Abstract
Sirtuins are NAD+-dependent deacylases that regulate numerous biological processes in response to the environment. SirT1 is the mammalian ortholog of yeast Sir2, and is involved in many metabolic pathways in somatic tissues. Whole body deletion of SirT1 alters reproductive function in oocytes and the testes, in part caused by defects in central neuro-endocrine control. To study the function of SirT1 specifically in the male germ line, we deleted this sirtuin in male germ cells and found that mutant mice had smaller testes, a delay in differentiation of pre-meiotic germ cells, decreased spermatozoa number, an increased proportion of abnormal spermatozoa and reduced fertility. At the molecular level, mutants do not have the characteristic increase in acetylation of histone H4 at residues K5, K8 and K12 during spermiogenesis and demonstrate corresponding defects in the histone to protamine transition. Our findings thus reveal a germ cell-autonomous role of SirT1 in spermatogenesis.
Keywords: SirT1, Male germ cell, Reproduction
INTRODUCTION
Sirtuins are a family of NAD+-dependent enzymes that exert many biological effects across various organisms (Haigis and Sinclair, 2010). Mammals possess seven Sir2 type genes (SirT), Sirt1 to Sirt7, which have different subcellular localizations, biological substrates and metabolic functions (Verdin et al., 2010). Sirt1 is the most similar in sequence to yeast Sir2, the gene found to regulate replicative aging in Saccharomyces cerevisiae, and is the most studied mammalian sirtuin protein to date. Whole body SirT1 knockout mice are born at sub-Mendelian ratios, have decreased body size and are infertile (McBurney et al., 2003). These mice have decreased levels of the gonadotropic hormones luteinizing hormone (LH) and follicle stimulating hormone (FSH), indicating a defect in the hypothalamic-pituitary-gonadotropin (HPG) axis, and accordingly fertility in SirT1 knockout females can be restored by administering these hormones (Kolthur-Seetharam et al., 2009; McBurney et al., 2003). Spermatozoa isolated from knockout males display abnormal morphology (McBurney et al., 2003) and are not as efficient in in vitro fertilization (IVF) experiments as wild-type sperm (Coussens et al., 2008). These latter deficiencies might result from a defect in the HPG axis, but might also presage an additional cell-autonomous defect in spermatogenesis.
Spermatogenesis comprises the division and differentiation of spermatogonial stem cells into mature spermatozoa in the testis. This process comprises two phases – the first includes mitotic and meiotic divisions, resulting in haploid round spermatids; and the second phase, termed spermiogenesis, features the differentiation of the round spermatids into mature spermatozoa (Russell et al., 1990). Differentiation takes place in the seminiferous tubules in testis, and proceeds from the outside to the inside of this structure, resulting in the release of mature spermatozoa into the lumen. Tubules are organized into segments of single cell associations, and cross-section analyses of tubules of adult animals reveals that a given segment exists at one of 12 morphologically distinguishable stages of differentiation (from spermatogonial stem cells to spermatozoa). A highly specialized program of chromatin remodeling and compaction characterizes the spermiogenesis program (Kimmins and Sassone-Corsi, 2005).
Proper DNA condensation during spermiogenesis requires that most histone-based nucleosomes be removed and replaced by nucleo-protamine structures covering the majority of the genome. Loss of protamines, or mutation of the testes-specific isoform of histone H2B that disrupts histone removal, render male mice infertile (Cho et al., 2001; Montellier et al., 2013). Histone removal is initiated by post-translation modification of the nucleosomes. Acetylation of histone H4 on residues K5 (H4K5) and K8 (H4K8) provide a binding site for the testis-specific bromodomain protein BRDT to bind to nucleosomes (Morinière et al., 2009). In the absence of BRDT-H4 binding, transition protein 2 (TP2) and protamines (PRM) are unable to properly localize within the nuclei of elongating and condensing spermatids, resulting in altered chromatin condensation and infertility (Gaucher et al., 2012).
Here, we investigated whether SirT1 has a cell-autonomous role in the male germ cell by specifically deleting it in pre-meiotic and post-meiotic cells. We find that deleting SirT1 in late stage post-meiotic cells has no effect, indicating that SirT1 is not expressed in these cells. However, deletion in the pre-meiotic cells results in multiple defects. Mice present with decreased testis size, a delay in pre-meiotic differentiation and abnormally shaped spermatozoa with increased levels of DNA damage. Furthermore, these mice exhibit a defect in the histone to protamine transition and altered chromatin condensation. Ultimately, these mice have decreased fecundity, providing evidence that SirT1 governs spermatogenesis through a male germ cell-autonomous role.
RESULTS
SirT1 is highly expressed in the testis and found in multiple types of germ cell
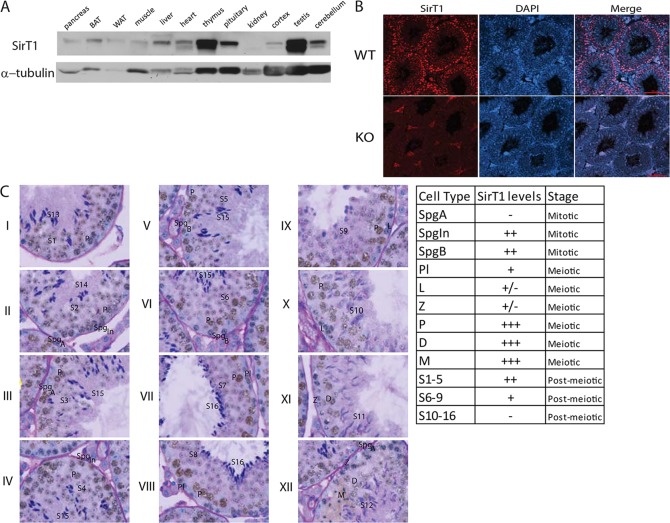
A comparison of SirT1 protein levels across various tissues, on a per microgram basis, indicates that it is highly expressed in the testes (Fig. 1A). Broad examination of SirT1 localization in the seminiferous tubules indicated that it is expressed in multiple cell types (Fig. 1B). A careful examination of the cell types in which SirT1 protein is present using immunohistochemistry staining demonstrated a biphasic expression pattern (Fig. 1C). SirT1 protein was not detectable in undifferentiated type A spermatogonia (stages II-III; SpgA), whereas intermediate spermatogonia (stage IV; SpgIn) and type B spermatogonia (stages V and VI; SpgB), as well as pre-leptotene cells (stage VII; Pl), contained moderate levels of SirT1 protein. Pachytene (stages I-X; P), diplotene (stages XI and XII; D) and meiotic spermatocytes (stage XII; M) contained the highest levels of SirT1 protein. Interestingly, SirT1 protein was evident in cells undergoing both mitotic (stage IV; SpgIn) and meiotic (stage XII; M) divisions. Some leptotene (stages IX and X; L) and zygotene (stage XI; Z) spermatocytes, but not all, demonstrated minimal staining for SirT1, whereas early round spermatids (stages I-V; S1-S5) displayed an intermediate signal for SirT1, which declined as they progressed through spermiogenesis (stage VI-IX; S6-S8). Elongating spermatids (stage X-XII; S9-S12) have very low levels of SirT1 protein, whereas condensing spermatids (stages I-VIII; S13-S16) noticeably lacked any SirT1 protein.
Fig. 1.
. SirT1 is highly expressed in the testis. (A) Immunoblot of SirT1 in mouse tissues. Each lane contains 50 µg of protein. (B) Immunofluorescence of SirT1 protein in cross sections of seminiferous tubules from wild-type and SirT1 whole body KO mice. (C) Immunohistochemistry for SirT1 counterstained with PAS/H in the XII stages of the seminiferous tubules. Representative images of stages I-XII taken with a 100× objective. The table on the right indicates the relative abundance in the different cell types, ± indicates that some of these cell populations demonstrate light staining. BAT, brown adipose tissue; WAT, white adipose tissue; SpgA, A type spermatogonia; SpgIn, Intermediate spermatogonia; SpgB, B type spermatogonia; Pl, pre-leptotene; L, leptotene; Z, zygotene; P, pachytene; D, diplotene; M, meiotic; S, spermatid.
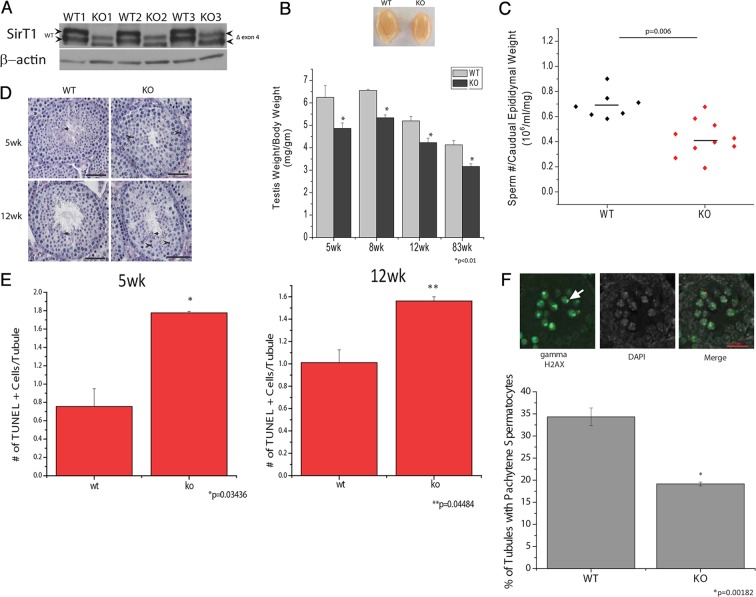
To determine whether SirT1 has a cell-autonomous role in testis, we deleted the catalytic exon 4 of SirT1 specifically in male germ cells. SirT1 floxed mice (f/f) (Cheng et al., 2003) were crossed to Stra8-iCre mice (Sadate-Ngatchou et al., 2008) or Prm1-Cre mice to generate a male germ cell-specific SirT1 knockout mouse that has SirT1 exon 4 deleted in pre-meiotic or post-meiotic germ cells, respectively. The Stra8-iCre; SirT1 f/f knockout mice demonstrated excision of exon 4 in immunoblot analyses, as evidenced by increased mobility in the protein when compared with SirT1 f/f (WT) mice (Fig. 2A). These mice did not have differences in body weight (supplementary material Fig. S1A) or epididymal weight (supplementary material Fig. S1B) when compared with SirT1 f/f (WT) controls. However, they did exhibit a decrease in testis weight as early as 5 weeks of age, which persists as the animals aged (Fig. 2B).
Fig. 2.
Deletion of SirT1 in pre-meiotic cells decreases testis weight and sperm number. (A) SirT1 immunoblotting on whole testis lysate from three biological replicates of 3-month-old SirT1f/f (WT) and Stra8-iCre; SirT1f/f (KO) mice. (B) Testis weight per gram of body weight in SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. Images are of 12-week-old WT and KO mice. (C) Number of sperm per caudal epididymis from 12-week-old SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. The line represents the mean sperm number. (D) PAS/H-stained cross sections of seminiferous tubules from 5-week- and 12-week-old SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. Black arrows indicate elongating and/or condensing spermatids, and arrowheads indicate dying and/or dead cells. (E) The number of TUNEL-positive cells per tubule cross section in SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice aged 5 weeks or 12 weeks. (F) Pachytene spermatocytes are identified by the γH2AX positive sex body (top; arrow). Quantification of tubules that contain pachytene spermatocytes in SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) 12 days post-partum mice (bottom). n=3 mice per group. Scale bars: 50 µm (D); 20 µm (F).
Analysis of protein extracts of whole testes from the Prm1-Cre; SirT1 f/f mice demonstrated the presence of full-length SirT1 without excision of exon 4. This could be due to SirT1 protein present in the stages that precede Prm1-Cre activation, or the removal of exon 4 by Prm1-Cre did not work (supplementary material Fig. S1D). To check that Prm1-Cre was active, we crossed Prm1-Cre; SirT1 f/f mice to the dTomato-green fluorescent protein (GFP) reporter mouse that normally expresses dTomato. However when Cre is expressed, dTomato is excised and the mouse then expresses GFP (Muzumdar et al., 2007). Analysis of these mice demonstrated extensive GFP expression in the post-meiotic cells, indicating that Prm1-Cre was indeed active (supplementary material Fig. S1E). Furthermore, genotyping of DNA isolated from the sperm showed that exon 4 had been deleted (supplementary material Fig. S1F), indicating that the Prm1-Cre was active. The Prm1 promoter expresses Cre in round spermatids at step 7, the point immediately before these cells begin to transition into spermatozoa (Mali et al., 1989). These data indicate that Sirt1 mRNA is not expressed in the late round and/or early elongated spermatids and that the SirT1 protein present in these cells is from mRNA transcribed in cells that precede these stages.
To determine whether deleting SirT1 in pre-meiotic germ cells with Stra8-Cre mice (herein referred to exclusively as KO) caused defects in the ability to produce sperm, we dissected the caudal epididymis and counted the number of sperm present. KO mice had significantly fewer sperm compared with WT littermate controls (Fig. 2C). The whole body SirT1 knockout mouse presented with deficits in Sertoli and Leydig cells (Kolthur-Seetharam et al., 2009). Therefore, we asked whether there is a difference in the population of these cells in our mouse. Analysis of mRNA transcripts specific for Leydig cells and Sertoli cells indicated that there was no difference in the abundance of these cells in the KO mouse compared with WT mice (supplementary material Fig. S1C). Importantly, there was no difference in Rhox5 mRNA levels, a testosterone responsive transcript, thus indicating that the KO mouse did not have altered levels of testosterone.
Spermatogenesis is delayed in the absence of SirT1
The first wave of spermatogonial stem cell differentiation into spermatozoa begins when mice are born, and by 5 weeks of age, the majority of seminiferous tubules contain both round spermatids and elongating and/or condensed spermatids (Fig. 2D upper left, arrows). Interestingly, the testes of 5-week-old SirT1 KO mice did not contain seminiferous tubules with elongating and/or condensing spermatids; the most advanced cell type present was round spermatids. However, at 12 weeks of age, elongating and/or condensing spermatids were present in the SirT1 KO mice (Fig. 2D bottom, arrows). At both ages, periodic acid-Schiff (PAS) staining indicated that SirT1 KO mice might have more dead cells than WT mice (Fig. 2D, arrowheads). Quantification of TUNEL-stained cross sections from these mice demonstrated that SirT1 KO mice indeed had more apoptotic cells per tubule at both 5 weeks and 12 weeks of age (Fig. 2E).
To determine whether the absence of elongating spermatids in the KO animals at 5 weeks of age is due to a delay in pre-meiotic and/or meiotic cells or post-meiotic cells, we examined cross sections of testes from 12-day-old mice. At 12 days of age, pachytene spermatocytes are the most advanced cell type present in the seminiferous tubules and were easily identified by the presence of γH2AX localization with unsynapsed sex chromosomes of the sex body (Fig. 2F top; arrow). Quantification of seminiferous tubules that contained a minimum of one pachytene spermatocyte demonstrated that SirT1 KO mice have a delay in differentiation of pre-meiotic and/or meiotic cells compared with WT mice (Fig. 2F bottom). This difference might be due to defects in synapsis of homologous chromosomes during meiosis. Therefore, we prepared meiotic spreads from WT and KO mice and stained with antibodies against SYCP1, SYCP3 and γH2AX to stage the cells of meiotic prophase. We did not observe any differences in γH2AX localization in leptotene, zygotene, pachytene and diplotene spermatocytes, or in the formation of the synaptonemal complex from SirT1 KO mice compared with those of WT mice (supplementary material Fig. S2), demonstrating that synapsis is normal in mutant mice. This indicates that loss of SirT1 delays spermatogenesis, most probably, in pre-meiotic cells.
Histone post-translational modifications are altered in the absence of SirT1
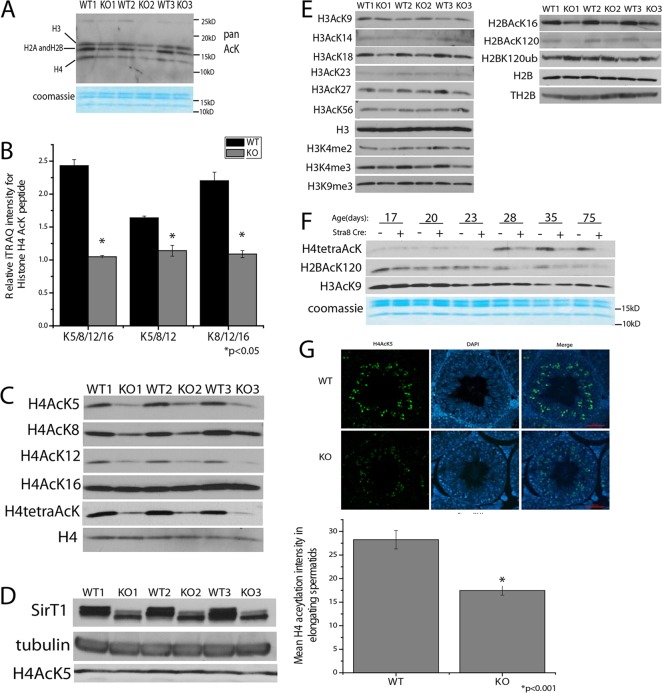
The histone to protamine transition is part of the morphological changes that are required to generate spermatozoa from haploid round spermatid cells. This process involves the removal of most histones from chromatin and the eventual deposition of protamines – highly charged proteins that facilitate and maintain the condensation of the sperm DNA (Balhorn, 1982). The initiation of this process is associated with hyper-acetylation of histones, H2A and H2B monoubiquitination, as well as H3 K4 tri-methylation (Baarends et al., 1999; Godmann et al., 2007; Hazzouri et al., 2000). To examine whether total histone acetylation is altered in the KO animals, we examined acid-extracted proteins from the chromatin of germ cell fractions. Using a pan-acetyl lysine antibody, we found that, paradoxically, histone acetylation was decreased in the SirT1 deacetylase KO mice compared with that of WT littermate controls (Fig. 3A). These changes occurred in the absence of differences in histone acetyltransferase activity between WT and KO animals (supplementary material Fig. S3A). To identify which lysine residues on the various histones were different in KO mice, we performed quantitative proteomics for acetylation on samples that had been generated from whole testes. We were only able to obtain quantification for histone H4 and found that KO mice had significantly lower levels of H4 acetylation (Fig. 3B). Correspondingly, antibodies specific for histone H4 acetyl lysine residues demonstrated that acetylation of H4K5, K8 and K12 was decreased, whereas acetylation of residue K16 remained unchanged in chromatin (Fig. 3C). Examination of the non-chromatin fraction of germ cells demonstrated that the levels of H4K5 acetylation were not different between KO mice and WT controls (Fig. 3D), suggesting that the deficit in H4 acetylation is not due to decreased acetylation of soluble histones before incorporation into chromatin.
Fig. 3.
Post-translational modifications of histones are altered in Stra8-Cre KO testes. (A) Pan-acetyl lysine (AcK) immunoblot of acid soluble proteins from the chromatin of germ cells isolated from three littermate pairs of 3-month-old SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. Coomassie Blue staining is shown at the bottom. (B) Quantification of iTRAQ abundance for histone H4 acetylated tryptic peptides from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) whole testes, n=3. (C) Immunoblots of histone H4 acetylation on acid soluble proteins isolated from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) germ cell chromatin. (D) Acetylated histone H4K5 (H4Ack5) levels in the non-chromatin NETN soluble fraction from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) germ cells. (E) Immunoblots of acid soluble proteins from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) germ cell chromatin for histone H3 and H2B post-translational modifications. me2, di-methylated, me3, tri-methylated. (F) Immunoblots of H4, H3, and H2B acetylation on acid extracts of SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) germ cell chromatin from mice of different ages (days postpartum). (G) Immunofluorescence in stage IX-X tubules from 12-week-old SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice for H4AcK5. Quantification of fluorescence intensity of images (n=9 total sections from three littermate pairs; three stage IX-X tubules per pair of mice). Scale bar: 50 µm.
The proteomic analysis did not detect acetylation of other histones; however, using antibodies for specific modifications, we determined that some other core histone post-translational modifications were changed in KO mice. Acetylation of H3K18 and K27 was decreased in mutant mice, whereas H3K9, K14, K23, and K56 acetylation levels were unchanged (Fig. 3E). Di- and tri-methylation of H3K4 were also decreased, whereas tri-methylation of H3K9 was unchanged. Moreover, H2B residue K16 and K120 acetylation was decreased in the KO mice, whereas there was an increase in H2B monoubiquitination at residue K120 (Fig. 3E). To determine which cell type(s) within the seminiferous tubules have altered levels of histone modification, we generated germ cell acid extracts from mice aged 17, 20, 23, 28, 35 and 75 days. Round spermatids begin to appear at around 20 days of age, and H4 acetylated on K5, K8, K12 and K16 (H4tetraAcK), a marker for elongating spermatids (Hazzouri et al., 2000), did not appear in the WT samples until day 28 (Fig. 3F). The differences in H4 acetylation between SirT1 WT and KO mice did not appear until 28 days of age and persisted until 75 days, suggesting that the observed difference was in elongating spermatids. Furthermore, acetylated H4K5 immunofluorescence on stage IX-X tubules, the stage at which round spermatids start to elongate, brightly stained elongating spermatids in cross sections from SirT1 WT mice, whereas the SirT1 KO mice demonstrated very low levels of staining (Fig. 3G), confirming that the defect is indeed in elongating spermatids.
Previous studies have indicated that a decrease in histone deacetylase (HDAC) activity in elongating spermatids is responsible for the increase in histone acetylation (Hazzouri et al., 2000), and that HDAC1 and HDAC2 protein is degraded at this point in spermiogenesis (Caron et al., 2003). Immunoblot analysis of germ cells from adult WT and KO mice demonstrated that there was not a global difference in HDAC1 or HDAC2 protein levels (supplementary material Fig. S3B). Analysis of a developmental time course similar to that shown in Fig. 3F indicated that there was not a difference in these HDACs between WT and mutant mice (supplementary material Fig. S3C). Furthermore, KO mice had similar levels of HDAC activity to WT mice (supplementary material Fig. S3D). Chromodomain-Y-like protein (CDYL) is expressed in elongating spermatids and recruits HDAC1 and HDAC2 to chromatin in order to induce deacetylation-mediated transcriptional silencing (Caron et al., 2003). Because there was no difference in HDAC protein levels or HDAC activity, we thought that maybe there would be increased levels of CDYL to facilitate the decrease in histone acetylation; however, there was no increase in CDYL protein (supplementary material Fig. S3B). These data indicate that SirT1 is necessary, through some unknown mechanism, for the burst of histone acetylation in elongating spermatids, which precedes histone removal.
Loss of SirT1 disrupts the histone to protamine transition
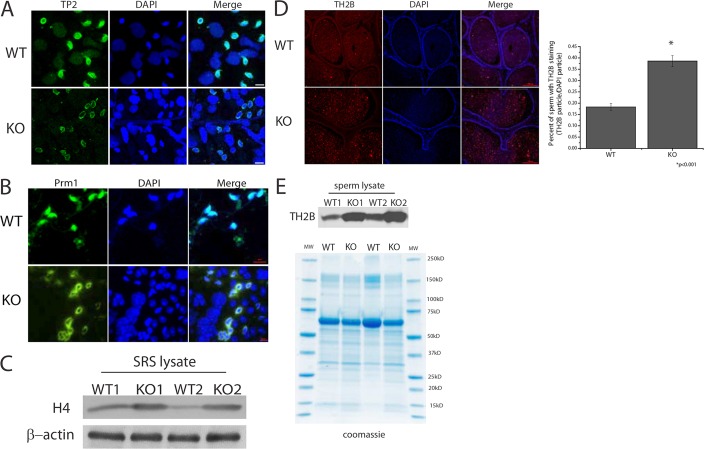
The histone to protamine transition starts with the hyper-acetylation of histone H4 in elongating spermatids, followed by removal of histones and the localization of transition proteins to DNA, and finally replacement of transition proteins by protamines that facilitate DNA condensation. Furthermore, without the removal of the testis-specific isoform of H2B (TH2B), this process is disrupted (Montellier et al., 2013). The transition proteins and protamines did not display differential expression between WT and KO mice (supplementary material Fig. S4A). To examine whether SirT1 KO mice exhibit functional defects in the histone to protamine transition, we examined the localization of TP2 in squash slide preparations. Elongating spermatids isolated from SirT1 WT seminiferous tubules demonstrated colocalization of TP2 and DNA, whereas in the SirT1 KO samples, we found that many elongating spermatids had marginalized TP2 localization, which did not overlap with DNA (Fig. 4A). Interestingly, previous reports have demonstrated that TP2 is acetylated in the C-terminus of the protein (Pradeepa et al., 2009). We were unable to detect acetylation of TP2 by immunoblot, but we were able to determine that residue K91 of TP2 is acetylated in vivo by using peptide liquid chromatography tandem mass spectrometry (LC-MS/MS) (supplementary material Fig. S4B). Quantification of TP2 residue K91 acetylation by peptide LC-MS/MS did not demonstrate a difference between SirT1 WT and KO mice (supplementary material Fig. S4C), indicating that SirT1 is not directly acting on TP2.
Fig. 4.
The histone to protamine transition is altered in Stra8-Cre KO testes. (A) Squash slides from stage IX-XII seminiferous tubules stained for transition protein 2 (TP2) and (B) protamine 1 (Prm1). Images are representative of multiple squash slides prepared from three sibling pairs of SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. (C) Immunoblot of histone H4 from sonication-resistant spermatid (SRS) lysates isolated from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. (D) Testis-specific histone H2B (TH2B) immunofluorescence on caudal epididymal cross sections (left) and quantification of sperm with staining of TH2B per nucleus (DAPI, blue) (right, n=4 mice; one section per mouse). (E) Immunoblot of TH2B from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) sperm lysates. Scale bars: 10 µm (A,B); 50 μm (D).
A similar pattern of TP2 has been reported in a mouse with mutated BRDT, the protein that recognizes and binds to hyper-acetylated histone H4 (Gaucher et al., 2012). This mutant mouse also displays defects in PRM1 localization, therefore we examined localization of PRM1. Similar to TP2, PRM1 demonstrated colocalization with DNA in the SirT1 WT samples, whereas SirT1 KO samples contained elongating spermatids with aberrant PRM1 localization that was not evident in WT samples (Fig. 4B). If there is a defective histone to protamine transition in the absence of SirT1, then we would expect sperm from SirT1 KO mice to have more histones than SirT1 WT mice. Immunoblot analysis on lysates from sonication-resistant spermatids (SRS), which contain only elongating and condensing spermatids, demonstrated that KO animals contained more histone H4 protein compared with WT animals (Fig. 4C). Furthermore, cross sections of caudal epididymis, which contain mature spermatozoa, from KO mice had more nuclei that were positive for TH2B compared with WT mice (Fig. 4D). Finally, immunoblotting of mature motile sperm biochemically corroborate the immunofluorescence data, indicating that KO mice contain more TH2B than WT mice (Fig. 4E). Combined, these data demonstrate that mutant mice have a defect in histone removal, potentially due to the failure in H4 acetylation and BRDT binding. This defect would then explain the presence of elongating spermatids with altered TP2 and PRM1 localization in the absence of SirT1.
Germ cell specific SirT1 is necessary for normal chromatin condensation and sperm morphology
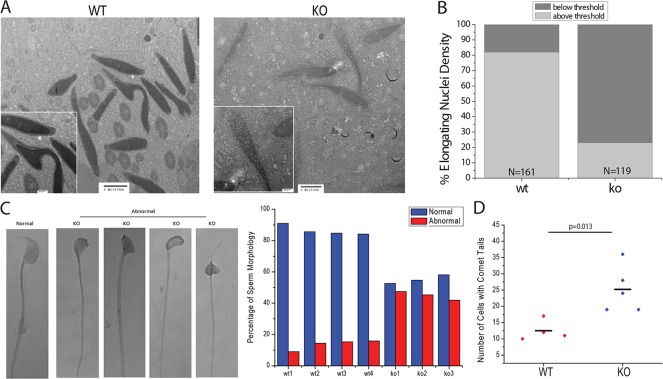
Chromatin condensation and sperm formation are events that succeed the histone to protamine transition. Therefore, we examined the extent of chromatin condensation in elongating spermatids by evaluating the density of nuclei in negatively stained electron micrographs of testes (Fig. 5A). Nuclei were scored blindly to be either above or below an arbitrary threshold. Approximately 20% of the nuclei scored from control mice were of below average density, whereas 80% of the KO nuclei were below density (Fig. 5B). Furthermore, enzymatic digestion of DNA with micrococcal nuclease indicated that chromatin from KO mice is more susceptible to digestion than chromatin from WT mice (supplementary material Fig. S5). Collectively, these data suggest that the elongating spermatids in the KO mice are defective, at least in part, because they are not able to fully condense their chromatin.
Fig. 5.
Pre-meiotic SirT1 KO mice have abnormal spermatozoa and decreased chromatin condensation in elongating spermatids. (A) Electron micrograph images of elongating spermatids from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice. (B) Quantification of relative electron density from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice, N=161 and 119 respectively. Insets in A and B show enlarged images of sections. (C) Phase-contrast images of normal and abnormal sperm (left) and the quantification of normal and abnormal sperm from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice (right). A minimum of 200 spermatozoa were counted. (D) Quantification of a comet assay on mature spermatozoa from SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) animals. Per mouse, 200 sperm cells were scored. Line represents mean number of comet tail-positive cells.
For spermatozoa to fertilize an oocyte they must have a properly formed head. To examine the role of SirT1 in the development of sperm morphology, we scored sperm as having normal or abnormal head morphology. Sperm isolated from control mice had approximately 85-90% normal head morphology. By contrast, sperm isolated from the KO mice had a significant decrease in normal sperm head morphology with a concomitant increase in the percentage of abnormal sperm (Fig. 5C). To further examine the integrity of the DNA in the mature spermatozoa, we performed a comet assay as an indicator of alkali labile lesions, representative of DNA breaks. KO spermatozoa had significantly more comet tail-positive nuclei than the control WT spermatozoa (Fig. 5D). These data indicate that the KO mice give rise to spermatozoa that are defective by morphological and molecular criteria.
Specific deletion of SirT1 in the male germ cell causes decreased fecundity
To test whether deletion of SirT1 activity in male germ cells affects fecundity, we paired 2-month-old WT or KO male mice with 2-month-old wild-type C57BL6/J female mice and monitored the number of pups born, as well as the total number of litters produced by these males over time. To ensure female fertility, we replaced female mice with new 2-month-old WT females every 3 months. SirT1 KO mice sired fewer pups per litter (Fig. 6A) and were unable to sire as many litters as the WT males (Fig. 6B). As a consequence of these defects, the KO mice produced far fewer progeny over their reproductive lifespan as WT littermate controls (Fig. 6C). In addition, the ability of KO males to produce any progeny ceased at a much earlier age than controls (Fig. 6D). These results show that the KO mice exhibit a large quantitative defect in progeny production and a premature aging phenotype with respect to fertility.
Fig. 6.
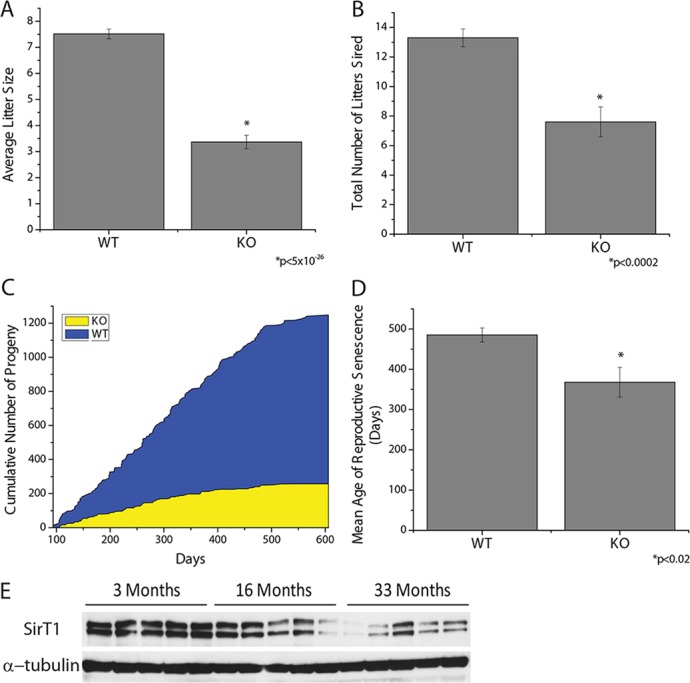
SirT1 is necessary to maintain male fecundity with age. (A-D) SirT1 f/f (WT) and Stra8-Cre; SirT1 f/f (KO) mice were mated to WT females and the number of pups per litter (A), total number of litters (B), cumulative number of pups (C) and mean age to reproductive senescence (D) were determined (n=10). (E) SirT1 protein levels, using immunoblotting, in the testes of mice aged 3, 16, or 33 months old.
The defect in fertility with age could be due an alteration in the abundance of germ stem cells and/or somatic cells. Histological analysis of testes from aged mice did not reveal a morphological difference between WT and KO mice (supplementary material Fig. S6A). Analysis of various cell-specific transcripts indicated that the abundance of Sertoli cells, Leydig cells or the different spermatogonia cell types do not differ between WT and KO mice with age (supplementary material Fig. S6B,C). Another possible explanation is that the function of the somatic cells could be changed due to impaired spermatogenesis. The expression of proteins involved in the formation of the blood testes barrier (supplementary material Fig. S6D), and a biotin tracer assay that assess the function of this barrier (supplementary material Fig. S6E), indicated the absence of SirT1 did not affect the blood testes barrier as a function of age. Sertoli cell-specific testosterone responsive genes Rhox5 (Bhardwaj et al., 2008) (supplementary material Fig. S6B) and claudin 3 (Meng et al., 2005) (supplementary material Fig. S6D) were not changed, providing evidence that differences in testosterone and Sertoli cell function are not causing decreased fecundity and early reproductive senescence in mutant mice. Interestingly, SirT1 protein declined in testis from WT mice aged 16 months and 33 months compared with that in mice at 3 months of age (Fig. 6E), indicating that SirT1 is probably required to stave off reproductive senescence.
DISCUSSION
SirT1 whole body KO mice have been shown to lack pituitary hormones for reproduction, and fertility in females can be rescued by administration of these hormones (Kolthur-Seetharam et al., 2009; McBurney et al., 2003). The present study demonstrates that SirT1 is necessary in male germ cells for normal spermatogenesis. Mice lacking SirT1 in male germ cells displayed decreased sperm counts, and many of their spermatozoa have aberrant morphology and increased DNA lesions. These defects explain why male germ cell SirT1 KO mice sire only ∼15% of the progeny number that are sired by control mice. Our findings indicate that SirT1 plays an additional essential role in male germ cells.
Cells in the mid-to-late stages of meiosis I contain the highest levels of SirT1, whereas it is present at moderate levels in differentiating spermatogonia and haploid round spermatids. Without SirT1, the appearance of pachytene cells in seminiferous tubules is delayed, demonstrating that SirT1 is required for pre-meiotic germ cell differentiation. Furthermore, the histone hyper-acetylation in haploid cells concomitant with chromatin condensation is decreased in KO mice; paradoxical to SirT1 enzymatic activity and its influence on histone acetyltransferase (HAT) and HDAC activity (Chen et al., 2012; Dobbin et al., 2013). This could be, in part, due to SirT1 regulating one or more processes in the generation, disassembly or substrate specificity of chromatin modifying complex(es) within round spermatids. Alternatively, the defect could be a result of SirT1 absence in meiotic cells, which persists to result in the incomplete removal of histones from elongating spermatids. Independent of the cell type in which these defects occur, they are not attributable to defects in somatic tissue.
Earlier studies have indicated that SirT1 is required for the hypothalamic-pituitary axis to produce hormones necessary for reproduction (Kolthur-Seetharam et al., 2009; McBurney et al., 2003). This raises the question why SirT1 also evolved an important cell-autonomous role in the male germ line. This function in testis might be analogous to the role of SirT1 in protecting peripheral tissues during calorie restriction – i.e. SirT1 might serve a quality control function in testis, in addition to its role in driving reproduction from the HPG axis. Interestingly, SirT1 levels did not change in response to either calorie restriction or high fat diet (supplementary material Fig. S7), but activity depends on NAD+, which might change with diet.
Aging is known to decrease fecundity in male humans (Ford et al., 2000). Older males exhibit decreased testis volume, decreased sperm counts, increased DNA damage of sperm, altered sperm morphology and decreased chromatin condensation (Giwercman et al., 2010; Pasqualotto et al., 2005; Rolf et al., 1996), all phenotypes reminiscent of male germ cell SirT1 KO mice. Indeed, we observed a decline in the ability of wild-type male mice to sire progeny with age, and this cessation occurred significantly earlier in mice lacking SirT1 in male germ cells. These data raise the possibility that decreased SirT1 levels are causally associated with decreased fecundity of aging males. Consistent with this idea, we observed a reduction in SirT1 levels in the testes of aging male mice.
Deletion of TAp73 (Inoue et al., 2014), GATA4 (Kyronlahti et al., 2011) and XPA (Nakane et al., 2008) results in an age-dependent deterioration of seminiferous tubules, primarily due to defects in germ cell proliferation (TAp73), Sertoli cell and Leydig cell maturation and dysfunction (TAp73 and GATA4) and a lack of post-meiotic cells (TAp73, GATA4 and Xpa). Deletion of SirT1 results in accelerated reproductive aging without a loss of post-meiotic cells or defects in somatic cell abundance or function, indicating that SirT1 plays a unique role in the aging of the testes. Our data indicates that SirT1 most probably operates, in part, to facilitate the proper packaging of chromatin, which is required to transmit the genome for fertilization and organismal propagation, reminiscent of the role that Sir2 plays in the maintenance of the genome in S. cerevisiae (Guarente, 1999). It will be interesting to test whether restoring SirT1 activity pharmacologically can prolong the reproductive lifespan of male mice, and, if so, whether human reproduction might be impacted by activating or inhibiting this sirtuin protein in the male germ line.
MATERIALS AND METHODS
Mice
All procedures were performed according to guidelines and under supervision of the Committee for Animal Care (CAC) of Massachusetts Institute of Technology. SirT1 flox/flox and Stra8iCre mice have been previously described (Cheng et al., 2003; Sadate-Ngatchou et al., 2008) and both the dTomato reporter and protamine-Cre mouse strains were obtained from Jackson Laboratories. For breeding studies, 3-month-old male SirT1 f/f or SirT1 f/f; Stra8-iCre mice were mated to 2-month-old wild-type C57BL/6J female mice (Jackson Laboratories). Every 2-3 months, the female was replaced with a 2-month-old female mouse. Cages were monitored daily for the presence and number of new-born mice. The study was terminated when all SirT1 f/f; Stra8-Cre mice failed to produce a litter for more than 60 days. Sperm were isolated from the caudal epididymis as described previously (Hazzouri et al., 2000). Briefly, the caudal epididymis was cut open with a razor blade in 2 ml of pre-warmed PBS in a 35-mm culture dish. The culture dish was placed at 37°C for 15 min to allow the sperm to swim out, after which 500 µl of sperm suspension was transferred to 2 ml of PBS and incubated at 60°C to inactivate swimming, and then the sperm were counted on a cellometer AUTO T4 (Nexcelom Biosciences) in quadruplicate. To assess sperm morphology, heat-killed sperm were spread onto glass slides, air dried and then PAS stained. Sperm were scored as normal or abnormal, and a minimum of 200 sperm were counted per sample.
Immunoblotting
All western blots were performed using Bio-Rad Criterion pre-cast gels. Samples were loaded in SDS-page buffer into either 4-15%, 10-20%, 7.5% or 18% precast PAGE gels (Bio-Rad) and run for 1-2 h at 100 V. The protein was transferred to PVDF (Millipore) membrane in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3) at 400 mA for 60-90 min at 4°C. Membranes were stained with Ponceau S [5% acetic acid,0.1% (w/v) Ponceau S], washed in dH2O and blocked in 5% milk in PBS with 0.1% Tween 20 (PBST) for 1 h at room temperature and then incubated with primary antibodies overnight at 4°C. Membranes were washed for 10 min in PBST and then 5% milk in PBST, incubated with either anti-mouse IgG horseradish peroxidase (Cell Signaling), anti-rabbit horseradish peroxidase (Cell Signaling) for 1 h at room temperature. Membranes were developed using enhanced chemiluminescence (Cell Signaling) or Immune-Star Western (Bio-Rad) after three washes in PBST. Germ cell fractions were prepared by mincing decapsulated whole testis in PBS and rotating for 15 min at 4°C. The cell mixture was filtered through a 70 µM nylon mesh at 1200 g for 10 min. Soluble fractions were generated by lysing the germ cell pellet or decapsulated testis in NETN (50 mM Tris-Cl pH 8, 100 mM NaCl, 2 mM EDTA, 0.5% NP-40) for 30 min on ice. The lysate was cleared by centrifuging at 12,000 g for 10 min and the supernatant was taken as the NETN soluble fraction. The pellet consisting of chromatin and insoluble material was washed with PBS, and then histones were extracted with 0.25 M HCl, precipitated with trichloroacetic acid and dissolved in water for the acid soluble chromatin fraction. Sperm lysates were generated by resuspending motile mature sperm that had been isolated, as described previously (Brykczynska et al., 2010), with RIPA buffer containing benzonase (Sigma-Aldrich). Briefly, motile mature spermatozoa were obtained by allowing spermatozoa to swim out of caudal epididymal tissue for 1 h at 37°C into sperm motility medium (135 mM NaCl, 5 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 30 mM Hepes pH 7.4; freshly supplemented with 10 mM lactate acid, 1 mM sodium pyruvate, 20 mg/ml BSA, 25 mM NaHCO3). To avoid contamination of somatic cells, only the top fractions were used. Sonication-resistant spermatids were isolated by homogenizing two testes from one mouse in a glass Teflon Dounce containing PBS 0.05% Triton X-100 and 7 mM BME, transferred to a 15 ml conical tube and centrifuged for 10 min at 5000 rpm (4750 g). The pellet was resuspended in PBS 0.05% Triton X-100 with 7 mM BME and sonicated three times for 30 s at approximately 40 W. Microscopic analysis indicated that >95% of the cells present were elongating spermatids. Cells were centrifuged and then lysed in RIPA buffer containing benzonase (Sigma-Aldrich).
The antibodies used were as follows: SirT1 (a gift from Shin Imai, Washington University, St Louis, MO, USA); α-tubulin and β-actin (Sigma-Aldrich); pan-AcK (ImmuneChem); H4AcK5, H4AcK8, H4AcK12, H4AcK16 and H3AcK14 were from Millipore; H4tetraAcK, H3AcK23, H2BAcK16, H2BAcK120 were from Active Motif; H4, H3, H3AcK9, H3AcK27, H3AcK56, H3K9me3, H3K4me2, H2B and TH2B were from Abcam; H3AcK18, H3K4me3 and H2BK120ub1 were from Cell Signaling Technologies.
Immunohistochemistry
Testes were fixed overnight in Bouin's solution and then transferred to 70% ethanol. Tissue was embedded, and 5-µM sections were made for staining. Slides were dehydrated and treated with sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6) for antigen retrieval. Slides were processed with the immunohistochemistry Autostainer 360 (Thermo Scientific) in the Histology Core Facility in the Koch Institute Swanson Biotechnology Center and then counterstained with periodic acid-Schiff and hematoxylin (PAS/H). SirT1 antibody (CST 2028) was used 1:200.
Immunofluorescence
Testis and caudal epididymis were fixed overnight in Bouin's solution and then transferred to 70% ethanol. Tissue was embedded and 5-µM sections were made for staining. Slides were dehydrated and treated with sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6) for antigen retrieval. Samples were washed twice for 5 min each time in TBS-Tx (TBS with 0.025% TX-100) and then blocked with 10% normal serum in 1% BSA in TBS for 1 h at room temperature before adding antibodies against H4AcK5 (1:500, Millipore), TH2B (1:3000, Abcam) or SirT1 (1:200, CST 2028) in 1% BSA in TBS overnight at 4°C. Slides were washed twice for 5 min each time in TBS-Tx before application of AlexaFluor 488 or 564 (Invitrogen) in 1% BSA in TBS for 1 h at room temperature. TBS was used to wash slides three times for 5 min each time; nuclei were stained with DAPI or DRAQ5 and imaged on either a LEICA TCS SP5 or Nikon A1R Ultra-Fast Spectral Scanning Confocal Microscope. Quantification of images was performed using ImageJ Fiji.
Squash slides were prepared as described previously (Kotaja et al., 2004). Slides were immediately placed into 4% paraformaldehyde in PBS upon removal from −80°C for 10 min, rinsed once in PBS and then incubated in 0.2% PBS-Tx at room temperature for 5 min. Slides were washed once in PBS and then blocked for 1 h at room temperature with 5% BSA in PBS. Antibodies against TP2 (Steve Kistler, University of South Carolina, Columbia, USA) or PRM1 (Shaltech) were applied in 5% BSA in PBS at 1:300, and slides were incubated overnight at 4°C. Slides were washed three times for 5 min per wash at room temperature in PBST (0.05%), incubated with AlexaFluor 488 (Invitrogen) in 5% BSA in PBS for 1 h at room temperature and washed again three times for 5 min per wash in PBST at room temperature. Nuclei were stained with DRAQ5, and slides were visualized using a Leica TCS SP5 confocal microscope or a Nikon A1R ultra-fast Spectral scanning confocal microscope.
Electron microscopy
Tissues were trimmed and fixed in 2.5% glutaraldehyde, 3% paraformaldehyde with 5% sucrose in 0.1 M sodium cacodylate buffer (pH 7.4), and post fixed in 1% osmium in veronal-acetate buffer. Tissues were stained in block overnight with 0.5% uranyl acetate in veronal-acetate buffer (pH 6.0), then dehydrated and embedded in Spurr’s resin. Sections were cut on a Leica Ultracut UCT microtome with a Diatome diamond knife at a thickness setting of 50 nm and then stained with uranyl acetate and lead citrate. The sections were examined using a FEI Tecnai spirit at 80 KV.
qPCR
Total testicular RNA was purified using Trizol and Nucleospin RNA II kit (Macherey-Nagel) after decapsulation of testis. cDNA was synthesized using the Retroscript kit according to the manufacturer's protocol (Ambion) and amplified and quantified using Sybr green (Bio-Rad) and the Light Cycler 480 (Roche) in the MIT BioMicro Center. Primers were from http://pga.mgh.harvard.edu/primerbank/.
LC-MS/MS
Whole testes were solubilized in 8 M urea, reduced, alkylated and digested with trypsin overnight, then desalted using Sep-Pak (Waters) and lyophilized. Samples were labeled with iTRAQ (AB Sciex) and dried. Peptides were fractionated into six fractions using isoelectric focusing (Invitrogen) and then enriched for acetylation by incubating with anti-acetyl lysine agarose (ImmuneChem). Peptides were eluted with 0.2 M glycine pH 2.0. Acetyl-lysine enriched peptides, as well as peptides in the supernatant of the immunoprecipitation, from each fraction were individually loaded onto a C18 pre-column and subsequently attached to a high-performance liquid chromatography (HPLC) instrument for LC-MS/MS analysis using nano electrospray on a Q Exactive (Thermo). The iTRAQ values were first corrected according to the manufacturer's instructions to account for isotopic overlap and then normalized to the non-acetylated peptides in the supernatant of the immunoprecipitation. Acetylated H4 peptides were present in fractions 1, 5 and 6.
HAT assay
HAT activity was determined using a HAT assay reagent kit (Millipore 17-284). Details are provided in the supplementary Materials and Methods.
HDAC assay
HDAC activity was determined with a colormetric HDAC assay kit (Active Motif). Further details are provided in the supplementary Materials and Methods.
Micrococcal nuclease digest
Chromatin from SirT1 f/f and SirT1 f/f; Stra8-Cre sonication-resistant spermatids was isolated and subjected to micrococcal nuclease digestion. DNA was subsequently purified and visualized with EtBR on an agarose gel. Details are provided in the supplementary Materials and Methods.
Blood testes barrier permeability
A biotin tracer was injected into the interstitium of exposed testes. Testes were removed and paraffin-embedded, and sections were incubated with streptavidin conjugated to AlexaFluor 555. Further details are provided in the supplementary Materials and Methods.
Statistical analysis
The data presented are means±s.e.m. Lines present in the box charts indicate the average of all data points, and each point or dot is a single mouse. Data were analyzed using two-way analysis of variance using Origin Pro 8. When the analysis of variance indicated a significant difference, individual differences were explored with two-tailed paired t-test. Statistical significance was determined at the 0.05 level.
Supplementary Material
Acknowledgements
We thank Laurie Boyer and Vidya Subramanian for their input, Robert Braun for the Stra8iCre mouse, Tsutomu Endo for helping stage SirT1 protein, Michelle Carmell and Kyomi Igarashi for help with meiotic spreads, reagents and input. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the Proteomics, Microscopy and the Histology core facilities. This work utilized equipment in the MIT BioMicro Center.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
E.L.B. and L.G. conceived the project. E.L.B., I.N., E.O.W., B.D.B., A.M.D.R. and N.W. performed the experiments and analyzed the data. L.G., F.M.W. and P.S.-C. provided reagents, equipment and guidance.
Funding
This work was supported by National Research Service Awards postdoctoral fellowship [F32 CA132358 for E.L.B.]; a California Institute for Regenerative Medicine postdoctoral fellowship to I.N.; the Anna Fuller, Pearl Staller, Krakauer and Hugh Hampton Young Memorial Graduate Fellowships for B.D.B.; National Institutes of Health (NIH) [U54 CA112967 for F.M.W.]; and grants from the NIH and a gift from the Glenn Foundation for Medical Research to L.G. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.110627/-/DC1
References
- Baarends, W. M., Hoogerbrugge, J. W., Roest, H. P., Ooms, M., Vreeburg, J., Hoeijmakers, J. H. J. and Grootegoed, J. A. (1999). Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207, 322-333 10.1006/dbio.1998.9155 [DOI] [PubMed] [Google Scholar]
- Balhorn, R. (1982). A model for the structure of chromatin in mammalian sperm. J. Cell Biol. 93, 298-305 10.1083/jcb.93.2.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, A., Rao, M. K., Kaur, R., Buttigieg, M. R. and Wilkinson, M. F. (2008). GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Mol. Cell. Biol. 28, 2138-2153 10.1128/MCB.01170-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska, U., Hisano, M., Erkek, S., Ramos, L., Oakeley, E. J., Roloff, T. C., Beisel, C., Schübeler, D., Stadler, M. B. and Peters, A. H. F. M. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679-687 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- Caron, C., Pivot-Pajot, C., van Grunsven, L. A., Col, E., Lestrat, C., Rousseaux, S. and Khochbin, S. (2003). Cdyl: a new transcriptional co-repressor. EMBO Rep. 4, 877-882 10.1038/sj.embor.embor917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Zhao, W., Yang, J. S., Cheng, Z., Luo, H., Lu, Z., Tan, M., Gu, W. and Zhao, Y. (2012). Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol. Cell. Proteomics 11, 1048-1062 10.1074/mcp.M112.019547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H.-L., Mostoslavsky, R., Saito, S. i., Manis, J. P., Gu, Y., Patel, P., Bronson, R., Appella, E., Alt, F. W. and Chua, K. F. (2003). Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 100, 10794-10799 10.1073/pnas.1934713100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y.-C., Hecht, N. B. and Eddy, E. M. (2001). Haploinsufficiency of protamine–1 or –2 causes infertility in mice. Nat. Genet. 28, 82-86 10.1038/ng0501-82 [DOI] [PubMed] [Google Scholar]
- Coussens, M., Maresh, J. G., Yanagimachi, R., Maeda, G. and Allsopp, R. (2008). Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS ONE 3, e1571 10.1371/journal.pone.0001571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin, M. M., Madabhushi, R., Pan, L., Chen, Y., Kim, D., Gao, J., Ahanonu, B., Pao, P.-C., Qiu, Y. and Zhao, Y.et al. (2013). SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 16, 1008-1015 10.1038/nn.3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, W. C. L., North, K., Taylor, H., Farrow, A., Hull, M. G. R. and Golding, J. and ALSPAC StudyTeam. (2000). Increasing paternal age is associated with delayed conception in a large population of fertile couples: evidence for declining fecundity in older men. Hum. Reprod. 15, 1703-1708 10.1093/humrep/15.8.1703 [DOI] [PubMed] [Google Scholar]
- Gaucher, J., Boussouar, F., Montellier, E., Curtet, S., Buchou, T., Bertrand, S., Hery, P., Jounier, S., Depaux, A. and Vitte, A.-L.et al. (2012). Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 31, 3809-3820 10.1038/emboj.2012.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giwercman, A., Lindstedt, L., Larsson, M., Bungum, M., Spano, M., Levine, R. J. and Rylander, L. (2010). Sperm chromatin structure assay as an independent predictor of fertility in vivo: a case-control study. Int. J. Androl. 33, e221-e227 10.1111/j.1365-2605.2009.00995.x [DOI] [PubMed] [Google Scholar]
- Godmann, M., Auger, V., Ferraroni-Aguiar, V., Sauro, A. D., Sette, C., Behr, R. and Kimmins, S. (2007). Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol. Reprod. 77, 754-764 10.1095/biolreprod.107.062265 [DOI] [PubMed] [Google Scholar]
- Guarente, L. (1999). Diverse and dynamic functions of the Sir silencing complex. Nat. Genet. 23, 281-285 10.1038/15458 [DOI] [PubMed] [Google Scholar]
- Haigis, M. C. and Sinclair, D. A. (2010). Mammalian sirtuins: biological insights and disease relevance. In Annual Review of Pathology-Mechanisms of Disease, Vol. 5, pp. 253-295 Palo Alto: Annual Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri, M., Pivot-Pajot, C., Faure, A.-K., Usson, Y., Pelletier, R., Sèle, B., Khochbin, S. and Rousseaux, S. (2000). Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone-deacetylases. Eur. J. Cell Biol. 79, 950-960 10.1078/0171-9335-00123 [DOI] [PubMed] [Google Scholar]
- Inoue, S., Tomasini, R., Rufini, A., Elia, A. J., Agostini, M., Amelio, I., Cescon, D., Dinsdale, D., Zhou, L. and Harris, I. S.et al. (2014). TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc. Natl. Acad. Sci. USA 111, 1843-1848 10.1073/pnas.1323416111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins, S. and Sassone-Corsi, P. (2005). Chromatin remodelling and epigenetic features of germ cells. Nature 434, 583-589 10.1038/nature03368 [DOI] [PubMed] [Google Scholar]
- Kolthur-Seetharam, U., Teerds, K., Rooij, D. G. d., Wendling, O., McBurney, M., Sassone-Corsi, P. and Davidson, I. (2009). The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol. Reprod. 80, 384-391 10.1095/biolreprod.108.070193 [DOI] [PubMed] [Google Scholar]
- Kotaja, N., Kimmins, S., Brancorsini, S., Hentsch, D., Vonesch, J.-L., Davidson, I., Parvinen, M. and Sassone-Corsi, P. (2004). Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat. Methods 1, 249-254 10.1038/nmeth1204-249 [DOI] [PubMed] [Google Scholar]
- Kyrönlahti, A., Euler, R., Bielinska, M., Schoeller, E. L., Moley, K. H., Toppari, J., Heikinheimo, M. and Wilson, D. B. (2011). GATA4 regulates Sertoli cell function and fertility in adult male mice. Mol. Cell. Endocrinol. 333, 85-95 10.1016/j.mce.2010.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P., Kaipia, A., Kangasniemi, M., Toppari, J., Sandberg, M., Hecht, N. B. and Parvinen, M. (1989). Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod. Fertil. Dev. 1, 369-382 10.1071/RD9890369 [DOI] [PubMed] [Google Scholar]
- McBurney, M. W., Yang, X., Jardine, K., Hixon, M., Boekelheide, K., Webb, J. R., Lansdorp, P. M. and Lemieux, M. (2003). The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23, 38-54 10.1128/MCB.23.1.38-54.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, J., Holdcraft, R. W., Shima, J. E., Griswold, M. D. and Braun, R. E. (2005). Androgens regulate the permeability of the blood-testis barrier. Proc. Natl. Acad. Sci. USA 102, 16696-16700 10.1073/pnas.0506084102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montellier, E., Boussouar, F., Rousseaux, S., Zhang, K., Buchou, T., Fenaille, F., Shiota, H., Debernardi, A., Hery, P. and Curtet, S.et al. (2013). Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 27, 1680-1692 10.1101/gad.220095.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinière, J., Rousseaux, S., Steuerwald, U., Sóler-Lopez, M., Curtet, S., Vitte, A.-L., Govin, J., Gaucher, J., Sadoul, K. and Hart, D. J.et al. (2009). Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461, 664-668 10.1038/nature08397 [DOI] [PubMed] [Google Scholar]
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. and Luo, L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nakane, H., Hirota, S., Brooks, P. J., Nakabeppu, Y., Nakatsu, Y., Nishimune, Y., Iino, A. and Tanaka, K. (2008). Impaired spermatogenesis and elevated spontaneous tumorigenesis in xeroderma pigmentosum group A gene (Xpa)-deficient mice. DNA Repair 7, 1938-1950 10.1016/j.dnarep.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualotto, F. F., Sobreiro, B. P., Hallak, J., Pasqualotto, E. B. and Lucon, A. M. (2005). Sperm concentration and normal sperm morphology decrease and follicle-stimulating hormone level increases with age. BJU Int. 96, 1087-1091 10.1111/j.1464-410X.2005.05806.x [DOI] [PubMed] [Google Scholar]
- Pradeepa, M. M., Nikhil, G., Hari Kishore, A., Bharath, G. N., Kundu, T. K. and Rao, M. R. S. (2009). Acetylation of Transition Protein 2 (TP2) by KAT3B (p300) Alters Its DNA Condensation Property and Interaction with Putative Histone Chaperone NPM3. J. Biol. Chem. 284, 29956-29967 10.1074/jbc.M109.052043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf, C., Behre, H. M. and Nieschlag, E. (1996). Reproductive parameters of older compared to younger men of infertile couples. Int. J. Androl. 19, 135-142 10.1111/j.1365-2605.1996.tb00451.x [DOI] [PubMed] [Google Scholar]
- Russell, L. D., Ettlin, R. A., Sinha Hikim, A. P. and Clegg, E. D. (1990). Histological and Histopathological Evaluation of The Testis, pp. 62-193 Clearwater, FL: Cache River Press. [Google Scholar]
- Sadate-Ngatchou, P. I., Payne, C. J., Dearth, A. T. and Braun, R. E. (2008). Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738-742 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin, E., Hirschey, M. D., Finley, L. W. S. and Haigis, M. C. (2010). Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669-675 10.1016/j.tibs.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.