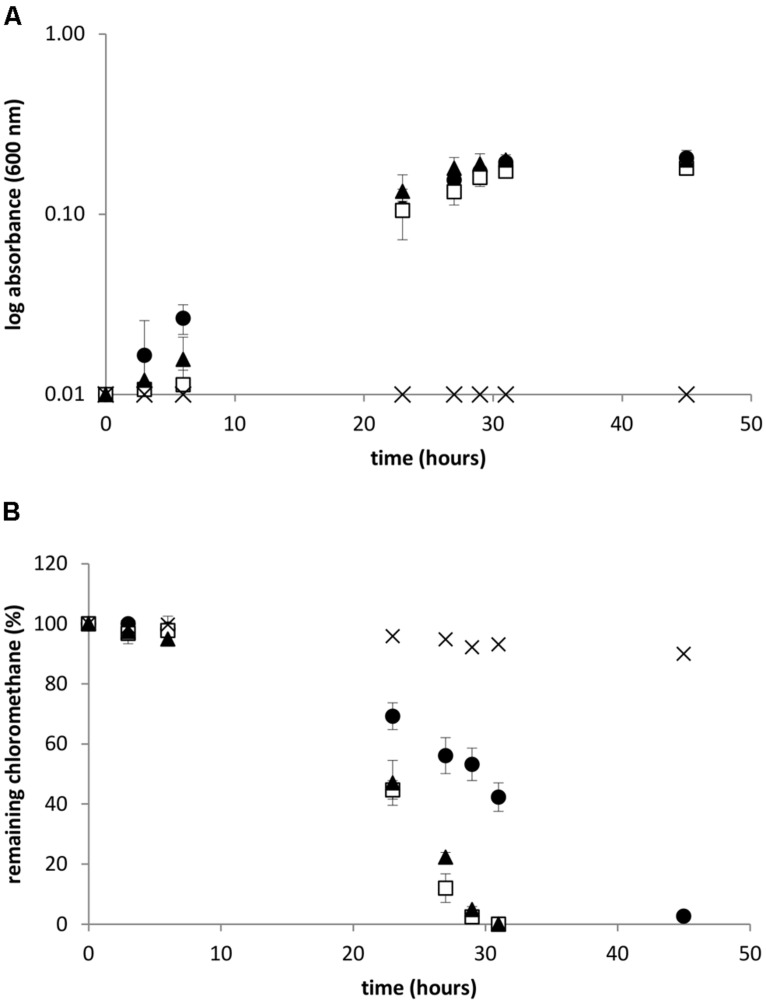
GROWTH OF STRAINS WITH CHLOROMETHANE AS SOLE CARBON AND ENERGY SOURCE
Methylobacterium extorquens CM4, Hyphomicrobium sp. MC1, and L. methylohalidivorans MB2 were cultivated with 10 mM CH3Cl as sole carbon and energy source in the recommended medium allowing fastest growth, i.e., minimal mineral medium for strains CM4 and MC1, and high-salt mineral medium for strain MB2 (Figure 2A). Chloromethane consumption during growth was measured in the gaseous phase by gas chromatography (Figure 2B). In cultures of M. extorquens CM4 and H. sp. MC1, CH3Cl was completely degraded after 30 h under the chosen growth conditions. In contrast, consumption of CH3Cl by the L. methylohalidivorans MB2 culture required a longer time (∼45 h) to proceed to completion, although its growth behavior was similar to that of the other two strains.
FIGURE 2.
Growth and chloromethane degradation during bacterial cultivation. Absorbance at 600 nm (A) and consumption of chloromethane (B). Leisingera methylohalidivorans MB2 (•), M. extorquens CM4 (□) Hyphomicrobium sp. MC1 ( ) and abiotic control (×). Error bars indicate the standard deviation of the mean of three biological replicates. Chloride concentration at the end of cultivation were 9.2 ± 0.2 and 9.2 ± 0.3 mol L-1 for strains CM4 and MC1, respectively, but could not be measured in the high-salt medium used for growth of strains MB2.
) and abiotic control (×). Error bars indicate the standard deviation of the mean of three biological replicates. Chloride concentration at the end of cultivation were 9.2 ± 0.2 and 9.2 ± 0.3 mol L-1 for strains CM4 and MC1, respectively, but could not be measured in the high-salt medium used for growth of strains MB2.
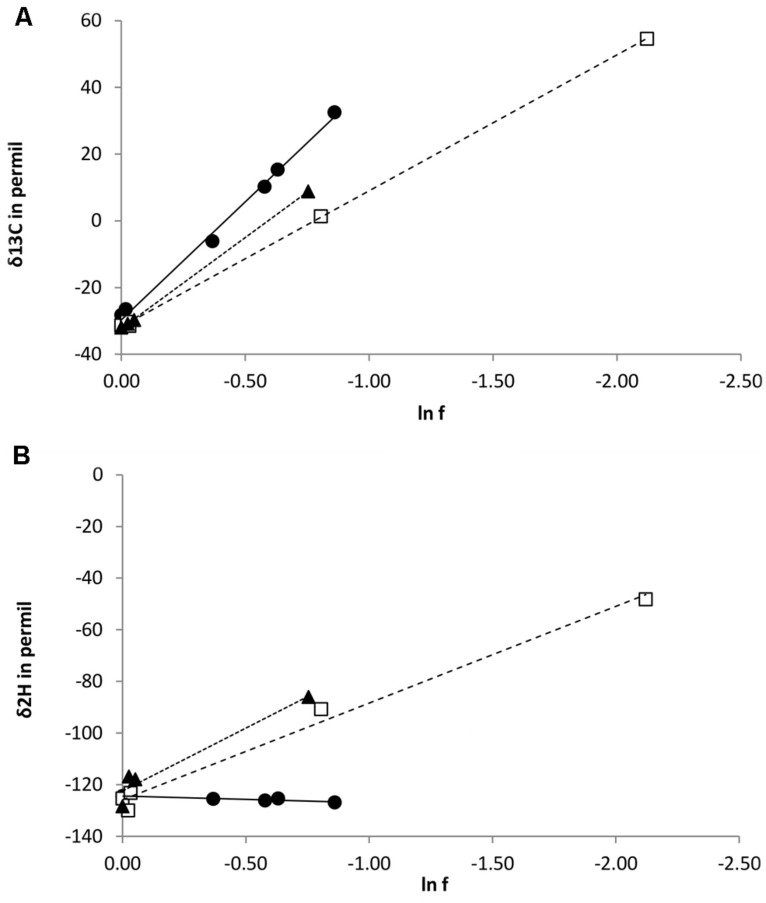
CARBON AND HYDROGEN ISOTOPE FRACTIONATION OF CHLOROMETHANE DURING GROWTH
During degradation of CH3Cl, δ13C values of residual chloromethane increased from approximately -32‰ (initial value) to 55, 9, and 33‰ for strains CM4, MC1, and MB2 respectively (Figure 3A). No carbon or hydrogen fractionation was observed in abiotic controls with media M3 and MAMS (data not shown). Derived values of isotope fractionation factor (αC) and of the corresponding enrichment factor were very similar for cmu pathway strains CM4 and MC1, and substantially larger for L. methylohalidivorans MB2 (Table 3).
FIGURE 3.
Isotope variations of chloromethane during bacterial cultivation with chloromethane. (A) δ13C and (B) δ2H in relation to the fraction of remaining chloromethane (f). Lines represent best-fit linear regressions. L. methylohalidivorans MB2 (•), M. extorquens CM4 (□), and Hyphomicrobium sp. MC1 ( ).
).
Table 3.
Isotopic enrichment (ε) and fractionation (α) factors for carbon and hydrogen during growth with chloromethane.
| εc(‰) | R2a | αC | εH(‰) | R2a | αH | |
|---|---|---|---|---|---|---|
| Methylobacterium extorquens CM4 | 42 | 0.9997 | 1.042 | 39 | 0.9886 | 1.039 |
| Hyphomicrobium sp. MC1 | 54 | 0.9999 | 1.054 | 51 | 0.9418 | 1.051 |
| Leisingera methylohalidivorans MB2 | 76 | 0.9951 | 1.076 | 0 | 0.8230 | 1.000 |
aQuality of fit to linear least-squares regression.
However, trends were markedly different for the three strains when considering the enrichment of 2H in residual CH3Cl during cultivation. For strains CM4 and MC1, δ2H values increased from approximately -124 at the start of the experiment to 16‰)and -12‰ for strains CM4 and MC1, respectively (Figure 3B). In marked contrast, however, no substantial change of δ2H was observed during degradation of CH3Cl by L. methylohalidivorans MB2 (Figure 3B). This resulted in large differences of hydrogen stable isotope fractionation factor (αH) and of the corresponding enrichment factor between strains CM4 and MC1 containing the cmu pathway for CH3Cl degradation on the one hand, and strain MB2 lacking the corresponding genes on the other hand (Table 3).
DISCUSSION
The presence of cmu genes in recently completed genome sequences was somewhat expected, but their detection in exclusively anaerobic bacteria came as a surprise considering that they had so far only been found in aerobic chloromethane-degrading bacteria. Anaerobic chloromethane-degrading bacteria reported so far use a different, although in one case at least also corrinoid-dependent, pathway (Traunecker et al., 1991; Messmer et al., 1993; Freedman et al., 2004). Worthy of note, CH3Cl dehalogenation by the cmu pathway does not require aerobic conditions and is actually sensitive to oxygen (Studer et al., 2001). It is thus possible that anaerobic bacteria with cmu genes identified here (Table 2) are actually able to use CH3Cl as a carbon and energy source, although this remains to be tested experimentally.
The conserved cmuBCA gene organization (Figure 1) in CH3Cl-degrading bacteria (Nadalig et al., 2011), and the high level of identity between the protein sequences encoded by cmu genes of Thermincola potens, Desulfurispora thermophila, Thermosediminibacter oceani, Desulfotomaculum alcoholivorax, and Desulfomonile tiejdei (>81, >74, and >58% for cmuA, cmuB, and cmuC, respectively), suggests a common evolutionary origin for these genes and their dissemination in the environment by horizontal gene transfer. The presence of an excisionase in the immediate environment of cmu genes in these five strains (Figure 1) further supports acquisition of cmu genes by horizontal transfer in these strains, as does the presence of two cmu gene clusters in Desulfomonile tiedjei whose sequences are not closer related to each other than to those of other chloromethane-degrading strains. To our knowledge, however, potential sources of CH3Cl that would support dissemination of cmu genes in anaerobic environments have not yet been reported.
Incidentally, our analysis also confirms the particular status in the cmu pathway of cmuC, a gene shown to be essential for growth of strain CM4 with CH3Cl (Studer et al., 2002; Roselli et al., 2013) but whose function remains elusive. Indeed, sequence conservation among the proteins encoded by genes cmuA, cmuB, and cmuC are lowest for the CmuC gene product (Table 2). Moreover, the probable loss of a cmuC homolog in one of the two cmu gene clusters of Desulfomonile tiedjei strain CM4 (Figure 1) also hints at its possibly lesser role in CH3Cl metabolism.
As to the chloromethane-degrading strain L. methylohalidivorans MB2, analysis of its genome (Buddruhs et al., 2013) confirms the initial report (Schäfer et al., 2007) that it lacks the cmu pathway. The best partial hit to CmuA (32% amino acid identity) is a 211-residue corrinoid protein, and cmuB or cmuC homologs were not detected in the L. methylohalidivorans MB2 genome (Table 2). However, downstream genes of the H4F-dependent cmu pathway (metF, folD, purU) were all found, so an H4F-dependent metabolic pathway for growth of L. methylohalidivorans MB2 with CH3Cl remains a possibility.
In our experiments, we showed that L. methylohalidivorans MB2, previously grown with CH3Cl (0.37 mM; Schaefer et al., 2002), is capable of using this one-carbon compound as sole carbon and energy source at an initial concentration of 10 mM. A direct comparison of its growth behavior with that of strains CM4 and MC1 is prevented by the fact that the latter two strains do not grow in high-salt mineral medium, whereas L. methylohalidivorans MB2 does not grow in the standard low-salt mineral medium used for strains CM4 and MC1. Incidentally, this suggests that salt adaptation may be unrelated to adaptation to intracellular chloride production during dehalogenation, as observed recently for bacteria growing with dichloromethane (Michener et al., 2014).
The differences in CH3Cl metabolism of L. methylohalidivorans MB2 suggested by comparative genomics were experimentally supported by isotope analysis (Figure 3; Table 3). For L. methylohalidivorans MB2, isotopic enrichment factor for carbon during growth was substantially larger than for CM4 and MC1, indicating a larger primary isotope effect and providing further evidence for operation of another pathway for utilization of CH3Cl in this strain. In contrast, a previous study on carbon isotopic fractionation of CH3Cl by cell suspensions of three bacterial strains, including L. methylohalidivorans MB2, gave similar isotopic enrichment values (ranging between 42 and 47‰; Miller et al., 2001). In particular, the value obtained for Aminobacter ciceronei strain IMB1, the only strain so far shown to possess cmuA but not cmuB (Woodall et al., 2001), was similar to those of strains CM4 and MC1 (Miller et al., 2001). This suggests that the corrinoid dehalogenase protein CmuA drives carbon isotopic fractionation in chloromethane-degrading strains with the cmu pathway. Moreover and unlike for carbon, a larger isotope effect than in previous resting cell experiments (Nadalig et al., 2013) was observed for hydrogen during growth in strains CM4 and MC1. However, the most striking finding of the present study was the lack of substantial hydrogen isotope enrichment upon CH3Cl degradation by L. methylohalidivorans MB2. This suggests that unlike CmuAB chloromethane dehalogenase, the unknown dehalogenase of this strain does not cause hydrogen fractionation during degradation of the chloromethane methyl group. Nevertheless and as a common denominator to all three chloromethane-degrading strains investigated here (Table 3), carbon isotope fractionation (the primary isotope effect in cleavage of the carbon-halogen bond) was more pronounced than hydrogen isotope fractionation (a secondary isotope effect in CH3Cl dehalogenation), as expected (Elsner et al., 2005).
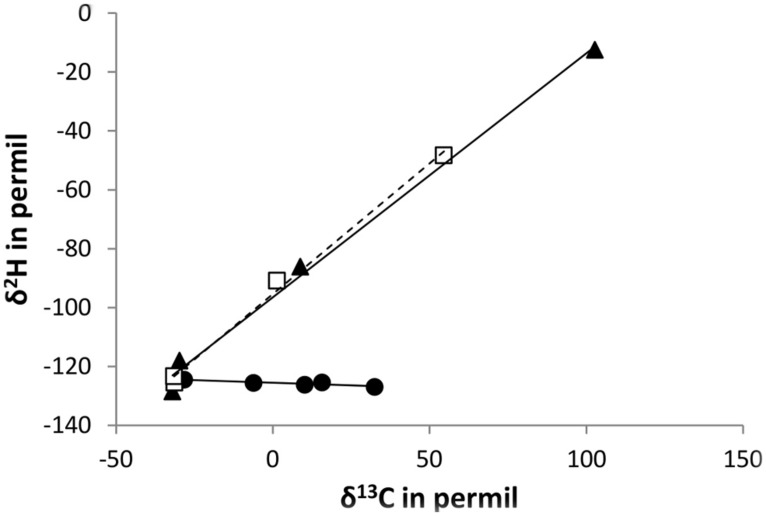
The observed differences in isotopic fractionation of CH3Cl carbon and hydrogen between the three strains CM4, MC1, and MB2 are best visualized in Figure 4, which shows the trends in enrichment of the heavier isotope of carbon and hydrogen for the different strains at different time points during growth. As proposed by Elsner et al. (2005), the slopes in these graphs constitute a clear indication that L. methylohalidivorans MB2 uses a different pathway for growth with CH3Cl than strains CM4 and MC1, which utilize the same pathway.
FIGURE 4.
Changes in carbon and hydrogen isotope ratios for degradation of chloromethane by L. methylohalidivorans MB2 (•), M. extorquens CM4 (□), and Hyphomicrobium sp. MC1 ( ) at different time points during growth (increase in time from left to right of the graph). Lines (slope = -0.03 (R2: 0.8014) for strain MB2, slope = 0.89 (R2: 0.9954) for strain CM4, and slope = 0.83 (R2: 0.9939) for strain MC1, respectively) represent best fit regressions.
) at different time points during growth (increase in time from left to right of the graph). Lines (slope = -0.03 (R2: 0.8014) for strain MB2, slope = 0.89 (R2: 0.9954) for strain CM4, and slope = 0.83 (R2: 0.9939) for strain MC1, respectively) represent best fit regressions.
Measurements of isotopic fractionation for a given environmental compartment will include the overall contribution of the metabolic diversity of chloromethane-degrading bacteria and their relative occurrence in that environment. It is tempting to speculate that chloromethane degradation in the soil environment, for which an isotopic fractionation of 49‰ similar to that found here for strains CM4 and MC1 was obtained in a previous study (Miller et al., 2004), is predominantly performed by bacteria with the cmu pathway. Our results on microbially driven hydrogen and carbon isotope fractionation suggest that using in a two-dimensional isotope scheme might help to confirm this hypothesis. Thus, a combination of genomic studies with physiological and isotopic characterisation of chloromethane-degrading bacterial strains, as performed here, will remain a major objective for the near future in order to constrain the bacterial sink strength of the atmospheric budget of CH3Cl.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial support for the acquisition of GC-FID equipment from REALISE (http://realise.unistra.fr), the Alsace network for research and engineering in the environmental sciences, is gratefully acknowledged. Frank Keppler is supported by the ESF (EURYI Award to Frank Keppler) and DFG (KE 884/2-1), and by the DFG research unit 763 “Natural Halogenation Processes in the Environment – Atmosphere and Soil” (KE 884/6-1; KE 884/7-1).
REFERENCES
- Buddruhs N., Chertkov O., Petersen J., Fiebig A., Chen A., Pati A., et al. (2013). Complete genome sequence of the marine methyl-halide oxidizing Leisingera methylohalidivorans type strain (DSM 14336T), a member of the Roseobacter clade. Stand. Genomic Sci. 9 128–141 10.4056/sigs.4297965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne-Bailey K. G., Wrighton K. C., Melnyk R. A., Agbo P., Hazen T. C., Coates J. D. (2010). Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J. Bacteriol. 192 4078–4079 10.1128/JB.00044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter C., Hamilton J. T. G., McRoberts W. C., Kulakov L., Larkin M. J., Harper D. B. (1999). Halomethane:bisulfide/halide ion methyltransferase, an unusual corrinoid enzyme of environmental significance isolated from an aerobic methylotroph using chloromethane as the sole carbon source. Appl. Environ. Microbiol. 65 4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czapiewski K., Czuba E., Huang L., Ernst D., Norman A., Koppmann R., et al. (2002). Isotopic composition of non-methane hydrocarbons in emissions from biomass burning. J. Atmos. Chem. 43 45–60 10.1023/A:1016105030624 [DOI] [Google Scholar]
- DeWeerd K. A., Mandelco L., Tanner R. S., Woese C. R., Suflita J. M. (1990). Desulfomonile tiedjei gen. nov., and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium. Arch. Microbiol. 154 23–30 10.1007/BF00249173 [DOI] [Google Scholar]
- Doronina N. V., Sokolov A. P., Trotsenko Y. A. (1996). Isolation and initial characterization of aerobic chloromethane-utilizing bacteria. FEMS Microbiol. Lett. 142 179–183 10.1111/j.1574-6968.1996.tb08427.x [DOI] [Google Scholar]
- Elsner M., Zwank L., Hunkeler D., Schwarzenbach R. P. (2005). A new concept linking observable stable isotope fractionation to transformation pathways of organic pollutants. Environ. Sci. Technol. 39 6896–6916 10.1021/es0504587 [DOI] [PubMed] [Google Scholar]
- Freedman D., Swamy M., Bell N., Verce M. (2004). Biodegradation of chloromethane by Pseudomonas aeruginosa strain NB1 under nitrate-reducing and aerobic conditions. Appl. Environ. Microbiol. 70 4629–4634 10.1128/AEM.70.8.4629-4634.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greule M., Huber S. G., Keppler F. (2012). Stable hydrogen-isotope analysis of methyl chloride emitted from heated halophytic plants. Atmos. Environ. 62 584–582 10.1016/j.atmosenv.2012.09.007 [DOI] [Google Scholar]
- Hamilton J. T. G., McRoberts W. C., Keppler F., Kalin R. M., Harper D. B. (2003). Chloride methylation by plant pectin: an efficient environmentally significant process. Science 301 206–209 10.1126/science.1085036 [DOI] [PubMed] [Google Scholar]
- Harper D. B. (2000). The global chloromethane cycle: biosynthesis, biodegradation and metabolic role. Nat. Prod. Rep. 17 337–348 10.1039/a809400d [DOI] [PubMed] [Google Scholar]
- Harper D. B., Hamilton J. T. G., Ducrocq V., Kennedy J. T., Downey A., Kalin R. M. (2003). The distinctive isotopic signature of plant-derived chloromethane: possible application in constraining the atmospheric chloromethane budget. Chemosphere 52 433–436 10.1016/S0045-6535(03)00206-6 [DOI] [PubMed] [Google Scholar]
- Harper D. B., Kalin R. M., Hamilton J. T. G., Lamb C. (2001). Carbon isotope ratios for chloromethane of biological origin: potential tool in determining biological emissions. Environ. Sci. Technol. 35 3616–3619 10.1021/es0106467 [DOI] [PubMed] [Google Scholar]
- Hartmans S., Schmuckle A., Cook A. M., Leisinger T. (1986). Methyl chloride: naturally occurring toxicant and C-1 growth substrate. Microbiology 132 1139–1142 10.1099/00221287-132-4-1139 [DOI] [Google Scholar]
- Jörg G., Bertau M. (2004). Thiol-tolerant assay for quantitative colorimetric determination of chloride released from whole-cell biodehalogenations. Anal. Biochem. 328 22–28 10.1016/j.ab.2004.01.027 [DOI] [PubMed] [Google Scholar]
- Kaksonen A. H., Spring S., Schumann P., Kroppenstedt R. M., Puhakka J. A. (2007). Desulfurispora thermophila gen. nov., sp. nov., a thermophilic, spore-forming sulfate-reducer isolated from a sulfidogenic fluidized-bed reactor. Int. J. Syst. Evol. Microbiol. 57 1089–1094 10.1099/ijs.0.64593-0 [DOI] [PubMed] [Google Scholar]
- Kaksonen A. H., Spring S., Schumann P., Kroppenstedt R. M., Puhakka J. A. (2008). Desulfotomaculum alcoholivorax sp. nov., a moderately thermophilic, spore-forming, sulfate-reducer isolated from a fluidized-bed reactor treating acidic metal- and sulfate-containing wastewater. Int. J. Syst. Evol. Microbiol. 58 833–838 10.1099/ijs.0.65025-0 [DOI] [PubMed] [Google Scholar]
- Keppler F., Harper D. B., Rockmann T., Moore R. M., Hamilton J. T. G. (2005). New insight into the atmospheric chloromethane budget gained using stable carbon isotope ratios. Atmos. Chem. Phys. 5 2403–2411 10.5194/acp-5-2403-2005 [DOI] [Google Scholar]
- Keppler F., Kalin R., Harper D., McRoberts W. C., Hamilton J. T. G. (2004). Carbon isotope anomaly in the major plant C1 pool and its global biogeochemical implications. Biogeosciences 1 393–412 10.5194/bgd-1-393-2004 [DOI] [Google Scholar]
- Marx C. J., Bringel F., Chistoserdova L., Moulin L., Farhan Ul Haque M., Fleischman D. E., et al. (2012). Complete genome sequences of six strains of the genus Methylobacterium. J. Bacteriol. 194 4746–4748 10.1128/JB.01009-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C. J., Van Dien S. J., Lidstrom M. E. (2005). Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 3:e16 10.1371/journal.pbio.0030016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnulla C., McDonald I. R., Murrell J. C. (2001). Methyl chloride utilising bacteria are ubiquitous in the natural environment. FEMS Microbiol. Lett. 201 151–155 10.1111/j.1574-6968.2001.tb10749.x [DOI] [PubMed] [Google Scholar]
- Messmer M., Wohlfarth. G., Diekert G. (1993). Methyl chloride metabolism of the strictly anaerobic, methyl chloride-utilizing homoacetogen strain MC. Arch. Microbiol. 160 383–387 10.1007/BF00252225 [DOI] [Google Scholar]
- Michener J., Vuilleumier S., Bringel F., Marx C. J. (2014). Phylogeny poorly predicts the utility of a challenging horizontally-transferred gene in Methylobacterium strains. J. Bacteriol. 196 2101–2107 10.1128/JB.00034-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L., Connell G., Guidetti J., Oremland R. (1997). Bacterial oxidation of methyl bromide in fumigated agricultural soils. Appl. Environ. Microbiol. 63 4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L., Kalin R. M., McCauley S. E., Hamilton J. T. G., Harper D. B., Millet D. B., et al. (2001). Large carbon isotope fractionation associated with oxidation of methyl halides by methylotrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 98 5833–5837 10.1073/pnas.101129798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L., Warner G., Baesman S., Oremland R., McDonald I. R., Radajewski S., et al. (2004). Degradation of methyl bromide and methyl chloride in soil microcosms: use of stable C isotope fractionation and stable isotope probing to identify reactions and the responsible microorganisms. Geochim. Cosmochim. Acta 68 3271–3283 10.1016/j.gca.2003.11.028 [DOI] [Google Scholar]
- Montzka S., Reimann S. (2011). “Ozone-depleting substances (ODSs) and related chemicals,” in Scientific Assessment of Ozone Depletion: 2010 Chap. 1 Global Ozone Research and Monitoring Project, Report No. 52. Geneva: World Meteorological Organization; 1–112 [Google Scholar]
- Nadalig T., Farhan Ul Haque M., Roselli S., Schaller H., Bringel F., Vuilleumier S. (2011). Detection and isolation of chloromethane-degrading bacteria from the Arabidopsis thaliana phyllosphere, and characterization of chloromethane utilization genes. FEMS Microbiol. Ecol. 77 438–448 10.1111/j.1574-6941.2011.01125.x [DOI] [PubMed] [Google Scholar]
- Nadalig T., Greule M., Bringel F., Vuilleumier S., Keppler F. (2013). Hydrogen and carbon isotope fractionation during degradation of chloromethane by methylotrophic bacteria. Microbiologyopen 2 893–900 10.1002/mbo3.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitluck S., Yasawong M., Munk C. (2010). Complete genome sequence of Thermosediminibacter oceani type strain (JW/IW-1228PT). Stand. Genomic Sci. 3 108–116 10.4056/sigs.1133078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker K. R., Kalin R. M. (2012). Methyl chloride isotopic signatures from Irish forest soils and a comparison between abiotic and biogenic methyl halide soil fluxes. Global Change Biol. 18 1453–1467 10.1111/j.1365-2486.2011.02600.x [DOI] [Google Scholar]
- Roselli S., Nadalig T., Vuilleumier S., Bringel F. (2013). The 380 kb pCMU01 plasmid encodes chloromethane utilization genes and redundant genes for vitamin B12- and tetrahydrofolate-dependent chloromethane metabolism in Methylobacterium extorquens CM4: a proteomic and bioinformatics study. PLoS ONE 8:e56598 10.1371/journal.pone.0056598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Yokouchi Y. (2008). Stable carbon isotope ratio of methyl chloride emitted from glasshouse-grown tropical plants and its implication for the global methyl chloride budget. Geophys. Res. Lett. 35:L08807 10.1029/2007GL032736 [DOI] [Google Scholar]
- Schaefer J. K., Goodwin K. D., McDonald I. R., Murrell J. C., Oremland R. S. (2002). Leisingera methylohalidivorans gen. nov., sp. nov., a marine methylotroph that grows on methyl bromide. Int. J. Syst. Evol. Microbiol. 52 851–859 10.1099/ijs.0.01960-0 [DOI] [PubMed] [Google Scholar]
- Schäfer H., McDonald I. R., Nightingale P. D., Murrell J. C. (2005). Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide-oxidizing bacteria. Environ. Microbiol. 7 839–852 10.1111/j.1462-2920.2005.00757.x [DOI] [PubMed] [Google Scholar]
- Schäfer H., Miller L. G., Oremland R. S., Murrell J. C. (2007). Bacterial cycling of methyl halides. Adv. Appl. Microbiol. 61 307–346 10.1016/S0065-2164(06)61009-5 [DOI] [PubMed] [Google Scholar]
- Studer A., McAnulla C., Büchele R., Leisinger T., Vuilleumier S. (2002). Chloromethane induced genes define a third C1 utilization pathway in Methylobacterium chloromethanicum CM4. J. Bacteriol. 184 3476–3482 10.1128/JB.184.13.3476-3484.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Stupperich E., Vuilleumier S., Leisinger T. (2001). Chloromethane: tetrahydrofolate methyl transfer by two proteins from Methylobacterium chloromethanicum strain CM4. Eur. J. Biochem. 268 2931–2938 10.1046/j.1432-1327.2001.02182.x [DOI] [PubMed] [Google Scholar]
- Traunecker J., Preuss A., Diekert G. (1991). Isolation and characterization of a methyl chloride utilizing, strictly anaerobic bacterium. Arch. Microbiol. 156 416–421 10.1007/BF00248720 [DOI] [Google Scholar]
- Vallenet D., Engelen S., Mornico D., Cruveiller S., Fleury L., Lajus A., et al. (2009). MicroScope: a platform for microbial genome annotation and comparative genomics. Database 2009 bap021. 10.1093/database/bap021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannelli T., Messmer M., Studer A., Vuilleumier S., Leisinger T. (1999). A corrinoid-dependent catabolic pathway for growth of a Methylobacterium strain with chloromethane. Proc. Natl. Acad. Sci. U.S.A. 96 4615–4620 10.1073/pnas.96.8.4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier S., Nadalig T., Farhan Ul Haque M., Magdelenat G., Lajus A., Roselli S., et al. (2011). Complete genome sequence of the chloromethane-degrading strain Hyphomicrobium sp. strain MC1. J. Bacteriol. 193 5035–5036 10.1128/JB.05627-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodall C. A., Warner K. L., Oremland R. S., Murrell J. C., McDonald I. R. (2001). Identification of methyl halide-utilizing genes in the methyl bromide-utilizing bacterial strain IMB-1 suggests a high degree of conservation of methyl halide-specific genes in Gram-negative bacteria. Appl. Environ. Microbiol. 67 1959–1963 10.1128/AEM.67.4.1959-1963.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Wang Y., Zeng T., Yantosca R. (2004). A three-dimensional global model study of atmospheric methyl chloride budget and distributions. J. Geophys. Res. 109:D24309 10.1029/2004JD004951 [DOI] [Google Scholar]