Abstract
Most eukaryotic genomes contain large regions of satellite DNA. These arrays are often associated with essential chromosomal functions, but remain largely absent from genome projects because of difficulties in cloning and sequence assembly. The numerous small chromosomes of the parasite Trypanosoma brucei fall into this category, yet are critical to understanding the genome because of their role in antigenic variation. Their relatively small size, however, makes them particularly amenable to physical mapping. We have produced fine-resolution maps of 17 complete minichromosomes and partial maps of two larger intermediate-sized chromosomes. This revealed a canonical structure shared by both chromosomal classes based around a large central core of 177-bp repeats. Around the core are variable-length genic regions, the lengths of which define chromosomal class. We show the core region to be a repetitive palindrome with a single inversion point common to all the chromosomes of both classes, suggesting a mechanism of genesis for these chromosomes. Moreover, palindromy appears to be a feature of (peri)centromeres in other species that can be easily overlooked. We propose that sequence inversion is one of the higher-order sequence motifs that confer chromosomal stability.
African trypanosomes of the species Trypanosoma brucei are extracellular protozoan parasites of the mammalian bloodstream. They survive for long periods in the host bloodstream through a process of antigenic variation involving periodic switching of the major cell surface protein, variable surface glycoprotein (VSG; see Barry and McCulloch 2001). To facilitate this important survival mechanism, there exists in T. brucei a highly specialized gene organization that includes a large number of small chromosomes—classified on the basis of electrophoretic mobility as either minichromosomes (MCs) or intermediate-sized chromosomes (ICs), as shown in Figure 1 (see also El-Sayed et al. 2000). The MCs are small (30–150 kb), linear, and very numerous. A population of ∼100 MCs, comprising ∼10% of the nuclear DNA, is maintained by T. brucei as a means of expanding the number of available telomeric VSG genes (VSGs). Loci from this repertoire of MC VSGs, despite being fewer in number than those on other chromosomes, are preferred genes for antigenic switching events early in parasitemia (Robinson et al. 1999). Such switching events proceed by duplicative recombination of VSGs of minichromosomal origin into an active VSG expression site on a larger chromosome, the MCs themselves being transcriptionally silent.
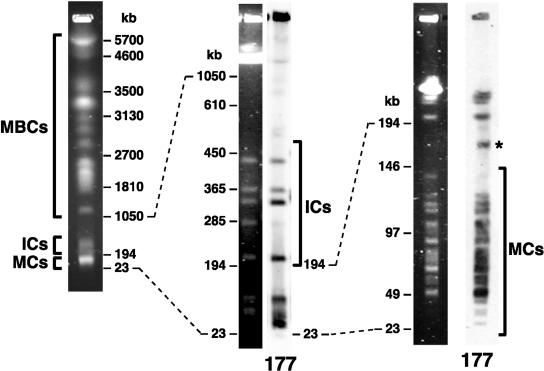
Figure 1.
The karyotype of Trypanosoma brucei as assessed by pulsed-field gel electrophoresis of whole-chromosome-sized DNAs. The chromosomes fall into three classes: megabase-sized chromosomes (MBCs), intermediate-sized chromosomes (ICs), and minichromosomes (MCs). ICs and MCs contain large quantities of a 177-bp satellite repeat sequence, as shown by hybridization to labeled repeat (177). This repeat is also found on a circular extrachromosomal element of ∼70 kb, which runs aberrantly on pulsed-field gels (star).
Central to the role of MCs in T. brucei pathogenesis is the diversity of VSGs they carry. During nuclear division, MCs are segregated via interactions with the mitotic spindle (Ersfeld and Gull 1997), and this segregation proceeds with considerable fidelity (Wickstead et al. 2003b). Such a situation allows the parasite to maintain a great diversity within the population of MCs (and hence within the library of VSGs it contains), which could never be achieved by random segregation. The discrepancy between the low number of microtubules in the mitotic spindle and high number of MCs has led to the proposal of a mechanism of segregation mediated by lateral interactions to the spindle (Gull et al. 1998).
T. brucei ICs are 200–700 kb in size and much less numerous than MCs, numbering between one and seven in most strains. They are of uncertain ploidy. ICs share repetitive elements with MCs (see Fig. 1) but, like the larger megabase-sized chromosomes (MBCs), contain expression sites for bloodstream VSGs. The MBCs are 0.9–6 Mb in size. They are diploid and form 11 homologous pairs that show considerable variation in size both between homologs and between strains (Melville et al. 1999). All of the housekeeping genes of the organism are contained on MBCs, and VSG expression sites are found at roughly half of the subtelomeres.
Given their central importance to the trypanosome's primary strategy for immune evasion, surprisingly little is known of the structure of the small chromosomes of T. brucei. The aim of the ongoing T. brucei genome sequencing project is a (near) complete sequence assembly for the MBCs only—reflected in the adoption of a chromosome-by-chromosome approach (involving isolation of DNA from individual MBCs by pulsed-field gel electrophoresis) for much of the sequencing (El-Sayed et al. 2003; Hall et al. 2003). The adoption of this strategy, and the lack of data on the small chromosomes more generally, is undoubtedly influenced by the presence on the MCs of a large quantity of a highly homogeneous 177-bp satellite repeat sequence (Sloof et al. 1983a).
In many organisms, regions of highly repetitive DNA present a huge hurdle to the generation of truly complete genome sequence data. Such regions are frequently difficult to map, problematic to clone, and extremely technically demanding to assemble. This led to the understandable exclusion of much of the satellite DNA from the “complete” higher eukaryote genome sequence assemblies produced in the last few years (The C. elegans Sequencing Consortium 1998; Adams et al. 2000; Arabidopsis Genome Initiative 2000; Lander et al. 2001). Yet such regions are frequently of great interest from the point of view of chromosome evolution and can possess essential functions for chromosome inheritance, such as centromeric activity (Sullivan et al. 2001).
Recent work has done much to push at the boundaries of these sequence gaps. Schueler et al. (2001) mapped and partially sequenced the region of transition between euchromatin and the centromere on the human X-chromosome, providing unprecedented information about the evolutionary landscape surrounding the centromere. However, the inferred assembly ends at the very start of the ∼3-Mb DXZ1 array that forms the centromeric core. In Arabidopsis also, although the heterochromatic pericentric regions on all five chromosomes have been largely sequenced (Arabidopsis Genome Initiative 2000; Haupt et al. 2001), there is little contiguous sequence data available from the satellites at the true centromeres. More recently, Sun et al. (2003) produced a detailed description of the 420-kb repetitive region that constitutes the centromere of the Drosophila minichromosome Dp1187. A combination of mapping and partial sequencing was used to infer a sequence assembly consisting of islands of complex DNA embedded in large regions of simple sequence repeat.
Here, we have used gene targeting to introduce exogenous plasmids at various positions along individual T. brucei MCs, ICs, and one MBC. We used these markers to create, for the first time, fine-resolution restriction maps of whole MCs, identifying regions of 177-bp repeat and complex DNA. In this way the structures of 17 independent MCs were elucidated, revealing a canonical organization for T. brucei MCs based around a large uninterrupted repetitive core with an inverted symmetry. This repetitive palindromic core seems to be common also to ICs, which differ from MCs only in the length of nonrepetitive subtelomeric sequences they possess. Moreover, sequencing of repeat inversion points suggests a single common ancestor for all the small chromosomes. Elucidation of MC structure may help in the identification of the, as yet undefined, centromere and origin of replication on these chromosomes. We also discuss the relevance of this unexpected palindromy to the analysis of larger satellite regions in other species.
RESULTS
Small Chromosome-Derived BACs
The bacterial artificial chromosome (BAC) library RPCI-93 (http://bacpac.chori.org/tbrucei93.htm) provides 84-fold haploid T. brucei genome coverage at an insert size of 50–200 kb. It was generated from non-chromosome-specific genomic DNA. The small chromosomes (MCs and ICs) of the organism are expected to be underrepresented in the library because they are: (1) effectively telomere-proximal at BAC resolution (average BAC insert size ∼140 kb), reducing their probability of being cloned; and (2) highly repetitive in nature owing to the presence of large numbers of the 177-bp satellite. However, given the size of BAC clone inserts, any insert of minichromosomal origin would cover nearly an entire MC and be extremely useful for analysis of MC structure.
Given the distribution of the 177-bp satellite (see Fig. 1), presence of the repeat is a good indication of BACs originating from small chromosomes in T. brucei. Screening of high-density filters has identified five of the 18,000 BACs in the library RPCI-93 as hybridizing to the 177-bp repeat (S. Melville, pers. comm.). Further analysis revealed only one of these, B36G19, to be a true positive for the 177-bp sequence (data not shown). We undertook physical mapping combined with limited sequencing to reconstruct the structure of the B36G19 BAC insert (Supplemental Fig. A). The BAC contains only ∼1.4 kb of the 177-bp repeat sequence (approximately eight repeat units), and the total insert size of 21 kb is well below the minimum size of DNA fragments selected during library construction (>50 kb). We believe this discrepancy in size to be the consequence of bacterial rearrangement of an originally much larger region of 177-bp repeats. Similar rearrangements have been reported in the products of attempted cloning of 177-bp satellite regions by plasmid rescue (Zomerdijk et al. 1992) and are our experience of handling the repeat (data not shown). The orientation of the repeats in the BAC insert is also suggestive of recombination (discussed further below).
Alongside 177-bp repeats, BAC B36G19 contains DNA homologous to genes found in subtelomeric VSG expression sites (ESAG3, ESAG4, and LRRP1) and also a non-LTR retroposon sequence, ingi, found throughout the T. brucei genome (∼400 copies per haploid genome). Expression-site-associated sequences are found on all three chromosomal classes in T. brucei. We attempted to determine the source of the B36G19 insert DNA by generating several small (0.5–2 kb) probes from the BAC. Unfortunately, the high degree of sequence identity shared by expression-site-associated sequences meant that no probe was specific to any one chromosome or, indeed, chromosomal class. However, sequence from the ESAG4 region hybridized to MBCs and ICs only (data not shown). Combined with the presence of the 177-bp satellite, this suggests that B36G19 is most likely derived from an IC. Hence, the 177-bp satellite, which constitutes >5% of the T. brucei nuclear DNA, makes up only 5 × 10–5% of the insert DNA in the BAC library RPCI-93. This comprises one highly compressed region on only 1 of 18,000 clones. The library appears devoid of minichromosomal DNA, and other approaches must be found to investigate MC and IC structure.
Ectopic Marking of Minichromosomes
At present, there are very few markers for individual chromosomes in the MC or IC populations. However, in Trypanosoma (Eid and Sollnerwebb 1991), as for Saccharomyces cerevisiae (Rothstein 1991), specific sequences in the parental genome can be targeted for plasmid insertion with relative ease because of a predominance of homologous recombination over nonhomologous end-joining. Moreover, trypanosomal chromatin is considerably looser than that of vertebrates (see Hecker et al. 1995), allowing transcription of (and hence selection for) transgenes embedded in “heterochromatic” satellite DNA (Wickstead et al. 2002). For these reasons, we chose to ectopically mark MCs by the targeted insertion of integrative vectors. Figure 2A illustrates the anatomy of the integrative vectors used in this study. The vectors contain the recognition sequence for the very rare-cutting endonuclease I-SceI. The probability of any one MC containing an innate recognition site for this enzyme is extremely small (p < 10–4), allowing the restriction site to be used for long-range mapping.
Figure 2.
(A) Anatomy of vectors used in this study to ectopically mark T. brucei chromosomes. Vectors were derived from pGad7 and differ only in targeting sequence: “forward” 177-bp repeat; “reverse” 177-bp repeat; VSG-G4 or VSG-S8. The recognition site for the extremely rare-cutting endonuclease I-SceI is shown. A unique ApaI site exists immediately adjacent to this site. (B) Integration into T. brucei small chromosomes by targeting the 177-bp repeat. The distribution of vector (pGad7) integration observed in a population of uncloned positive transformants (popn.) closely matches the distribution of the 177-bp repeat (see Fig. 1) and is the result of single integration events in many cells, as seen when the population is cloned. The pGad7 probe also hybridizes to pGEM-derived sequence on an MBC in the parental PTP cells. (C) Mapping 177-bp repeats regions on ectopically marked MCs. Genomic DNA from a clone marked at a 177-bp repeat was digested to completion with either I-SceI or ApaI endonucleases, both of which cut uniquely within the exogenous plasmid sequence. DNA digested with ApaI was then digested with 0, 0.08, 0.4, or 2 units of HpaI. Hybridization of PFGE-separated DNAs to either GFP and Hygr probes or pGEM probe is shown. Double stars indicate DNA uncut by HpaI. Arrows denote the extent of the 177-bp repeat regions. Note that there is cross-hybridization of the probes to sequences in the parental PTP cell line, particularly pGEM, which is present in PTP cells. The migration profile of a 4.9-kb ladder (left panel) under the same conditions gives an indication of gel resolution.
Initially, we targeted integrative vectors to the 177-bp satellite repeat sequence (Sloof et al. 1983a). This repeat is found on all, or nearly all, MCs and is also a marker of ICs (Fig. 1), and vectors targeted to the repeat showed integration into both chromosomal classes (Fig. 2B). The distribution of integration events in a population of transformants conforms closely to the distribution of the 177-bp repeat, showing no apparent bias for particular chromosomes (note that insertion of plasmid increases the size of MCs by ∼6 kb). The population represents many transformants resulting from single integration events, rather than a few transformants with integration at multiple sites: clones taken from the population had plasmid sequence on only one chromosome in all cases tested (n = 34; illustrated in Fig. 2B). We also found no evidence of tandem integration events into the same MC (see below).
The 177-bp repeat was targeted in both orientations. Because the 177-bp satellite is not detectably transcribed, we know of no intrinsic directionality to the repeat. However, for clarity, in this study we have adopted the following nomenclature: vectors in which the 177-bp repeat as published by Sloof et al. (1983a) runs in the same direction as plasmid GFP and Hygr transcription are said to contain a “forward” orientation repeat; those in which the Sloof et al. (1983a) sequence runs antisense to GFP transcription contain a “reverse” repeat.
We also targeted integration to two minichromosomal VSG genes: VSG-G4 and VSG-S8 (accession nos. AF294806 and AF294807). These genes were identified previously in a search for MC-specific DNA elements and occur on four and one MCs, respectively (Alsford et al. 2001); vector integration is consistent with this (data not shown).
Fine-Resolution Mapping of Minichromosomes
We used integrated markers to subject individual MCs to fine-resolution restriction endonuclease mapping. I-SceI was used to cleave chromosomes uniquely within the vector sequence. Then, to map the extent of the 177-bp satellite arrays, these cut chromosomes were subjected to partial digestion with HpaI, which has one recognition site per 177-bp repeat unit. The sizes of partially digested DNAs were measured by PFGE separation and hybridization to probes occurring on either side of the I-SceI recognition site (exemplified in Fig. 2C). Whenever possible, DNA manipulation was eased by the replacement of I-SceI for a restriction enzyme with a shorter recognition sequence that also cut uniquely within the vector sequence (ApaI in the case shown). The positions of nonrepetitive DNA on marked MCs were mapped in a similar way with the 4-cutters Sau3AI and MspI (exemplified in Supplemental Fig. B). These enzymes are expected to cut random DNA sequence on average every ∼250 bp, but do not cut the 177-bp repeat, telomeric repeats, or the T. brucei 70-bp repeat satellite that is found upstream of VSG genes and is known to occur on MCs (Sloof et al. 1983b).
We subjected 16 marked MCs to fine-resolution restriction mapping: seven randomly selected MCs targeted by a “forward” orientation 177-bp repeat; seven randomly selected MCs targeted by a “reverse” repeat; one MC marked at VSG-G4; and one MC marked at VSG-S8. Figure 3 shows the structures of these MCs. It is immediately clear from these maps that the 177-bp repeats form a large 20–80-kb central core to MCs. This core contributes ∼55% of the total minichromosomal DNA. In only one case was this core region interrupted by other sequence—in this case the non-177-bp sequence (∼6.5 kb) was also refractory to 4-cutting endonucleases, suggesting that it is a different simple sequence DNA (Fig. 3A).
Figure 3.
(A–D) Physical maps of 20 T. brucei chromosomes deduced with the aid of ectopic plasmid insertions. Here 17 complete minichromosomes were mapped from integration events targeted to (A) “forward” 177-bp repeat; (B) “reverse” 177-bp repeat; and (C) VSG-G4, VSG-S8, and an orphan rDNA promoter (Zomerdijk et al. 1992). Partial maps were also produced from integration events into 177-bp repeats occurring on two ICs and one MBC (D). The IC of 345 kb was targeted using “forward” repeats, whereas the IC of 300 kb and MBC of ∼1.7 Mb were targeted using “reverse” repeats. Partial maps only cover DNA up to ∼100 kb from the site of integration. All chromosomes are arranged such that vector transcription runs left to right. The resolution of the maps decreases with increasing distance from the integrated vector (see Fig. 2C). Combining data from several gels, we estimate the standard error in measurements of size around the site of insertion to be <20 bp, whereas that 50 kb–100 kb away is ∼2 kb. (E) Position of the integration events along the 177-bp core when targeting “forward” or “reverse” 177-bp repeats. Positions are normalized with respect to the total length of repetitive core region (excluding vector sequence) for each individual chromosome.
The identification of an uninterrupted repetitive core to MCs is at odds with the report of Zomerdijk et al. (1992) of a centrally located island of ∼5 kb of complex DNA. In that work— the only previous example of physical mapping of whole MCs—a single 60-kb MC was ectopically marked at an orphan rRNA gene promoter found on four MCs and one or two ICs (van Leeuwen et al. 2000). The positions of several 6-bp endonuclease recognition sites were then mapped using complete (rather than partial) digestions. A clustering of sites to a region in the center of the chromosome was observed. We obtained the cell line carrying this marked MC (T20(+)2; kind gift of P. Borst) and subjected it to our partial mapping approach (Fig. 3C). This demonstrated that this marked MC does not contain an island of complex DNA in the repeat core, but an unusually large (for an MC) nonrepetitive subtelomeric region. The 177-bp repeat core of this chromosome is uninterrupted.
Regions of nonrepetitive DNA were found at most, but not all, subtelomeres. It is unclear how many of these sequences are VSG genes. Undoubtedly some are VSGs (e.g., VSG-G4 and -S8), although other nonrepetitive DNAs have been described at MC subtelomeres (Weiden et al. 1991; Zomerdijk et al. 1992). The remaining ∼35% of MC DNA is simple sequence consisting of telomeric and subtelomeric repeats, and other repetitive elements such as the 70-bp repeat. Of these classes, the most common seems to be telomeric repeats, which appear to start close to the end of nonrepetitive regions (confirmed by the limited sequence data available for these regions) and show considerable heterogeneity. For example, the chromosome-end proximal to the VSG-S8 locus can be as little as ∼1.5 kb or at least 25 kb from the end of the gene in isolated clones (data not shown). Interestingly, MCs did not seem to possess the long tracts of 70-bp repeats apparent in VSG expression sites on MBCs (see Lips et al. 1993). Indeed, several nonrepetitive regions (including a known VSG) abut directly onto the 177-bp core region with few or no intervening repetitive regions.
The 177-bp Palindrome
The vast majority of 177-bp repeat sequence exists in blocks of direct tandem repeats. This can be seen in data from a number of sources: (1) Complete digestion of genomic DNA with endonucleases that cut once per repeat unit results in fragments that are n × 177 bp in size. When hybridized to the 177-bp repeat, only integer values of n are seen (Sloof et al. 1983a). Gaps or inversions would result in noninteger values for at least some endonucleases. (2) Partial digestion of marked MCs with endonucleases that cut within the repeat gives a ladder with a step size of n × 177 bp (see Fig. 2C). (3) Selecting at random >50 end sequences containing 177-bp repeats generated by the T. brucei genome sequencing project (http://www.sanger.ac.uk/Projects/T_brucei/; http://www.tigr.org/tdb/mdb/tbdb/) we found all contained direct repeats. Each of these data sets is limited but, taken together, we estimate the number of nondirect repeats to be <1 in every 100, equating to at least 18 kb of direct head-to-tail repeats for each nondirect repeat unit.
Marking MCs by targeting the 177-bp repeat in the “forward” orientation produces asymmetric integration along the chromosomes. Integration events are biased such that ectopic transcription runs toward the center of the chromosome and away from the closest telomere (Fig. 3A). Such a situation could be caused by selective pressure for this transcriptional orientation. Alternatively, it could result from a higher-order organization to the 177-bp repeat core. The former proposition was disproved by targeting the 177-bp repeat in the “reverse” orientation; in this case, integration occurred such that ectopic transcription ran toward the closest telomere (Fig. 3B). The distribution of integration into the 177-bp satellite core reflects the intrinsic orientation of the repeats. As can be clearly seen in Figure 3E, the minichromosomal repetitive core region has a palindromic organization consisting of direct head-to-tail 177-bp repeats running in from both telomeres to a point of inversion around the center of the repetitive region (p < 0.001). In agreement with this, the 177-bp repeat region on the BAC B36G19, although highly compressed, still displays an inverted symmetry.
The 177-bp Repeat Inversion Point
The fine-resolution mapping of the satellite core regions presented here refutes the presence of large (>3 kb) sections of unique DNA lying at the center of most MCs in T. brucei. However, the existence of small elements in the arrays cannot be excluded on the basis of these data. We isolated DNA from the inversions in 177-bp repeat orientation using a single-primer PCR approach. Three separate primers directed toward the “forward” orientation of the 177-bp repeat resulted in ladders of PCR products with step sizes of 177 bp (data not shown). Analogous primers to the “reverse” orientation repeat gave no PCR product for two out of three primers. Product from the third reverse primer was sequenced and established to have arisen from mispriming occurring in the widespread retroposon ingi.
We cloned and sequenced 31 PCR products comprising 16 nonredundant (i.e., not identical) sequences that were unambiguously repeat inversions, as shown by the alignment in Figure 4A. These were isolated using templates of total genomic DNA (15 sequences) or size-selected whole chromosomes of ∼37–50 kb (six sequences) and ∼75–87 kb (three sequences). Although several variants are apparent, these sequences are clearly all related—in each, the repeats meet in the same way (shown schematically in Fig. 4B). None of the repeat inversions contained any unique DNA, but an asymmetric meeting of the repeats results in a central region of asymmetry (CRA). In 15 out of 16 sequences, this CRA is ∼70 bp, because of the presence of a 20-bp subrepeat duplication in the repeat on one side of the inversion (Fig. 4B). Although relatively rare, similar duplications are also found in head-to-tail repeat units. This can be attributed to the 177-bp repeat having, in common with many satellite repeats, a suprarepeat structure—in this case, of diverged 20-bp subrepeats (see Sloof et al. 1983a).
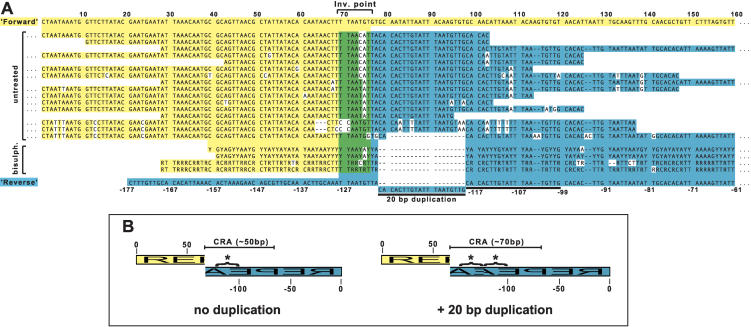
Figure 4.
The structure of minichromosomal 177-bp repeat inversions. (A) Sequence of 16 nonredundant repeat inversions isolated from untreated genomic DNA and four nonredundant inversions from bisulfite-treated DNA. Because bisulfite treatment introduces genetic code degeneracy, these sequences consist of residues {A, G, Y} for “A” strand amplicons or {C, T, R} for “B” strand amplicons. Sequences are aligned against “forward” (yellow) or “reverse” (blue) orientations of the 177-bp repeat. Around the inversion point are nucleotides that match both orientations (green). A 20-bp subrepeat duplication found in some inversion point sequences is marked with a solid line. (B) Schematic representations of the isolated repeat inversions highlighting the central region of asymmetry (CRA). This CRA is ∼50 bp or ∼70 bp depending on duplication of the 20-bp subrepeat (starred).
Palindrome amplification by PCR is technically challenging, largely because of foldback of single-stranded DNA (Devine et al. 1997). The problem is exacerbated as the proximity and identity of the inverted sequences increase. Palindromic DNA is also poorly propagated in bacteria (e.g., Sucgang et al. 2003). We were concerned that these factors might bias our results toward isolation of repeat inversions with lower degrees of palindromy then was representative. To verify the results from the single-primer PCR, we isolated inversion points from DNA from which the capacity for self-complementation had been removed. This was achieved by chemical conversion of cytosine residues to uracil by bisulfite-mediated deamination (Frommer et al. 1992). Eight clones comprising four nonredundant sequences all contained the same inversions as isolated from untreated genomic DNA, but lacking the 20-bp subrepeat duplication (Fig. 4). Isolation of variants with a smaller central region of asymmetry (50 bp) demonstrates the bias inherent in the single-primer approach. However, isolation of the same kind of repeat inversion validates the results obtained from untreated DNA.
It is worthy of note that the efficiency of cloning palindromic sequences amplified from untreated DNA was ∼100-fold less than that of cloning nonpalindromic DNA. Also, clones containing >30 bp of palindromic DNA could not be sequenced as an entity because Taq failed to process through the inversion point. Full sequence for these clones had to be attained by careful assembly of sequence from subcloned DNA. We believe that these cloning and sequencing biases are also manifest in the sequence produced during the genome sequencing effort. Sequencing of the T. brucei sheared DNA library has generated ∼44,000 end sequences (∼19 Mb of sequence) of which 1246 (∼3%; ∼500 kb of sequence) contain the 177-bp satellite (expectation value <10–10). This is approximately half the number that would be expected in an entirely unbiased library. More significantly, none of the sequence produced contains the repeat inversion isolated here.
We also isolated and sequenced the repeat inversion region occurring on the 177-bp repeat-containing BAC. This inversion is not of the type isolated more “directly” from T. brucei DNA above, as would be expected for sequence arising from bacterial rearrangement.
Partial Maps of Other Chromosomes
As well as generating physical maps of 17 MCs, we have generated partial maps from integration events into 177-bp repeat arrays occurring on larger chromosomes (Fig. 3D). These maps extend ∼100 kb on either side of the integration events. Mapping of two ICs, of 300 kb and 345 kb, demonstrated the existence of uninterrupted 177-bp repeat satellite arrays on these chromosomes akin to those found on MCs. Moreover, the orientation of integration events into these arrays was also consistent with the palindromic organization seen on MCs (Fig. 3E). This strengthens the evidence from the organization of 177-bp repeats in the IC-derived BAC B36G19 already discussed. The major difference between the structure of these ICs and the mapped MCs was the presence of much larger tracts of nonrepetitive DNA at one subtelomere. These regions could not be completely mapped by the technique used in this study.
In contrast to the situation in MCs and ICs, mapping of the ectopically marked MBC revealed a very small array of 177-bp repeats (nine repeat units in total). These are situated very close to a telomere, separated from it by only 2 kb of nonrepetitive DNA (Fig. 3D). Integration of plasmid targeted to the “reverse” orientation repeat into the most telomere-distal repeat argues against this array being palindromic. Hybridization to whole-chromosome-sized DNAs separated by pulsed-field gel electrophoresis showed the marked chromosome to be of ∼1.7 Mb (data not shown). These data demonstrate that the 177-bp repeat is found on MBCs in T. brucei, if only in very small quantities.
DISCUSSION
The data presented here reveal for the first time a canonical structure for the minichromosomes of T. brucei (Fig. 5). This structure consists of a central core of 177-bp repeats, shorter subtelomeric regions containing nonrepetitive DNA, and telomeres of variable length. The 177-bp repeat core region makes up ∼55% of total MC DNA (20–80 kb per chromosome). Although this is a large portion of the MCs, it is considerably less than the estimate of >90% made by Weiden et al. (1991). Part of the discrepancy is probably owing to the subset of MCs on which the earlier estimate was made. Also, the minichromosomal repeat core is uninterrupted by intervening sequences. MCs do not generally (if ever) contain the centrally located island of complex DNA proposed by Zomerdijk et al. (1992), which we show to be instead an unusually long nonrepetitive subtelomeric region occurring on the chromosome mapped.
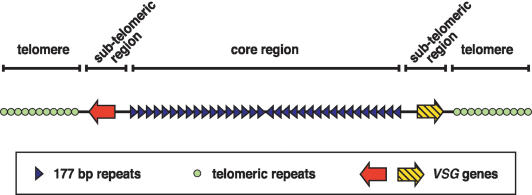
Figure 5.
A canonical structure for the MCs of T. brucei. The core region is a large repetitive palindrome produced by direct head-to-tail repeats running from both subtelomeres to a central inversion point. Subtelomeric regions contain nonrepetitive regions, at least some of which are VSG genes. Short subtelomeric repeats are capped by telomeres of equivalent size and sequence to those of the large housekeeping chromosomes. The structure of ICs is analogous, but contains larger, more complex subtelomeric regions.
An examination of integration events along the MC core region showed it to be a repetitive palindrome—tandem 177-bp repeats run in from both telomeres to a point of inversion around the center of the repeat region. This observation was strengthened by the isolation of several variants of the repeat inversion, each of which is in the orientation predicted by the mapping data. Furthermore, our mapping data demonstrate that the 177-bp repeat palindrome is also a feature of the other small chromosomal class in T. brucei (the ICs, also associated with antigenic variation). The identification of an IC-derived BAC possessing a highly compressed remnant of the repeat palindrome supports this. The independent description recently of a BAC containing 140 kb of an IC expression site but no 177-bp satellite sequence led the authors to speculate that ICs are segregated in a different manner to MCs (Berriman et al. 2002). Although this may still be proved correct, we suggest that it cannot be inferred from the paucity of 177-bp repeats occurring on BACs, because the repeats are hugely excluded from this BAC library (occurring in only 1 of 18,000 clones) and appear subject to bacterial rearrangement. We propose, instead, that the distinction between ICs and MCs is an artificial one. Both chromosomal classes share a common organization around the 177-bp repeat core and differ only in the quantity of subtelomeric expression-site-associated sequence acquired. We expect that recombinational activity will promote or demote the same chromosomal core between the two classes, and may account for the variability in the number of ICs observed between different strains.
Palindrome Formation
Large inverted-duplication, or palindrome, formation is often associated with gene amplification. In the slime moulds Physarum and Dictyostelium, and in the ciliate Tetrahymena, programmed amplification of the ribosomal RNA genes (rDNA) proceeds by the formation of numerous extrachromosomal dimers of the rDNA locus in the form of head-to-head palindromes (Engberg et al. 1976; Karrer and Gall 1976; Vogt and Braun 1976; Cockburn et al. 1978). Palindrome formation of a more stochastic nature is also observed in Saccharomyces, mammalian, and Leishmania cell lines that have been subjected to selection for drug resistance (Beverley et al. 1984; Ford and Fried 1986; Walton et al. 1986). Large inverted duplications are also found in human tumors and in cells transfected with viral oncogenes (see Tlsty 1997).
Work in Tetrahymena (Yasuda and Yao 1991), Saccharomyces (Butler et al. 1996) and, recently, mammalian cells (Tanaka et al. 2002) shows that large palindrome formation is readily seeded by double-stranded chromosome breaks near short inverted repeats in a non-sequence-specific manner. Moreover, inverted repeats are able to induce double-strand breaks and are hotspots for recombination (Lobachev et al. 2000). The self-complementary nature of palindromic DNA imparts the potential for intrastrand base pairing leading to extrusion of cruciform secondary structure. Several models can then be invoked to explain DNA instability around the inverted repeat (see Leach 1994): (1) Hairpin structures might block the replication fork, promoting template switching. (2) Secondary structures may be substrates for conformation-specific nucleases, such as the SbcCD complex in Escherichia coli, producing double-strand breaks and leading to rearrangements. (3) The cruciform structure of an extruded inversion resembles a Holliday junction that can be a substrate for the homologous recombination machinery.
The palindromy of the central region of T. brucei MCs may be indicative of their derivation (Fig. 6). An inversion in a region of tandem repeats occurring upstream of a telomeric VSG could extrude to a cruciform and induce a DNA double-strand break. Self-ligation could create a telomeric fragment with a hairpin cap that, upon replication, forms an MC with two functioning telomeres and an inverted symmetry. MC numbers could build up either through many such palindrome formation events or chromosome missegregation. In the light of this proposed descent, the identification of 177-bp repeats at the subtelomeres of an MBC is interesting, but is as likely to represent sequence acquired from MCs as it is to be a progenitor of MCs. More significantly, our isolation of several variants of essentially the same repeat inversion argues that all the small chromosomes (MCs and ICs) of T. brucei arose from a single ancestral palindrome formation event. The generation of the diverse library of subtelomeres observed today is thus a post hoc adaptation.
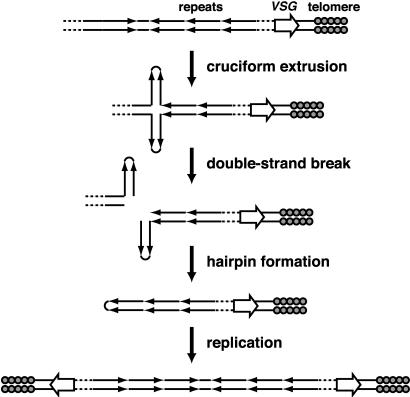
Figure 6.
A model for the generation of palindromic MCs from larger chromosomes containing inverted repeats. Extrusion of inverted repeats into a cruciform structure induces a chromosome double-strand break. Interstrand ligation creates a hairpin that becomes a large palindrome upon DNA replication.
Minichromosomal Structure and Origins of Replication
The stable propagation of MCs requires an origin of replication and a centromere. Neither structure has yet been defined in trypanosomatid nuclear DNA. Patnaik et al. (1993) have demonstrated that a region encompassing the procyclin gene promoter confers a partial stability to plasmids introduced into T. brucei, indicating an autonomously replicating sequence (ARS) element within this region. In Leishmania, targeted deletions of a stable linear extrachromosome have implicated an ∼30-kb region in mitotic stability (Dubessay et al. 2001).
The reduced size and complexity of MCs compared with MBCs represents a considerable advantage in the search for elements involved in chromosome stability. Weiden et al. (1991) have previously shown that replicating MCs exhibit single bidirectional replication bubbles located near the chromosome center. The data presented here now demonstrate that the 177-bp satellite core is the prime candidate for the minichromosomal origin of replication. In S. cerevisiae, replication origins consist of an AT-rich conserved sequence and two or three nonconserved elements (see Francon et al. 1999). One of the crucial roles for these sequences is in influencing higher-order DNA structures, a DNA bend that recruits ARS binding factor 1, and an antibent configuration induced by homopolymer A≥3 and T≥3 tracts (Eckdahl and Anderson 1990; Marilley 2000). Homopolymers also play a role in Schizosaccharomyces pombe ARS function (Okuno et al. 1999). The T. brucei 177-bp repeat contains alternating A3 and T3 tracts with a periodicity of ∼30 bp. However, this periodicity is much larger than observed in antibent S. cerevisiae or bacterial replication origins (∼12 bp and ∼8 bp, respectively; Eckdahl and Anderson 1990), and the number of homopolymer tracts is no greater than would be expected in random DNA sequence of the same base composition. Perhaps, even if each repeat was an inefficient initiator of replication, the high copy number of the repeat might ensure MC stability.
Alternatively, the inverted symmetry of the 177-bp repeat region may play a role in the origin of replication function. The use of inverted repeats in replication origins has been perfected by viruses (see Boulikas 1996). The HSV-1 oriL is a near perfect 144-bp palindrome, and the potential for cruciform formation at origins of replication may be more general. Origins of replication in monkey have inverted repeats as well as AT-rich regions (Landry and Zannis-Hadjopoulos 1991). In Tetrahymena, however, replication of the palindromic rDNA minichromosome is sequence-specific (Blomberg et al. 1997) and does not require palindromy (Reischmann et al. 1999). Furthermore, deletions in a Leishmanial linear palindrome that removed A≥6 homopolymer tracts but preserved inverted symmetry dramatically reduced chromosome stability (Dubessay et al. 2001). At present, although the data presented here implicate the 177-bp repeat region in replication function, no good model for its mode of action can be made on the basis of primary sequence alone.
A Role for Palindromy in Centromere Identity?
The best-defined centromeres are those of S. cerevisiae (see Sullivan et al. 2001). These simple “point” centromeres are determined by primary sequence alone. This elementary sequence–function relationship is appealing, but is a misleading model for other centromeres, even for other fungi. Despite centromeric function being conserved throughout evolution, centromeric sequences are diverse and rapidly evolving (see Henikoff et al. 2001). However, some structural themes can be seen at centromeres. Generally, centromeres are gene-poor, heterochromatic, and rich in satellite repeats. One of the striking features of the centromeric repeats from many species is that, although they are very divergent in terms of primary sequence, the repeat unit lengths are frequently remarkably similar—typically in the range of 150–180 bp or 300–360 bp (with some notable exceptions). These lengths correspond closely to either one or two nucleosomal units, suggesting an interaction between the repeats and centromeric histone H3. It is clear, however, that the presence of a particular repeat is neither necessary nor sufficient to determine centromere identity in most species (see Choo 2000; Sullivan et al. 2001). This has led to suggestions that either secondary sequence characteristics or some higher-order repeat organization may be required for centromeric function (Murphy and Karpen 1998; Koch 2000; Blower 2002).
The centromeres of T. brucei have yet to be defined. However, the telling repeat unit length, tandem organization, and commonality to most, if not all, of the small chromosomes implicates the 177-bp satellite in chromosome stability. In the context of putative centromeric function, the inverted structure of the 177-bp repeat region is particularly interesting. Recently, Dubessay et al. (2002) described an artificial extrachromosome in Leishmania, whose mitotic stability they assigned to the presence of 272-bp subtelomeric satellite sequence. However, this stability could as easily be assigned to the chromosomal core that contained tandemly repeated exogenous DNA arranged in a large palindrome, especially as subtelomeric repeats (although not necessarily 272-bp repeats) are insufficient for mitotic stability of nonpalindromic chromosomes in the same species (Tamar and Papadopoulou 2001).
The centromeres of S. pombe consist of nonhomologous ∼5-kb islands of complex DNA that are flanked by repetitive elements common to all three centromeres (see Sullivan et al. 2001). These repetitive flanks have an inverted symmetry and are themselves mirrored across the core region. The centromeres of humans and Arabidopsis are frequently referred to as consisting of arrays of satellite repeats oriented in a head-to-tail fashion, but it is still largely unclear as to how repeats are organized across the centromeres of these organisms. The centromeric core of the human Y-chromosome at least is not grossly palindromic (Cooper et al. 1993). However, several inversions in the repeat arrays can be found in the partial pericentric sequence assembled by the respective genome sequencing projects (Arabidopsis Genome Initiative 2000; Lander et al. 2001).
Palindromy can also be seen in centromeres lacking nucleosome-length satellites. In humans, neocentromeres can arise on chromosomes in regions devoid of centromeric α-satellite DNA (see Amor and Choo 2002). Interestingly, it has been recently shown that one of these neocentromeres maps to an ancestral centromere containing inverted duplications (Ventura et al. 2003). The functional centromere on the Drosophila minichromosome Dp1187 contains no satellite sequence of nucleosomal length, containing instead pentameric simple sequence repeats and some complex DNA (Sun et al. 2003). It does, however, retain several inverted duplications in the Maupiti region. Moreover, the simple sequence repeats themselves form stable foldback structures caused by G · A mismatch pairing on the GA-rich strand (Ortiz-Lombardía et al. 1998).
It has been proposed that inverted repeats may cause secondary structure formation at centromeres that is necessary to “present” the centromeric chromatin to the mitotic machinery (Blower 2002). If so, how ubiquitous might such inversions be? Could higher-order sequence organization be going undetected in complex centromeres? Satellite repeats on the small chromosomes of T. brucei present many of the same problems to sequence assembly as those seen at regional centromeres. However, several technical factors (not least of which is their relatively small size) allowed us to reveal a higher-order structure to the repeat regions that was unlikely to emerge from many other approaches. We propose that similar inversions associated with satellites, and in particular centromeric satellites, may be as yet undiscovered in other organisms because of the technical difficulties in characterizing these regions.
METHODS
Targeting Constructs
All plasmids used in this study (Fig. 2A) were derived from the vector pGad7 (Wickstead et al. 2003a). This root vector encodes GFP and the hygromycin-resistance marker under the control of a tetracycline-inducible procyclin gene promoter (Wirtz and Clayton 1995). Downstream from the genes is a recognition site for the meganuclease I-SceI. The pGad7 derivatives used here differ only in the sequence used for targeting their integration into the trypanosomal genome. All have been described previously (Wickstead et al. 2003a). Briefly, the “forward” orientation of the 177-bp repeat was targeted using pGad8-177t1. The repeat in pGad8-177t1 is orientated such that the 177-bp sequence, as originally described by Sloof et al. (1983a), runs in the same direction as transcription of the GFP-Hygr polycistron. pGad8-rev177 contains the same targeting sequence, but in the opposite (“reverse”) orientation. Constructs pGad8-VSG-G4 and pGad8-VSG-S8 contain targeting sequences derived from the minichromosomal genes VSG-G4 and VSG-S8, respectively. Both genes are orientated in the same direction as plasmid transcription.
Cell Lines and Transformations
Cells were cultured at 28°C in SDM-79 with 10% (v/v) fetal calf serum (Brun and Schönenberger 1979). Cell lines containing ectopically marked chromosomes were generated by stable transformation of the T. brucei “PTP” cell line (Wickstead et al. 2003a). Transformations were achieved by electroporation of cells in the presence of linearized vector as described elsewhere (Wickstead et al. 2003a). The T20(+)2 cell line was a gift of P. Borst. It was generated from 427 strain procyclic cells by the integration of a neomycin-resistance gene at an orphan ribosomal RNA gene promoter occurring on a minichromosome, as described (Zomerdijk et al. 1992).
Pulsed-Field Gel Electrophoresis
Whole-chromosome-sized DNAs were prepared in agarose plugs as described elsewhere (Melville et al. 1998). For fine-resolution mapping, DNA up to ∼200 kb in size was isolated as follows: ∼5 × 108 cells were harvested from actively dividing culture by centrifugation (1500g, 10 min). Cells were washed in 140 mM NaCl, 3 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate, (pH 7.4; PBS), and resuspended in 0.5 mL of PBS. Cells were lysed by addition of 6 mL of 2.5 M LiCl, 4% (v/v) Triton X-100, 50 mM Tris-HCl, 63 mM EDTA (pH 8), mixed by inversion, and incubated for 5 min at room temperature. The lysate was extracted against 1:1 phenol:chloroform and nucleic acids were precipitated from the aqueous fraction with ethanol. The pellet was dissolved in 10 mM Tris-HCl and 50 mM EDTA (pH 8) and digested with 150 μg/mL RNaseA for 1 h at 37°Cthen 300 μg/mL proteinase K for 2 h at 50°C. Finally, the solution was extracted against 1:1 phenol:chloroform, then DNA precipitated and redissolved in 10 mM Tris-HCl (pH 8).
PFGE was carried out in a contour-clamped homogeneous electric field electrophoresis apparatus (CHEF Mapper; Biorad). DNA separation was performed in 1% agarose (SeaKem Gold; FMC BioProducts) in either 90 mM Tris-borate, 0.2 mM EDTA (pH 8.2; 1× TB[0.1]E) or 45 mM Tris-borate, 1 mM EDTA (pH 8; 0.5× TBE) with an included angle of 120°. Specific conditions were (1) MBC separation (Fig. 1) 142 h at 2 V/cm in 1× TB(0.1)E at 12°C, switching time ramped linearly 18–42 min; (2) IC separation (Fig. 1) 37 h at 5 V/cm in 1× TB(0.1)E at 12°C, switching time ramped linearly 20–80 sec; (3) MC separation (Figs. 1 and 2B) 42 h at 4.6 V/cm in 1× TB(0.1)E at 12°C, switching time ramped linearly 8–15 sec; (4) mapping (Fig. 2C) 15.5 h at 5.3 V/cm in 0.5× TBE at 14°C, switching time ramped linearly 1–8 sec.
Southern Blot Analysis
DNA gels stained in 1 μg/mL ethidium bromide were prepared for transfer by UV nicking (80 mJ, 250 nm UV). Gels were equilibrated for 15 min in 0.4 M NaOH, 1.5 M NaCl and then transferred to positively charged nylon membrane (Roche) by overnight capillary transfer in the same solution. After transfer, membranes were neutralized with 0.5 M Tris-HCl (pH 7). Fluoroscein-labeled probes were generated by a random priming (Gene Images kit; Amersham) from the following unlabeled DNA: pGad7 uncut plasmid; GFP ORF; Hygr ORF; pGEM-4Zf+ uncut plasmid; and 177-bp repeat DNA.
Hybridization was performed overnight in 0.1% (w/v) SDS, 5% (w/v) dextran sulfate, 5% (v/v) blocking solution (Amersham), 750 mM NaCl, 75 mM sodium citrate (pH 7) at 60°C. Blots were washed to a stringency of 0.1% (w/v) SDS, 30 mM NaCl, 3 mM sodium citrate (pH 7) at 62°C. Hybridized probe was detected by treating membranes with an anti-fluorescein alkaline phosphatase-conjugated antibody followed by thorough washing and addition of the chemiluminescent substrate CPD-star (Amersham). If reprobing was desired, membranes were stripped by washing twice for 10 min with just-boiled 0.3% (w/v) SDS, 0.3 M NaOH, followed by neutralization with 0.5 M Tris-HCl (pH 7) and rinsing several times with 300 mM NaCl, 30 mM sodium citrate (pH 7).
Isolation of Repeat Inversions
Genomic DNA for PCR was isolated from strain 427 T. brucei procyclic cells as described previously (Sambrook and Russell 2001). Whole-chromosome-sized DNAs of 25–125 kb were isolated from 1% low-melting temperature agarose pulsed-field gels (Macro-Sieve Low Melt; Flowgen) run for MC separation (see above). Agarose was divided into eight blocks each containing minichromosomal DNA over a window of ∼12.5 kb (i.e., 25–37.5 kb, 37.5–50 kb, etc.). DNA was recovered from agarose by digestion with β-agaraseI (New England Biolabs) followed by ethanol precipitation. Single-primer PCR for the isolation of repeat inversions used the “forward” orientation primers ATTAAACAATGC GCAGTTAACG, GTGTGCAACATTAAATAC, or TTAATTACAAGT GTGCAACA. “Reverse” orientation primers were GTGTATAA TAGCGTTAACTGCG, GTAATTAATATGGCACAC, or TATTGCA CACATTAAAAGTT. All primers specifically amplified 177-bp satellite sequence when used in conjunction with a primer of the opposite orientation (data not shown). PCR amplicons were ligated into pGEM-T Easy (Promega) and transformed into recombination-deficient bacteria (SURE2; Stratagene) to reduce the rearrangement of sequences during propagation.
Bisulfite Treatment
Chemical conversion of cytosine residues in genomic DNA to uracil residues was achieved by bisulfite-mediated hydrolytic deamination (Frommer et al. 1992), following the improved method of Paulin et al. (1998). DNA was initially prepared by hydrodynamic shearing (passing 20 times through a 30-gauge needle). The resultant fragments had an average size of ∼20 kb as assessed by electrophoretic mobility. Converted palindromes were amplified using the primers ATTAAATAATGTGTAGT TAATG and ATTAAACAATACACAATTAACA. Primers in the opposite orientation gave no product (data not shown).
Database Mining
Genome sequence data were produced by the Institute for Genomic Research (http://www.tigr.org/tdb/mdb/tbdb/) and the Pathogen Sequencing Group at the Sanger Institute (http://www.sanger.ac.uk/Projects/T_brucei/) and can be obtained from ftp://ftp.tigr.org/pub/data/ and ftp://ftp.sanger.ac.uk/pub/pathogens. The T. brucei sheared DNA library (average insert size 4 kb) was created at the Institute for Genomic Research. End sequences were generated by the Pathogen Sequencing Group at the Sanger Institute and are available at ftp://ftp.sanger.ac.uk/pub/databases/T.brucei_sequences/GSS/. BLAST homology searches were enacted by a standalone local host using executables from NCBI (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/).
Acknowledgments
We thank Sara Melville (Department of Pathology, University of Cambridge, UK) for information on the BAC clone hybridizations. The T20(+)2 cell line was a kind gift of Piet Borst (Division of Molecular Biology, Netherlands Cancer Institute). This work was supported by a BBSRC postgraduate studentship to B.W. and a Wellcome Trust Programme and Equipment Grant to K.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2227704.
Footnotes
[Supplemental material is available online at www.genome.org. The following people kindly provided reagents, samples, or unpublished information as indicated in the paper: S. Melville and P. Borst.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Alsford, S., Wickstead, B., Ersfeld, K., and Gull, K. 2001. Diversity and dynamics of the minichromosomal karyotype in Trypanosoma brucei. Mol. Biochem. Parasitol. 113: 79–88. [DOI] [PubMed] [Google Scholar]
- Amor, D.J. and Choo, K.H.A. 2002. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71: 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Barry, J.D. and McCulloch, R. 2001. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49: 1–70. [DOI] [PubMed] [Google Scholar]
- Berriman, M., Hall, N., Sheader, K., Bringaud, F., Tiwari, B., Isobe, T., Bowman, S., Corton, C., Clark, L., Cross, G.A.M., et al. 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122: 131–140. [DOI] [PubMed] [Google Scholar]
- Beverley, S.M., Coderre, J.A., Santi, D.V., and Schimke, R.T. 1984. Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization. Cell 38: 431–439. [DOI] [PubMed] [Google Scholar]
- Blomberg, P., Randolph, C., Yao, C.H., and Yao, M.C. 1997. Regulatory sequences for the amplification and replication of the ribosomal DNA minichromosome in Tetrahymena thermophila. Mol. Cell. Biol. 17: 7237–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas, T. 1996. Common structural features of replication origins in all life forms. J. Cell. Biochem. 60: 297–316. [DOI] [PubMed] [Google Scholar]
- Brun, R. and Schönenberger, M. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36: 289–292. [PubMed] [Google Scholar]
- Butler, D.K., Yasuda, L.E., and Yao, M.C. 1996. Induction of large DNA palindrome formation in yeast: Implications for gene amplification and genome stability in eukaryotes. Cell 87: 1115–1122. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. 2000. Centromerization. Trends Cell Biol. 10: 182–188. [DOI] [PubMed] [Google Scholar]
- Cockburn, A.R., Taylor, W.C., and Firtel, R.A. 1978. Dictyostelium rDNA consists of nonchromosomal palindromic dimers containing 5S and 36S coding regions. Chromosoma 70: 19–29. [DOI] [PubMed] [Google Scholar]
- Cooper, K.F., Fisher, R.B., and Tyler-Smith, C. 1993. The major centromeric array of alphoid satellite DNA on the human Y chromosome is non-palindromic. Hum. Mol. Genet. 2: 1267–1270. [DOI] [PubMed] [Google Scholar]
- Devine, S.E., Chissoe, S.L., Eby, Y., Wilson, R.K., and Boeke, J.D. 1997. A transposon-based strategy for sequencing repetitive DNA in eukaryotic genomes. Genome Res. 7: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubessay, P., Ravel, C., Bastien, P., Lignon, M.F., Ullman, B., Pagès, M., and Blaineau, C. 2001. Effect of large targeted deletions on the mitotic stability of an extra chromosome mediating drug resistance in Leishmania. Nucleic Acids Res. 29: 3231–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubessay, P., Ravel, C., Bastien, P., Stuart, K., Dedet, J.-P., Blaineau, C., and Pagès, M. 2002. Mitotic stability of a coding DNA sequence-free version of Leishmania major chromosome 1 generated by targeted chromosome fragmentation. Gene 289: 151–159. [DOI] [PubMed] [Google Scholar]
- Eckdahl, T.T. and Anderson, J.N. 1990. Conserved DNA structures in origins of replication. Nucleic Acids Res. 18: 1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid, J. and Sollnerwebb, B. 1991. Stable integrative transformation of Trypanosoma brucei that occurs exclusively by homologous recombination. Proc. Natl. Acad. Sci. 88: 2118–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed, N.M., Hegde, P., Quackenbush, J., Melville, S.E., and Donelson, J.E. 2000. The African trypanosome genome. Int. J. Parasit. 30: 329–345. [DOI] [PubMed] [Google Scholar]
- El-Sayed, N.M.A., Ghedin, E., Song, J.M., MacLeod, A., Bringaud, F., Larkin, C., Wanless, D., Peterson, J., Hou, L.H., Taylor, S., et al. 2003. The sequence and analysis of Trypanosoma brucei chromosome II. Nucleic Acids Res. 31: 4856–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg, J., Anderson, P., and Leick, V. 1976. Free ribosomal DNA molecules form Tetrahymena pyriformis GL are giant palindromes. J. Mol. Biol. 104: 455–470. [DOI] [PubMed] [Google Scholar]
- Ersfeld, K. and Gull, K. 1997. Partitioning of large and minichromosomes in Trypanosoma brucei. Science 276: 611–614. [DOI] [PubMed] [Google Scholar]
- Ford, M. and Fried, M. 1986. Large inverted duplications are associated with gene amplification. Cell 45: 425–430. [DOI] [PubMed] [Google Scholar]
- Francon, P., Maiorano, D., and Mechali, M. 1999. Initiation of DNA replication in eukaryotes: Questioning the origin. FEBS Lett. 452: 87–91. [DOI] [PubMed] [Google Scholar]
- Frommer, M., McDonald, L.E., Millar, D.S., Collis, C.M., Watt, F., Grigg, G.W., Molloy, P.L., and Paul, C.L. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. 89: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull, K., Alsford, S., and Ersfeld, K. 1998. Segregation of minichromosomes in trypanosomes: Implications for mitotic mechanisms. Trends Microbiol. 6: 319–323. [DOI] [PubMed] [Google Scholar]
- Hall, N., Berriman, M., Lennard, N.J., Harris, B.R., Hertz-Fowler, C., Bart-Delabesse, E.N., Gerrard, C.S., Atkin, R.J., Barron, A.J., Bowman, S., et al. 2003. The DNA sequence of chromosome I of an African trypanosome: Gene content, chromosome organisation, recombination and polymorphism. Nucleic Acids Res. 31: 4864–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, W., Fischer, T.C., Winderl, S., Fransz, P., and Torres-Ruiz, R.A. 2001. The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27: 285–296. [DOI] [PubMed] [Google Scholar]
- Hecker, H., Betschart, B., Burri, M., and Schlimme, W. 1995. Functional morphology of trypanosome chromatin. Parasitol. Today 11: 79–83. [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Karrer, K.M. and Gall, J.G. 1976. The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J. Mol. Biol. 104: 421–453. [DOI] [PubMed] [Google Scholar]
- Koch, J. 2000. Neocentromeres and a satellite: A proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 9: 149–154. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Landry, S. and Zannis-Hadjopoulos, M. 1991. Classes of autonomously replicating sequences are found among early-replicating monkey DNA. Biochim. Biophys. Acta 1088: 234–244. [DOI] [PubMed] [Google Scholar]
- Leach, D.R. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893–900. [DOI] [PubMed] [Google Scholar]
- Lips, S., Revelard, P., and Pays, E. 1993. Identification of a new expression site-associated gene in the complete 30.5 kb sequence from the Antat 1.3a variant surface protein gene-expression site of Trypanosoma brucei. Mol. Biochem. Parasitol. 62: 135–137. [DOI] [PubMed] [Google Scholar]
- Lobachev, K.S., Stenger, J.E., Kozyreva, O.G., Jurka, J., Gordenin, D.A., and Resnick, M.A. 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19: 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marilley, M. 2000. Structure–function relationships in replication origins of the yeast Saccharomyces cerevisiae: Higher-order structural organization of DNA in regions flanking the ARS consensus sequence. Mol. Gen. Genet. 263: 854–866. [DOI] [PubMed] [Google Scholar]
- Melville, S.E., Leech, V., Gerrard, C.S., Tait, A., and Blackwell, J.M. 1998. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 94: 155–173. [DOI] [PubMed] [Google Scholar]
- Melville, S.E., Gerrard, C.S., and Blackwell, J.M. 1999. Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Res. 7: 191–203. [DOI] [PubMed] [Google Scholar]
- Murphy, T.D. and Karpen, G.H. 1998. Centromeres take flight: A satellite and the quest for the human centromere. Cell 81: 139–148. [DOI] [PubMed] [Google Scholar]
- Okuno, Y., Satoh, H., Sekiguchi, M., and Masukata, H. 1999. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol. Cell. Biol. 19: 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lombardía, M., Cortés, A., Huertas, D., Eritja, R., and Azorín, F. 1998. Tandem 5′-GA:GA-3′ mismatches account for the high stability of the fold-back structures formed by the centromeric Drosophila dodeca-satellite. J. Mol. Biol. 277: 757–762. [DOI] [PubMed] [Google Scholar]
- Patnaik, P.K., Kulkarni, S.K., and Cross, G.A.M. 1993. Autonomously replicating single-copy episomes in Trypanosoma brucei show unusual stability. EMBO J. 12: 2529–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin, R., Grigg, G.W., Davey, M.W., and Piper, A.A. 1998. Urea improves efficiency of bisulfite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res. 26: 5009–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischmann, K.P., Zhang, Z., and Kapler, G.M. 1999. Long range cooperative interactions regulate the initiation of replication in the Tetrahymena thermophila rDNA minichromosome. Nucleic Acids Res. 27: 3079–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, N.P., Burman, N., Melville, S.E., and Barry, J.D. 1999. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell. Biol. 19: 5839–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue—Integrative DNA transformation in yeast. Methods Enzymol. 194: 281–301. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115. [DOI] [PubMed] [Google Scholar]
- Sloof, P., Bos, J.L., Konings, A., Menke, H.H., Borst, P., Gutteridge, W.E., and Leon, W. 1983a. Characterization of satellite DNA in Trypanosoma brucei and Trypanosoma cruzi. J. Mol. Biol. 167: 1–21. [DOI] [PubMed] [Google Scholar]
- Sloof, P., Menke, H.H., Caspers, M.P.M., and Borst, P. 1983b. Size fractionation of Trypanosoma brucei DNA: Localization of the 177 bp repeat satellite DNA and a variant surface glycoprotein gene in a minichromosomal DNA fraction. Nucleic Acids Res. 11: 3889–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucgang, R., Chen, G.K., Liu, W., Lindsay, R., Lu, J., Muzny, D., Shaulsky, G., Loomis, W., Gibbs, R., and Kuspa, A. 2003. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res. 31: 2361–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., Blower, M.D., and Karpen, G.H. 2001. Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2: 584–596. [DOI] [PubMed] [Google Scholar]
- Sun, X.P., Le, H.D., Wahlstrom, J.M., and Karpen, G.H. 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamar, S. and Papadopoulou, B. 2001. A telomere-mediated chromosome fragmentation approach to assess mitotic stability and ploidy alterations of Leishmania chromosomes. J. Biol. Chem. 276: 11662–11673. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Tapscottt, S.J., Trask, B.J., and Yao, M.C. 2002. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. 99: 8772–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlsty, T.D. 1997. Genomic instability and its role in neoplasia. Curr. Top. Microbiol. Immunol. 221: 37–46. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., Kieft, R., Cross, M., and Borst, P. 2000. Tandemly repeated DNA is a target for the partial replacement of thymine by β-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol. 109: 133–145. [DOI] [PubMed] [Google Scholar]
- Ventura, M., Mudge, J.M., Palumbo, V., Burn, S., Blennow, E., Pierluigi, M., Giorda, R., Zuffardi, O., Archidiacono, N., Jackson, M.S., et al. 2003. Neocentromeres in 15q24–26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 13: 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, V.M. and Braun, R. 1976. Structure of ribosomal DNA in Physarum polycephalum. J. Mol. Biol. 106: 567–587. [DOI] [PubMed] [Google Scholar]
- Walton, J.D., Paquin, C.E., Kaneko, K., and Williamson, V.M. 1986. Resistance to antimycin-A in yeast by amplification of Adh4 on a linear, 42 kb palindromic plasmid. Cell 46: 857–863. [DOI] [PubMed] [Google Scholar]
- Weiden, M., Osheim, Y.N., Beyer, A.L., and Van der Ploeg, L.H.T. 1991. Chromosome structure: DNA nucleotide-sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 11: 3823–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead, B., Ersfeld, K., and Gull, K. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125: 211–216. [DOI] [PubMed] [Google Scholar]
- Wickstead, B., Ersfeld, K., and Gull, K. 2003a. The frequency of gene targeting in Trypanosoma brucei is independent of target-site copy number. Nucleic Acids Res. 31: 3993–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead, B., Ersfeld, K., and Gull, K. 2003b. The mitotic stability of the minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 132: 97–100. [DOI] [PubMed] [Google Scholar]
- Wirtz, E. and Clayton, C. 1995. Inducible gene-expression in trypanosomes mediated by a prokaryotic repressor. Science 268: 1179–1183. [DOI] [PubMed] [Google Scholar]
- Yasuda, L.F. and Yao, M.C. 1991. Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell 67: 505–516. [DOI] [PubMed] [Google Scholar]
- Zomerdijk, J.C.B.M., Kieft, R., and Borst, P. 1992. A ribosomal RNA gene promoter at the telomere of a minichromosome in Trypanosoma brucei. Nucleic Acids Res. 20: 2725–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nlm.nih.gov/blast/executables/; NCBI.
- ftp://ftp.sanger.ac.uk/pub/databases/T.brucei_sequences/GSS/; Sanger Institute T. brucei GSS reads databank.
- ftp://ftp.sanger.ac.uk/pub/pathogens; download genome sequence data.
- ftp://ftp.tigr.org/pub/data/; download genome sequence data.
- http://bacpac.chori.org/tbrucei93.htm; bacterial artificial chromosome (BAC) library RPCI-93.
- http://www.sanger.ac.uk/Projects/T_brucei/; Sanger Institute T. brucei genome sequencing project homepage.
- http://www.tigr.org/tdb/mdb/tbdb/; The Institute for Genomic Research T. brucei genome sequencing project homepage.