Abstract
Coordinated gene expression controlled by long-distance enhancers is orchestrated by DNA regulatory sequences involving transcription factors and layers of control mechanisms. The Shh gene and well-established regulators are an example of genomic composition in which enhancers reside in a large desert extending into neighbouring genes to control the spatiotemporal pattern of expression. Exploiting the local hopping activity of the Sleeping Beauty transposon, the lacZ reporter gene was dispersed throughout the Shh region to systematically map the genomic features responsible for expression activity. We found that enhancer activities are retained inside a genomic region that corresponds to the topological associated domain (TAD) defined by Hi-C. This domain of approximately 900 kb is in an open conformation over its length and is generally susceptible to all Shh enhancers. Similar to the distal enhancers, an enhancer residing within the Shh second intron activates the reporter gene located at distances of hundreds of kilobases away, suggesting that both proximal and distal enhancers have the capacity to survey the Shh topological domain to recognise potential promoters. The widely expressed Rnf32 gene lying within the Shh domain evades enhancer activities by a process that may be common among other housekeeping genes that reside in large regulatory domains. Finally, the boundaries of the Shh TAD do not represent the absolute expression limits of enhancer activity, as expression activity is lost stepwise at a number of genomic positions at the verges of these domains.
Keywords: Sonic hedgehog (Shh), Enhancers, Long-range regulation, Topological domains, Sleeping beauty transposon, Mouse
INTRODUCTION
The regulatory architecture of highly regulated genes such as those involved in controlling developmental processes has been particularly difficult to define. Identification of regulatory elements by sequence alone has proven difficult; most progress has been made using multispecies conservation and functional analysis such as transgenic mice and DNase sensitivity. In general the regulatory composition of a single gene may consist of multiple elements that reside within introns of the gene and extend to large distances at either end occupying positions in gene deserts and even neighbouring genes. This composition orchestrates layers of control mechanisms, posing a number of questions about the capacity of the regulatory components within these complex regulatory domains.
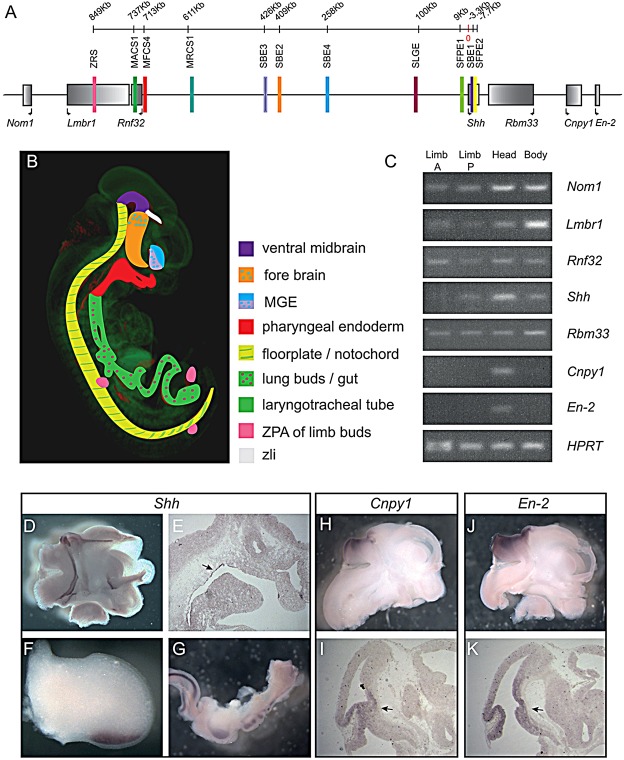
Sonic hedgehog (Shh) is an example of a developmental gene dependent on long-range gene regulatory mechanisms for its full spatiotemporal pattern of expression. With the coding region lying adjacent to a large gene desert, the expression is controlled by a group of cis-regulators, many of which were identified by mouse transgenic reporter assays. Epstein et al. (1999) identified two intronic enhancers and one outside the gene, lying ∼9 kb upstream of Shh. These potential enhancers of Shh activate lacZ reporter expression within the ventral midline of the spinal cord and hindbrain and the ventral midbrain and caudal region of the diencephalon. A further study, using comparative sequence analysis and mouse reporter assays, uncovered three forebrain enhancers located 300-450 kb upstream of Shh (Jeong et al., 2006). Comparative genomics was also used to identify another more distal cluster of three cis-regulatory elements ranging approximately 600-900 kb upstream of Shh, one of which lies within an intron of the Rnf32 gene. This cluster of regulatory elements directs regional expression of Shh in a co-linear pattern along the anteroposterior body axis within the epithelial linings of the oral cavity to the hindgut (Sagai et al., 2009). Shh initiation and spatial expression control within the posterior margin of the limb bud called the zone of polarizing activity (ZPA) is also regulated by a cis-regulatory element designated the ZRS (Lettice et al., 2002, 2003; Sagai et al., 2004, 2009). The ZRS is another intragenic regulator and is found within Lmbr1, a gene that lies within a cluster of genes flanking the desert. ZRS is the farthest known enhancer for Shh, acting over a distance of nearly 900 kb. Overall the Shh genomic regulatory domain comprises nearly 900 kb, containing a number a regulators extending into two unrelated genes, the Lmbr1 and Rnf32 genes. (The entire locus and Shh expression pattern are summarised diagrammatically in Fig. 1A,B.)
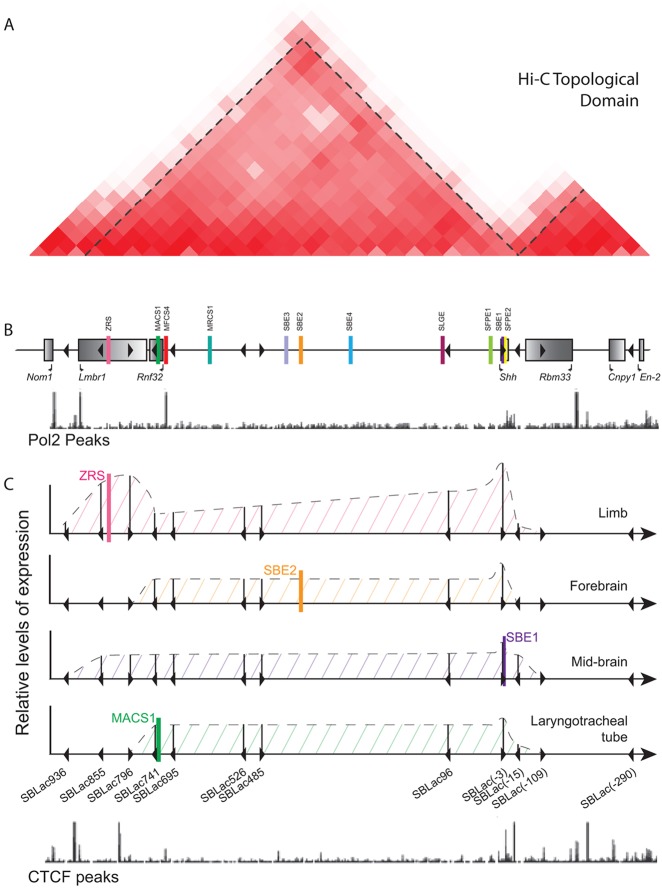
Fig. 1.
Expression of genes within the Shh regulatory locus. (A) The genes within the interval from Nom1 to En2 are marked by the grey rectangles, shaded from dark to light in the 5′ to 3′ orientation. Known enhancers are shown as coloured bars. (B) Schematic illustration of the multiple sites of Shh expression in the E11.5 embryo; the colours used match the relevant enhancers shown in their genomic context in A. Expression in the ZLI is driven by an unknown enhancer. (C) Results of a study of the embryonic expression at E11.5 of those genes conducted by RT-PCR [in anterior and posterior halves of limb buds, in carcass (body) and isolated heads]. In situ hybridisation, in E11.5 embryos are shown for Shh in whole mount in a bisected head (D), limb (F) and isolated gut (G), and in tissue sections showing expression in the epithelial lining oral cavity (E). Expression of Cpny1 and En2 are shown in whole-mount bisected heads (H,J) and in sections (I,K). Expression within the midbrain-hindbrain boundary is marked by arrows.
Disruption of the long-range cis-regulation of Shh causes human congenital defects. Chromosomal rearrangements that disrupt cis-regulators act as autosomal dominant mutations to cause a highly variable holoprosecencephaly phenotype (Belloni et al., 1996), whereas chromosomal rearrangements that cause duplications of the ZRS are associated with triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV (Klopocki et al., 2008; Sun et al., 2008). Additionally, point mutations in the ZRS result in ectopic anterior expression of Shh, which is a major cause of preaxial polydactyly type 2 (PPD2) (Lettice and Hill, 2005). Furthermore, a large-scale intrachromosomal rearrangement that places Shh in a novel regulatory environment, called ‘enhancer adoption’, has been demonstrated to result in a severe limb phenotype in humans (Lettice et al., 2011). A similar gain of regulatory information may explain the brachydactyly phenotype observed in the mouse Dsh (short digit) mutant (Niedermaier et al., 2005).
In order to understand coordinated regulatory events responsible for Shh expression in the embryo, we modified the endogenous locus. By exploiting the enhancer monitoring aspect of the Local Hopping Enhancer Detection System (LHED) (Kokubu et al., 2009) based on the Sleeping Beauty (SB) transposon, we specifically engineered the chromosomal region containing the Shh gene. In this study, we targeted the transposon vector containing the lacZ reporter gene into the locus then utilised its transposition capacity to insert extensively throughout the region. This approach enabled us to map enhancer activity throughout the Shh regulatory domain to determine a potential relationship between enhancer activity and its position within the locus and elucidate on a chromosomal scale the regulatory events manifested over a long range.
RESULTS
Surveillance of the Shh regulatory domain
In order to examine the activity of the Shh regulatory complex and the long-range activity of cis-acting enhancers, we chose to insert reporter genes interspersed throughout the Shh regulatory domain on chromosome 5. Vectors were designed based on the LHED strategy (Kokubu et al., 2009), which carries the SB inverted repeats (IRs)/direct repeats (DRs) and combines standard knock-in technology, and transposon based enhancer detection. As our initial concern was to investigate the activity of the most distal enhancer, the ZRS, we inserted the local hopping transposon on either side of this cis-regulator. Embryonic stem (ES) cells were generated that contained a targeted integration of the LHED vector within either of the two positions. One, called 5′ Insert, lies 5′ of the ZRS (orientation based on position relative to the Shh coding region) about 860 kb from Shh and the second, called 3′ Insert, was located on the 3′ side of the ZRS, 781 kb from Shh.
Transposon ‘hopping’ was induced in the ES cell lines by transfection of the pCMV-SB100x encoded transposase (Mates et al., 2009). Mobilisation of the transposon confers puromycin resistance, and approximately 200 puromycin-resistant colonies originating from the 5′ Insert ES cells were picked, and 400 colonies from the 3′ Insert ES cells. Reinsertion of the transposon occurred in 60% of 5′-Insert-derived colonies and in 50% of the 3′-Insert-derived colonies. Of the 325 clones where the insertion site was mapped, 50% of the insertion sites were found within chromosome 5, and of these more than 40% were within 1 Mb of the site of origin (summarised in supplementary material Table S2). In addition, the transposon insertion showed no bias in orientation in relation to the site of origin. Sequence of the insertion site from 50 ES cell clones (including those discussed in the text) were analysed for indels generated during the transposition event (data not shown) but no significant insertions or deletions were identified; thus, reinsertion of the transposon appears to be a highly accurate event.
As heterozygous embryos carrying pLHED insertions were sufficient to analyse cis-regulatory reporter function, a number of ES cell clones carrying a single insert were selected and injected into tetraploid blastocysts, which generate embryos derived wholly from the ES cells. At E11.5, these embryos were analysed for enhancer activity by examining lacZ expression from the transposed transgene.
Relative expression activity within the gene desert
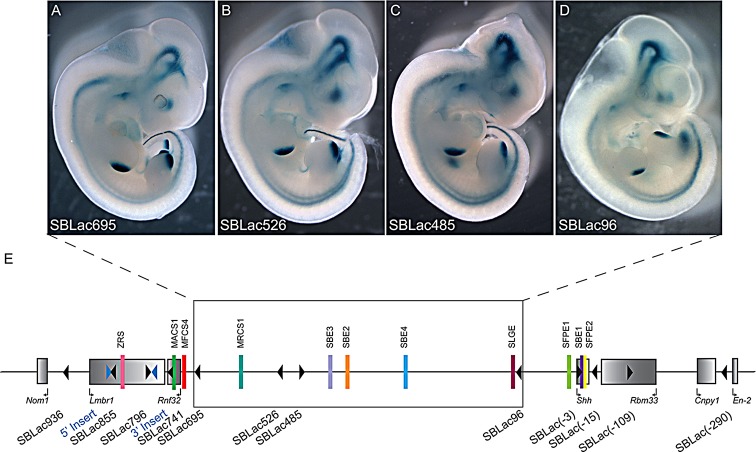
Initially, an ES cell clone was chosen if the SB transposon insertion site was situated within the gene desert, between the Shh and Rnf32 genes. The gene desert covers ∼730 kb of DNA containing a number of well-characterised Shh enhancers responsible for expression in the brain, the floor plate of the neural tube and the epithelial lining of the pharynx and oral cavity. To systematically analyse regulatory activity across this large intergenic region, four ES cell clones were selected that carried reporter genes inserted within the gene desert. Each insert is called SBLac (using the convention of Ruf et al., 2011) and is distinguished by their distance from the transcriptional start of the Shh gene; for example, the SBLac96 (n=9) lies 96 kb from Shh and lies between SFPE1 and SBE4. The other three inserts within the desert that were analysed were SBLac485 (n=6), SBLac526 (n=7) and SBLac695 (n=7), the last sits within the cluster of gut epithelial enhancers between MRCS1 and MFCS4 (Fig. 2E). These clones enabled us to sample enhancer activity across the gene desert as a measure of both the state of the chromatin and the susceptibility of enhancers to genomic distances.
Fig. 2.
Expression of SBLac insertions within the gene desert. (A-D) Embryos derived from the ES cells containing the SBLac re-insertions within the intergenic desert depicted in E, which were harvested at E11.5 and stained for expression of β-gal. The staining observed reflects the Shh expression pattern. The diagram in E depicts the genomic interval from Nom1 to En2. In addition to the genes (grey rectangles) and known enhancers (coloured bars), the sites of the original pHLED insertions are marked by the blue triangles and the positions of the mobilised and re-inserted SBLac insertions are marked with black arrowheads. The direction in which the arrowhead points depicts the 5′-to-3′ orientation of the reporter gene. Those SBLac insertions shown in A-D have been boxed.
Tetraploid complementation embryos at E11.5 were analysed, and all four reporter insertions showed similar, complex expression patterns (Fig. 2A-D), which compared well to the major sites of Shh expression (revealed by in situ hybridisation, Fig. 1D-G and previously reported by Echelard et al., 1993; Riddle et al., 1993; Epstein et al., 1999; Chuong et al., 2000). There were no apparent omissions in the patterns of expression. Expression analyses of the genes within the Shh interval from Nom1 to En2 have been examined by RT-PCR (Fig. 1C), and in situ hybridisation in whole mounts and on sections. Specific expression was detected for Shh within the brain, pharyngeal endoderm, limb and gut, and for Cnpy1 and En2 within the midbrain-hindbrain junction. However, the nearest neighbours to Shh (Nom1, Lmbr1, Rnf32 and Rbm33) all appear to be expressed widely in the embryo (Fig. 1C and data not shown). This widespread expression is supported by embryonic day 14.5 (E14.5) RNA-seq data from the mouse ENCODE project (see webpage, http://genome.ucsc.edu/ENCODE/dataSummaryMouse.html) (The ENCODE Consortium Project, 2011).
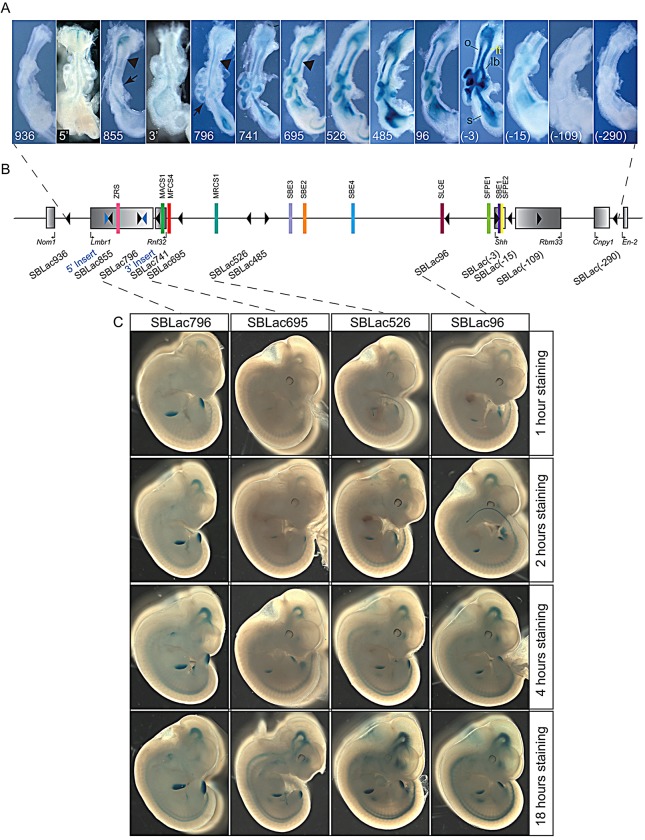
The analysis for lacZ reporter gene activity was done for each embryo under similar β-galactosidase (β-gal) staining conditions such that expression levels were comparable among the different insertion sites. All embryos were stained for 18 h to ensure all sites of expression are detected by the β-gal enzyme reaction. (The consistency of the β-gal staining for similarly staged embryos for two different insertion sites is shown in supplementary material Fig. S1.) To ensure that the final staining pattern reflected the levels of expression, we followed the timecourse of staining for four different insertions within the Shh regulatory domain; SBLac 796, SBLac695, SBLac526 and SBLac96 (Fig. 3C). The timecourse included embryos at 1, 2, 4 and 18 h and showed that qualitative comparison of expression levels is possible between the insertion sites. The progression of staining in the early time points reflects the relative pattern at the 18 h time end point.
Fig. 3.
Expression within guts and a timecourse for β-gal staining. (A) A series of guts, dissected at E11.5, from embryos derived from cells carrying the initial insertions called 5′ Insert (5′) and 3′ Insert (3′) and the mobilised SBLac containing ES cells throughout the entire interval and stained for β-gal expression. Insertions in the middle of the interval, from SBLac741 to SBLac(-15), reflect the Shh expression pattern, with expression being detected in the laryngotracheal tube [in panel labelled (-3)], oesophagus, the lung buds and stomach. The embryonic gut from either end of the interval, SBLac936, SBLac(-109) and SBLac(-290), show no detectable staining. Expression in the laryngotracheal tube, driven by the MACS1 enhancer, is not detected in SBLac855 and SBLac796 (arrowheads). MACS1 shows a directional preference within the regulatory domain, because SBLac695, which lies a similar distance 3′ of MACS1 as SBLac796 lies 5′, shows more activity (expression marked by arrowhead). SBLac855 also shows no expression in the developing lung buds (arrow). (B) The genomic interval, as in Fig. 2E. (C) The timecourse for β-gal staining in embryos from the SBLac796, SBLac695, SBLac526 and SBLac96 insertions, dissected at E11.5 and stained for 1, 2, 4 and 18 h. lb, lung buds; lt, laryngotracheal tube; o, oesophagus; s, stomach.
Analysis of the expression patterns for the gene desert inserts suggest that the desert is in an open conformation over its length and each reporter is receptive to all the known enhancer activities. In addition, the insert position did not affect the pattern of expression and none of the reporters showed unusual or unexpected additions to the expression pattern as would be expected for a holo-enhancer (Marinic et al., 2013).
Distance as a modulator of enhancer activity
Although the pattern of expression was not affected by position of the reporter within the regulatory domain, the levels of expression showed positional differences. The question of how location of an SB reporter insert pertains to expression levels was addressed by focusing on the activity of two of the enhancers, ZRS and MACS1, which are at the extreme 5′ end of the regulatory domain. These two enhancers afford the opportunity to examine enhancer activity over a large genomic distance.
The MACS1 enhancer is responsible for widespread expression in the epithelium of the gut, stomach, alveoli and laryngotracheal tube, and lies inside the Rnf32 gene (Fig. 1A). A nonconserved secondary enhancer, called SLGE, lies within 100 kb 5′ of the Shh gene and drives expression that overlaps the MACS1 pattern in all tissues except the laryngotracheal tube [labelled in yellow in Fig. 3A, panel marked (-3)] which has no known secondary enhancer (Tsukiji et al., 2014). Analysis of the four reporters between MACS1 and the Shh gene lying within the gene desert (Fig. 3) showed levels of expression within the laryngotracheal tube, which appeared to be highest in SBLac485. Surprisingly, expression of the two nearest reporter genes, SBLac695 and SBLac741, which lie within the Rnf32 gene and at 5 kb away are the closest insertions to MACS1, showed that proximity to the MACS1 enhancer has no influence on expression activity. The MACS1 enhancer also has an apparent directional preference within the regulatory domain; the two reporter insertions on the 5′ side (SBLac796 and SBLac855) show very low activity in the laryngotracheal tube. SBLac796, which is 60 kb on the 5′ side of MACS 1, is a similar distance to the more active SBLac695 (∼40 kb), which lies on the opposite side (compare the expression marked by the black arrowheads in SBLac796 and SBLac695 in Fig. 3). Additionally, no activity is detectable in the SBLac855 (arrowhead), suggesting a gradual loss of MACS1 activity at this side of the enhancer.
The ZRS is the enhancer that maps the furthest distance from the Shh gene, lying in intron 5 of the Lmbr1 gene. The spatiotemporal information that drives the limb expression is contained in this single enhancer; a previous report shows that deletion of this region results in loss of limb expression (Sagai et al., 2005). The two insertions, SBLac796 (n=12) and SBLac855 (n=15), sit to either side of the ZRS and are the nearest reporter genes to this enhancer. In contrast to the low expression of these two SBLac insertions in the gut, and laryngotracheal tube (Fig. 3A), the expression in the limb is appreciable, confirming that this region of the domain is open for enhancer activity (Fig. 4D-F). Other insertions between the ZRS and Shh also showed expression, but differences were difficult to discern, as the limb buds stain appreciably at the 18 h time point. To compare expression across the regulatory domain, expression in the limb buds was analysed over the timecourse in Fig. 3C. The limb bud expression in SBLac796 was compared to the three insertions within the gene desert, SBLac695, SBlac526 and SBLac96. SBLac796 staining is readily seen in the limb buds at 1 h and appears to approach saturation by 4 h of staining, whereas the insertions within the gene desert lying at a greater distance lag behind. SBlac695, SBLac526 and SBlac96 show increasing limb staining (see Fig. 3) the closer the reporter lies to the Shh gene. In contrast to the MACS1 enhancer, the ZRS exhibits appreciably higher activity with reporters that are located nearby. This activity falls dramatically in the proximal gene desert and then increases gradually again with proximity to Shh.
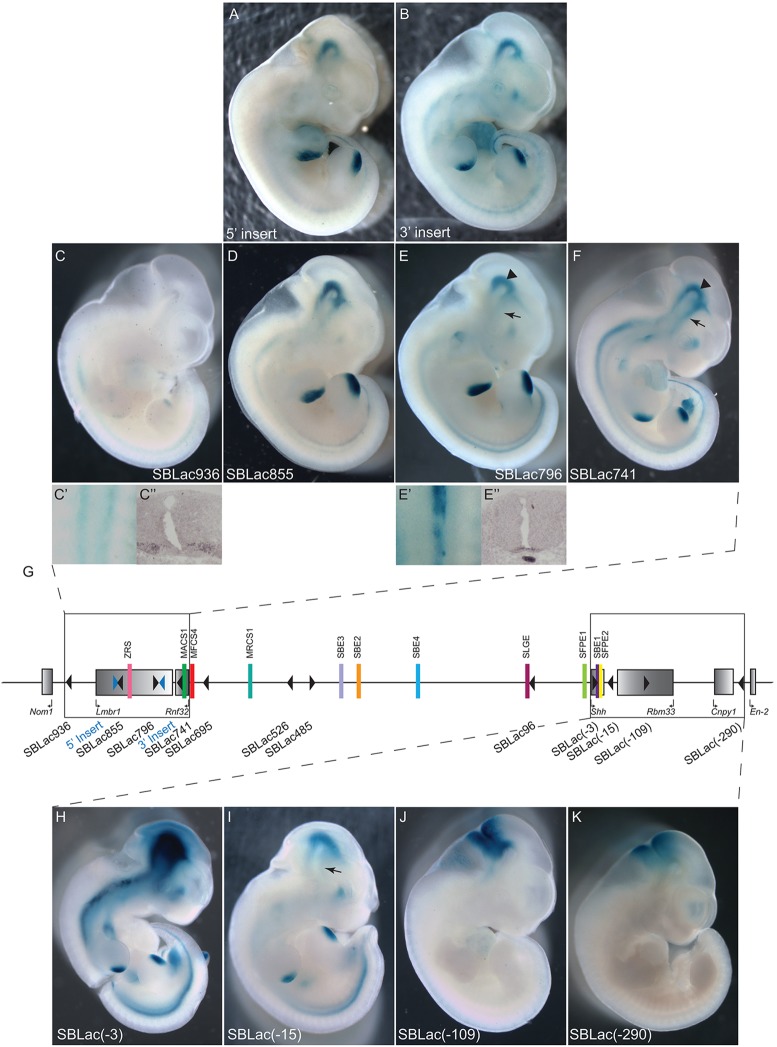
Fig. 4.
Expression of SBLac insertions at the extreme ends of the Shh regulatory locus. (A,B) Embryos derived from ES cells carrying the initial insertion sites and (C-F) from SBLac-carrying ES cells from the 5′ end of the regulatory domain. SBLac936 shows faint Shh like expression in the limb buds but the staining down the back is detected in two stripes (C′), which does not reflect the Shh pattern in the floor plate and notochord (E′), but more likely reflects expression of Mnx1, the next gene past Nom1. Expression of Mnx1 is shown by in situ hybridisation (C″). In situ hybridisation for Shh expression in the floor plate and notochord is shown in E″. Expression in SBLac855, SBLac796 and SBLac741 does reflect the Shh pattern; however, expression is lost preferentially from the developing forebrain in SBLac855 and SBLac796, whereas expression in the midbrain is maintained. [Compare expression marked by arrows (forebrain), with that marked by arrowheads (midbrain) in E,F.] (G) The regulatory locus with the relevant SBLac insertions (black triangles) boxed. At the 3′ end of the region (H-K), highest levels of expression are detected in SBLac(-3), which has integrated within the coding region of Shh. Expression in the next insertion, SBLac(-15), is maintained in most of the sites of Shh expression but at much lower levels, and expression is completely missing from the forebrain (arrow). However, in the last two insertions, SBLac(-109) and SBLac(-290), expression is detected only at the midbrain-hindbrain junction, mirroring expression of Cnpy1 and En2, and suggesting these SBLac insertions have integrated within a different topological domain.
Expression susceptibility inside an ubiquitously expressed gene
The Rnf32 gene is expressed widely throughout the embryo, but no developmental role has, thus far, been assigned. The Rnf32 gene has Shh enhancers upstream, (the ZRS limb-specific enhancer resides in intron 5 of the Lmbr1 gene) downstream and residing inside the gene itself (the MACS1 enhancer, discussed above, resides inside intron 8). However, the Rnf32 gene expression is not detectably upregulated in the gut or any other regions that reflect the Shh pattern (Fig. 1C and data not shown). To determine whether there is a property integral to the Rnf32 gene that renders it unresponsive to the local regulatory landscape, we selected an insertion, SBLac741, which is located in intron 6 of the Rnf32 gene. SBLac741 is expressed throughout the central nervous system (CNS), brain, gut and limb in a typical Shh spatial pattern (Fig. 4F). Therefore Rnf32, which resides inside the regulatory domain and carries an active promoter (data from ENCODE summarised in Fig. 6B), is resistant to outside enhancer influence; however, the body of the gene is accessible to the surrounding expression activity. Although the Shh enhancers are not limited to the Shh promoter, as evidenced by the activity of the heterologous promoter of the reporter genes, it appears that the Rnf32 promoter is refractory to these activities.
Fig. 6.
Summary of the limits of topological domains and enhancer activity. (A) The Hi-C analysis from mouse ES cells (taken from the Mouse ENCODE website), and the boundaries of the topological domain determined by the directionality index and marked by dotted lines. The relationship between the positions of these features and the genes and insertions within the Nom1 to En2 region are indicted in B. Below the genes is the track showing the positions of RNA polymerase II (Pol2 peaks marked with black lines) in E14.5 limb buds taken from ENCODE. (C) A summary of relative expression activity driven by individual enhancers in particular tissues (ZRS driving expression in the limb bud, SBE2 in the forebrain, SBE1 in the midbrain and MACS1 in the laryngotracheal tube) at each of the SBLac insertions. The solid vertical lines show an estimate of the relative expression levels and the dotted lines the predicted levels throughout the genomic interval. Expression at the 5′ end shows that reduction in expression occurs over an interval of greater than 100 kb, whereas limb bud expression is still detectable at the furthest insertion site. Positions of CTCF peaks (bright blue lines) in E14.5 limb buds taken from ENCODE are shown below. There are no appreciable peaks in the middle of the interval, the majority lying at either end; however, there is no relationship between position of the peaks and the position at which expression is reduced.
Activity of regulators that reside within the Shh gene
Two enhancers reside within the introns of the Shh gene. These enhancers work at the shortest distance and, by operating within the context of the gene, avoid the need for any participation with the upstream regulatory domain. This gave us the opportunity to assay the intragenic enhancer activity to determine whether intronic enhancers have similar characteristics to long-range-acting enhancers. The floor plate enhancer SFPE2 in exon 3 overlaps the activity of a similar enhancer in the gene desert at E11.5, the SFPE1, and therefore activity was difficult to specifically attribute to SFPE2 (Fig. 1B). We therefore focused on the SBE1 enhancer, which is located in intron 2 of the Shh gene and is responsible for Shh expression in the ventral midline of the rostral midbrain and caudal diencephalon (Fig. 1B). Mice carrying a 525 bp targeted deletion of the SBE1 enhancer initiate Shh expression at E8.5 in these brain regions but are unable to maintain it after E10.0 (Jeong et al., 2011). By contrast, Shh expression within the forebrain tissue, the zona limitans intrathalamica (ZLI), is unaffected. These data suggest that SBE1 is required for expression in the ventral midbrain at E11.5 and thus presents a distinctive expression pattern that can be used to analyse enhancer function. We found that all SBLac reporters in the gene desert (Fig. 2A-D) and residing in the Rnf32 and Lmbr1 genes (Fig. 4A,B,D-F) are under the influence of the ventral midbrain SBE1 regulator, as reporter gene expression is present within the midbrain (although not the ZLI in some instances), showing that this enhancer, although near the Shh promoter, has the information to act at a very long distance of over 700 kb. Therefore, enhancer proximity to the promoter and residence inside the gene does not indicate a different class of regulator and underscores the idea that mechanisms are shared by intragenic enhancers and long-distance enhancers.
Defining the boundaries of Shh enhancer activity
We observed that reporter gene activity within Lmbr1 decreased differentially in a subset of Shh-expressed tissues. For example, between SBLac741 and SBLac796 there is little change in production in the ventral midbrain (driven by the SBE1 enhancer) (indicated by arrowheads in Fig. 4E,F), whereas the forebrain expression (SBE2 enhancer) is noticeably reduced (indicated by arrows in Fig. 4E,F). Also the laryngotracheal expression (MACS1 enhancer) is drastically reduced between the same two insertion sites (arrowheads in Fig. 3A). The lung expression dependent on the MACS1 and SLGE enhancers is dramatically reduced between SBLac796 and SBLac855 (arrows in Fig. 3A).
To examine the possibility that orientation of the insertion, especially those located inside active genes, may affect the relative expression of reporters, we compared five different insertion sites within genes at the Lmbr1 end of the regulatory domain. SBLac741in the Rnf32 gene and in the Lmbr1 gene, SBLac796 and SBLac855, and the insertion sites of the initial transposon constructs, 5′ Insert and 3′ Insert were examined. SBLac 855 and the 5′ Insert and SBLac796 and the 3′ Insert are nearest neighbours and are situated in opposite orientations (Fig. 3B). Relative expression appears to be dependent on position and not on orientation (compare gut expression in Fig. 3A and expression in whole embryos in Fig. 4A,D and Fig. 4B,E). For example, the laryngotracheal tract and forebrain expression is present in SBLac741 but not in the inserts in the Lmbr1 gene, regardless of orientation.
In order to determine how far Shh enhancer activities persist within the chromosomal domain, embryos were generated containing an SBLac insertion located 24 kb upstream of the Lmbr1 gene (SBLac936). The SBLac936 insertion was situated between Lmbr1 and the neighbouring Nom1 gene. In situ hybridisation and RT-PCR (Fig. 1C and data not shown) showed that Nom1 was expressed widely throughout the embryo with no discernible Shh pattern. SBLac936 (n=5) expression was undetectable throughout most of the embryo, except within the neural tube and the limb. The limb expression mirrors that of Shh in the ZPA and is probably due to residual activity from the ZRS (Fig. 4C). However, dissected embryos showed that the neural tube expression was unlikely to be Shh related given that expression was found in two lateral stripes corresponding to the motoneurons (Fig. 4C′), rather than within the Shh midline domain in the floor plate and notochord (Fig. 4E′). This probably reflects expression of Mnx1, which is the next gene past Nom1. (For comparison, expression of Mnx1 by in situ hybridisation is shown in Fig. 4C″, whereas Shh in situ is shown in Fig. 4E″.) No other expression resembling the Shh pattern was found.
SBLac insertions were also examined downstream of Shh (Fig. 4G): firstly, SBLac[-15] (n=10) within the intergenic region between Shh and the nearest neighbouring gene Rbm33, approximately 15 kb from the Shh 5′ end; and secondly, an insertion within an intronic region of the Rbm33 gene SBLac[-109] (n=7). Finally, the furthest 3′ insertion studied, SBLac[-290] (n=6), lies between Cnpy1 and En2 (Fig. 4H-K). The expression pattern of SBLac[-15] was shown to reflect that of the Shh pattern within the CNS, gut and limbs; however, levels were appreciably lower than the adjacent insertion, SBLac[-3]. SBLac[-3] (n=5) shows the highest levels of expression observed, but this insertion resides within the Shh gene and lies in the orientation of transcription. In addition, the forebrain expression driven by the SBE2 enhancer was undetectable (arrow in Fig. 4I). We used optical projection tomography (OPT) (Sharpe et al., 1999) to examine this expression in SBLac[-15] compared to SBLac96 and SBLac526 to gain a 3D view of the expression (Fig. 5B-D,F-H), which agreed with the relative loss of expression in the forebrain. The lack of forebrain expression observed in SBLac[-15] (Fig. 5D,H) is also very similar to that at the opposite 5′ end of the regulator domain in SBLac855 (Fig. 5A,E), suggesting that a similar mechanism might be at play at both extreme ends. Focusing on the ventral midbrain enhancer (SBE1) that resides inside the Shh gene, which has no promoter lying between it and the reporter in SBLac[-15], also shows lower activity (Fig. 4I), suggesting that downregulation of activity within the Shh regulatory domain is not due to the promoter generating an interfering boundary; however, promoter preference cannot be ruled out. The SBE1 enhancer is capable of working bidirectionally; however, action in the opposite direction from the target gene is severely limited. The more distal insertions SBlac[-109] and SBLac[-290] showed no Shh related expression; instead expression was found within the midbrain-hindbrain junction. Thus a distinctive boundary exists between SBLac[-15] and SBLac[-109] but similar to loss of activity at the 5′ end of the regulatory domain, expression is initially reduced before becoming undetectable.
Fig. 5.
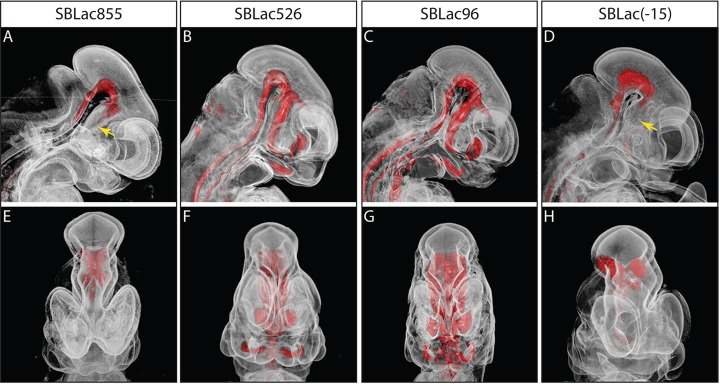
OPT analysis of SBLac insertions. Outputs of OPT analysis of E11.5 embryos derived from SBLac insertions across the regulatory locus [SBLac855 (A,E), SBLac526 (B,F), SBLac96 (C,G) and SBLac(-15) (D,H)]. Sagittal views are shown in A-D and frontal views in E-H. The anatomy of the samples is translucent, whereas the β-gal expression is coloured red. Expression is confirmed to be missing from the forebrains of SBLac8555 and SBLac(-15) (yellow arrows), whereas midbrain expression is maintained.
The midbrain/hindbrain expression patterns observed in SBlac[-109] and SBLac[-290] support the notion that these two reporter genes were influenced by the same regulatory element but within a domain distinct from that of Shh. Downstream of Shh lie three genes: Rbm33, Cnpy1 and En2. Very little is known about the first gene other than sequence composition suggesting an RNA-binding activity. Cnpy1 and En2, however, are expressed within the midbrain-hindbrain boundary region (Fig. 1H-K) and En2 plays a role in restricting the fate of progenitor cells to a midbrain or hindbrain lineage (Davis et al., 1988; Paek et al., 2012). The expression of SBlac[-109] and SBLac[-290] appears to be under the control of the midbrain-hindbrain regulatory element. SBLac[-109] lies inside the Rbm33 gene, presenting a second example of a gene resistant to outside regulatory influence, similar to the observations made for the Rnf32 gene. This raises the possibilities that promoter resistance plays a general role within large regulatory domains that encompass multiple genes.
DISCUSSION
The analysis of the Shh regulatory locus took advantage of the previous transgenic and genetic analyses that established the contribution of the highly conserved elements on the spatial expression activities (Epstein et al., 1999; Lettice et al., 2002, 2003; Sagai et al., 2004, 2009; Jeong et al., 2006). Most of these Shh regulators operate at mid-gestation; thus, generation of tetraploid complementation embryos focusing on a single stage (∼E11.5) in development proved to be an effective assay for enhancer activity across a large chromosomal domain.
Coordinated gene expression controlled by long-distance enhancers is recognised to fall into a number of different, but possibly overlapping, mechanistic classes. Regulatory landscapes that control the output of the HoxD gene cluster rely on multiple inputs from regulatory elements scattered across hundreds of kilobases (called regulatory archipelagos) that control sequential expression of genes in the HOX cluster as well as the co-expression of the ‘bystander’ genes Lnp and Evx2 (Spitz et al., 2003; Montavon et al., 2011). The holo-enhancer of FGF8 requires multiple regulatory elements, many of which lie inside genes acting together to generate a complex developmental expression pattern. The enhancer activity and the expression pattern are highly dependent on the position of the gene relative to the regulatory elements, suggesting a complex interaction of the regulatory elements. These regulators do not activate neighbouring genes (Marinic et al., 2013). By contrast, Shh developmental activity may represent a more common genomic composition (Symmons and Spitz, 2013) relying on a regulatory domain primarily composed of regulators that contribute to the spatiotemporal expression pattern as a summation of the individual enhancer activities. The enhancers that regulate Shh expression populate a gene desert but extend past this desert into two genes of a neighbouring gene-rich region. As with holo-enhancers, neighbouring gene expression is unaffected.
Gene desert is open to enhancer activity
Reporter genes inserted into any of the four positions of the 729 kb gene desert displayed spatial patterns that reflected Shh expression upon examination of the whole embryo and the dissected gut. Long-range activity is inbuilt into the enhancers assayed in the Shh region. The gene desert is broadly open to transcriptional activity; and the spatial pattern of expression was independent of the position of the reporters within the desert, suggesting that there is no segmentation of the regulatory terrain into open and closed chromatin and that the chromatin within the gene desert is functionally indistinguishable.
Long-range acting enhancers are believed to have adopted mechanisms that enable the interaction of enhancers with their respective promoters by a looping mechanism, which is cytologically visible by 3D fluorescence in situ hybridisation (FISH) (Bickmore, 2013). In support of these mechanisms the most distal Shh regulator, the ZRS enhancer, was shown to employ these interactions (Amano et al., 2009) and these were related to gene activity (Lettice et al., 2014). If these mechanisms are crucial for long-range activity, the question arises as to how these relate to enhancers acting over much shorter distances. Notably, the ventral midbrain enhancer that resides within the Shh gene and operates on the target promoter only 6 kb away has full capacity to recognise reporter genes up to a distance of ∼800 kb. This relationship of enhancer and promoter interactions to gene activity suggest uniformity in enhancer mechanisms across the whole regulatory domain and indicates that proximity does not necessarily require the implementation of different mechanisms. Although this approach does not indicate if there is a promoter preference, it does show that heterologous promoters are accessible by the Shh enhancers. These enhancers are fully capable of recognising promoters in numerous positions within the regulatory domain, suggesting that enhancers possess a surveillance activity that bidirectionally scans across broad regions of the regulatory domain.
Activity level differs dependent on position
Although the majority of the gene desert appears to be open to enhancer activity, the activity levels are not equivalent across the domain. We examined the most distal enhancer, the ZRS limb-specific enhancer lying in the Lmbr1 gene, which showed a trend in which the reporter closest to the enhancer was high with a decrease in expression toward the middle of the domain and an increase with the reporters near the Shh gene. This increase in pHLED-derived reporter activity nearer the gene was also reported for the Pax1 gene (Kokubu et al., 2009). Alternatively, the MACS1 gut enhancer is found within the Rnf32 gene (Tsukiji et al., 2014) and expression driven by MACS1 in the laryngotracheal tube suggests a mechanism more focused in the direction of the gene but does not increase with proximity to the Shh gene.
Resistance to enhancers by widely expressed genes
The Rnf32 gene lies fully within the Shh regulatory domain and carries one of the known enhancers within an intron. Expression of Rnf32, however, does not reflect the Shh expression pattern but is expressed broadly throughout the embryo at E11.5. One possible explanation for this resistance to the Shh regulatory domain is that the gene is outside regulatory influence; however, the insertion inside the gene has the full capacity to generate the Shh expression pattern. In accordance, this regulatory evasion may be a common mechanism, as a similar resistance to nearby enhancer activity was shown for the Rbm33 gene. Both the Rbm33 and Rnf32 genes appear to be fully within range of enhancer influence, and we suggest that the promoters are refractory to enhancer activity, perhaps highlighting a common event within large regulatory domains.
Topological domains and the limits of enhancer activity
We have mapped the extent of the regulatory domain responsible for the spatiotemporal expression of the Shh gene. The Shh enhancers were shown to operate within a single long-range chromosomal domain with boundaries that limit enhancer activity. On a large genomic scale, chromatin interaction studies using Hi-C techniques suggest that the genome is arranged into large megabase-sized ‘topological domains’ or ‘topological associated domains’ (TADS), the boundaries of which are expected to correspond to insulator or barrier elements, which prevent the spread of heterochromatin (Smallwood and Ren, 2013). Furthermore, a genome-wide functional study examining randomly integrated reporters shows that large regulatory domains are confined to TADs and that enhancers generally act pervasively throughout these regulatory domains (Symmons et al., 2014). The Shh regulatory domain agrees with the TAD previously predicted (Fig. 6A) (Smallwood and Ren, 2013). However, distinct boundaries that block enhancer activity within these domains are not absolute. At the Lmbr1 end of the domain enhancer, function is found to tail off in a stepwise fashion within the gene (depicted in Fig. 6C). The laryngotracheal expression is drastically reduced between SBLac741 and SBLac796, whereas the expression in the lung primordial is lost between SBLac796 and SBLac855. Finally, the ventral midbrain and the limb expression levels are drastically reduced between SBLac855 and SBLac936, but low levels of Shh in the limb are still detectable. At the Shh end of the regulatory domain, enhancer activity is reduced 15 kb downstream of the gene promoter, whereas the forebrain expression is undetectable. All expression is lost at the next insert within the Rbm33 gene. These data suggest that there are no absolute limits to enhancer function and a number of barriers exist that downregulate expression.
TAD boundary regions are postulated to be enriched for the insulator binding protein CTCF (Dixon et al., 2012) that prevent illegitimate enhancer-promoter interactions. The major CTCF sites identified in the analysis of E14.5 limbs (Shen et al., 2012) are shown in Fig. 6C. Within the Shh and Lmbr1 region such sites were identified (ENCODE), and these sites correspond well with the end of the TAD mapped by Hi-C (Smallwood and Ren, 2013). Notably, no CTCF sites lie within the gene desert (Fig. 6C). The incremental loss of expression toward the limits of the Shh topological domain tallies with multiple CTCF sites; however, there is no direct relationship between the position of the CTCF sites and the loss of enhancer activity, suggesting that additional mechanisms limit the activity of enhancers within a regulatory domain.
MATERIALS AND METHODS
Targeting vector construction
Two vectors were designed for targeted integration into the Lmbr1 locus: the first a 7.7 kb homology arm at the 5′ end of the Lmbr1 gene, 7.5 kb upstream of the ZRS; and the second a 7.6 kb arm 74 kb downstream of the ZRS. Mini targeting arms were generated by PCR (the primers are listed in supplementary material Table S1) and cloned using the underlined restriction sites into pBluescript (Invitrogen). Bacterial recombineering (Liu et al., 2003) was then employed to retrieve the homology arms from the Pac RCPI-21 542n10 (Osoegawa et al., 2000). These homology arms were subcloned (using the bold restriction sites) into the pLHED vector (Kokubu et al., 2009). The constructs had been designed with gaps, of 600 and 666 bp, respectively, flanked by HindIII sites that were used to linearise the vectors for targeting.
Cell culture and gene targeting
E14TG2a ES cells were cultured and targeted using standard techniques (Lettice et al., 1999). 100 µg of each linearised vector were electroporated into 1×107 ES cells. After 10 days of selection with G418, individual clones were picked, screened for correct targeting by PCR (primers are listed in supplementary material Table S1) and the insertion position confirmed by DNA FISH.
In vitro mobilisation of transposon
LHED targeted cells were plated at densities of 1×106 per 10 cm plate, transiently transfected with 20 µg of the transposase vector pCMV-SB100x (Mates et al., 2009), using TransFast (Promega) and cultured for 48 h before the addition of puromycin (2 µg/ml). After 10 days of selection, resistant colonies were picked into 96-well plates, grown and DNA produced for analysis. Excision of the transposon was confirmed by PCR using the primers for Puromycin gene and the PGK promoter. Reinsertion of transposon was detected using lacZ-specific primers.
Identifying transposon insertion sites
To identify transposon insertion sites, a nested asymmetric PCR strategy was applied as described by Ruf et al. (2011) (primers are listed in supplementary material Table S1).
Embryo production, whole-mount X-gal staining and in situ hybridisation
Tetraploid blastocysts were produced by electrofusion, and ES cells injected to make entirely ES-cell-derived embryos by tetraploid complementation (Nagy et al., 1993). Whole-mount X-gal staining of embryos was performed at E11.5 as described previously (Lettice et al., 2003), but on this occasion the staining was allowed to proceed at room temperature for between 1 and 18 h in a concentration of 300 µg/ml X-gal.
Wild-type mouse embryos were harvested at E11.5 and in situ hybridisation was performed with DIG-labelled gene-specific antisense probes as previously described (Hecksher-Sorensen et al., 1998).
Probes were generated for Lmbr1, Rnf32, Nom1 and Rbm33 by RT-PCR and cloned into the pBluescript vector (Agilent Technologies) (primers are listed in supplementary material Table S1). The Shh probe was kindly provided by Andy McMahon (Echelard et al., 1993) and the Cnpy1 and En2 probes by Jean M Herbert (Paek et al., 2012).
Gene expression analysis by RT-PCR
RNA was extracted from E11.5 mouse embryos by using RNAbee (AMSBio) and cDNA was made using a First-Strand cDNA Synthesis Kit (Roche). Primers (listed in supplementary material Table S1) were exon specific.
OPT analysis
OPT imaging was performed (Sharpe et al., 2002). Briefly, PFA-fixed embryos were embedded in 1% low-melting-point agarose and then immersed in methanol for 24 h to remove all water. The embryo was then cleared for 24 h in BABB (one part benzyl alcohol/two parts benzyl benzoate). The sample was then scanned using a Bioptonics 3001 scanner (www.biooptonics.com) with an image taken every 0.9° (of a 360° rotation). Upon completion, images are reconstructed using Bioptonics proprietary software with the outputs then being viewed with Dataviewer (Bioptonics) and Bioptonics Viewer. Additional, 3D outputs were produced using Drishti rendering software (Ajay Limaye-Volume Exploration and Presentation Tool).
Supplementary Material
Acknowledgements
We thank Ben Moore for advice on the Shh TAD; professors Nick Hastie and David Fitzpatrick for carefully reading the manuscript; Chikara Kokubu for the pHLED vector and for advice on its transposition; Andy McMahon and Jean Herbert for the Shh, Cnpy1 and En2 in situ probes; Zsuzsanna Izsvak for the transposase vector pCMV-SB100x; Harris Morrison for OPT skills and the staff in the MRC Evan's Building for expert technical assistance.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
E.A. performed the majority of experiments and helped with the interpretation of the results. P.S.D. produced the genetic and embryological resources. L.A.L. and R.E.H. supervised the study, and analysed and interpreted the data. E.A., L.A.L. and R.E.H. prepared the manuscript.
Funding
This work was supported by a Medical Research Council Core Grant. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.108480/-/DC1
References
- Amano, T., Sagai, T., Tanabe, H., Mizushina, Y., Nakazawa, H. and Shiroishi, T. (2009). Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47-57 10.1016/j.devcel.2008.11.011 [DOI] [PubMed] [Google Scholar]
- Belloni, E., Muenke, M., Roessler, E., Traverso, G., Siegel-Bartelt, J., Frumkin, A., Mitchell, H. F., Donis-Keller, H., Helms, C., Hing, A. V.et al. (1996). Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat. Genet. 14, 353-356 10.1038/ng1196-353 [DOI] [PubMed] [Google Scholar]
- Bickmore, W. A. (2013). The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14, 67-84 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Chuong, C.-M., Patel, N., Lin, J., Jung, H.-S. and Widelitz, R. B. (2000). Sonic hedgehog signaling pathway in vertebrate epithelial appendage morphogenesis: perspectives in development and evolution. Cell. Mol. Life Sci. 57, 1672-1681 10.1007/PL00000650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C. A., Noble-Topham, S. E., Rossant, J. and Joyner, A. L. (1988). Expression of the homeo box-containing gene En-2 delineates a specific region of the developing mouse brain. Genes Dev. 2, 361-371 10.1101/gad.2.3.361 [DOI] [PubMed] [Google Scholar]
- Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., Hu, M., Liu, J. S. and Ren, B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard, Y., Epstein, D. J., St-Jacques, B., Shen, L., Mohler, J., McMahon, J. A. and McMahon, A. P. (1993). Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417-1430 10.1016/0092-8674(93)90627-3 [DOI] [PubMed] [Google Scholar]
- Epstein, D. J., McMahon, A. P. and Joyner, A. L. (1999). Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development 126, 281-292. [DOI] [PubMed] [Google Scholar]
- Hecksher-Sorensen, J., Hill, R. E. and Lettice, L. (1998). Double labeling for whole-mount in situ hybridization in mouse. Biotechniques 24, 914–916, 918. [DOI] [PubMed] [Google Scholar]
- Jeong, Y., El-Jaick, K., Roessler, E., Muenke, M. and Epstein, D. J. (2006). A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761-772 10.1242/dev.02239 [DOI] [PubMed] [Google Scholar]
- Jeong, Y., Dolson, D. K., Waclaw, R. R., Matise, M. P., Sussel, L., Campbell, K., Kaestner, K. H. and Epstein, D. J. (2011). Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development 138, 531-541 10.1242/dev.058917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki, E., Ott, C.-E., Benatar, N., Ullmann, R., Mundlos, S. and Lehmann, K. (2008). A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J. Med. Genet. 45, 370-375 10.1136/jmg.2007.055699 [DOI] [PubMed] [Google Scholar]
- Kokubu, C., Horie, K., Abe, K., Ikeda, R., Mizuno, S., Uno, Y., Ogiwara, S., Ohtsuka, M., Isotani, A., Okabe, M.et al. (2009). A transposon-based chromosomal engineering method to survey a large cis-regulatory landscape in mice. Nat. Genet. 41, 946-952 10.1038/ng.397 [DOI] [PubMed] [Google Scholar]
- Lettice, L. A. and Hill, R. E. (2005). Preaxial polydactyly: a model for defective long-range regulation in congenital abnormalities. Curr. Opin. Genet. Dev. 15, 294-300 10.1016/j.gde.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Lettice, L. A., Purdie, L. A., Carlson, G. J., Kilanowski, F., Dorin, J. and Hill, R. E. (1999). The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc. Natl. Acad. Sci. USA 96, 9695-9700 10.1073/pnas.96.17.9695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice, L. A., Horikoshi, T., Heaney, S. J. H., van Baren, M. J., van der Linde, H. C., Breedveld, G. J., Joosse, M., Akarsu, N., Oostra, B. A., Endo, N.et al. (2002). Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl. Acad. Sci. USA 99, 7548-7553 10.1073/pnas.112212199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice, L. A., Heaney, S. J. H., Purdie, L. A., Li, L., de Beer, P., Oostra, B. A., Goode, D., Elgar, G., Hill, R. E. and de Graaff, E. (2003). A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725-1735 10.1093/hmg/ddg180 [DOI] [PubMed] [Google Scholar]
- Lettice, L. A., Daniels, S., Sweeney, E., Venkataraman, S., Devenney, P. S., Gautier, P., Morrison, H., Fantes, J., Hill, R. E. and FitzPatrick, D. R. (2011). Enhancer-adoption as a mechanism of human developmental disease. Hum. Mutat. 32, 1492-1499 10.1002/humu.21615 [DOI] [PubMed] [Google Scholar]
- Lettice, L. A., Williamson, I., Devenney, P. S., Kilanowski, F., Dorin, J. and Hill, R. E. (2014). Development of five digits is controlled by a bipartite long-range cis-regulator. Development 141, 1715-1725 10.1242/dev.095430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Jenkins, N. A. and Copeland, N. G. (2003). A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476-484 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinić, M., Aktas, T., Ruf, S. and Spitz, F. (2013). An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell 24, 530-542 10.1016/j.devcel.2013.01.025 [DOI] [PubMed] [Google Scholar]
- Mates, L., Chuah, M. K. L., Belay, E., Jerchow, B., Manoj, N., Acosta-Sanchez, A., Grzela, D. P., Schmitt, A., Becker, K., Matrai, J.et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753-761 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- Montavon, T., Soshnikova, N., Mascrez, B., Joye, E., Thevenet, L., Splinter, E., de Laat, W., Spitz, F. and Duboule, D. (2011). A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132-1145 10.1016/j.cell.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. and Roder, J. C. (1993). Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90, 8424-8428 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermaier, M., Schwabe, G. C., Fees, S., Helmrich, A., Brieske, N., Seemann, P., Hecht, J., Seitz, V., Stricker, S., Leschik, G.et al. (2005). An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J. Clin. Invest. 115, 900-909 10.1172/JCI200523675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Tateno, M., Woon, P. Y., Frengen, E., Mammoser, A. G., Catanese, J. J., Hayashizaki, Y. and de Jong, P. J. (2000). Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10, 116-128. [PMC free article] [PubMed] [Google Scholar]
- Paek, H., Antoine, M. W., Diaz, F. and Hébert, J. M. (2012). Increased beta-catenin activity in the anterior neural plate induces ectopic mid-hindbrain characteristics. Dev. Dyn. 241, 242-246 10.1002/dvdy.22787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, R. D., Johnson, R. L., Laufer, E. and Tabin, C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401-1416 10.1016/0092-8674(93)90626-2 [DOI] [PubMed] [Google Scholar]
- Ruf, S., Symmons, O., Uslu, V. V., Dolle, D., Hot, C., Ettwiller, L. and Spitz, F. (2011). Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat. Genet. 43, 379-386 10.1038/ng.790 [DOI] [PubMed] [Google Scholar]
- Sagai, T., Masuya, H., Tamura, M., Shimizu, K., Yada, Y., Wakana, S., Gondo, Y., Noda, T. and Shiroishi, T. (2004). Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh). Mamm. Genome 15, 23-34 10.1007/s00335-033-2317-5 [DOI] [PubMed] [Google Scholar]
- Sagai, T., Hosoya, M., Mizushina, Y., Tamura, M. and Shiroishi, T. (2005). Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132, 797-803 10.1242/dev.01613 [DOI] [PubMed] [Google Scholar]
- Sagai, T., Amano, T., Tamura, M., Mizushina, Y., Sumiyama, K. and Shiroishi, T. (2009). A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development 136, 1665-1674 10.1242/dev.032714 [DOI] [PubMed] [Google Scholar]
- Sharpe, J., Lettice, L., Hecksher-Sørensen, J., Fox, M., Hill, R. and Krumlauf, R. (1999). Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr. Biol. 9, 97-100 10.1016/S0960-9822(99)80022-0 [DOI] [PubMed] [Google Scholar]
- Sharpe, J., Ahlgren, U., Perry, P., Hill, B., Ross, A., Hecksher-Sørensen, J., Baldock, R. and Davidson, D. (2002). Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296, 541-545 10.1126/science.1068206 [DOI] [PubMed] [Google Scholar]
- Shen, Y., Yue, F., McCleary, D. F., Ye, Z., Edsall, L., Kuan, S., Wagner, U., Dixon, J., Lee, L., Lobanenkov, V. V.et al. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116-120 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood, A. and Ren, B. (2013). Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 25, 387-394 10.1016/j.ceb.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz, F., Gonzalez, F. and Duboule, D. (2003). A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405-417 10.1016/S0092-8674(03)00310-6 [DOI] [PubMed] [Google Scholar]
- Sun, M., Ma, F., Zeng, X., Liu, Q., Zhao, X.-L., Wu, F.-X., Wu, G.-P., Zhang, Z.-F., Gu, B., Zhao, Y.-F.et al. (2008). Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long range, limb-specific SHH enhancer. J. Med. Genet. 45, 589-595 10.1136/jmg.2008.057646 [DOI] [PubMed] [Google Scholar]
- Symmons, O. and Spitz, F. (2013). From remote enhancers to gene regulation: charting the genome's regulatory landscapes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120358 10.1098/rstb.2012.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons, O., Uslu, V. V., Tsujimura, T., Ruf, S., Nassari, S., Schwarzer, W., Ettwiller, L. and Spitz, F. (2014). Functional and topological characteristics of mammalian regulatory domain. Genome Res. 24, 390-400 10.1101/gr.163519.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Consortium Project. (2011). A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiji, N., Amano, T. and Shiroishi, T. (2014). A novel regulatory element for Shh expression in the lung and gut of mouse embryos. Mech. Dev. 131, 127-136 10.1016/j.mod.2013.09.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.