Abstract
Purpose.
The CRYAAN101D transgenic mouse model expressing deamidated αA-crystallin (deamidation at N101 position to D) develops cortical cataract at the age of 7 to 9 months. The present study was carried out to explore the molecular mechanism that leads to the development of cortical opacity in CRYAAN101D lenses.
Methods.
RNA sequence analysis was carried out on 2- and 4-month-old αA-N101D and wild type (WT) lenses. To understand the biologic relevance and function of significantly altered genes, Ingenuity Pathway Analysis (IPA) was done. To elucidate terminal differentiation defects, immunohistochemical, and Western blot analyses were carried out.
Results.
RNA sequence and IPA data suggested that the genes belonging to gene expression, cellular assembly and organization, and cell cycle and apoptosis networks were altered in N101D lenses. In addition, the tight junction signaling and Rho A signaling were among the top three canonical pathways that were affected in N101D mutant. Immunohistochemical analysis identified a series of terminal differentiation defects in N101D lenses, specifically, increased proliferation and decreased differentiation of lens epithelial cells (LEC) and decreased denucleation of lens fiber cells (LFC). The expression of Rho A was reduced in different-aged N101D lenses, and, conversely, Cdc42 and Rac1 expressions were increased in the N101D mutants. Moreover, earlier in development, the expression of major membrane-bound molecular transporter Na,K-ATPase was drastically reduced in N101D lenses.
Conclusions.
The results suggest that the terminal differentiation defects, specifically, increased proliferation and decreased denucleation are responsible for the development of lens opacity in N101D lenses.
Keywords: alpha A crystallin, denucleation, cortical opacity, deamidation
Previously we have shown that the lenses of mice expressing CRYAAN101D transgene develop mild cortical lens opacity at the age of 7 months. In continuation of that work, the present study elucidates a potential molecular mechanism of formation of cortical opacity in CRYAAN101D transgenic mice (N101D mutant).
Introduction
The ocular lens has a unique cellular architecture consisting of a single layer of epithelial cells at the anterior surface and concentric layers of highly specialized, terminally differentiated fiber cells. Epithelial cells in the germinative zone proliferate continuously to replicate throughout the life of an organism. The newly formed epithelial cells migrate to the lens equator in the transitional zone, where they elongate and differentiate into secondary fiber cells. The coordinated proliferation, migration, and elongation of lens epithelial cells is required for the lens transparency and refraction.1–4 Differentiation of lens epithelial cells (LEC) into lens fiber cells (LFC) is a complex process involving changes at the molecular and morphologic levels. The morphologic changes are associated with membrane cytoskeleton remodeling, actin filament assembly, and cell adhesion turnover.5–8 The molecular changes include the synthesis and short-range, ordered packing of crystallins and the degradation of organelles to create an organelle-free zone (OFZ). Together, these changes make the lens transparent and refractive, allowing light to pass its way to the retina.9,10 Disruption in any of these processes has been known to cause defects in LEC proliferation, elongation, and differentiation, resulting in persistence of organelles in LFC, perturbance of OFZ formation, and lens opacity.11 Besides proper execution of the terminal differentiation process, lens transparency is also dependent on many other factors, including regulated interactions of crystallins with membrane and cytoplasmic proteins, proper maintenance of cellular ionic concentration, and cellular homeostasis. Any perturbation in these cellular processes also leads to lens opacity. For example, transgenic mice expressing αAR116C transgene have been reported to develop lens opacity in which mutant crystallin showed an approximate 10-fold increased affinity for membrane.12,13 Membrane defects have also been reported to be associated with lens opacity induced by dexamethasone or mechanical stress in cultured lenses where the cells had lost cadherin junctions.14 Mutations on the fiber cell-specific proteins, such as aquaporin and connexin also caused lens opacity mediated by inhibition of Na,K-ATPase, which led to an imbalance of Na+ and K+ concentration in the cell.15,16 Abnormally high sodium content has been reported in lenses of many cataractous patients.17 Na,K-ATPase maintains ion concentration in the lens by pumping out three sodium ions in exchange for two potassium ions by using the energy from ATP hydrolysis.
Alpha crystallin, a major protein component of the lens, plays a vital role in the prevention of aggregation of misfolded proteins during aging, under heat, or in other cellular stress conditions.18–20 In addition, αA crystallin has been shown to prevent apoptosis induced by various factors such as staurosprine,21 UVA irradiation,22,23 and TNF.24
The limited protein turnover of lens crystallins makes them susceptible to a wide variety of posttranslational modifications (PTMs), including proteolysis, glycation, disulfide bonding, oxidation, and deamidation.25 These PTMs destabilize the crystallin structure and cause aggregation, crosslinking, and precipitation of lens proteins, which lead to lens opacity.26
Among various PTM, deamidation is the most common PTMs among the crystallins, accounting for greater than 60% of total reported modifications. The deamidation process adds a negative charge to aspargine and glutamine residues, thereby inducing structural changes and affecting protein–protein interactions.27–29 In αA-crystallin, the occurrence of deamidation at the Gln-50 and Asn-101 positions was more frequent than at Gln-6 and Asn-123,27 suggesting that Gln-50 and Asn-101 residues are more susceptible to deamidation. Our in vitro analysis suggested that a deamidation at the N101 position has a more pronounced effect on the structure and function of αA-crystallin than deamidation at the N123 position.30,31 Nevertheless, both the mutants showed reduced chaperone activities, larger protein aggregates, and altered secondary structures compared with the wild-type (WT) αA-crystallin. To explore the in vivo effect of αA crystallin deamidation, we previously generated a transgenic mouse model expressing deamidated αA-crystallin,32 CRYAAN101D transgenic mouse (transgenic line 1.B2, in which 28.2% of total αA protein is deamidated [αAN101D]; Fig. 1). Compared with WT, lenses from CRYAAN101D transgenic mice showed altered packing of fiber cells, persistence of nuclei in the deeper cortical region, increased insolubilization of crystallin, and mild cortical opacity at the age of 7 months. In the present study, using various molecular approaches, we elucidated the molecular events that lead to the development of cortical opacity in lenses of N101D transgenic mice. Based on the results, we propose that the greater association of deamidated αA-crystallin (αAN101D) with the membrane may be responsible for the disruption of Na,K-ATPase expression and function, which, in turn, alters Rho A GTPase expression and activities. The altered Rho A GTPases cause a series of terminal differentiation defects, including increased proliferation and decreased differentiation of lens epithelial cells (LEC) and delayed denucleation in lens fiber cells (LFC), which eventually lead to lens opacity in N101D mutants.
Figure 1.
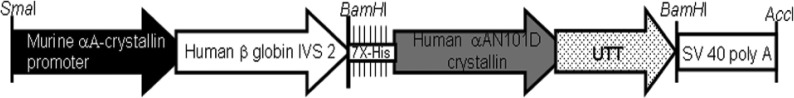
Graphical representation of deamidated αA crystallin transgenic construct used for generation of CRYAAN101D transgenic mice. Reprinted with permission from Gupta R, Asomugha CO, Srivastava OP. The common modification in alphaA-crystallin in the lens, N101D, is associated with increased opacity in a mouse model. J Biol Chem. 2011;286:11579–11592. Copyright 2011 American Society for Biochemistry and Molecular Biology. Transgene construct contain αA crystallin promoter, human β-globin intervening sequence, the coding region of αA crystallin that has deamidation at 101 position, with an in-frame epitope tag (7X-His) at the N-terminus of αA crystallin and untranslated sequences (UTT) and polyadenylatation SV40 sequence at the C-terminus.
Materials and Methods
Ethics Statement
All animal experiments were performed in accordance with ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. In addition, they were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Alabama at Birmingham (Protocol no. 130208393). Mice were housed in specific pathogen-free facilities at the University of Alabama at Birmingham. Isofluorane was used (in accordance to the IACUC-approved animal research protocols) to perform humane euthanasia of mice, and all efforts were made to minimize suffering of animals.
Materials
Unless stated otherwise, the molecular biology-grade chemicals were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA) or Fisher (Atlanta, GA, USA) companies. The polyclonal anti-human aquaporin-0 (AQP0) antibody was purchased from Alpha Diagnostics (San Antonio, TX, USA). Additional commercial sources of various antibodies used in the study are described throughout the text.
RNA Extraction
After euthanizing the mice, lenses from the mice of desired ages were extracted under dissecting microscope and immediately placed in 500 μL of Trizol, and total RNA was extracted using the illustra RNAspin Mini RNA isolation kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) as per the manufacturer's protocol. The concentration and the quality of RNA was determined using a Nanodrop spectrophotometer (Thermo Scientific, Pittsburgh, PA, USA).
Whole-Genome Transcryptome Analysis
The mRNA sequencing was performed on the Illumina HiSeq2000 (paired end 2 × 50 bp sequencing runs; Illumina, San Diego, CA, USA) using the latest versions of the sequencing reagents and flow cells providing up to 300 Gb of sequence information per flow cell. The quality of the total RNA isolated from lenses of 2- and 4-month-old N101D transgenic and WT mice was briefly assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) followed by 2-rounds of poly A+ selection and conversion to cDNA. The TruSeq library generation kits were used as per the manufacturer's instructions (Illumina). Library construction consisted of random fragmentation of the polyA mRNA followed by cDNA production using random primers. The ends of the cDNA were repaired, A-tailed, and adaptors ligated for indexing (up to 12 different barcodes per lane) during the sequencing runs. The cDNA libraries were quantified using qPCR in a Roche LightCycler 480 (Roche Diagnostics, Indianapolis, IN, USA) with the Kapa Biosystems kit for library quantification (Kapa Biosystems, Woburn, MA, USA) prior to cluster generation. Clusters were generated to yield approximately 725 to 825 K clusters/mm.33 Cluster density and quality was determined during the run after the first base addition parameters were assessed.
The raw data files were converted to the fastq file format using Illumina's CASAVA version 1.8.1.TopHat version 1.2.0 (Illumina, San Diego, CA, USA) was used to align the raw RNA-Seq fastq reads to the mouse mm9 reference genome using the short read aligner Bowtie (Johns Hopkins University, Baltimore, MD, USA).34,35 TopHat also analyzes the mapping results to identify splice junctions between exons. Cufflinks version 1.0.30 (University of California, Berkeley, CA, USA and Johns Hopkins University, Baltimore, MD, USA) was used to align the readings from TopHat to assemble transcripts, estimate their abundances, and test for differential expression and regulation.36 Cuffcompare, which is part of Cufflinks, merged the assembled transcripts to a reference annotation and is capable of tracking Cufflinks transcripts across multiple experiments. Finally, Cuffdiff found significant changes in transcript expression, splicing, and promoter use.
The above datasets were analyzed using Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, [in the public domain] www.ingenuity.com). The data set contained gene identifiers and corresponding expression values and was uploaded into the application. Each identifier was mapped to its corresponding object in Ingenuity's Knowledge Base. A fold-change cutoff of ±2 was set to identify target molecules whose expression was significantly upregulated or downregulated. These Network Eligible Molecules were overlaid onto a Global Molecular Network developed from information contained in Ingenuity's Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity. The Functional Analysis identified the biological functions that were most significant to the dataset. Molecules from the dataset that met the ±2-fold change cutoff and were associated with biologic functions in Ingenuity's Knowledge Base were considered for analysis. Right-tailed Fisher's exact test was used to calculate a P value determining the probability that each biologic function assigned to that data set is due to chance alone (this experiment was done at the Gene Expression Shared Facility located in the Heflin Center for Genomic Sciences at the University of Alabama at Birmingham, Birmingham, AL, USA).
Immunohistochemistry
Paraffin-embedded sections (5-μM thick axial and equatorial sections) were obtained from whole eyes of WT and N101D mutant transgenic mice. Sections were deparaffinized and rehydrated using PBS, followed by heat-induced epitope retrieval in 10 mM sodium citrate and 0.2% Tween 20. The sections were rinsed with water and blocked in 3% (wt/vol) BSA in PBS for 30 minutes, and incubated at 4°C overnight with the individual primary polyclonal antibodies raised against aquaporin-0, RhoA, Cdc42, Rac1, and NaK+ATPase (Cell Signaling, Danvers, MA, USA). Next, the sections were washed with PBS and incubated with Alexa fluor 488-conjugated anti-rabbit secondary antibodies (Invitrogen, Grand Island, NY, USA), at 1:1000 dilution in PBS, for 1 hour at room temperature. The sections were then washed in PBS and stained with Hoechst 33342 nuclear stain (1:100 dilution) for 1 minute and rinsed with PBS. For the negative controls, the individual primary antibodies were omitted. The sections were mounted with aqueous mounting media and examined with an SP-2 confocal microscope (at UAB High Resolution Imaging Core facility, Birmingham, AL, USA), or, in some cases, with a Zeiss Axioplan 2 fluorescence microscope equipped with a CCD camera (Hercules, CA, USA, at Vision Science Research Center Core facility). For staining the membrane with Tetramethyl rhodamine (TRITC), the BSA-incubated sections were incubated with TRITC-labeled Lectin from Triticum vulgaris (Sigma-Aldrich Corp.).
Western Blot Analysis
To quantify the total amount of Rho GTPase protein in both WT and N101D transgenic mice, four to six lenses were homogenized in Radioimmunoprecipitation assay buffer (RIPA buffer; Sigma-Aldrich Corp.) and centrifuged at 800g for 10 minutes at 4°C. Protein concentration in supernatant fraction was determined, and equal amounts of protein from WT and N101D transgenic mice lenses were separated by SDS PAGE, transferred to a PVDF membrane (using BioRad Trans-Blot Turbo Transfer System, City, State, Country), and subsequently probed with antibodies against Rho A, Rac1, or Cdc42. A horse raddish peroxidase (HRP)-conjugated goat anti-rabbit (1:1000 dilution; Cell Signaling) was added, and the immunoreactive bands were then detected through the LI-COR odyssey Fc dual mode imaging system using enhanced chemiluminescence substrate (LI-COR, Lincoln, NE, USA). β-actin was used as a loading control.
Results
RNA Sequence Analysis of Lenses of N101D and Age-Matched WT Mice
To understand the global view of dysregulated genes and pathways affected in the N101D mutants, we investigated gene expression in 2- and 4-month-old WT and N101D mutants. RNA sequencing data have been submitted to the Gene Expression Omnibus under accession number GSE57959 ([in the public domain] http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57959)
The only gene expressions that were considered significant were those, which showed at least a 2-fold changes (with P ≤ 0.05) in N101D compared with WT mice. Overall, compared with WT, 611 genes were differentially expressed in the 2-month-old N101D mutant, and among those, 311 genes were upregulated and 300 genes were downregulated. In 4-month-old N101D lenses, the expressions of 331 genes were altered, of which, 160 genes were upregulated and 171 genes were downregulated. Interestingly, 303 genes that were altered in 2-month-old N101D lenses were also altered in 4-month-old in N101D lenses. The complete list of altered genes is reported as Supplementary Excel File S1.
To further understand the biologic relevance of the differentially expressed genes in lenses of N101D mice, the IPA was used to explore their relationships and known annotations. Networks affected in 2- and 4-month-old N101D lenses relative to WT mice are listed in Table 1. Notably, gene expression was the most affected network with the highest scores of 29 and 35 in 2- and 4-month-old lenses, respectively. Other top networks affected were found to be similar in 2- and 4-month-old lenses. In 2- and 4-month-old N101D lenses, the top three biofunction networks affected are listed in Table 2. Interestingly, the common biofunction networks affected in 2- and 4-month-old lenses were associated with overall cell structure and assembly. This is consistent with our earlier phenotypic characterization of N101D lenses showing disrupted cell structure and disordered packing of fiber cells.32 The top canonical pathways affected in 2- and 4-month-old N101D lenses that were significant to the selected data set are listed in Table 3. In accordance with the above observations, the top affected biofunction network in N101D lenses was cellular assembly and organization; nearly all top canonical pathways affected were related to cellular assembly and organization. In 2-month-old lenses, the tight junction was the top canonical pathway with a 31/161 molecule ratio (molecule ratio was determined by dividing the number of differentially expressed genes from the data set that map to that pathway by the total number of molecules that exist in that canonical pathway). Again, this observation is consistent with our earlier SEM results,32 in which fiber cells from 5-months and older N101D lenses exhibited altered membranes, disorganized fiber cells in parallel arrays, and loss of ball-and-socket interdigitation among fiber cells. In addition to tight junctions, the other top two canonical pathways affected in N101D were actin cytoskeleton signaling with 34/233 molecules and Rho A signaling with 23/111 molecules. Interestingly, the Rho A signaling pathway was also found among the top three canonical pathways affected in 4-month-old lenses with 18/111 molecules affected. The other top two canonical pathways affected in 4-month-old lenses were the role of Nuclear Factor of Activated T cells (NFAT) in cardiac hypertrophy with 28/196 molecules and Thrombin-signaling with 26/199 affected molecules (Table 3).
Table 1.
Top Networks Affected in Lenses of a 2-Month-Old and 4-Month-Old CRYAAN101D Relative to CRYAAWT Mice
|
Associated Network Functions |
Score |
Affected Genes |
|
| 2 Months Old | |||
| 1 | Gene expression, decreased levels of albumin, embryonic development | 29 | 33 |
| 2 | Cell morphology, cellular development, nervous system development and function | 29 | 33 |
| 3 | Nervous system development and function, cellular assembly and organization, infectious disease | 27 | 32 |
| 4 | Gene expression, cellular development, cellular growth and proliferation | 26 | 31 |
| 5 | Cell-to-cell signaling and interaction, cellular assembly and organization, cellular function and maintenance | 26 | 31 |
| 4 Months Old | |||
| 1 | Gene expression, cardiovascular system development and function, cellular development | 35 | 33 |
| 2 | Decreased levels of albumin, gene expression, cellular assembly and organization | 30 | 31 |
| 3 | Cellular assembly and organization, cellular function and maintenance, cancer | 30 | 31 |
| 4 | Gene expression, cellular assembly and organization, cellular compromise | 29 | 30 |
| 5 | Cell cycle, cell death, cancer | 23 | 27 |
Networks significantly affected in 2- and 4-month-old CRYAAN101D lenses compared as determined by IPA. The score is based on the P value of the affected network. The network with a score of 15 or greater is considered significant.
Table 2.
Top Molecular and Cellular Functions Significantly Affected in 2- and 4-Month-Old N101D Lenses Relative to WT Lenses as Determined by Ingenuity Pathway Analysis
|
P
Value |
Number of Molecules |
|
| 2 Months Old | ||
| Cellular assembly and organization | 8.50E-07–1.99E-02 | 292 |
| Cell morphology | 1.04E-10–1.62E-02 | 187 |
| Cellular function and maintenance | 1.33E-10–1.62E-02 | 183 |
| 4 Months Old | ||
| Cellular assembly and organization | 8.50E-07–1.99E-02 | 211 |
| Molecular transport | 1.97E-06–1.99E-02 | 159 |
| Cellular function and maintenance | 1.04E-09–1.62E-02 | 153 |
Table 3.
Top Canonical Pathway Significantly Affected In 2- and 4-Month-Old N101D Lenses as Determined by Ingenuity Pathway Analysis
|
P
Value |
Ratio |
|
| 2 Months Old | ||
| Tight junction signaling | 7.77E-06 | 31/161 |
| Rho A signaling | 5.66E-05 | 23/11 |
| Actin cytoskeleton signaling | 3.02E-04 | 34/233 |
| 4 Months Old | ||
| Role of NFAT in cardiac hypertrophy | 3.77E-06 | 28/196 |
| Thrombin signaling | 7.82E-05 | 26/199 |
| Rho A signaling | 1.05E-04 | 18/111 |
Pathways with P value less than 0.05 defined as significant.
The Expression of αAN101D Transgene Affects Proliferation Differentiation and Denucleation of LEC in N101D Lenses
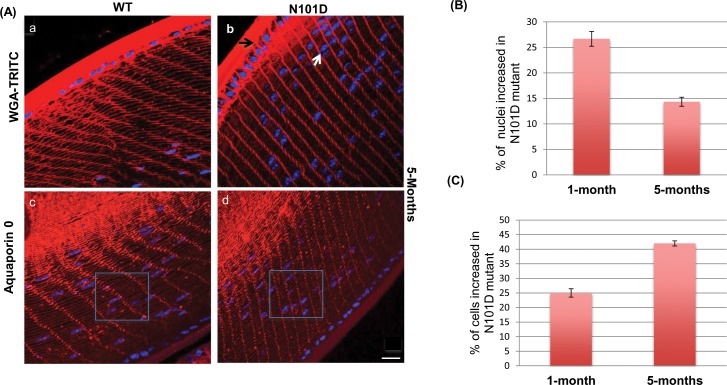
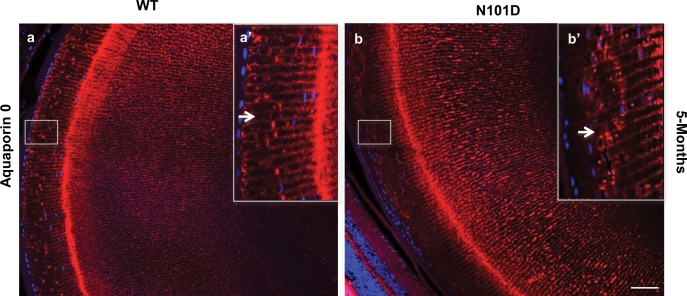
To further characterize the cellular assembly and organization defects in N101D lenses during development, eye sections from 1- and 5-month-old N101D and WT mice lenses were stained with wheat germ agglutinin-conjugated to TRITC, and also immunostained with antiaquaporin-0. Since LECs constantly divide, elongate, migrate, and terminally differentiate into LFCs during the development, immunohistochemical analysis should able to identify any defects in these processes. The results showed that in N101D mouse lenses the terminal differentiation processes were disturbed (Fig. 2). The defects included: (1) compared to WT, the thickness of the anterior epithelium increased in N101D (black arrow in Fig. 2A, b), (2) the nuclei in LEC and LFC were narrow and elongated in WT lenses, whereas in N101D lenses the nuclei in LEC and LFC were bigger, round, and more compact, (3) the number of fiber cells that retained nuclei in the outer cortex of N101D was increased (Fig. 2A, b, Supplementary Fig. S1B). In 1- and 5-month-old lenses, there were 26% and 15% more fiber cells that retained nuclei, respectively, compared with the fiber cells in WT lenses (Figs. 2A, b and 2B), (4) the number of fiber cells in N101D lenses was increased compared with WT lenses. At 1- and 5-months of age there were 25% and 42% increases, respectively, in cell number in N101D lenses compared to the WT lenses (Figs. 2A, d, and 2C, Supplementary Fig. S1A), (5) At the transition zone, the expression of aquaporin-0, a marker for LFC differentiation37 was slightly decreased in N101D lenses relative to WT lenses (Fig. 3, b). Wild-type lenses showed strong aquaporin-0 immunostaining from the equator to the interior of the lens. In contrast, in N101D lenses, the aquaporin-0 immunostaining was very weak at the equator below the epithelium (Fig. 3, compare inset a' and b', indicated by arrows), but at the interior of the lens, aquaporin-0 immunostaining in N101D was comparable to immunostaining in WT lenses (Fig. 3), and (6) in addition, the lens-remodeling zone (RZ,38 a zone consisting of matured fiber cells with convoluted membranes that lack regular hexagonal shape) in the WT lenses spanned closer to the epithelium at both 1- (Supplementary Fig. S2, RZ in a and c) and 5-months of age (Supplementary Fig. S2, RZ in e and g) relative to a farther shift toward the center of the lenses in N101D at 1- (Supplementary Fig. S2, RZ in b and d) and 5-months of age (Supplementary Fig. S2, RZ in f and h). Together, these results suggest that N101D lenses have LEC proliferation, differentiation, and denucleation defects.
Figure 2.
Visualization of cellular assembly and organization in WT and N101D lenses by membrane labeling with WGA-TRITC and aquaporin-0 (A). Five-month-old equatorial sections were stained with WGA-TRITC (a, b), antiaquaporin 0 antibody (c, d), counterstained the nuclei with DAPI (blue). The black arrow in (b) shows increased thickness of epithelium in N101D compared to WT (a). The white arrow in N101D shows differences in nuclei in N101D compared to WT. Inset square in (c, d) demonstrate an increased cell number in N101D compared with WT lenses. Scale bar: 60 μm. (B) Quantification of increased number of nuclei in N101D relative to WT. Total number of nuclei in WT versus N101D was counted and quantified. Results are based on four lens sections from two mice for each age. (C) Quantification of increased number of cells in N101D relative to WT. Total number of cells inside the square were counted and quantified. The represented data are from four lens sections of two mice of each age.
Figure 3.
Expression of aquaporin-0 is delayed in N101D mutant lenses. In WT lenses, the aquaporin-0 staining with antiaquaporin antibody was observed in transitional zone just bellow epithelium layer (a) and in (Inset a': enlarged area, indicated by an arrow in a'. In contrast, in the N101D mutant lenses, aquaporin-0 staining (b) and (Inset [b']: enlarged area) was observed farther away from the epithelial layer. Nuclei were counterstained with DAPI (blue). The results shown are from lens sections from two animals per age group. Scale bar: 80 μm.
Rho A GTPases Expression Is Altered in N101D Mutant Lenses
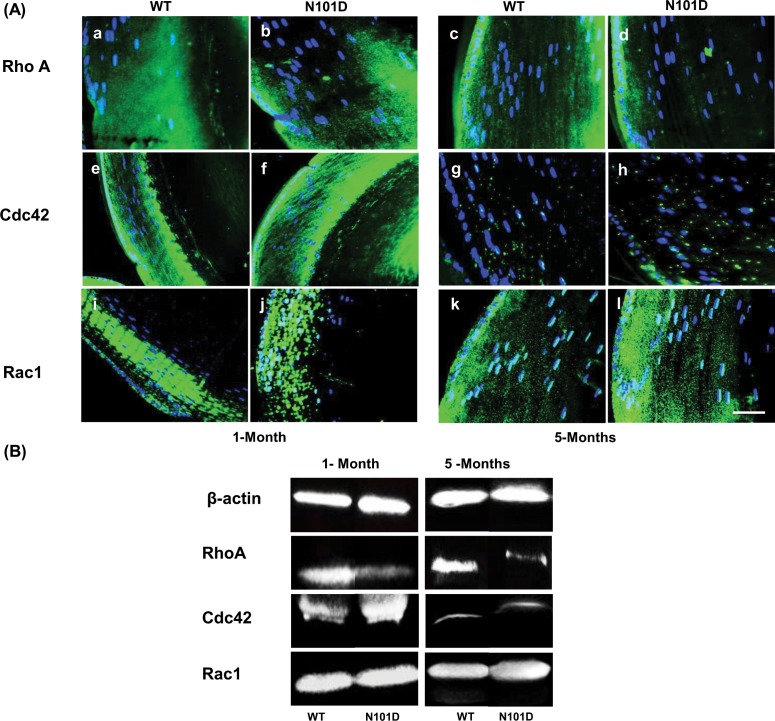
Rho A GTPases are GTP-binding proteins that play an important role in a number of cellular processes including regulation of actin cytoskeletal organization, cell–cell adhesion, cell polarity, morphogenesis, migration, vesicle trafficking, cell-cycle progression, and transcriptional activity.39–41 The previous studies on Rho A GTPases implicated the role of Rho GTPases in lens development and structural integrity and noted that their defects lead to lens abnormalities, including LEC proliferation, differentiation and elongation, polarity, shape, and packing defects.42–44 Therefore, because our whole-genome transcriptome results showed that Rho A signaling was affected in N101D mutant relative to WT lenses, and their potential role in the LEC terminal differentiation process, we analyzed the expression of the Rho A GTPases (Rho A, Cdc42, and Rac1) in 1- and 5-month-old lenses.
Rho A Immunostaining
In 1-month-old WT lenses, the epithelium and newly differentiated secondary fiber cells in the outer cortical region showed strong Rho A immunoreactivity (Fig. 4A, a). In contrast, the 1-month-old N101D mutant lenses showed a patchy and reduced Rho A immunoreactivity at the transitional zone where cells were differentiating (Fig. 4A, b). Additionally, the Rho A immunostaining in 5-months-old WT lenses was substantially reduced compared with 1-month-old WT lenses, suggesting an age-dependent reduction in the Rho A expression. Similar to 1-month-old results, 5-month-old N101D lenses also showed a weaker Rho A immunostaining (Fig. 4A, d) compared with 5-month-old WT lenses (Fig. 4A, c). Quantification of intensity by ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA) suggested that, compared with WT lenses, N101D lenses showed approximately 2-fold decrease in Rho A expression in 1- and 5-month-old lenses, respectively.
Figure 4.
Expressions of Rho A GTPases (Rho A, Cdc42, and Rac1) are altered in lenses of N101D mutant compared with WT. (A) Immunostaining of Rho A in 1- and 5-month-old N101D lenses (b, d), was decreased relative to 1- and 5-month-old WT lenses, (a, c). In contrast, the Cdc42 immunostaining in 1-month-old N10D lenses (f) was increased compared with 1-month-old WT lenses (e). Slight increase in Cdc42 immunostaining in 5-month N101D lenses (h) compared to 5-month WT lenses (g). Similarly, Rac1 immunostaining in, 1- and 5-month-old N101D lenses (j, l) increased relative to 1-and 5-month-old WT lenses (i, k). Nuclei were counter-stained with DAPI. Scale bar: 60 μm. (B) Western blot analysis was used to determine levels of Rho A, Cdc42, and Rac1 in WT and N101D lenses. The 1- and 5-month-old WT and N101D lenses were homogenized in RIPA buffer as described in Materials and Methods and probed with anti-Rho A, Cdc42, and Rac1 antibodies. Equal amounts of protein loading was confirmed by probing for anti–β-actin antibody. Four lenses from two animals for each age were used in the experiment.
Cdc42 Immunostaining
The immunostaining of Cdc42 was very strong in lens epithelium and cortical fiber cells of 1-month-old WT lenses (Fig. 4A, e), specifically at the basal end of elongating secondary fiber cells. In 1-month-old N101D lenses, the Cdc42 immunoreactivity was relatively stronger (Fig. 4A, f), and the staining region was much wider relative to the WT lens. Similar to Rho A expression, the total amount of Cdc42 protein expression in 5-month-old WT and N101D lenses was reduced. In 5-month-old N101D lenses, the expression of Cdc42 was slightly increased (Fig. 4A, h) compared with WT (Fig. 4A, g). ImageJ quantification of fluorescence intensity indicated that, compared with WT, the N101D lenses show approximately 2- and 1.5-fold increases in Cdc42 staining in 1- and 5- month-old lenses, respectively.
Rac1 Immunostaining
In 1-month-old WT lenses, the epithelium and secondary fiber cells in the outer cortical region showed uniform Rac1 immunostaining (Fig. 4A, i). In contrast, the lenses of 1-month-old N101D mutant showed increasing, but patchy, Rac1 staining compared to WT lenses (Fig. 4A, j). Similarly, in 5-month-old N101D lenses, Rac1 immunostaining was much stronger (Fig. 4A, l) compared with 5-month-old WT lenses (Fig. 4A, k). ImageJ quantification of Rac1 staining intensity indicated that compared to WT, N101D lenses showed approximately 2- and 4-fold increases in Rac1 staining in 1-and 5-month-old lenses, respectively.
To quantify the altered expression of the Rho GTPases proteins (Rho A, and Cdc42 and Rac1) in N101D lenses, proteins from 1- and 5-month-old lenses were analyzed by Western blot (Fig. 4B). Compared with age-matched WT lenses, the N101D lenses showed approximately 2 ± 0.5-fold reduction in Rho A protein expression, 3- and less than 2-fold increase in Cdc42 expression, and 2- and 4-fold increases in Rac1 expression in 1- and 5-month-old lenses, respectively. These results were mostly consistent with the above immunohistochemical data.
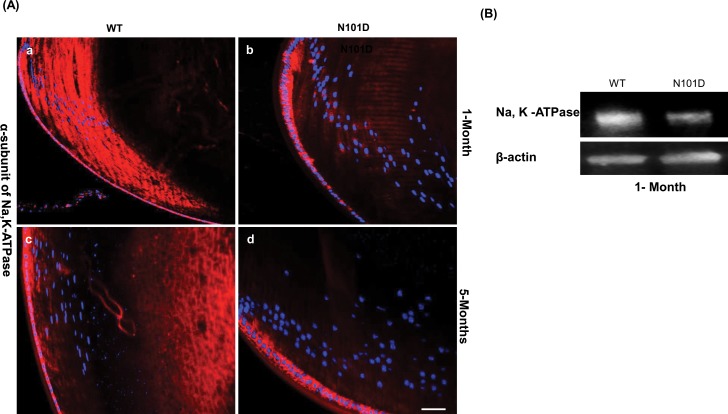
The Expression of Na,K-ATPase Is Impaired in Lenses of N101D Mutant
Next, we wanted to find out how the expression of αAN101D transgene alters Rho A GTPase expression and function. It has been shown in MDCK cells that the Rho A GTPases are essential for the assembly and function of tight junction and that activity of Rho A is modulated by Na,K-ATPase.45 Interestingly, our present results also suggest that tight junction and Rho GTPases activity are affected in N101D lenses. Additionally, our RNA sequence data indicated that the expression of α2-subunit of Na,K-ATPase is one of the most affected molecular transporter genes (333-fold downregulated) in 2-month-old N101D lenses compared with age-matched WT lenses. To further confirm altered Na,K-ATPase expression at the protein level, immunohistchemical analysis was carried out using an antibody against the α- and β-subunits of Na,K-ATPase on 1-and 5-month-old N101D and WT lenses. The results showed a large difference in intensity of immunostaining of the α-subunit Na,K-ATPase in 1-month-old N101D lenses compared with 1-month-old WT lenses (Fig. 5A). Quantification of immunostaining intensity by Image quant suggested that the expression of α-subunits of Na,K-ATPase in 1-month-old N101D was decreased by approximately 6-fold (Fig. 5A, b) compared with 1-month-old WT lenses (Fig. 5A, a). Additionally, Western blot analysis of 1-month-old lens homogenates confirmed a decrease in α-subunits of Na,K-ATPase expression N101D lenses compared with WT lenses (Fig. 5B). Furthermore, the expression of the β-subunit of Na,K-ATPase was also decreased (~3-fold) in 1-month-old N101D lenses (Supplementary Fig. S3). Consistent with RNA sequence data at 5-month-old lenses, there was no significant difference in immunostaining of Na,K-ATPase in N101D (Fig. 5A, d versus WT lenses Fig. 5A, c).
Figure 5.
Expression of Na,K-ATPase is downregulated in N101D lenses. (A) Immunofluorescent staining of α-subnit of Na,K-ATPase (red) in 1-month-old N101D lenses (b) was drastically reduced compared with 1-month-old WT lenses (a). No significant difference in immumostaining of Na,K-ATPase in 5-month-old WT (c) and N101D lenses (d) was observed. Nuclei were counterstained with DAPI. Scale bar: 60 μm. (B) Western blot analysis confirming the decreased expression of Na, K-ATPase in 1-month-old N101D compared with 1-month-old WT lenses. Lens homogenates from 1-month-old N101D and 1-month-old WT mice (n = 2) were used for the immunoblot. β-actin was used as a loading control.
Discussion
In this study, we demonstrated that the expression of αAN101D transgene results in disruption of lens morphogenesis, which is characterized by defective LEC proliferation, elongation, and lens fiber cell denucleation. The most notable defects of expression of αAN101D transgene during development is that αAN101D transgene renders LECs to become hyperproliferative, leading to an increased cell number in the outer cortex of N101D lenses compared with the number of cells in the corresponding region of WT lenses. The increased proliferation delays LEC differentiation and impairs the denucleation process in N101D lenses. The undegraded nuclei and accumulated nuclear debris in fiber cells in the inner cortex and in the nuclear region of lens might, therefore, responsible for development of opacity in N101D lenses.
RNA sequence data indicated that a large number of genes were differentially expressed in N101D lenses compared with WT lenses. Analysis of differentially expressed genes in 2- and 4-month-old N101D lenses by Ingenuity Pathway suggested that most of the genes affected were from gene expression, cellular assembly, and organization networks. The biofunctions affected were cellular structures and maintenance and molecular transport (Tables 1, 2). The top canonical pathways affected were tight junction signaling and Rho A signaling (Table 3). Thus, RNA sequence results were largely consistent with earlier phenotypic and SEM analysis of LEC and LFC from 5-month and older N101D mutant lenses.32
The Expression of αAN101D Transgene Induce Proliferation, Differentiation, and Denucleation Defects
The characteristic shape and organization of a lens mainly depends on synchronized proliferation and differentiation of LECs. Any abnormality in proliferation, migration, and elongation of LECs disrupts the symmetry and alters the development and synchronized differentiation.1,2 Once the self-renewing LECs exit from the cell cycle at the lens equator, they commit to becoming LFCs, which intercalate to form a highly ordered meridional row of elongating fibers.11 These cells then migrate and differentiate into mature fiber cells by eliminating light-scattering nuclei and other organelle and intracellular structures. In N101D mutants, these processes were disrupted, as evident from the labeling of the membrane with WGA-TRITC and antiaquaporin 0 antibody (Figs. 2, 3). The equatorial LFCs retained their proliferative ability and did not commit to the transition zone to become fiber cells; and as a result, they lacked the expression of the fiber cell marker aquaporin 0. Compared with WT, the transition zone in N101D lenses was shifted more interior from the epithelium. The increased proliferation delayed the LEC differentiation process and resulted in a relatively larger outer cortical region with a greater number of cells in the outer cortex. In addition, our preliminary results from cell culture demonstrate that LECs derived from N101D transgenic mice were found to be hyperproliferative (unpublished data). Taken together, these data confirm altered cell-cycle progression of LEC in N101D mutants. Interestingly, previous reports have also shown the hyperproliferative nature of LEC derived from αB-knockout mice.46 These results suggest an important role of αA- and αB-crystallins in cell-cycle regulation. However, the LEC derived from the αA-knockout mice show opposite results (i.e., 50% slower growth compared with LEC derived from WT mice).22 Therefore, it is unclear how a αAN101D mutation would induce hyperproliferation of LECs, whereas a loss of αA crystallin reduces LEC proliferation. It is possible that in N101D mutants the hyperproliferation of LEC is not due to a direct effect of αA mutation on cell cycle per se, but rather due to an effect of the mutation on upstream molecular players such as Rho A GTPases, which are the master switches that control the cell-cycle progression. In support, our results suggest that the expressions of Rho A GTPases are altered in N101D mutants. Furthermore, RNA sequencing data indicated that in 2-month-old N101D lenses, there was a 3.9-fold decrease in Rho A expression, and a 4.7-fold increase in Rac1 expression. Consistent with RNA sequencing data, the immunohistochemical and immunoblot results data also indicated an approximate 2-fold downregulation of Rho A and 2- to 4-fold upregulation of Rac1 protein in N101D lenses relative to WT lenses (Fig. 4).
Rho A GTPases (Rho A, Cdc42, and Rac1) are crucial regulators of actin cytoskeletal reorganization, actin polymerization, cell adhesion, and cell-cycle progression. The importance of Rho GTPases in LEC differentiation has been shown in Sfrp2 transgenic mice47 and in bovine Rho GDP dissociation inhibitor (RhoGDIα) studies.43 Some of the terminal differentiation defects of N101D lenses were similar to bovine RhoGDIα transgenic mice, such as the hyperproliferative nature of LEC.43 In bovine RhoGDIα transgenic mice, the active Rho GTPases including Rho A, Cdc42, and Rac1 activities were reduced; however, in N101D transgenic mice the expression of Rho A is reduced, whereas the expressions of Cdc42 and Rac1 were increased relative to WT lenses (Figs. 4A, 4B). This suggests an antagonistic regulation between Rho A and Cdc42/Rac1 in N101D lens fibers. Such a regulatory relationship was also documented in other systems such as N1E-115 neuroblastoma, MCDK, NIH 3T3, and SV80 cells,48–50 suggesting a balance between activities among Rho A GTPases. Although, we have not determined the Rho A GTPases activity in N101D lenses, however, since the activity of a protein is also regulated by controlling its amount, the altered Rho A GTPase protein expressions in N101D lenses likely to alter their GTPase activities.
An overexpression of Rho GTPases has been demonstrated in a number of human tumors including lung, colon, advanced breast cancer, upper and lower urinary tract, and pancreatic adeno carcinoma.51–53 However, inhibition of Rho GTPase activity is also shown to promote progenitor cell proliferation.54 The balanced activity between Rho A and Rac was shown to exist in pancreatic cancer.55 Taken together, these results suggest that a proper balance of Rho GTPase expression and activities is crucial for cellular functions.
Increased Association of αAN101D Crystallin With the Membrane May Be Responsible for the Decrease in Expression of the Na,K-ATPase in N101D Mutant Lenses
αA-crystallin is known to associate with the LFC membrane, and its association increases in aging, or in diseased conditions.56 In our previous study, based on increased association of αAN101D mutant crystallin with membrane and loss of interdigitation among fiber cells in N101D mutants, we proposed that circulation of fluid might be perturbed in N101D lenses.32 Consistent with that observation, our present RNA sequence data also implicate that the circulation of fluid is most likely affected in N101D, since genes involved in molecular transport and tight junction were highly affected in these lenses compared with WT lenses. Moreover, RNA sequence data also indicated that the expression of Na,K-ATPase (ATP1A2) mRNA was highly downregulated (333-fold) in 2-month-old N101D lenses. Immunohistochemical analysis by antibody against the α-subunit of Na,K-ATPase further confirmed a 6-fold decreased expression of the pump in N101D lenses (Fig. 5).
Na,K-ATPase is an important regulator of intracellular electrolyte levels in all mammalian cells. It is one of the well-characterized transporters that regulate tight junction functions.57 Various mechanisms have been shown to be involved in regulation of Na,K-ATPase function. However, long-term regulation mainly involves changes in gene expression.58 Na,K-ATPase has been shown to interact with a multitude of proteins, from ion transporters to structural and cell signaling molecules. The physical and functional coupling between Na,K-ATPase and Na+/Ca2+ is important for regulation of intercellular Ca2+. Moreover, in MDCK cells it has been shown that inhibition of Na,K-ATPase activity prevented the formation of tight junction and also inhibited Rho A GTPase activity.59–61
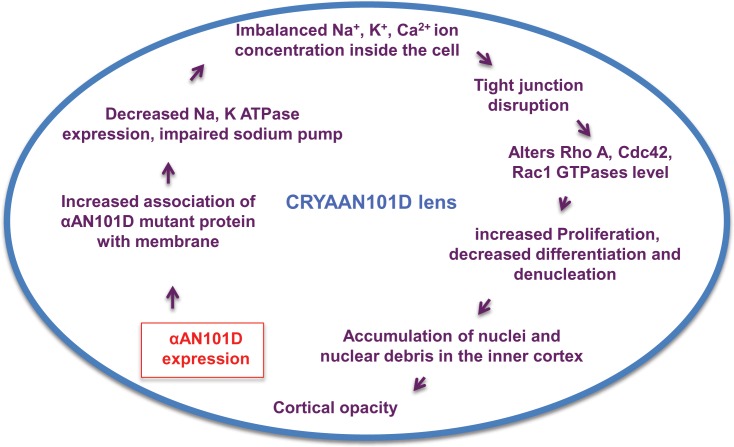
Based on the above observations and our results from N101D lenses, we propose a mechanistic model (Fig. 6) to explain the development of opacity in lenses of N101D mutants. According to this model, an increased association of αAN101D crystallin with the membrane may be responsible for the decreased expression of the major membrane ion-transporter, Na,K-ATPase. The decreased expression and activity of this pump may affects tight junction assembly, and therefore would alter intercellular ion homeostasis and Rho A GTPases expression. The consequence of altered Rho GTPases is a series of terminal differentiation defects including altered proliferation and cell-cycle progression of LEC and LFCs. The delayed LEC differentiation and denucleation ultimately lead to cortical lens opacity in N101D lenses. As described above, since Na,K-ATPase has been shown to interact with a multitude of proteins including signaling molecules, it will be interesting to investigate in the future whether the expression of αAN101D transgene impairs only Na,K pump-related activities, or if it also impairs the interaction with signaling molecules. Focused study on LEC derived from N101D transgenic mice should be able to differentiate the defects derived from impaired interaction of Na,K-ATPase with signaling molecules from defects associated with impaired pump activities.
Figure 6.
Schematic representation of a molecular mechanism involved in development of lens opacity in N101D transgenic mice. In N101D lenses, an increased association of αAN101D mutant crystallin with the membrane may responsible for the decreased expression of membrane bound transporter Na,K-ATPase. The impaired Na,K-ATPase pump activity is most likely to imbalance the ion concentration inside the cell and affects tight junction assembly and Rho A GTPases expression. The altered Rho A GTPase expression in turn causes a series of terminal differentiation defects, such as increased proliferation and decreased differentiation of LEC and decreased denucleation of LFC. This defective terminal differentiation process leads to accumulation of nuclei and nuclear debris in the inner cortex of the lens that eventually causes lens opacity.
We strongly believe that the observed defects in N101D mutant lenses are mainly due to the expression of deamidated alpha A crystallin. However, one of the caveat of our study is that we have analyzed only (1.B2) transgenic lines. To establish the relationship between the ratio of deamidated alpha A crystalline and the changes observed, and to rule out the possibility that those observed defects in N101D are not due to the site of transgene insertion, but, rather, are specific to the expression of deamidated alpha A crystallin, investigation on other N101D transgenic lines (either 1.1, 2.21, or 3.42) is required.
Acknowledgments
The authors thank the technical assistance received from core facilities of the Transgenic Mouse high Resolution Imaging and Heflin Genomics (David Crossman, PhD) at the University of Alabama at Birmingham (Birmingham, AL, USA).
Supported by National Institutes of Health (Bethesda, MD, USA) Grants EY06400 and P30-EY03039.
Disclosure: S.M. Hegde, None; K. Srivastava, None; E. Tiwary, None; O.P. Srivastava, None
References
- 1. Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001; 17: 255–296 [DOI] [PubMed] [Google Scholar]
- 2. Piatigorsky J. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation. 1981; 19: 134–153 [DOI] [PubMed] [Google Scholar]
- 3. Lovicu FJ, McAvoy JW. Growth factor regulation of lens development. Dev Biol. 2005; 280: 1–14 [DOI] [PubMed] [Google Scholar]
- 4. Kuszak JR, Zoltoski RK, Sivertson C. Fibre cell organization in crystalline lenses. Exp Eye Res. 2004; 78: 673–687 [DOI] [PubMed] [Google Scholar]
- 5. Lee A, Fischer RS, Fowler VM. Stabilization and remodeling of the membrane skeleton during lens fiber cell differentiation and maturation. Dev Dyn. 2000; 217: 257–270 [DOI] [PubMed] [Google Scholar]
- 6. Rao PV, Maddala R. The role of the lens actin cytoskeleton in fiber cell elongation and differentiation. Semin Cell Dev Biol. 2006; 17: 698–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weber GF, Menko AS. Actin filament organization regulates the induction of lens cell differentiation and survival. Dev Biol. 2006; 295: 714–729 [DOI] [PubMed] [Google Scholar]
- 8. Beebe DC, Vasiliev O, Guo J, Shui YB, Bassnett S. Changes in adhesion complexes define stages in the differentiation of lens fiber cells. Invest Ophthalmol Vis Sci. 2001; 42: 727–734 [PubMed] [Google Scholar]
- 9. Delaye M, Tardieu A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature. 1983; 302: 415–417 [DOI] [PubMed] [Google Scholar]
- 10. Wride MA. Lens fibre cell differentiation and organelle loss: many paths lead to clarity. Philos Trans R Soc Lond B Biol Sci. 2011; 366: 1219–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp Eye Res. 2009; 88: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu CD, Kymes S, Petrash JM. A transgenic mouse model for human autosomal dominant cataract. Invest Ophthalmol Vis Sci. 2006; 47: 2036–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cobb BA, Petrash JM. Structural and functional changes in the alpha A-crystallin R116C mutant in hereditary cataracts. Biochemistry. 2000; 39: 15791–15798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lyu J, Kim JA, Chung SK, Kim KS, Joo CK. Alteration of cadherin in dexamethasone-induced cataract organ-cultured rat lens. Invest Ophthalmol Vis Sci. 2003; 44: 2034–2040 [DOI] [PubMed] [Google Scholar]
- 15. Yu XS, Jiang JX. Interaction of major intrinsic protein (aquaporin-0) with fiber connexins in lens development. J Cell Sci. 2004; 117 (pt 6): 871–880 [DOI] [PubMed] [Google Scholar]
- 16. Chung J, Berthoud VM, Novak L, et al. Transgenic overexpression of connexin50 induces cataracts. Exp Eye Res. 2007; 84: 513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duncan G, Bushell AR. Ion analyses of human cataractous lenses. Exp Eye Res. 1975; 20: 223–230 [DOI] [PubMed] [Google Scholar]
- 18. Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci U S A. 1992; 89: 10449–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993; 268: 1517–1520 [PubMed] [Google Scholar]
- 20. Aoyama A, Frohli E, Schafer R, Klemenz R. Alpha B-crystallin expression in mouse NIH 3T3 fibroblasts: glucocorticoid responsiveness and involvement in thermal protection. Mol Cell Biol. 1993; 13: 1824–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mao YW, Liu JP, Xiang H, Li DWC. Human [alpha]A- and [alpha]B-crystallins bind to Bax and Bcl-XS to sequester their translocation during staurosporine-induced apoptosis. Cell Death Differ. 2004; 11: 512–526 [DOI] [PubMed] [Google Scholar]
- 22. Andley UP, Song Z, Wawrousek EF, Bassnett S. The molecular chaperone alphaA-crystallin enhances lens epithelial cell growth and resistance to UVA stress. J Biol Chem. 1998; 273: 31252–31261 [DOI] [PubMed] [Google Scholar]
- 23. Lee JS, Liao JH, Wu SH. Chiou SH: alpha-crystallin acting as a molecular chaperonin against photodamage by UV irradiation. J Protein Chem. 1997; 16: 283–289 [DOI] [PubMed] [Google Scholar]
- 24. Mehlen P, KretzRemy C, Preville X, Arrigo AP: Human hsp27, Drosophila hsp27 and human alpha B-crystallin expression-mediated increase in glutathione is essential for the protective activity of these proteins against TNF alpha-induced cell death. Embo J. 1996; 15: 2695–2706 [PMC free article] [PubMed] [Google Scholar]
- 25. Wilmarth PA, Tanner S, Dasari S, et al. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: Does deamidation contribute to crystallin insolubility? J Proteome Res. 2006; 5: 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma Z, Hanson SR, Lampi KJ, David LL, Smith DL, Smith JB. Age-related changes in human lens crystallins identified by HPLC and mass spectrometry. Exp Eye Res. 1998; 67: 21–30 [DOI] [PubMed] [Google Scholar]
- 27. Takemoto L, Boyle D: Deamidation of alpha-A crystallin from nuclei of cataractous and normal human lenses. Mol Vis. 1999; 5: 2 [PubMed] [Google Scholar]
- 28. Takata T, Oxford JT, Demeler B, Lampi KJ. Deamidation destabilizes and triggers aggregation of a lens protein, beta A3-crystallin. Protein Sci. 2008; 17: 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takata T, Woodbury LG, Lampi KJ. Deamidation alters interactions of beta-crystallins in hetero-oligomers. Mol Vis. 2009; 15: 241–249 [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R, Srivastava OP. Deamidation affects structural and functional properties of human alphaA-crystallin and its oligomerization with alphaB-crystallin. J Biol Chem. 2004; 279: 44258–44269 [DOI] [PubMed] [Google Scholar]
- 31. Gupta R, Srivastava OP. Effect of deamidation of asparagine 146 on functional and structural properties of human lens alphaB-crystallin. Invest Ophthalmol Vis Sci. 2004; 45: 206–214 [DOI] [PubMed] [Google Scholar]
- 32. Gupta R, Asomugha CO, Srivastava OP. The common modification in alphaA-crystallin in the lens, N101D, is associated with increased opacity in a mouse model. J Biol Chem. 2011; 286: 11579–11592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012; 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010; 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grey AC, Li L, Jacobs MD, Schey KL, Donaldson PJ. Differentiation-dependent modification and subcellular distribution of aquaporin-0 suggests multiple functional roles in the rat lens. Differentiation. 2009; 77: 70–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim JC, Walker KL, Sherwin T, Schey KL, Donaldson PJ. Confocal microscopy reveals zones of membrane remodeling in the outer cortex of the human lens. Invest Ophthalmol Vis Sci. 2009; 50: 4304–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002; 420: 629–635 [DOI] [PubMed] [Google Scholar]
- 40. Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004; 116: 167–179 [DOI] [PubMed] [Google Scholar]
- 41. Ridley AJ. Rho GTPases and cell migration. J Cell Sci. 2001; 114 (pt 15): 2713–2722 [DOI] [PubMed] [Google Scholar]
- 42. Rao V, Wawrousek E, Tamm ER, Zigler S Jr. Rho GTPase inactivation impairs lens growth and integrity. Lab Invest. 2002; 82: 231–239 [DOI] [PubMed] [Google Scholar]
- 43. Maddala R, Reneker LW, Pendurthi B, Rao PV. Rho GDP dissociation inhibitor-mediated disruption of Rho GTPase activity impairs lens fiber cell migration, elongation and survival. Dev Biol. 2008; 315: 217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maddala R, Chauhan BK, Walker C, et al. Rac1 GTPase-deficient mouse lens exhibits defects in shape, suture formation, fiber cell migration and survival. Dev Biol. 2011; 360: 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jou TS, Schneeberger EE, Nelson WJ. Structural and functional regulation of tight junctions by RhoA and Rac1 small GTPases. J Cell Biol. 1998; 142: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andley UP, Song Z, Wawrousek EF, Brady JP, Bassnett S, Fleming TP. Lens epithelial cells derived from alphaB-crystallin knockout mice demonstrate hyperproliferation and genomic instability. FASEB J. 2001; 15: 221–229 [DOI] [PubMed] [Google Scholar]
- 47. Chen YJ, Stump RJW, Lovicu FJ, Shimono A, McAvoy JW. Wnt signaling is required for organization of the lens fiber cell cytoskeleton and development of lens three-dimensional architecture. Dev Biol. 2008; 324: 161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997; 17: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leeuwen FN, Kain HE, Kammen RA, Michiels F, Kranenburg OW, Collard JG. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J Cell Biol. 1997; 139: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sander EE, Collard JG. Rho-like GTPases: their role in epithelial cell–cell adhesion and invasion. Eur J Cancer. 1999; 35: 1905–1911 [DOI] [PubMed] [Google Scholar]
- 51. Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci U S A. 2000; 97: 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fritz G, Just I, Kaina B. Rho GTPases are over-expressed in human tumors. Int J Cancer. 1999; 81: 682–687 [DOI] [PubMed] [Google Scholar]
- 53. Kamai T, Yamanishi T, Shirataki H, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004; 10: 4799–4805 [DOI] [PubMed] [Google Scholar]
- 54. Ghiaur G, Lee A, Bailey J, Cancelas JA, Zheng Y, Williams DA. Inhibition of RhoA GTPase activity enhances hematopoietic stem and progenitor cell proliferation and engraftment. Blood. 2006; 108: 2087–2094 [DOI] [PubMed] [Google Scholar]
- 55. Guo X, Wang M, Jiang J, et al. Balanced Tiam1-rac1 and RhoA drives proliferation and invasion of pancreatic cancer cells. Mol Cancer Res. 2013; 11: 230–239 [DOI] [PubMed] [Google Scholar]
- 56. Su SP, McArthur JD, Friedrich MG, Truscott RJ, Aquilina JA. Understanding the alpha-crystallin cell membrane conjunction. Mol Vis. 2011; 17: 2798–2807 [PMC free article] [PubMed] [Google Scholar]
- 57. Rajasekaran SA, Palmer LG, Moon SY, et al. Na, K-ATPase activity is required for formation of tight junctions, desmosomes, and induction of polarity in epithelial cells. Mol Biol Cell. 2001; 12: 3717–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Therien AG, Blostein R. Mechanisms of sodium pump regulation. Am J Physiol Cell Physiol. 2000; 279: C541–C566 [DOI] [PubMed] [Google Scholar]
- 59. Kometiani P, Li J, Gnudi L, Kahn BB, Askari A, Xie Z. Multiple signal transduction pathways link Na+/K+-ATPase to growth-related genes in cardiac myocytes. The roles of Ras and mitogen-activated protein kinases. J Biol Chem. 1998; 273: 15249–15256 [DOI] [PubMed] [Google Scholar]
- 60. Tian J, Cai T, Yuan Z, et al. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol Biol Cell. 2006; 17: 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dostanic I. Schultz Jel J, Lorenz JN, Lingrel JB. The alpha 1 isoform of Na, K-ATPase regulates cardiac contractility and functionally interacts and co-localizes with the Na/Ca exchanger in heart. J Biol Chem. 2004; 279: 54053–54061 [DOI] [PubMed] [Google Scholar]