Abstract
We describe the identification of zyxin as a regulator of synapse maintenance in mechanosensory neurons in C. elegans. zyx-1 mutants lacked PLM mechanosensory synapses as adult animals. However, most PLM synapses initially formed during development but were subsequently lost as the animals developed. Vertebrate zyxin regulates cytoskeletal responses to mechanical stress in culture. Our work provides in vivo evidence in support of such a role for zyxin. In particular, zyx-1 mutant synaptogenesis phenotypes were suppressed by disrupting locomotion of the mutant animals, suggesting that zyx-1 protects mechanosensory synapses from locomotion-induced forces. In cultured cells, zyxin is recruited to focal adhesions and stress fibers via C-terminal LIM domains and modulates cytoskeletal organization via the N-terminal domain. The synapse-stabilizing activity was mediated by a short isoform of ZYX-1 containing only the LIM domains. Consistent with this notion, PLM synaptogenesis was independent of α-actinin and ENA-VASP, both of which bind to the N-terminal domain of zyxin. Our results demonstrate that the LIM domain moiety of zyxin functions autonomously to mediate responses to mechanical stress and provide in vivo evidence for a role of zyxin in neuronal development.
Keywords: Axon retraction, Cytoskeleton, Stress fiber, Synaptogenesis
INTRODUCTION
During neuronal development, the formation and maintenance of synapses are highly regulated to ensure the proper function of both pre- and postsynaptic partners. The outgrowth of axons to their targets and the formation and stabilization of synaptic contacts are critically dependent on adhesion of the neuron to extracellular substrates during outgrowth and subsequently to target cells (Raper and Mason, 2010; Benson and Huntley, 2012). These sites of adhesion provide the linkages to transduce forces generated by the cytoskeleton that drive axonal outgrowth and shape, developing synaptic specializations (Suter and Miller, 2011). However, neurons do not grow in isolation, but rather in the context of the growth of the entire organism. This environment is the source of many extrinsic forces that also impact the cell and thus must be sensed and accounted for during this process (Franze, 2013). The influence of extrinsic mechanical forces on neuronal development has long been recognized. For example, Weiss (1941) described the process of stretch growth of axons, and recent experiments have documented the extensive influence of stretch forces on axon growth rates (Pfister et al., 2004). Likewise, muscle contractions are crucial for the axonal guidance of Rohon-Beard cells in developing zebrafish (Paulus et al., 2009). Although the extensive influence of extrinsic mechanical forces on neuronal development is well documented, the cellular mechanisms by which these forces are detected and harnessed are poorly understood.
Zyxin is one protein implicated in regulating cellular responses to mechanical and stress forces applied to sites of adhesion. Zyxin was originally purified from avian smooth muscle as a basic phosphoprotein (Beckerle, 1986). Zyxin localizes to focal adhesion plaques as well as adherens junctions, and is implicated in regulating cell adhesion, migration, mechanotransduction and actin-based morphological remodeling in normal and cancerous cells (Drees et al., 1999; Yoshigi et al., 2005; Hoffman et al., 2006; Yu and Luo, 2006; Sy et al., 2006; Colombelli et al., 2009; Mori et al., 2009; Sperry et al., 2010). Zyxin contains an N-terminal proline-rich region and three tandemly arrayed zinc-finger LIM domains at the C-terminus (Sadler et al., 1992). Biochemical studies revealed that the N-terminal region of zyxin binds to the actin-remodeling proteins α-actinin and ENA-VASP (Crawford et al., 1992; Drees et al., 2000); the LIM domains interact with a variety of structural and signaling proteins including transcription factors (Degenhardt and Silverstein, 2001; Martynova et al., 2008). This supports an important role of zyxin in relaying signals from actin-rich cell adhesion sites to the nucleus. The LIM domain is sufficient to localize zyxin to actin stress fibers (Smith et al., 2013) and to the leading edge of migrating cells in response to force (Uemura et al., 2011) and is required for stretch-induced actin remodeling (Hoffman et al., 2012), thus also implicating zyxin in more local cytoskeletal remodeling. Notably, zyxin is often found to be enriched in epithelial and muscular tissues, but is expressed at relatively low levels in the nervous system (Macalma et al., 1996). Using cultured neurons, several groups reported zyxin localization at nerve growth cones, suggesting that zyxin might be involved in regulating actin dynamics in extending axons (Gomez et al., 1996; Jay, 2000). However, the role of zyxin in the nervous system has not been carefully examined, and thus our understanding of its role in neural development remains limited.
Vertebrates express multiple proteins with a similar domain organization to zyxin, including LPP (Petit et al., 2000), TRIP6 (Yi and Beckerle, 1998) and the more distantly related proteins WTIP (Srichai et al., 2004), Ajuba (Goyal et al., 1999) and LimD1 (Sharp et al., 2004). Each contains a poorly conserved N-terminal domain and three C-terminal LIM domains. Many members of this family have been demonstrated to localize to sites of cell adhesion and also to translocate from these sites to the nucleus under a variety of conditions (Petit et al., 2003; Srichai et al., 2004). Various family members have been implicated in diverse processes including microRNA-mediated gene silencing (James et al., 2010), responses to hypoxia (Foxler et al., 2012), hippo signaling (Das Thakur et al., 2010), PIP2 signaling (Kisseleva et al., 2005) and stress fiber maintenance (Smith et al., 2010; Hoffman et al., 2012). Molecular genetic analysis in mouse suggests that vertebrate zyxin family members assume functionally redundant cellular roles, since mouse knockouts of LPP and zyxin exhibit few overt phenotypic abnormalities (Hoffman et al., 2003; Vervenne et al., 2009), whereas Drosophila mutants are inviable (Das Thakur et al., 2010; Renfranz et al., 2010).
Here we report that zyx-1 acts to stabilize synaptic contacts during synapse development in mechanosensory neurons. In contrast to vertebrates and Drosophila, the C. elegans genome encodes only one zyxin homologue (Smith et al., 2002). We recovered a zyx-1 mutant in a genetic screen for mutants that disrupted synapse formation in the touch receptor mechanosensory neurons. Timecourse imaging analysis revealed that zyx-1 animals retained the ability to form synapses during development, but that the synapses were often lost. At least some of the synaptic loss was due to breakage and retraction of the neuronal processes that formed synapses. In addition, we discovered that PLM synapses were preserved in immobilized mutants, suggesting that zyx-1 protects PLM mechanosensory processes and synapses from locomotion-induced mechanical stress forces. C. elegans ZYX-1, like the vertebrate homologue, contains an N-terminal proline-rich region and three tandemly arrayed C-terminal LIM domains. Unexpectedly, we found that the neuronal function of ZYX-1 was mediated through an isoform that contains only the LIM domains. We conclude that the LIM domain module constitutes not only a domain of zyxin used to target stress fibers, but also a fully functional protein capable of mediating mechanical responses completely independently of the N-terminal domain.
RESULTS
zyx-1 mutants lack PLM mechanosensory synapses
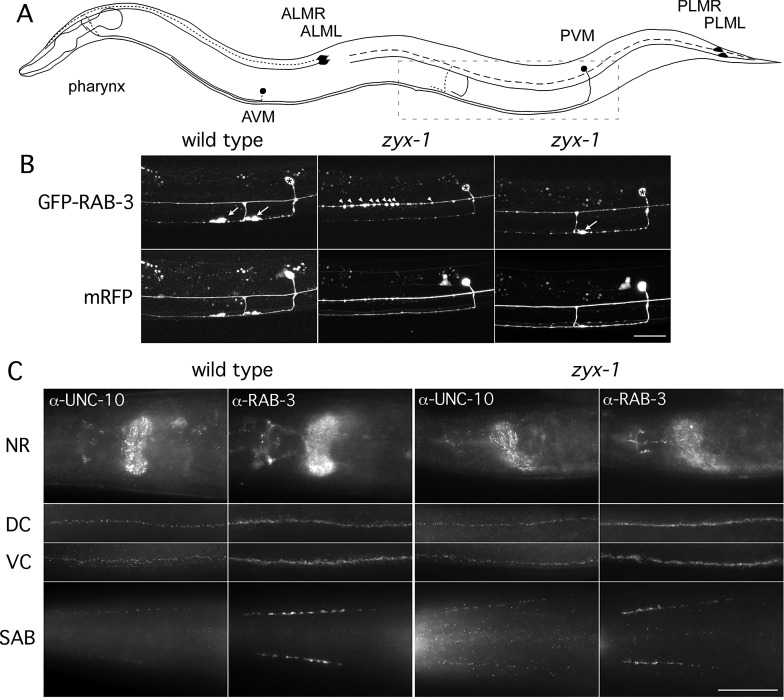
The touch receptor neurons of C. elegans consist of six neurons that sense gentle touch (Chalfie et al., 1985). Three cells (PLML, PLMR and PVM) mediate touch responses in the posterior, and three others (ALML ALMR and AVM) mediate touch responses in the anterior of the animal (Fig. 1A). These cells extend sensory processes along the body just under the cuticle, where specialized mechanosensory receptors are localized and form synapses with interneurons that coordinate locomotion (Chalfie et al., 1985; Emtage et al., 2004). We have focused on the two posterior lateral mechanosensory PLM (L and R) neurons and the synapses that they form in the ventral nerve cord (VNC) just posterior of the vulva (Fig. 1A,B). The synapses of the PLMs were visualized using GFP synaptic vesicle (SV)-localized tags and neuronal morphology with cytosolic mRFP, both of which were specifically expressed in the mechanosensory neurons (Nonet, 1999; Bounoutas et al., 2009). To identify genes that regulate PLM synapse development, we performed ethyl methanesulfonate mutagenesis and screened for disruption of GFP accumulations at the PLM synapses. We isolated sam mutants (SV tag abnormal in mechanosensory neurons) including sam-6(js417) (A. M. Schaefer, PhD Thesis, Washington University, 2001). Single nucleotide polymorphism mapping, transgenic rescue and sequencing (see supplementary material Methods for details) revealed that sam-6(js417) is allelic to zyx-1, which encodes the sole C. elegans homolog of the zyxin family of cytoskeleton-associated proteins.
Fig. 1.
zyx-1 mutants lack PLM mechanosensory synapses. (A) Schematic depiction of the touch neuron circuit in C. elegans. Two PLM neurons are located on the left (PLML) and right (PLMR) side of the tail ganglia, and each sends out a process anteriorly and branch into the VNC, where they form a large synaptic varicosity. Two ALM neurons are located on the left (ALML) and right (ALMR) side in the mid-body, and each extends a process anteriorly and branch into the nerve ring. PVM is located on the left side in the posterior and extends a single process to the VNC, where it turns and extends far anteriorly. AVM is located on the right side in the anterior and extends a single process to the VNC, where it turns anteriorly and extends to the nerve ring. Our studies focus on the PLM branches and synapses found in the boxed region, as shown in B. (B) Morphology and presynaptic specializations of PLM neurons visualized with cytosolic mRFP and GFP::RAB-3 expressed under the mec-7 promoter. Confocal images of wild-type animals revealed the two PLM synaptic varicosities labeled by GFP::RAB-3 in the VNC (arrows). By contrast, most zyx-1 young adults lacked PLM synapses; rather, the GFP::RAB-3 signal accumulated as puncta in the PLM processes, forming a beads-on-a-thread-like pattern (arrowheads). Occasionally, PLM synapses were observed in zyx-1; however, these usually were of reduced size and GFP::RAB-3 intensity (arrow). The morphology of the PVM neuron was not affected in zyx-1 mutants (asterisks indicate the PVM cell body). Strains used were NM3361 and NM3410. (C) zyx-1 mutants have otherwise grossly normal synaptic structures. Wild-type and zyx-1(gk190) mutant animals were fixed, permeabilized and immunostained for the synaptic vesicle-associated protein RAB-3 and the presynaptic active zone protein UNC-10. No significant differences were observed between wild-type and zyx-1 animals in the nerve ring (NR), VNC, DNC or in the head cholinergic SAB motor neurons. Strains used were N2 and VC299. Scale bars: 20 μm.
We analyzed the mechanosensory neurons of wild-type and zyx-1 mutant animals. In wild-type animals, the SV marker GFP::RAB-3 was predominantly localized to two large accumulations in the VNC, as formed by the two PLM neurons (Fig. 1A,B). These correspond to the synaptic varicosities observed by electron microscopy (Chalfie et al., 1985). By contrast, in over 80% of zyx-1(js417) and zyx-1(gk190) mutants, the GFP::RAB-3 accumulations were absent from the VNC, and in an additional 15% of mutants one of the two varicosities was absent (Fig. 1B). Using a cytosolic mRFP marker to visualize the entire neuronal process, we observed that in late L4 zyx-1 animals the synaptic branch was absent or terminated prematurely before reaching the VNC. Moreover, GFP::RAB-3 accumulated in closely spaced puncta in the distal region of the PLM anterior-posterior-directed processes (Fig. 1B). In the animals that formed synapses, the varicosities were often smaller than in the wild type (Fig. 1B) and were occasionally mispositioned posteriorly (discussed in detail below).
To assess if the reduction of zyx-1 function affects more general aspects of nervous system development, we determined the localization of the SV protein RAB-3 and the active zone-localized protein UNC-10 (homologous to vertebrate Rim1). RAB-3 and UNC-10 immunostaining in the nerve ring, the VNC, the dorsal nerve cord (DNC) and the SAB head cholinergic neurons of zyx-1 mutants was indistinguishable from that of the wild type (Fig. 1C), indicating that zyx-1 worms have a grossly normal organization of synaptic structures. Furthermore, we observed no changes in the gross morphology of cholinergic, GABAergic or GLR-1-expressing neurons in zyx-1 animals (supplementary material Fig. S1). Likewise, PVD neurons, which branch extensively, were also normal, showing a comparable density of branches to the wild type (supplementary material Fig. S2). Furthermore, ALM touch neurons formed morphologically normal synapses in the nerve ring (supplementary material Fig. S3). Therefore, we conclude that the function of zyx-1 in regulating synapse development is restricted to subsets of neurons and in particular to the PLM neurons of the mechanosensory system.
Delayed synapse formation in zyx-1 mutants
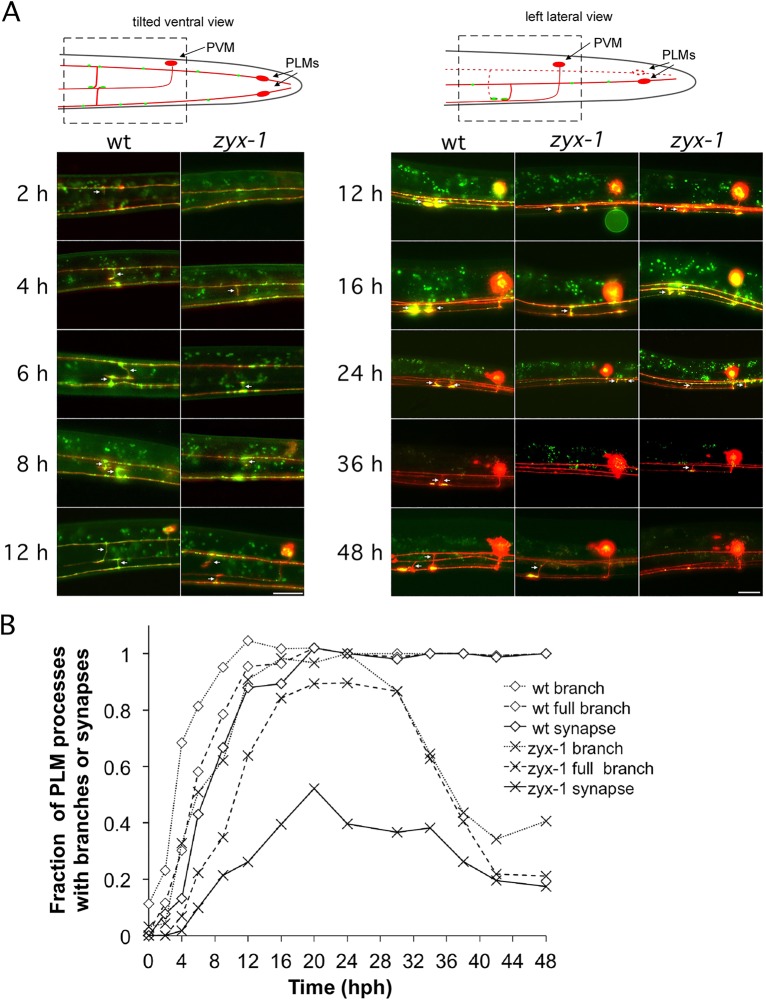
To assess whether the apparent absence of synapses in zyx-1 mutants reflects a failure of synapse formation or of synaptic maintenance, we analyzed the GFP::RAB-3 localization pattern in mechanosensory neurons of both wild-type and mutant animals throughout larval development. At hatching, PLM neurons in wild type had undergone outgrowth along the anterior-posterior axis, with GFP::RAB-3 puncta scattered throughout the PLM processes and the soma (Fig. 2A). In wild-type animals, PLM neurons began extending a ventral branch ∼2 h post hatching (hph) and, by the end of the L1 stage (14 hph), 85% of PLM neurons had extended a branch to the VNC and terminated in bright GFP::RAB-3-filled varicosities. These nascent ‘synapses’ were stable and grew in size throughout larval development (Fig. 2A,B; supplementary material Fig. S4).
Fig. 2.
zyx-1 mutants are unable to maintain mechanosensory synapses. (A) Timecourse of PLM synapse development in wild-type and zyx-1(gk190) animals at 22.5°C. The mechanosensory neurons were double labeled with GFP::RAB-3 and cytosolic mRFP. Images are maximal projections of multiple image planes taken in widefield epifluorescence configuration. The left panels show animals in a roughly ventral orientation, whereas the larger animals in the right panels are in a lateral orientation. Arrows mark the position of PLM branches. PVM is born at ∼8 hph, and thus was absent from the earlier time points. Scale bars: 10 µm for 2-16 hph; 20 µm for 24-48 hph. (B) Quantification of PLM processes that had initiated branches, formed full branches reaching the VNC, and formed synapses at various time points after hatching. n=24-192 for individual time points. A detailed breakdown of this dataset is given in supplementary material Fig. S4. Strains used were NM3361 and NM3413.
Our developmental studies suggest that zyx-1 mutants are able to form PLM synapses, although they form more slowly and at a lower frequency than in the wild type. In early L1 zyx-1 animals, branch formation was delayed by several hours, but nevertheless in 88% of animals at least one PLM had extended a branch to the VNC by the end of the L1 stage (Fig. 2A; supplementary material Fig. S4). At the middle of the L2 stage, 77% of zyx-1 animals had formed at least one PLM synapse (Fig. 2B; supplementary material Fig. S2). Although branch initiation, branch extension and GFP::RAB-3 varicosity growth were delayed, the growth rate (time from hatching to adulthood) of the zyx-1 animals was comparable to that of the wild type, indicating that this delay did not reflect an overall delay in the development of the animal.
zyx-1 PLM synaptic varicosities are unstable
The GFP::RAB-3 accumulations that formed in zyx-1 mutants often failed to expand in size and were observed less frequently later in larval development. This was revealed by the decreasing number of PLM branches and synapses after 24 hph (Fig. 2B; supplementary material Fig. S4). By the time zyx-1 animals were young adults, only 34% of zyx-1 PLM synaptic branches still showed accumulations in the VNC. Two other developmental timecourse experiments performed at room temperature with a less frequent sampling protocol showed a similar timecourse of PLM branch formation and loss (supplementary material Fig. S5). Thus, zyx-1 animals form PLM synaptic branches and synapses, but many of these are not stably maintained through development.
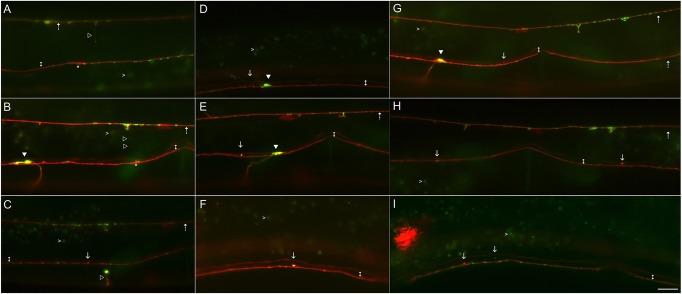
Evidence that zyx-1 processes break
What happened to the PLM branches that were lost from L2 to adult stages in zyx-1 animals? Several lines of evidence indicate that zyx-1 animals continue a repeated cycle of branch formation and breakage. First, we saw evidence of broken PLM branches with ‘retraction bulb’-like structures in which the PLM process was broken and a large bright spherical accumulation of both mRFP and GFP::RAB-3 was located at the end of the process (Fig. 3). These types of structure are associated with axon retraction in vertebrates (Balice-Gordon et al., 1993; Gan and Lichtman, 1998). In association with these structures, we also observed mRFP-positive (and sometimes GFP::RAB-3-positive) spherical ‘remnants’ in positions where mature synapses usually reside (Fig. 3). Although the retraction bulbs were rare (presumably because they are highly transient structures), the remnants were commonly observed in L3, L4 and young adult, but were only rarely observed in the wild type (supplementary material Fig. S4). Furthermore, these remnants were not observed in zyx-1 animals that had formed two mature PLM neurons, and cases of two remnants were only observed in animals without PLM synapses. These data are consistent with these structures representing the degrading distal portions of PLM synapses after PLM branch breakage.
Fig. 3.
Breakage and retraction of PLM collateral branches. Epifluorescence images of live L4 zyx-1 animals. (A-C) Examples of retracting branches (open arrowheads) that sometimes included a retraction bulb (see C). (D-I) mRFP-positive PLM remnants (arrows) were often present in L3, L4 and young adult zyx-1 animals. They usually consist of one very bright spot (but occasionally multiple closely spaced spots, see D) that were positioned between the processes of PVM (lower brighter process marked by double arrow) and AVG (upper brighter process marked by double arrow), which demarcate the most ventral and most dorsal regions of the VNC. Most remnants were mRFP positive and GFP negative, but a minority were double positive (see E,F). Occasionally, two distinct widely spaced remnants were seen (H,I; the less bright remnant is out of the plane of focus). Dashed arrows mark PLM processes; white arrowheads label large PLM synapses; and asterisks label presumed distal ends of severed PLM branches (in A,B). Gut granules (examples marked with chrevrons) are also visible in many images. Strains used were NM3361 and NM3413. Scale bar: 10 µm.
We also observed that as zyx-1 animals developed they formed supplementary branches in addition to the normal two (one for each PLM) found in the wild type. First, we observed numerous larval zyx-1 animals with three and occasionally four or five collateral branches (16% of L2 stage animals; supplementary material Fig. S4). The presence of three complete branches was exceedingly rare in the wild type (1% in L2 and not observed at later stages; supplementary material Fig. S4). Furthermore, we observed branches in positions where branches were virtually never observed in the wild type; 16% of zyx-1 L3 animals, 9% of L4 animals and 7% of adult animals had collateral branches extending ventrally between the PLM soma and the PVM process, whereas these posterior branches were never observed in the wild type (Fig. 2A; supplementary material Fig. S4). In many cases these branches formed varicosities (which could theoretically be functional synapses, as the postsynaptic partners of PLM are present in the VNC in this area). These branches were never observed in L1 and early L2 zyx-1 or wild-type animals. We interpret these as additional late-initiated attempts to form synaptic collateral branches.
In summary, we conclude from our observations that in zyx-1 animals synaptic collateral branches are initiated, but the branches only inefficiently transition into mature synaptic branches due to catastrophic events, which are likely to comprise axon breakage. In response to these failures, zyx-1 animals initiate additional branch formation events, and many of these additional branches share the same fate as the original branches.
Developmental studies confirm synaptic loss
To confirm that zyx-1 mutants fail to maintain presynaptic specializations, we followed individual animals by imaging them multiple times during development. In one experiment we imaged animals three times: at 12 hph (late L1), at 24 hph (∼L2/L3 molt) and at 48 hph (just after the L4/adult molt). GFP::RAB-3 accumulations in wild-type animals were stable in these time intervals (Fig. 4A). By contrast, of the 12 hermaphrodite animals we successfully triply imaged, seven showed apparent synaptic retractions: five began with two accumulations and lost one, and two animals began with one accumulation that later disappeared. In addition, we also quantified branch and synaptic loss in a set of three experiments in which 15-20 animals were examined in the early L2 larval stage and then again in the L4 larval stage. In these experiments (summarized in supplementary material Fig. S5), 42% of the animals lost or retracted one branch, 17% lost or retracted two branches, 32% lost one GFP::RAB-3 accumulation, and 15% lost two accumulations. Together, our results indicate that synapse formation is slightly delayed in PLM neurons and that most synapses formed in zyx-1 animals are unstable. Thus, ZYX-1 participates in the cellular process of PLM synapse formation, maintenance and growth during larval development.
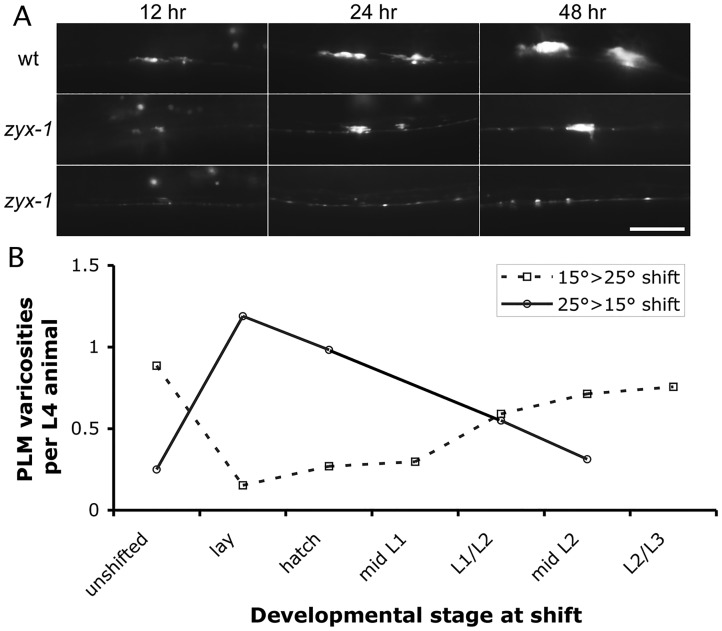
Fig. 4.
zyx-1 mutant synapses retract during development. (A) Timecourse of PLM synapse structure of representative animals. Individual wild-type or zyx-1(gk190) animals were immobilized in 10 mM sodium azide at 12 hph, imaged at 100 ×, recovered on an NGM plate, and then reimaged in similar fashion at 24 hph and 48 hph. The zyx-1 examples illustrate that PLM synapse loss is stochastic. In the top example, one synapse is lost while the other grows. Furthermore, they illustrate that PLM synapses can be lost both early (bottom) as well as late (top) in development. Scale bar: 10 µm. (B) Temperature shift of zyx-1(gk190) animals showing the PLM synapses in L4 larvae as a function of the time of shift (from permissive to non-permissive temperature, or vice versa). n=127-253 animals for shifted time points and n=95-173 for unshifted controls. Strains used were NM3361 and NM3413.
We also observed that the ability of zyx-1 mutants to form synaptic varicosities was greatly influenced by temperature. When zyx-1 mutants were raised at 15°C, synapses appeared to form much more reliably than when animals were raised at 25°C (supplementary material Fig. S6; Fig. 4B). Since both the zyx-1 alleles that we examined (including a deletion allele) exhibited similar temperature sensitivity, we presume this is the result of a temperature sensitivity in the requirement for zyx-1 in the process of PLM synaptogenesis, rather than a temperature sensitivity of the ZYX-1 mutant protein. We used this temperature effect to determine when during development zyx-1 gene function is crucial for synaptic development. When zyx-1 embryos were fertilized and grown at 15°C, but shifted to 25°C in mid-L1 or earlier, animals formed few GFP::RAB-3 accumulations. By contrast, animals shifted after the end of L2 formed GFP::RAB-3 accumulations much more robustly. In downshift experiments, mutant animals initially raised at 25°C formed accumulations robustly if shifted after hatching and before the synaptic branch initiated, but formed synapses progressively less effectively when shifting occurred later in the L1 or L2 larval stage. These observations indicate that zyx-1 contributes to synapse formation and maintenance throughout the period of L1 and L2, and thus there is not just a single time point at which it is needed to initiate branches or to stabilize a branch upon reaching the VNC.
ZYX-1B functions cell-autonomously in PLMs to regulate synapse development
zyx-1 is a complex gene that is expressed as multiple different isoforms in both muscle and neurons (Lecroisey et al., 2013) (supplementary material Fig. S7). The most abundant (at the RNA level) are a long isoform, ZYX-1A, and a short isoform, ZYX-1B. The ZYX-1A isoform, which is analogous to vertebrate zyxin in structure, consists of a poorly conserved domain-less region of 405 amino acids and a C-terminal 198 amino acid region with three highly conserved LIM domains (supplementary material Fig. S8). The 200 amino acid ZYX-1B isoform consists of only the three LIM domains. The zyx-1(gk190) 777 bp deletion allele, which was used in most of our experiments, removes the coding exons for the first two LIM domains and disrupts all protein isoforms of ZYX-1 (supplementary material Fig. S8A), whereas the js417 allele that we recovered contains a splice site lesion in a ZYX-1B-specific exon and thus selectively disrupts ZYX-1B (supplementary material Fig. S8A), suggesting that C-terminal LIM domains are important for mechanosensory synapse development.
We examined zyx-1 expression during development using N- and C-terminal tagged genomic constructs (supplementary material Fig. S7). We found that GFP-ZYX-1B and ZYX-1-GFP were both expressed broadly in neurons and muscle throughout development, while mCherry-ZYX-1 was expressed at low levels throughout development (supplementary material Figs S7 and S9). Furthermore, we examined the expression patterns of a YFP reporter driven by distinct promoters of the zyx-1 gene. The promoter upstream of the js417 lesion drove expression broadly in both neurons and muscle and was specifically expressed in the PLMs (supplementary material Figs S7 and S10). We then directed the expression of ZYX-1::GFP in mechanosensory neurons, in postsynaptic command interneurons and in muscle of zyx-1 mutants to examine which cell types were required for ZYX-1 function. The PLM synaptogenesis defects were rescued only when ZYX-1 was expressed in the PLMs, indicating that ZYX-1 functions cell-autonomously in the presynapse (Fig. 5A).
Fig. 5.
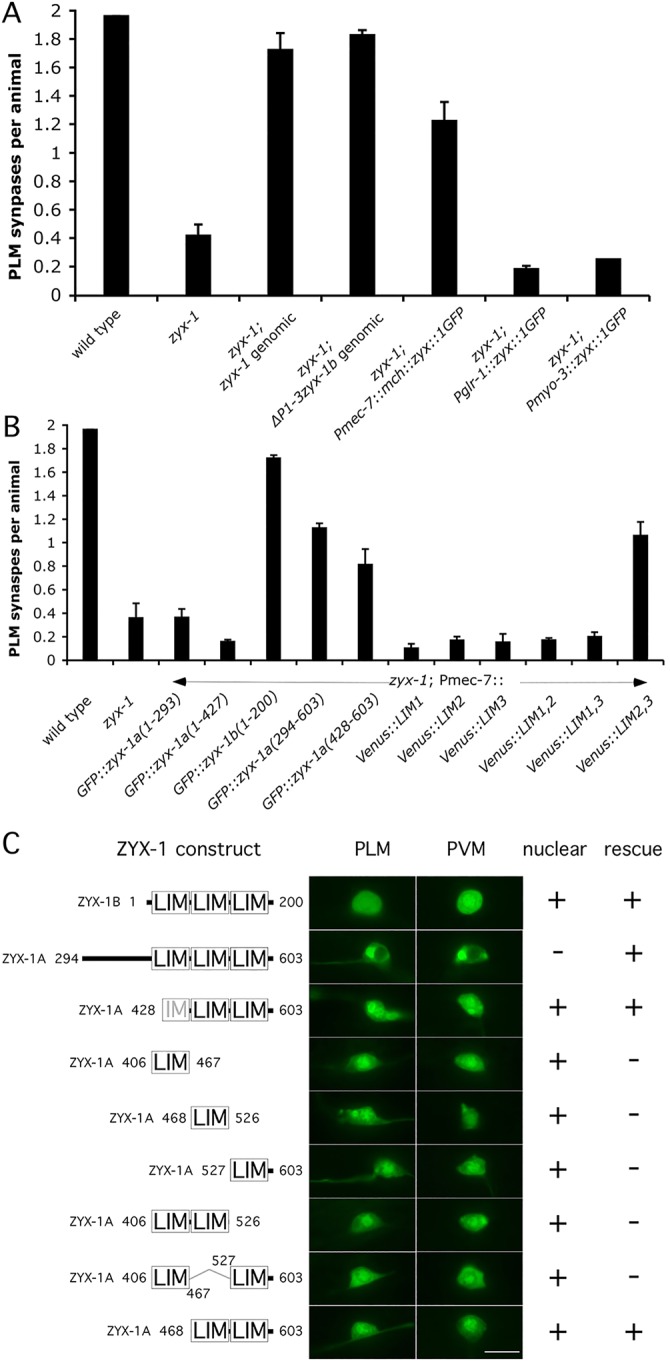
ZYX-1 functions cell-autonomously to regulate mechanosensory synapse development. (A) The expression of full-length ZYX-1A cDNA in presynaptic mechanosensory neurons (Pmec-7::mCherry::zyx-1::GFP) rescues the zyx-1 mutant phenotype, whereas muscular or interneuronal expression does not. PLM synapses formed per animal are displayed for wild type, zyx-1, and zyx-1 mutants expressing zyx-1::GFP fusions under the control of distinct promoters. Also shown are rescues by a complete zyx-1 genomic construct and a genomic construct only capable of expressing zyx-1b from the P4 promoter. (B) The expression of ZYX-1 C-terminal LIM domains in mechanosensory neurons rescues the PLM synapse defects of zyx-1 mutants. PLM synapses formed per animal are displayed for wild type, zyx-1, and zyx-1 mutants expressing various domains of ZYX-1 under the control of the mec-7 promoter. (A,B) Error bars indicate mean ± s.e.m. n=58-897. (C) Subcellular localization of ZYX-1 LIM domain fusions. The structure of various ZYX-1 domain constructs is depicted alongside the localization pattern observed in PLM and PVM, and the phenotypic consequences of expression of the construct in touch receptor neurons. Many LIM domain constructs were able to traffic to the nucleus, but the rescuing activity of the constructs was independent of subcellular localization. Scale bar: 10 μm.
To assess which domains of ZYX-1 are required for PLM development, we expressed the N- and C-terminal portions in mechanosensory neurons of zyx-1 mutants and assayed for the number of GFP accumulations in the VNC of transgenic progeny. Expression of the ZYX-1 N-terminal domain failed to rescue the zyx-1 synaptic phenotype (Fig. 5B). However, expression of the ZYX-1B isoform containing only the three LIM domains and ZYX-1 fragments harboring only the second and third LIM domains fully rescued the zyx-1 synaptic defect. Each of the smaller rescuing LIM domain constructs localized to both the cytosol and nucleus (Fig. 5C). Since vertebrate zyxin has both nuclear and cytoplasmic functions, we expressed a larger fragment of ZYX-1 that contains a region N-terminal to the LIM domains where a nuclear export signal (NES) is located in vertebrate zyxin (Fig. 5A). This larger construct retained the rescuing activity but was excluded from the nucleus, suggesting that ZYX-1 function in PLM synaptogenesis might not depend on its nuclear localization. Together, our results show that the second and third ZYX-1 LIM domains act cell-autonomously to regulate mechanosensory synapse development.
Worm ZYX-1 functions independently of α-actinin and other ZYX-1-interacting proteins
Extensive analysis of several members of the zyxin family has defined a set of molecular partners that mediate interactions of zyxin family members with the cytoskeleton both in vertebrates (Wang et al., 2003; Hirata et al., 2008) and in worms (Smith et al., 2002; Lecroisey et al., 2008, 2013). To define potential mechanisms of action for zyx-1 in mediating PLM synapse development, we examined C. elegans mutants in which these components are disrupted. Mutants in C. elegans homologs of the vertebrate zyxin-interacting proteins nectin and ENA-VASP, and mutants disrupting seven previously defined C. elegans ZYX-1-interacting proteins, all formed synapses robustly, in contrast to zyx-1 mutants (Table 1). Zyxin has been demonstrated to interact with focal adhesions via α-actinin (Reinhard et al., 1999). Furthermore, prior studies revealed that zyx-1 muscle phenotypes (in a dys-1 hlh-1 mutant background) are mediated via the worm α-actinin ATN-1 (Lecroisey et al., 2013). However, atn-1 mutants formed PLM synapses normally (Table 1), indicating that ZYX-1 functions via different interacting proteins in muscle versus PLM neurons.
Table 1.
PLM synapse phenotype of selected mutants
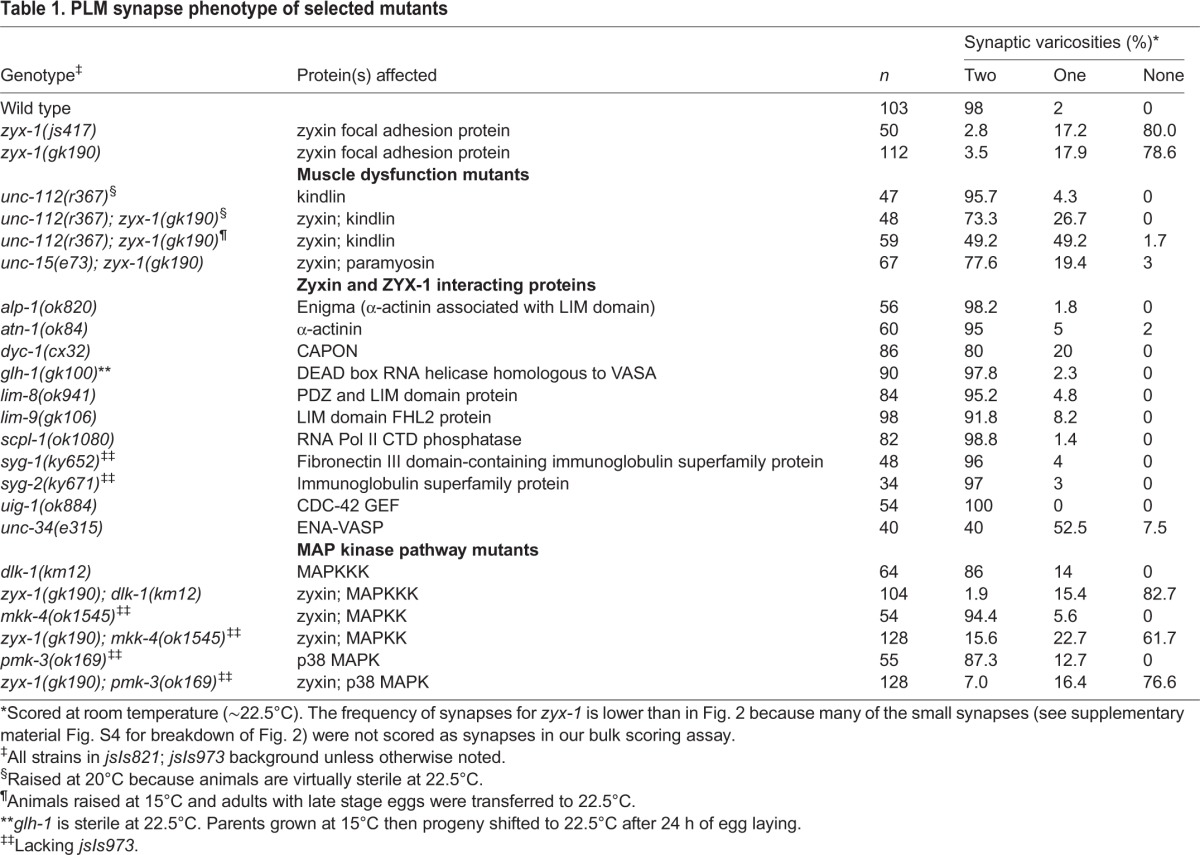
Since the PLM synaptic retraction phenotype of zyx-1 animals resembles that of rpm-1 E3 ubiquitin ligase mutants (Schaefer et al., 2000) and ptrn-1 patronin mutants (Marcette et al., 2014), we examined whether disruptions of the dlk-1 mkk-4 pmk-3 MAP kinase pathway could suppress zyx-1 defects in a similar manner to their suppression of rpm-1 and ptrn-1 defects (Nakata et al., 2005; Marcette et al., 2014). However, PLM synapse assembly was not perturbed in the MAP kinase mutants, and zyx-1 mutant defects were not altered in any of the three MAP kinase double mutants (Table 1). We conclude that the zyx-1 retraction defects are likely to be mediated by a pathway that is independent of the rpm-1/dlk-1 pathway that initiates axon regeneration in response to injury.
zyx-1 stabilizes synapses but is not essential for PLM synaptogenesis
One possible explanation for the delayed PLM synapse formation in zyx-1 mutants is that the absence of functional ZYX-1 impairs the stabilizing influence of adhesion complexes during PLM synaptic branch extension and mechanosensory synapse formation and growth. Synapses in the VNC and DNC of C. elegans are under constant mechanical strain due to the locomotion pattern of the animal. This was exemplified by the constant deformation of the PLM synaptic varicosities during locomotion, which occurred without long-term changes in overall synaptic morphology (Fig. 6A). Thus, wild-type PLM presynaptic varicosities are sufficiently adherent to endure these repetitive locomotion-induced mechanical forces.
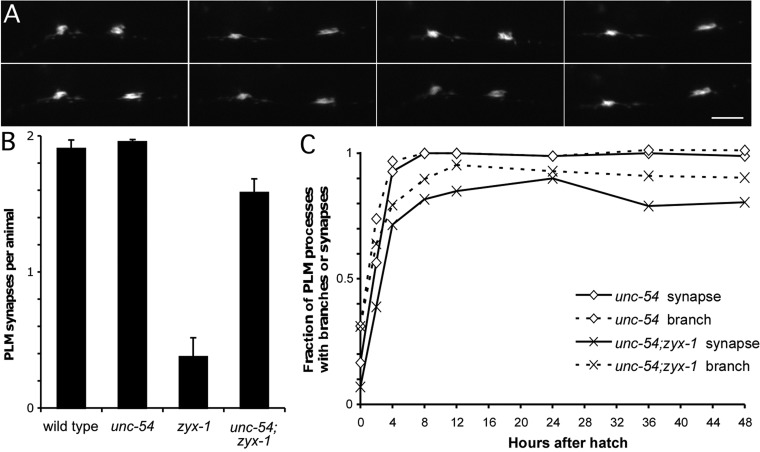
Fig. 6.
PLM synapses are preserved in zyx-1 mutants when locomotion is decreased. (A) Live imaging of PLM synapses during sinusoidal locomotion of an L4 animal. Images taken over the course of 5 min are aligned to show the stability of the morphology of synapses during the physical strain of locomotion. Scale bar: 10 µm. (B) Disruption of the muscle structural gene for myosin heavy chain, unc-54, led to immobilization of zyx-1 animals and restoration of PLM synapses. PLM synapses per animal are plotted as a function of genotype. n=69-140. Error bars indicate ± s.e.m. (C) Disruption of unc-54 suppresses the delay in branch outgrowth and synapse formation caused by loss of zyx-1. The fraction of PLM processes with branches or synapses is shown as a function of time after hatching for unc-54 and unc-54; zyx-1 animals. n=78-136 axons for post-hatching times and n=48-58 axons at hatching.
To examine whether zyx-1 synaptic defects are the result of deficiencies in the ability to respond to mechanical forces acting on the neuronal process, we utilized mutants that disrupt muscle function to test whether reducing movements of the VNC during PLM synaptic development could ameliorate zyx-1 synapse defects. unc-54 animals lack a functional myosin heavy chain and are virtually immobile on agar plates (Waterston et al., 1982). In an unc-54 background, zyx-1 mutants were able to form and maintain PLM synapses at rates approaching those of wild-type animals (Fig. 6B). Similarly, immobile paramyosin unc-15; zyx-1 mutant animals also exhibited robustly restored PLM synaptic structures in the VNC (Table 1). Mutants in unc-112 have late-onset paralysis, with animals moving at hatching but becoming progressively less mobile during development, and are completely paralyzed as adults. Consistent with a major role for zyx-1 during larval stages, unc-112(r367) was much less effective in suppressing the PLM synaptogenesis defect of zyx-1 than the other muscle mutants examined (Table 1).
Lastly, we examined the timecourse of branch and synapse formation in the unc-54; zyx-1 animals. Branch initiation was restored in early L1 larval stages and synapse formation occurred at a comparable time in L1 development as in unc-54 control animals (Fig. 6C). This strongly suggests that zyx-1 functions both during branch outgrowth and during synapse growth to stabilize the extending processes and newly formed presynaptic varicosities in an environment of constantly varying mechanical forces.
DISCUSSION
Previous studies have established a role for cell adhesion molecules in regulating synapse development. However, how these cell adhesion molecules transmit the signals in response to mechanical forces in the cell remains poorly understood. Our work provides the first whole-animal in vivo evidence that zyxin mediates cytoskeletal responses to mechanical forces. Our findings suggest that the C. elegans homolog of the focal adhesion-associated protein zyxin is a likely candidate to transmit cell adhesion tension signals during synaptogenesis and to stabilize synaptic varicosities in mechanosensory neurons during their growth at larval stages. Several observations support this notion. First, zyx-1 functions cell-autonomously in mechanosensory neurons to regulate PLM synapse development. Second, PLM synapses are well preserved in immobilized zyx-1 animals, suggesting that zyx-1 regulates the adherent properties of PLM synaptic contacts. Third, temperature-shift experiments suggest that zyx-1 is required not only during initial synapse formation in the L1 larvae, but also after these synapses are formed. Fourth, the C-terminal LIM domains of ZYX-1 are evolutionarily conserved and these domains of zyxin have been implicated in the response to stress forces. Fifth, in C. elegans the LIM domains are necessary and sufficient to maintain PLM synapses, suggesting that ZYX-1 LIM domains interact with cytoskeletal or cell adhesion components. Our results suggest that zyxin might provide a crucial link between force-bearing cellular structures and their cytoplasmic signaling function in modulating cytoskeletal and cell adhesion properties to maintain PLM synapses in a force-bearing environment.
Mechanistically, how does ZYX-1 function to regulate process outgrowth and synapse formation and maintenance? To address this issue, we examined several classes of candidates based on known biological functions and/or protein interactions with mammalian zyxin family members (Table 1; supplementary material Table S1). Two confounding factors limited our ability to test comprehensively the potential role of some of the proteins in PLM development. In particular, although ZYX-1 is clearly associated with focal adhesions, it is difficult to assess directly whether focal adhesion proteins are involved from the analysis that we performed (supplementary material Table S1). First, the α-integrin pat-2, the β-integrin pat-3, the integrin-linked kinase pat-4 and the PINCH protein unc-97 mutants arrest as twofold paralyzed embryos without completing elongation during embryogenesis before PLM synaptogenesis occurs. In these cases, we examined hypomorphic alleles or null alleles rescued by partially functional transgenes to circumvent the lethality, which nevertheless also confounded the interpretation. Second, we have shown that zyx-1 function is dispensable for PLM synapse formation in the absence of movement, and many focal adhesion mutants have severe locomotory defects due to requirements for the affected genes in muscle. The muscle contraction defects in the candidate mutants that we examined could suppress their requirement for PLM development, much as unc-54 suppresses zyx-1. Ideally, muscle-specific rescue lines or PLM-specific knockdown lines could be examined to test for neuron-specific function, but this was not performed as creating such lines is an extensive experimental undertaking. Thus, although our data did not reveal a role for focal adhesion components in PLM synaptogenesis (supplementary material Table S1), they do not eliminate the possibility that ZYX-1 functions to regulate synaptogenesis through focal adhesions. Our data do provide more forceful conclusions regarding other components (the null mutants of which are viable and motile), such as eliminating the possibility of an essential role for focal adhesion kinase and α-actinin in mediating ZYX-1 actions in synaptogenesis.
Prior work in C. elegans has examined the role of zyx-1 in muscle (Lecroisey et al., 2008, 2013). As mentioned earlier, ZYX-1 is localized to dense bodies in muscle and this localization is ATN-1 dependent. zyx-1 mutants showed no muscle phenotypes on their own, but in a dys-1 hlh-1 double mutant that exhibits 12-18% muscle cell degeneration the introduction of zyx-1(gk190) to create zyx-1 dys-1 hlh-1 triple-mutant animals reduced this degeneration by 60%. atn-1 mutants showed similar effects to zyx-1 on muscle cell degeneration, and this suppression phenotype was partially reversed by expression of the ZYX-1A isoform. Thus, ZYX-1 function in regulating muscle cell degeneration is mediated primarily by ZYX-1A (the ZYX-1B isoform acted as a dominant-negative in their assay) and is ATN-1 dependent. By contrast, ZYX-1 function in PLM synaptic development is mediated entirely by ZYX-1B and is independent of ATN-1. Thus, the roles of ZYX-1 in muscle versus PLM neurons are likely to be mediated by distinct molecular mechanisms.
Our work provides the first in vivo evidence for a role of zyxin in nervous system development. However, the zyx-1 mutant phenotypes appear to be specific to the PLM mechanosensory neurons. There are several possible reasons why the defect is so selective. First, PLMs have 15 specialized protofilament microtubules (MTs), which contain MEC-7 β-tubulin and MEC-12 α-tubulin, and these unusual MTs interact differently with adhesion complexes than typical neuronal MTs, making them more sensitive to applications of strong force. Second, from prior imaging analysis of PLM synaptic branch processes during outgrowth, we concluded that this process is unlikely to result from a growth cone-mediated process (M.L.N., A.M.S. and S.L., unpublished), but more likely from a filopodial-initiated process similar to that observed in collateral branching in vertebrate systems (Gallo, 2011). This filopodial-based process extension might employ fundamentally different adhesion mechanisms than growth cone extensions and could explain why many other processes (e.g. GABAergic commissural processes) are unaffected in zyx-1 animals. Lastly, it is possible that we have identified a specialized cellular feedback mechanism in the mechanosensory cells, which is sensing mechanical forces during the formation of mechanosensory synapses to regulate the relative strength of chemical versus electrical connections that mechanosensory neurons make with postsynaptic partners. Further dissection of the molecular mechanism underlying the zyx-1 phenotype will be required to understand the basis of this selective sensitivity.
We observed growth and subsequent retraction of PLM synaptic branches at high frequency in zyx-1 mutants. In addition, we also observed small globular synaptic marker-positive structures in the VNC (posterior to the vulva) that were no longer connected by a synaptic branch. These very likely represent the remnants of synaptic varicosities that remained after the breakage of the synaptic branches. Surprisingly, in zyx-1 mutants we did not see evidence of cellular attempts to remodel the PLM neurons, as previously described in rpm-1 and ptrn-1 mutants (Schaefer et al., 2000; Zhen et al., 2000; Nakata et al., 2005; Marcette et al., 2014). Thus, these breakage events did not appear to induce strong dlk-1 activation, which is necessary for axonal regrowth (Nix et al., 2011) and is characterized by exuberant overgrowth of touch neuron processes. Perturbations of MTs are known to activate dlk-1 and induce such remodeling (Valakh et al., 2013; Marcette et al., 2014). Perhaps loss of zyx-1 leads primarily to perturbations of the actin cytoskeleton network (such as actin stress fibers), and such disruptions are sensed differently than MT perturbation by the axonal damage cellular surveillance systems.
The classic zyxin protein contains a long ‘pre’-LIM domain with binding sites for α-actinin, ENA/VASP and adhesion molecules, a nuclear export sequence, and a C-terminal moiety with three LIM motifs. Most studies of this family treat the protein as one entity. However, C. elegans expresses two isoforms of zyxin (1B and 1C) that consist of only the LIM domains. Furthermore, our work demonstrates that the LIM domain-only isoform ZYX-1B functions independently of the pre-LIM domain in mediating the cellular synaptogenesis-promoting functions of ZYX-1 in PLM neurons. Our data also indicate that this isoform of ZYX-1 is intimately involved in sensing forces. Although an analogous LIM domain-only isoform of zyxin has not been characterized in vertebrates, Uemura et al. (2011) have demonstrated the ability of a LIM domain-only fragment to localize at adhesion sites in response to applied mechanical force. Furthermore, several mouse cDNAs consistent with the presence of such an isoform in vivo are present in GenBank (supplementary material Fig. S8). The low expression of zyxin in brain could explain the rarity of such transcripts in the public DNA sequence databases (Macalma et al., 1996). In addition, short isoform cDNAs for mouse Ajuba and WTIP are also present in sequence databases (supplementary material Fig. S8), and a short isoform of TRIP6 has also been characterized in detail (Kassel et al., 2004). Thus, we propose that the neuronal functions of vertebrate zyxin (and perhaps other family members) might be mediated by isoforms that have thus far been poorly defined at the molecular level.
Our work implies that the LIM domains of zyxin are capable not only of detecting forces, as has been previously demonstrated, but also of mediating the cellular responses to such forces. However, the mechanism by which the LIM domains mediate these responses remains elusive. LIM domains are known to mediate the translocation of both zyxin and paxillin to stress fibers. An attractive model is that the LIM domains bind directly to actin stress fibers that are being stretched and provide mechanical support to these fibers. Such a mechanism could provide a first line of cytoskeletal support that acts before the recruitment of other cytoskeletal remodeling factors by interaction via the N-terminal domain of zyxin. Alternatively, the LIM domains might be recruited to sites bearing mechanical force by as yet undefined proteins. Further genetic and biochemical studies should refine our understanding of the contribution of the LIM domains in the response to mechanical perturbations of the cytoskeleton.
MATERIALS AND METHODS
C. elegans strains
All C. elegans strains (listed in supplementary material Table S4) were maintained on E. coli OP50-spotted NGM plates using standard procedures (Sulston and Hodgkin, 1988). Alleles used are listed in supplementary material Table S2. Transgenic markers are listed in supplementary material Table S3.
Molecular biology
Cloning of zyx-1(js417) is described in the supplementary material Methods; SNP markers used in cloning and mapping are detailed in supplementary material Table S6. Characterization of gk442 is described in supplementary material Fig. S11. A detailed description of plasmid constructions is provided in the supplementary material Methods. Oligonucleotides used in these constructions are listed in supplementary material Table S5.
Immunohistochemistry
Animals were fixed and permeabilized as previously described using anti-UNC-10 (Ab5431, 1:20,000) and anti-RAB-3 (Ab326, 1:2000) (Koushika et al., 2001).
Temperature-shift experiments
Wild-type and mutant young adult animals were allowed to lay eggs at 15°C or 25°C on a fresh plate for 1 h and then the adults were removed. Plates were shifted to the alternate temperature at various times after the lay, and then scored for PLM synaptic defects when the animals reached the L4 larval stage.
Scoring for PLM synapses
Bulk scoring of L4 animals (Table 1) was performed by mounting animals on 2% agarose or by placing animals in 8 µl diluted 25 µm solid polystyrene Polybeads (Polysciences) with 10 mM sodium azide. Animals were imaged on an Olympus BX60 or a Zeiss Akioskop microscope and scored for the presence of GFP::RAB-3 accumulations in the VNC just posterior to the vulva. A synaptic varicosity was defined as a GFP::RAB-3 accumulation at the end of the ventral collateral branch extending from the lateral process of the PLMs that was brighter than the GFP::RAB-3 puncta presented in the PVM process in the VNC.
Analysis of neuronal morphology of other cell types and PLM neurons expressing other markers is described in supplementary material Methods.
Quantification of PLM synapse development
Wild-type and zyx-1 animals were grown for various times after hatching from synchronized populations of eggs. Animals were either placed on 2% agarose plates, or placed in diluted 10 or 25 µm polystyrene Polybeads on a slide and covered with a coverslip and anesthetized using 10 mM levamisole or 10 mM sodium azide. Animals were photographed using a Retiga EXi CCD camera (Q-imaging) using OpenLab (PerkinElmer) on an Olympus BX60 in brightfield at 4, 8 or 20 ×, and at several different focal planes in green and red epifluorescence channels using either a 60 × NA 1.4 (L1 and L2 larvae) or a 40 × NA 0.75 (all stages), or a 20 × NA 0.5 (L4 and adults) objective. Length of animals was measured using ImageJ (NIH). Animals were positioned on a developmental timeline based on both length and morphological characteristics (presence of PVM, position of the PVM axon and germ line size).
A branch was defined as a ventral or dorsal protrusion in the mRFP channel extending at least 2.5 µm. A full branch was defined as a branch extending into the VNC. A synapse was defined as a complete branch that was positive for GFP::RAB-3 in the VNC at levels brighter than the average punctum in the PVM process. A large synapse was defined as a VNC GFP::RAB-3 accumulation in a full branch that was brighter than any GFP::RAB-3 signal in the PLM lateral branch (trafficking SV and SV precursors). Remnants were defined as RFP-positive spherical structures that were brighter than the PVM process, between the PVM process and AVG process, and were not connected by a process to the PLM lateral process.
Multifocal plane images were produced by creating a maximal projection of individual planes, and green and red channel images were merged in Photoshop (Adobe), and levels were adjusted to equalize the background of different images.
Germ line transformation
The transgenic lines were constructed using standard germ line transformation procedures (Mello et al., 1991). All the zyx-1 rescue plasmids were injected at a final concentration of 30 ng/μl, while the co-injection marker Pmyo-2::GFP and the plasmid vector pcDNA3 were injected at 5 ng/μl and 100 ng/μl, respectively. At least two independent transgenic lines of each injection were analyzed for the rescue of PLM synapse development phenotypes.
Supplementary Material
Acknowledgements
We thank Dr A. Francis Stewart for providing reagents for recombineering cloning and Rose Vincent for technical assistance in mapping and transformation rescue.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
All authors contributed to performing the experiments and data analysis. M.L.N., S.L. and A.M.S. prepared the manuscript.
Funding
This work was supported by a grant [NS040094] from the National Institutes of Health to M.L.N. Some strains used in this work were obtained from the Caenorhabditis elegans Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources. Images for this work were obtained using the Bakewell Imaging Facility, which is supported by an NIH Neuroscience Blueprint Core Grant [NS057105] to Washington University. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.108217/-/DC1
References
- Balice-Gordon, R. J., Chua, C. K., Nelson, C. C. and Lichtman, J. W. (1993). Gradual loss of synaptic cartels precedes axon withdrawal at developing neuromuscular junctions. Neuron 11, 801-815 10.1016/0896-6273(93)90110-D [DOI] [PubMed] [Google Scholar]
- Beckerle, M. C. (1986). Identification of a new protein localized at sites of cell-substrate adhesion. J. Cell Biol. 103, 1679-1687 10.1083/jcb.103.5.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D. L. and Huntley, G. W. (2012). Synapse adhesion: a dynamic equilibrium conferring stability and flexibility. Curr. Opin. Neurobiol. 22, 397-404 10.1016/j.conb.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas, A., Zheng, Q., Nonet, M. L. and Chalfie, M. (2009). mec-15 encodes an F-box protein required for touch receptor neuron mechanosensation, synapse formation and development. Genetics 183, 607-617 10.1534/genetics.109.105726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie, M., Sulston, J. E., White, J. G., Southgate, E., Thomson, J. N. and Brenner, S. (1985). The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 5, 956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombelli, J., Besser, A., Kress, H., Reynaud, E. G., Girard, P., Caussinus, E., Haselmann, U., Small, J. V., Schwarz, U. S. and Stelzer, E. H. K. (2009). Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J. Cell Sci. 122, 1665-1679 10.1242/jcs.042986 [DOI] [PubMed] [Google Scholar]
- Crawford, A. W., Michelsen, J. W. and Beckerle, M. C. (1992). An interaction between zyxin and alpha-actinin. J. Cell Biol. 116, 1381-1393 10.1083/jcb.116.6.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur, M., Feng, Y., Jagannathan, R., Seppa, M. J., Skeath, J. B. and Longmore, G. D. (2010). Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20, 657-662 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt, Y. Y. and Silverstein, S. (2001). Interaction of zyxin, a focal adhesion protein, with the e6 protein from human papillomavirus type 6 results in its nuclear translocation. J. Virol. 75, 11791-11802 10.1128/JVI.75.23.11791-11802.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B. E., Andrews, K. M. and Beckerle, M. C. (1999). Molecular dissection of zyxin function reveals its involvement in cell motility. J. Cell Biol. 147, 1549-1560 10.1083/jcb.147.7.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B., Friederich, E., Fradelizi, J., Louvard, D., Beckerle, M. C. and Golsteyn, R. M. (2000). Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 275, 22503-22511 10.1074/jbc.M001698200 [DOI] [PubMed] [Google Scholar]
- Emtage, L., Gu, G., Hartwieg, E. and Chalfie, M. (2004). Extracellular proteins organize the mechanosensory channel complex in C. elegans touch receptor neurons. Neuron 44, 795-807 10.1016/j.neuron.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Foxler, D. E., Bridge, K. S., James, V., Webb, T. M., Mee, M., Wong, S. C. K., Feng, Y., Constantin-Teodosiu, D., Petursdottir, T. E., Bjornsson, J.et al. (2012). The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat. Cell Biol. 14, 201-208 10.1038/ncb2424 [DOI] [PubMed] [Google Scholar]
- Franze, K. (2013). The mechanical control of nervous system development. Development 140, 3069-3077 10.1242/dev.079145 [DOI] [PubMed] [Google Scholar]
- Gallo, G. (2011). The cytoskeletal and signaling mechanisms of axon collateral branching. Dev. Neurobiol. 71, 201-220 10.1002/dneu.20852 [DOI] [PubMed] [Google Scholar]
- Gan, W.-B. and Lichtman, J. W. (1998). Synaptic segregation at the developing neuromuscular junction. Science 282, 1508-1511 10.1126/science.282.5393.1508 [DOI] [PubMed] [Google Scholar]
- Gomez, T. M., Roche, F. K. and Letourneau, P. C. (1996). Chick sensory neuronal growth cones distinguish fibronectin from laminin by making substratum contacts that resemble focal contacts. J. Neurobiol. 29, 18-34 [DOI] [PubMed] [Google Scholar]
- Goyal, R. K., Lin, P., Kanungo, J., Payne, A. S., Muslin, A. J. and Longmore, G. D. (1999). Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 19, 4379-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, H., Tatsumi, H. and Sokabe, M. (2008). Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun. Integr. Biol. 1, 192-195 10.4161/cib.1.2.7001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. M., Nix, D. A., Benson, B., Boot-Hanford, R., Gustafsson, E., Jamora, C., Menzies, A. S., Goh, K. L., Jensen, C. C.et al. (2003). Targeted disruption of the murine zyxin gene. Mol. Cell. Biol. 23, 70-79 10.1128/MCB.23.1.70-79.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. M., Jensen, C. C., Kloeker, S., Wang, C.-L. A., Yoshigi, M. and Beckerle, M. C. (2006). Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 172, 771-782 10.1083/jcb.200512115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. M., Jensen, C. C., Chaturvedi, A., Yoshigi, M. and Beckerle, M. C. (2012). Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol. Biol. Cell 23, 1846-1859 10.1091/mbc.E11-12-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, V., Zhang, Y., Foxler, D. E., de Moor, C. H., Kong, Y. W., Webb, T. M., Self, T. J., Feng, Y., Lagos, D., Chu, C.-Y.et al. (2010). LIM-domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA-mediated gene silencing. Proc. Natl. Acad. Sci. USA 107, 12499-12504 10.1073/pnas.0914987107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay, D. G. (2000). The clutch hypothesis revisited: ascribing the roles of actin-associated proteins in filopodial protrusion in the nerve growth cone. J. Neurobiol. 44, 114-125 [DOI] [PubMed] [Google Scholar]
- Kassel, O., Schneider, S., Heilbock, C., Litfin, M., Gottlicher, M. and Herrlich, P. (2004). A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-kappaB-regulated promoters. Genes Dev. 18, 2518-2528 10.1101/gad.322404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva, M., Feng, Y., Ward, M., Song, C., Anderson, R. A. and Longmore, G. D. (2005). The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKI alpha. Mol. Cell. Biol. 25, 3956-3966 10.1128/MCB.25.10.3956-3966.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika, S. P., Richmond, J. E., Hadwiger, G., Weimer, R. M., Jorgensen, E. M. and Nonet, M. L. (2001). A post-docking role for active zone protein Rim. Nat. Neurosci. 4, 997-1005 10.1038/nn732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey, C., Martin, E., Mariol, M.-C., Granger, L., Schwab, Y., Labouesse, M., Segalat, L. and Gieseler, K. (2008). DYC-1, a protein functionally linked to dystrophin in Caenorhabditis elegans is associated with the dense body, where it interacts with the muscle LIM domain protein ZYX-1. Mol. Biol. Cell 19, 785-796 10.1091/mbc.E07-05-0497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey, C., Brouilly, N., Qadota, H., Mariol, M.-C., Rochette, N. C., Martin, E., Benian, G. M., Ségalat, L., Mounier, N. and Gieseler, K. (2013). ZYX-1, the unique zyxin protein of Caenorhabditis elegans, is involved in dystrophin-dependent muscle degeneration. Mol. Biol. Cell 24, 1232-1249 10.1091/mbc.E12-09-0679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalma, T., Otte, J., Hensler, M. E., Bockholt, S. M., Louis, H. A., Kalff-Suske, M., Grzeschik, K.-H., von der Ahe, D. and Beckerle, M. C. (1996). Molecular characterization of human zyxin. J. Biol. Chem. 271, 31470-31478 10.1074/jbc.271.49.31470 [DOI] [PubMed] [Google Scholar]
- Marcette, J. D., Chen, J. J. and Nonet, M. L. (2014). The Caenorhabditis elegans microtubule minus-end binding homolog PTRN-1 stabilizes synapses and neurites. Elife 3, e01637 10.7554/eLife.01637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynova, N. Y., Eroshkin, F. M., Ermolina, L. V., Ermakova, G. V., Korotaeva, A. L., Smurova, K. M., Gyoeva, F. K. and Zaraisky, A. G. (2008). The LIM-domain protein Zyxin binds the homeodomain factor Xanf1/Hesx1 and modulates its activity in the anterior neural plate of Xenopus laevis embryo. Dev. Dyn. 237, 736-749 10.1002/dvdy.21471 [DOI] [PubMed] [Google Scholar]
- Mello, C. C., Kramer, J. M., Stinchcomb, D. and Ambros, V. (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, M., Nakagami, H., Koibuchi, N., Miura, K., Takami, Y., Koriyama, H., Hayashi, H., Sabe, H., Mochizuki, N., Morishita, R.et al. (2009). Zyxin mediates actin fiber reorganization in epithelial-mesenchymal transition and contributes to endocardial morphogenesis. Mol. Biol. Cell 20, 3115-3124 10.1091/mbc.E09-01-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata, K., Abrams, B., Grill, B., Goncharov, A., Huang, X., Chisholm, A. D. and Jin, Y. (2005). Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120, 407-420 10.1016/j.cell.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Nix, P., Hisamoto, N., Matsumoto, K. and Bastiani, M. (2011). Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc. Natl. Acad. Sci. USA 108, 10738-10743 10.1073/pnas.1104830108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet, M. L. (1999). Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein-GFP fusions. J. Neurosci. Methods 89, 33-40 10.1016/S0165-0270(99)00031-X [DOI] [PubMed] [Google Scholar]
- Paulus, J. D., Willer, G. B., Willer, J. R., Gregg, R. G. and Halloran, M. C. (2009). Muscle contractions guide rohon-beard peripheral sensory axons. J. Neurosci. 29, 13190-13201 10.1523/JNEUROSCI.2179-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, M. M. R., Fradelizi, J., Golsteyn, R. M., Ayoubi, T. A. Y., Menichi, B., Louvard, D., Van de Ven, W. J. M. and Friederich, E. (2000). LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11, 117-129 10.1091/mbc.11.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, M. M. R., Meulemans, S. M. P. and Van de Ven, W. J. M. (2003). The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J. Biol. Chem. 278, 2157-2168 10.1074/jbc.M206106200 [DOI] [PubMed] [Google Scholar]
- Pfister, B. J., Iwata, A., Meaney, D. F. and Smith, D. H. (2004). Extreme stretch growth of integrated axons. J. Neurosci. 24, 7978-7983 10.1523/JNEUROSCI.1974-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper, J. and Mason, C. (2010). Cellular strategies of axonal pathfinding. Cold Spring Harb. Perspect. Biol. 2, a001933 10.1101/cshperspect.a001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard, M., Zumbrunn, J., Jaquemar, D., Kuhn, M., Walter, U. and Trueb, B. (1999). An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment. J. Biol. Chem. 274, 13410-13418 10.1074/jbc.274.19.13410 [DOI] [PubMed] [Google Scholar]
- Renfranz, P. J., Blankman, E. and Beckerle, M. C. (2010). The cytoskeletal regulator zyxin is required for viability in Drosophila melanogaster. Anat. Rec. (Hoboken) 293, 1455-1469 10.1002/ar.21193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler, I., Crawford, A. W., Michelsen, J. W. and Beckerle, M. C. (1992). Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J. Cell Biol. 119, 1573-1587 10.1083/jcb.119.6.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, A. M., Hadwiger, G. D. and Nonet, M. L. (2000). rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26, 345-356 10.1016/S0896-6273(00)81168-X [DOI] [PubMed] [Google Scholar]
- Sharp, T. V., Munoz, F., Bourboulia, D., Presneau, N., Darai, E., Wang, H.-W., Cannon, M., Butcher, D. N., Nicholson, A. G., Klien, G.et al. (2004). LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc. Natl. Acad. Sci. USA 101, 16531-16536 10.1073/pnas.0407123101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P., Leung-Chiu, W.-M., Montgomery, R., Orsborn, A., Kuznicki, K., Gressman-Coberly, E., Mutapcic, L. and Bennett, K. (2002). The GLH proteins, Caenorhabditis elegans P granule components, associate with CSN-5 and KGB-1, proteins necessary for fertility, and with ZYX-1, a predicted cytoskeletal protein. Dev. Biol. 251, 333-347 10.1006/dbio.2002.0832 [DOI] [PubMed] [Google Scholar]
- Smith, M. A., Blankman, E., Gardel, M. L., Luettjohann, L., Waterman, C. M. and Beckerle, M. C. (2010). A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev. Cell 19, 365-376 10.1016/j.devcel.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. A., Blankman, E., Deakin, N. O., Hoffman, L. M., Jensen, C. C., Turner, C. E. and Beckerle, M. C. (2013). LIM domains target actin regulators paxillin and zyxin to sites of stress fiber strain. PLoS ONE 8, e69378 10.1371/journal.pone.0069378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry, R. B., Bishop, N. H., Bramwell, J. J., Brodeur, M. N., Carter, M. J., Fowler, B. T., Lewis, Z. B., Maxfield, S. D., Staley, D. M., Vellinga, R. M.et al. (2010). Zyxin controls migration in epithelial-mesenchymal transition by mediating actin-membrane linkages at cell-cell junctions. J. Cell Physiol. 222, 612-624. [DOI] [PubMed] [Google Scholar]
- Srichai, M. B., Konieczkowski, M., Padiyar, A., Konieczkowski, D. J., Mukherjee, A., Hayden, P. S., Kamat, S., El-Meanawy, M. A., Khan, S., Mundel, P.et al. (2004). A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J. Biol. Chem. 279, 14398-14408 10.1074/jbc.M314155200 [DOI] [PubMed] [Google Scholar]
- Sulston, J. and Hodgkin, J. (1988). Methods. In The Nematode Caenorhabditis elegans (ed. Wood W. B.), pp. 587-606 Cold Spring Harbor, New York: Cold Spring Harbor Laboratory. [Google Scholar]
- Suter, D. M. and Miller, K. E. (2011). The emerging role of forces in axonal elongation. Prog. Neurobiol. 94, 91-101 10.1016/j.pneurobio.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy, S. M., Lai, P. B., Pang, E., Wong, N. L., To, K. F., Johnson, P. J. and Wong, N. (2006). Novel identification of zyxin upregulations in the motile phenotype of hepatocellular carcinoma. Mod. Pathol. 19, 1108-1116. [DOI] [PubMed] [Google Scholar]
- Uemura, A., Nguyen, T.-N., Steele, A. N. and Yamada, S. (2011). The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys. J. 101, 1069-1075 10.1016/j.bpj.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valakh, V., Walker, L. J., Skeath, J. B. and DiAntonio, A. (2013). Loss of the spectraplakin short stop activates the DLK injury response pathway in Drosophila. J. Neurosci. 33, 17863-17873 10.1523/JNEUROSCI.2196-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervenne, H. B. V. K., Crombez, K. R. M. O., Delvaux, E. L., Janssens, V., Van de Ven, W. J. M. and Petit, M. M. R. (2009). Targeted disruption of the mouse Lipoma Preferred Partner gene. Biochem. Biophys. Res. Commun. 379, 368-373 10.1016/j.bbrc.2008.12.074 [DOI] [PubMed] [Google Scholar]
- Wang, C.-T., Lu, J.-C., Bai, J., Chang, P. Y., Martin, T. F. J., Chapman, E. R. and Jackson, M. B. (2003). Different domains of synaptotagmin control the choice between kiss-and-run and full fusion. Nature 424, 943-947 10.1038/nature01857 [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., Smith, K. C. and Moerman, D. G. (1982). Genetic fine structure analysis of the myosin heavy chain gene unc-54 of Caenorhabditis elegans. J. Mol. Biol. 158, 1-15 10.1016/0022-2836(82)90447-8 [DOI] [PubMed] [Google Scholar]
- Weiss, P. (1941). Nerve patterns: the mechanics of nerve growth. Growth: Third Growth Symposium 5, 163-203. [Google Scholar]
- Yi, J. and Beckerle, M. C. (1998). The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics 49, 314-316 10.1006/geno.1998.5248 [DOI] [PubMed] [Google Scholar]
- Yoshigi, M., Hoffman, L. M., Jensen, C. C., Yost, H. J. and Beckerle, M. C. (2005). Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171, 209-215 10.1083/jcb.200505018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. P. and Luo, J.-H. (2006). Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 66, 7414-7419 10.1158/0008-5472.CAN-06-0227 [DOI] [PubMed] [Google Scholar]
- Zhen, M., Huang, X., Bamber, B. and Jin, Y. (2000). Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26, 331-343 10.1016/S0896-6273(00)81167-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.