Abstract
Differences in gene repertoire among bacterial genomes are usually ascribed to gene loss or to lateral gene transfer from unrelated cellular organisms. However, most bacteria contain large numbers of ORFans, that is, annotated genes that are restricted to a particular genome and that possess no known homologs. The uniqueness of ORFans within a genome has precluded the use of a comparative approach to examine their function and evolution. However, by identifying sequences unique to monophyletic groups at increasing phylogenetic depths, we can make direct comparisons of the characteristics of ORFans of different ages in the Escherichia coli genome, and establish their functional status and evolutionary rates. Relative to the genes ancestral to γ-Proteobacteria and to those genes distributed sporadically in other prokaryotic species, ORFans in the E. coli lineage are short, A+T rich, and evolve quickly. Moreover, most encode functional proteins. Based on these features, ORFans are not attributable to errors in gene annotation, limitations of current databases, or to failure of methods for detecting homology. Rather, ORFans in the genomes of free-living microorganisms apparently derive from bacteriophage and occasionally become established by assuming roles in key cellular functions.
Bacterial genomes display variation in size, even among strains of the same species. And because these microorganisms have very little noncoding or repetitive DNA, the variation in genome size usually reflects differences in gene repertoire. Some species, particularly bacterial parasites and symbionts, have undergone massive genome reduction and simply contain a subset of the genes present in their ancestors (Moran 1996). However, in free-living bacteria, such gene loss cannot explain the observed disparities in genome size because ancestral genomes would have had to contain improbably large numbers of genes. Surprisingly, a substantial fraction of the difference in gene contents in free-living bacteria is due to the presence of ORFans, that is, open reading frames (ORFs) that have no known homologs and are consequently of no known function (Fischer and Eisenberg 1999). The high numbers of ORFans in bacterial genomes indicate that, with the exception of those species with highly reduced genomes, much of the observed diversity in gene inventories does not result from either the loss of ancestral genes or transfer from well-characterized organisms (processes that result in a patchy distribution of orthologs but not in unique genes) or from recent duplications (which would likely yield homologs within the same or closely related genome).
The high frequencies of ORFans detected in bacterial genomes were originally attributed to the limited set of sequenced genomes then available for comparison, and it was predicted that this category of genes would dwindle as databases expanded. Nevertheless, the number of ORFans in databases has grown despite an increase in the number and diversity of complete genome sequences. A recent survey estimated their frequency to be 14% of the total genes from 60 completely sequenced genomes (Siew and Fischer 2003b).
The existence of ORFans in virtually every genome has been termed a “mystery” (Dujon 1996), and numerous explanations have been offered to account for their occurrence and for the inability to classify these genes into existing protein families (Fischer and Eisenberg 1999). The most common explanation is that ORFans represent very rapidly evolving genes, or possibly pseudogenes, undergoing rates of substitution and rearrangements that obscure their similarity to known proteins (Domazet-Loso and Tautz 2003). Alternatively, these genes could be produced de novo from noncoding sequences, which are more highly diverged between taxa. Furthermore, they could represent genes transferred from organisms that have no representatives in the databases such that no significant similarity can be detected. Then again, ORFans might not be real genes, but simply artifacts resulting from the algorithms used to recognize coding sequences in genomes. It is possible that the factors responsible for the presence of ORFans will differ among taxomonic groups. For example, in eukaryotes, the sparse sampling of complete genome sequences may account for a large proportion of ORFans, whereas in many bacterial groups, this explanation is less likely because of the large numbers of genome sequences already available.
Previous analyses of the species or strain-specific genes in bacteria showed that such sequences tend to have lower G+C contents than genes with a wider distribution among species. Charlebois et al. (2003), noting that intergenic regions are often A+T rich in bacterial genomes, interpreted the biased base composition of species-specific ORFans as indicating that they might be either annotation artifacts or fast-evolving genes. In contrast, Daubin et al. (2003) viewed the existence of unique ORFans in very closely related genomes as evidence that these sequences arose by lateral gene transfer.
Although long ORFans are likely to be actual coding sequences, short hypothetical ORFs must be viewed with caution (Ochman 2002; Siew and Fischer 2003a). Unfortunately, the uniqueness of ORFans is the very feature that prevents use of a comparative approach to examine sequence function and evolution. However, if ORFans arise throughout the history of a bacterial lineage, they will exist at every phylogenetic level, such that each clade of bacteria contains ORFans that are unique to that particular group. Considering sets of genes that are restricted to monophyletic groups as well as those confined to individual genomes allows us to examine the properties of ORFans across organisms and to answer questions about their origins in bacterial genomes.
RESULTS
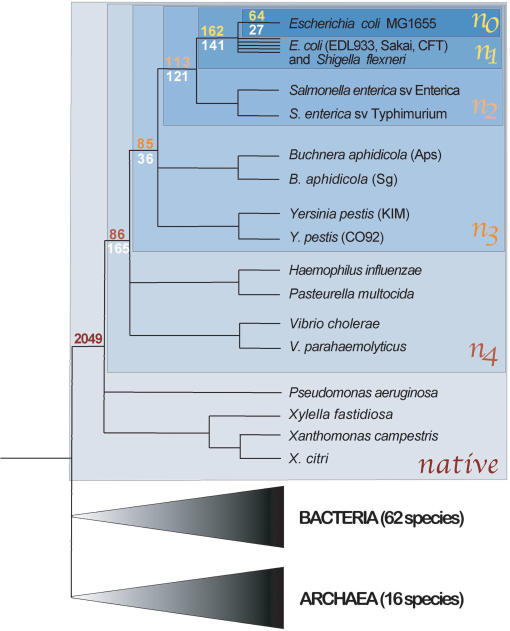
The phylogenetic distributions of annotated genes in the Escherichia coli MG1655 genome are highly variable. At each node of the phylogeny, two classes of clade-specific genes are evident: those with sporadic matches in distantly related prokaryotic species (HOPs for heterogeneous occurrence in prokaryotes) and those with no detectable match to any sequences in the databases (ORFans). Based on this distinction, the E. coli MG1655 genome contains >500 genes that have no homologs outside of the γ-Proteobacteria, with 64 ORFans that are unique to this genome. The close relationship and relatively recent divergence of the sequenced E. coli strains considered in this analysis imply a rapid mechanism for the generation of ORFans in a genome. In addition to the ORFans found only in the E. coli MG1655 genome (n0), 162 ORFans are restricted to the clade including all sequenced representatives of E. coli (n1), an additional 113 ORFans are confined to the E. coli–Salmonella enterica clade (n2), and 85 ORFans are specific to the enteric bacteria (n3). Moreover, there are numerous HOPs that are both confined to particular clades and detected in some distantly related prokaryotic genome as well as >2000 native genes ancestral to all γ-proteobacteria (Fig. 1). Given the criteria that we applied for classifying homologs, including the requirement of conserved gene context, these are conservative estimates of the numbers of genes specific to each clade.
Figure 1.
Distribution of clade-specific genes at different phylogenetic depths within the γ-Proteobacteria. The topology of the tree is based on Lerat et al. (2003), and successive blue boxes (n0–n4, native) encompass the clades considered in the present study. Numbers of ORFans (yellow/red) and HOPs (white) in the E. coli MG1655 genome specific to n0–n4 are shown at the basal nodes of each clade. The number of native genes (n = 2049) corresponds to genes in the E. coli MG1655 genome that are present in at least one member of each clade. Species numbers of Bacteria and Archaea denote all those included in BLASTP searches.
Features of ORFans
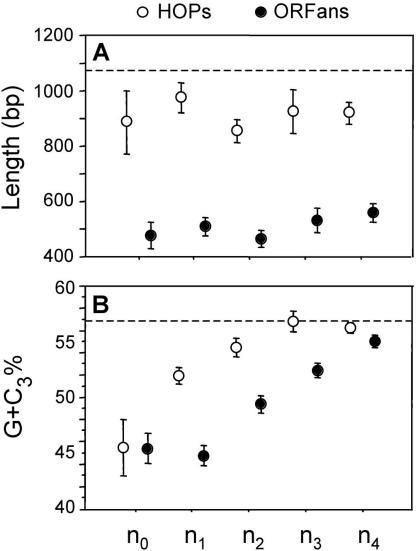
ORFans have some peculiar characteristics when compared with other genes. Within all clades, the ORFans spanned a similar size distribution and were significantly shorter than either HOPs or native genes (Fig. 2A). In addition, the ORFans from each clade are A+T rich, with those restricted to younger clades (n0 and n1) showing the most extreme biases in base composition (Fig. 2B). It is interesting to note that, within each clade, the G+C contents of HOPs and ORFans, although biased toward A+T relative to native genes, are distinct, suggesting separate histories for these two classes of genes.
Figure 2.
Characteristics of ORFans (black circles) and HOPs (open circles) in γ-Proteobacterial clades of increasing phylogenetic depth. Clade designations (n0-n4) follow those shown in Figure 1, and dashed lines denote values for native genes. (A) Average size (in base pairs). (B) Average %G+C content at the third position of codons (G+C3). Bars represent one standard error.
ORFans Encode Functional Proteins
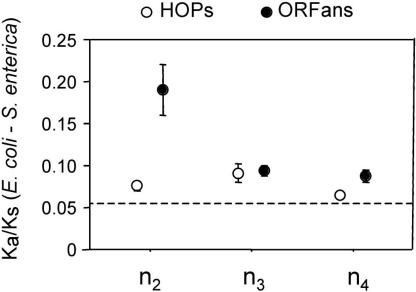
ORFans, particularly those that are short, have been attributed to errors in gene annotation or possibly pseudogenes (Fischer and Eisenberg 1999; Amiri et al. 2003; Charlebois et al. 2003). One method for determining the functional status of a sequence is by a comparative approach. Because selective constraints differ between synonymous and nonsynonymous sites, ORFs that specify functional proteins tend to have much higher divergence at synonymous sites (Ks) than at nonsynonymous sites (Ka). Similarly, in sequences that lack functional constraints, such as pseudogenes or misannotated regions, values of Ka and Ks are expected to be the same, and for protein-coding regions undergoing adaptive evolution, Ka/Ks ratios will exceed one. This Ka/Ks test requires comparisons of orthologs from lineages that are sufficiently divergent to have accumulated numerous changes, but similar enough such that alignments are not confounded by multiple substitutions; and therefore, we adopted this approach to examine genes restricted to the E. coli–S. enterica clade (n2). The average Ka/Ks ratio of the 113 ORFans unique to this clade is 0.19 ± 0.030 (with only five having Ka/Ks ratios not significantly differing from one), denoting that the vast majority of ORFans encode functional proteins (Fig. 3). Furthermore, the 121 HOPs confined to this clade have an average Ka/Ks = 0.08 ± 0.005; and among native genes, the average Ka/Ks = 0.05 ± 0.001. Similar Ka/Ks values have been reported for the ORFans in Drosophila (Domazet-Loso and Tautz 2003). Both in bacteria and in eukaryotes, the ORFans encode functional proteins, and these proteins evolve faster than more widely distributed genes. The increase in rates of protein evolution among ORFans is, in fact, more pronounced than that divulged by Ka/Ks ratios: because the average synonymous substitution rate of ORFans (Ks = 1.28) is significantly greater than that of native genes (Ks = 0.99), there is an even larger disparity between the amino acid substitution rates of ORFans and that of other genes.
Figure 3.
Average Ka/Ks ratios for ORFans (black circles) and HOPs (open circles) restricted to clades of increasing phylogenetic depth (n2-n4). All calculations of Ka and Ks are based on E. coli and S. enterica orthologs. The dashed line corresponds to the average Ka /Ks value for native genes of E. coli and S. enterica. Bars represent one standard error.
The Recognition and Origination of Fast-Evolving Sequences
The small size and rapid substitution rates of ORFans suggest that the lack of detectable homologs might result from artifacts inherent to the methods used to infer similarity among sequences. However, the presence of genes unique to E. coli MG1655 (and absent from very closely related strains) indicate that these ORFans did not originate from ancestral genes with enhanced evolutionary rates. Similarly, a re-examination of genes restricted to the E. coli–S. enterica clade (n2) reveals that heightened rates of evolution do not affect our ability to recognize orthologs and identify true ORFans. Although ORFans evolve, on average, more quickly than native genes, nearly 80% of the ORFans shared by E. coli and S. enterica have a Ka < 0.2. When native genes having similar or greater levels of divergence (i.e., equal or faster evolutionary rates) were subjected to BLAST similarity searches, homologs could be detected in all γ-Proteobacteria as well as several more distantly related genomes at our E-value threshold. Although the number of genes incorrectly assigned as ORFans is expected to be higher in deeper clades, Fischer and Eisenberg (1999) have shown that only a small proportion of ORFans can be attributed to limitations of sequence similarity algorithms. Restricting our analyses to genes having Ka < 0.1 between Escherichia and Salmonella (which would remove any falsely identified ORFans due to increased substitution rates) yielded the same results in terms of the differences in the lengths and G+C contents between ORFans and other genes.
Genomic Context of ORFans and HOPs
Nearly 40% of the ORFans and HOPs reside together in clusters (ranging up to 18 genes), which are randomly distributed around the E. coli MG1655 chromosome. This clustering differs significantly from a random distribution (p < 0.001), and those ORFans and HOPs restricted to more recent clades are more clustered. For instance, 54% of the ORFans and HOPs from the n0 clade are found in clusters of two or more genes, as compared with 29% for n1 and 14% for n2. Many clusters are in close proximity to regions associated with the integration of alien DNA, with ∼25% adjacent to tRNAs, IS elements or prophages. The fact that ORFans and HOPs restricted to the same clade often reside within the same cluster suggests that, despite a separate origin for each of these two classes of genes, certain events involve both types of sequences. This implies a common mechanism for introducing ORFans and HOPs into a genome, possibly involving bacteriophages.
DISCUSSION
ORFans are annotated open reading frames with no homologs in current databases. This might suggest that many of them are attributable to errors in annotation, to the failure of methods for detecting homology, or to inadequacies of the databases. The first two factors apparently play little role in the generation of ORFans in bacteria. Our analyses indicate that the majority of ORFans confined to γ-Proteobacterial clades are functional proteins (rather than annotation artifacts), and that sequence alignment algorithms combined with the analysis of genome context would likely recognize homologs of ORFans, if present, in other prokaryotes. With regard to the contents of current databases, recent increases in the quantity and diversity of sequenced genomes have not reduced the total number of documented ORFans (Siew and Fischer 2003b). Although sequencing additional genomes that are virtually identical to those already available would have the trivial consequence of masking ORFans by recovering homologs, the genomic sequences of closely related strains of E. coli have had quite the opposite effect. Despite a very recent divergence, each of the sequenced strains of E. coli harbors hundreds of kilobases of unique DNA as well as a large number of strain-specific ORFans (Welch et al. 2002). Therefore, not only are ORFans a common feature of bacterial genomes but they can also originate very quickly.
Because species sampling can influence the recognition of the ORFans unique to a genome, we analyzed ORFans appearing over the evolutionary history of a lineage by identifying sequences unique to monophyletic groups containing E. coli MG1655 at increasing phylogenetic depths (Fig. 1). Whereas most previous studies have focused on the ORFans confined to individual genomes, our approach allows direct comparisons of the numbers and characteristics of ORFans of different ages in the E. coli genome, and yields information about their functional status and substitution rates.
Taken together, ORFans in the E. coli lineage are short, functional, A+T rich, and quickly evolving, and can be differentiated based on their sequence properties both from those laterally acquired genes that are distributed in other bacteria (HOPs) and from those genes ancestral to all γ-Proteobacteria. Whereas the degenerative or accelerated evolution of ancestral genes can serve as a mechanism for generating ORFans both in bacteria species undergoing genome reduction (Amiri et al. 2003) and in eukaryotes (Domazet-Loso and Tautz 2003), the extreme differences in base composition of ORFans and native genes indicate that this is not the case for ORFans in E. coli. Because all bacterial genomes with G+C contents >30% have intergenic regions that are relatively A+T rich, it has also been suggested that ORFans arise from errors in annotating noncoding regions (Charlebois et al. 2003). But the fact that most of the intergenic regions are homologous among strains of E. coli indicates that these regions are not the source of unique, strain-specific sequences. And in addition, the Ka/Ks tests in older ORFans demonstrate that the vast majority encode functional proteins.
Despite their distinguishing features, ORFans in E. coli do not comprise a static group: the older ORFans (i.e., those present in deeper clades) approach characteristics of native genes in terms of base composition and evolutionary rates, whereas the younger ORFans tend to be clustered and adjacent to laterally transferred sequences. Together with their fast rate of origination in bacterial genomes, this chromosomal distribution suggests that, in bacteria such as E. coli, ORFans do not arise from the degradation of ancestral coding regions or from intergenic sequences, but rather by lateral gene transfer. Moreover, genes that originate together might be expected to become dispersed over time owing to rearrangements, insertions, and deletions, which accounts for the fact that ORFans restricted to shallower clades are more typically found in larger gene clusters.
Similar to what has been observed for bacteria (Charlebois et al. 2003; Daubin et al. 2003), the ORFans in Drosophila are short, AT rich, and quickly evolving relative to ancestral genes (Domazet-Loso and Tautz 2003). Unfortunately, the sparse sampling of insect genomes raises the possibility that the ORFans identified in Drosophila include genes that originated recently as well as those lost by (or not detected in) other lineages. Therefore, the populations of ORFans recognized in bacterial and eukaryotic genomes likely result from very different mechanisms and are not yet directly comparable.
A Role for Phage in Generating ORFans
Although several features of ORFans suggest that they originate through lateral gene transfer, it is unlikely that other bacteria serve as the major source of these genes. The compositional properties of genes native to bacteria differ from those observed in ORFans (Daubin et al. 2003), and even those genes with patchy distributions among prokaryotes (HOPs) have sequence characteristics that distinguish them from ORFans. Several lines of evidence implicate phage as the principal candidates for introducing ORFans into bacterial genomes: phage—particularly ssDNA— are A+T rich (Rocha and Danchin 2002), their genes have compositional characteristics that match recent ORFans (Daubin et al. 2003), and their raison d'être is to proliferate alien sequences in bacterial genomes. And because phage diversity has been so poorly sampled, it is not surprising that genes that originate from phage are rarely seen as having homologs, even when querying all-inclusive databases.
Many phages are known to encode short A+T-rich genes, a high proportion of which are ORFans (Pedulla et al. 2003). Although the function of these genes remains obscure, it has been proposed that they play a role in the retention of prophages by supplying properties that benefit the bacterial host (Hendrix et al. 2000; Juhala et al. 2000). This helps explain the persistence of ORFans in bacterial genomes, even after the disappearance of the neighboring phage sequences.
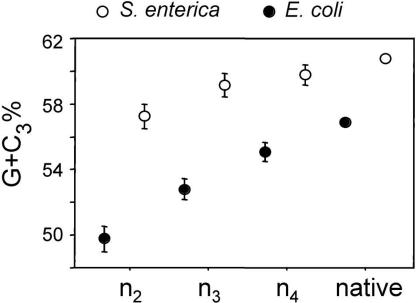
The introduction of ORFans by an A+T-rich donor population has been occurring throughout the evolutionary history of the γ-Proteobacterial lineage leading to E. coli. Because bacterial genomes manifest distinct mutational patterns, atypical sequences are expected to ameliorate and eventually resemble the base composition of native genes (Sueoka 1988; Lawrence and Ochman 1997). This process accounts for the higher G+C contents of ORFans from deeper clades (n2 and n3): these relatively ancient ORFans are still A+T rich, which links them to the younger ORFans in shallower clades, but their average base composition is closer to that of native genes. Finally, examining the sequence features of ORFans that originate over the history of a lineage also allows us to monitor the process of amelioration of these ancestrally A+T-rich genes toward their new genomic environments. ORFans restricted to E. coli and S. enterica (clade n2) still display significant differences in their G+C contents when compared with native genes, suggesting that the assimilation of genes to the sequence characteristics of their new host genome is, indeed, slow. It is interesting to note that although the G+C content of native genes is higher in Salmonella than in E. coli, the difference between ORFans and native genes is smaller, suggesting that the amelioration process is faster in Salmonella (Fig. 4).
Figure 4.
Average G+C at the third position of codons (G+C3) for orthologous ORFans of different classes of genes (ORFans, HOPs, native) in E. coli (black) and S. enterica (open). Bars represent one standard error.
If ORFans originate in phages, it is anticipated that their sequences will harbor additional characteristics of bacteriophage genes. Because dinucleotide frequencies can provide signatures that discriminate among sequences from different organisms and have been used to identify alien genes within genomes (Karlin 1998), we compared the dinucleotide frequencies from native genes, ORFans, and bacteriophages known to infect E. coli (see Supplemental material). Recent ORFans and phages are similarly biased for CpG, TpC, ApG, CpC, and ApA, whereas the older ORFans progressively approach the dinucleotide compositions of native genes. These results provide further support of an ancestral relationship between the ORFans present in the E. coli genomes and bacteriophages.
The Function of ORFans
Most ORFans, although under selective constraints, are of unknown function. However, the identification and function of a few of the ORFans in the E. coli lineage have been established; the most surprising of these is rpsV, the gene encoding ribosomal protein S22, which is restricted to the n2 clade. (A ribosomal protein called S22 has been described in chloroplast and mitochondria, but it is not homologous to the one found in E. coli; Keiper and Wormington 1990; Bubunenko and Subramanian 1994.) Despite its relatively low evolutionary rate, the rpsV gene of E. coli, like other ORFans, is short and A+T rich. The RpsV protein is associated with the small subunit of ribosomes during the early stages of the stationary phase (Wada 1998), and based on a genome-wide survey using E. coli microarrays, rpsV expression increases substantially in nongrowing cells (Selinger et al. 2000). Surprisingly, among the 25 genes showing the greatest increase in expression in stationary phase (Selinger et al. 2000), we found that, in addition to rpsV, 10 were ORFans (mostly n2) and five were HOPs. Because the stationary phase in bacteria often corresponds to cell stress or starvation, this may reveal an ancestral link between these genes and selfish elements that are mobilized under stress conditions.
When they originate, ORFans are unlikely to encode essential functions; but if maintained, ORFans can become incorporated into cellular processes and take on roles more crucial to cell survival. This could occur either by assuming the function of an ancestral gene or by conferring a new property that is integral to the host cell. As evidence of these processes, some of the ORFans restricted to the n3 clade are conserved in the highly reduced genome of the aphid symbiont Buchnera aphidicola. Because Buchnera has eliminated the vast majority of genes that were present in its free-living ancestor, its genome is thought to encode very few accessory functions and to have retained a minimal set of required genes. Among the ORFans conserved by E. coli and Buchnera is dnaT, which encodes a primosome assembly protein responsible for loading the replicative helicase DnaB onto DNA. Its immediate neighbor in both Escherichia and Buchnera is the gene specifying DnaC, another primosome assembly protein, which was classified as an HOP specific to the n3 clade because of its weak similarity to a protein in Gram-positive bacteria. Therefore, both dnaT and dnaC were likely acquired together in the ancestor of enteric bacteria and have since taken on a role in DNA replication previously performed by nonorthologous genes. This hypothesis is further supported by the detection of genes with similarity to dnaC and dnaT in two bacteriophages (Epsilon NC_004775 and P27 NC_003356, respectively). An additional 46 ORFans (n0–n4) show significant similarity to genes in sequenced phage genomes (Supplemental Table 1). Although the contribution of bacteriophage to the evolution of pathogenicity in bacteria has been well documented, these results suggest a more profound role of phage in bacterial evolution.
Two mechanisms, based on separate findings and having different evolutionary implications, could account for the existence of ORFans in bacterial genomes. In the first, ORFans are the remnants of ancestral sequences that result from the erosion and degradation of previously functional genes, and their presence is viewed as an indicator of genome dynamics (Amiri et al. 2003). In the second, ORFans elaborate new functions that are potentially beneficial to the organism and represent a means of bacterial adaptation. Whereas the first mechanism is certainly operating in host-associated bacteria undergoing massive genome reduction, our estimates from E. coli suggest that ORFans constitute a substantial fraction of the genomes of free-living, nonpathogenic bacteria. We have shown that the majority of ORFans encode functional proteins and display numerous features that indicate acquisition from phage. These results show that ORFan genes can become established in bacterial genomes and assume key roles essential to the cell, and support the argument for an unprecedented role of phage in bacterial long-term evolution.
METHODS
Delimiting ORFans in Clades of Different Phylogenetic Depths
We initially queried all completely sequenced prokaryotic genomes (n = 94) available in the EMGlib database (Perriere et al. 2000; release February 26, 2003) with the annotated ORFs of E. coli MG1665 using BLASTP (Altschul et al. 1997; applying the BLOSUM62 matrix). To identify those ORFs of E. coli MG1655 that are restricted to clades of different phylogenetic depths, we narrowed our analysis to the gene contents of sequenced enteric bacteria, currently the best represented bacterial group, including five strains of E. coli (Blattner et al. 1997; Hayashi et al. 2001; Perna et al. 2001; Welch et al. 2002; Wei et al. 2003), two subspecies/serovars of S. enterica (McClelland et al. 2001; Parkhill et al. 2001a), two species of Buchnera (Shigenobu et al. 2000; Tamas et al. 2002), and two strains of Yersinia pestis (Parkhill et al. 2001b; Deng et al. 2002) as well as those of several other γ-Proteobacteria, including Vibrio cholerae (Schoolnik and Yildiz 2000), Vibrio parahaemolyticus (Makino et al. 2003), Haemophilus influenzae (Fleischmann et al. 1995), Pasteurella multocida (May et al. 2001), Pseudomonas aeruginosa (Stover et al. 2000), Xylella fastidiosa (Simpson et al. 2000), Xanthomonas campestris, and Xanthomonas citri (da Silva et al. 2002). (Species are listed in order of increasing genetic distance to E. coli.) We defined monophyletic clades within the sequenced γ-Proteobacteria based on their genetic relationships, as established by phylogenetic analysis of the ∼200 genes conserved among all taxa considered (Lerat et al. 2003). From this phylogeny, we made use of all clades that included E. coli MG1655, except those that placed highly reduced genomes (e.g., Buchnera, Haemophilus) as the outside reference taxa because this would restrict the identification of clade-specific genes.
We first considered the ORFs restricted to the E. coli MG1665 genome (n0) and then to four key clades (Fig. 1), corresponding to the E. coli species (n1), the Escherichia–Salmonella group (n2), the enterics (n3), and the group including Vibrio spp., Haemophilus, Pasteurella, and the enterics (n4). To minimize the inclusion of genes expected to have sporadic distributions among bacteria, we removed all recognized IS elements as well as sequences associated with known prophages from the E. coli MG1655 gene set. Then, we defined clade-specific ORFans as those having no detectable homologs outside of a specific clade. Because of deletion events (particularly in reduced genomes), it is possible that some clade-specific ORFs are missing in certain members of a clade. Thus, for ORFans specific to n0, we retain all ORFs from E. coli MG1655 that have no match (E-value < 0.01) in any other genome considered. Similarly, the ORFans specific to clade n1 are those present in E. coli MG1655 and at least one of the other sequenced E. coli strains (E-value < 10–5) but absent (E-value > 0.01) from the other genomes. The ORFans specific to the deeper clades n2, n3, and n4 were similarly defined, but with the additional requirement that ORFs be present in at least one genome from each clade subsumed by the deeper clade. Those ORFs present in each clade and in at least one of the other γ-Proteobacterial genomes were considered ancestral (designated “native”). Our method thus allows us to identify ORFans that arise at different phylogenetic depths, and consequently to study their evolution using comparative analyses.
Genes With Heterogeneous Occurrence in Prokaryotes (HOPs)
To determine if ORFans share features with either laterally transferred or native genes, we performed BLAST searches to obtain sets of genes that were acquired before the divergence of each of the clades considered. Genes that are restricted to a particular clade within the γ-Proteobacteria but are distributed sporadically among more distantly related taxa are considered likely candidates of lateral gene transfer, and we refer to these genes as HOPs, for Heterogeneous Occurrence in Prokaryotes. For example, the HOPs in E. coli MG1655 (n0) were those ORFs with no homologs (E-value < 0.01) in other genomes included in clade n2 but showing some similarity (E-value < 10–5) to ORFs in more distantly related species. Similarly, HOPs in clade n1 were those ORFs from E. coli MG1655 also present in another E. coli, but absent from other species of clade n3 and having a match in a distantly related species. The procedure was repeated to identify the HOPs in clades n0 through n4.
Confirmation of ORFans and HOPs
Because the similarity among ORFs from more distantly related clades may be overlooked by BLAST—a particular consideration for quickly evolving genes—we verified the authenticity of each of the clade-specific ORFans and HOPs by considering their genomic context and locations. For example, if an ORF was detected via BLAST in multiple strains of E. coli but not S. enterica, we visually searched the alignments of the Escherichia and the Salmonella genomes for sequences showing any trace of similarity using the Percent Identity Plots (PIPs; Florea et al. 2003) provided on the EnteriX server (http://globin.cse.psu.edu/enterix). When the corresponding regions displayed PIPs >50%, the candidate ORFan was excluded from further analysis. Furthermore, because it is possible for taxa within a clade to have acquired homologous genes independently, analysis of gene context allowed us to determine if clade-specific ORFans and HOPs had retained their positions within genome, thus confirming their orthology. This was conducted for all genes common to E. coli and Salmonella; and those ORFs in different locations, or having different neighboring genes, were eliminated. Although the stringency of the criteria resulted in the removal of nearly 40% of the annotated E. coli MG1655 ORFs from further analyses, this process allowed us to identify clade-specific and native ORFs with a high degree of certainty.
Compositional Features and Substitution Rates
For each class of genes (i.e., ORFans and HOPs in clades n0,n1,n2,n3, and n4, and native ORFs ancestral to all γ-Proteobacteria), base composition was calculated over the entire gene and at each codon position in E. coli MG1655, and for the orthologous genes from S. enterica. For all E. coli and S. enterica orthologs, we calculated Ka and Ks by the method of Li (1997).
Acknowledgments
This work was funded by grants from the NIH (GM56120) and the DOE (DEFG0301ER63147).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2231904.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri, H., Davids, W., and Andersson, S.G. 2003. Birth and death of orphan genes in rickettsia. Mol. Biol. Evol. 20: 1575–1587. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M.G. and Subramanian, A.R. 1994. Recognition of novel and divergent higher plant chloroplast ribosomal proteins by Escherichia coli ribosome during in vivo assembly. J. Biol. Chem. 269: 18223–18231. [PubMed] [Google Scholar]
- Charlebois, R.L., Clarke, G.D., Beiko, R.G., and St Jean, A. 2003. Characterization of species-specific genes using a flexible, web-based querying system. FEMS Microbiol. Lett. 225: 213–220. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C., Ferro, J.A., Reinach, F.C., Farah, C.S., Furlan, L.R., Quaggio, R.B., Monteiro-Vitorello, C.B., Van Sluys, M.A., Almeida, N.F., Alves, L.M., et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417: 459–463. [DOI] [PubMed] [Google Scholar]
- Daubin, V., Lerat, E., and Perriere, G. 2003. The source of laterally transferred genes in bacterial genomes. Genome Biol. 4: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W., Burland, V., Plunkett III, G., Boutin, A., Mayhew, G.F., Liss, P., Perna, N.T., Rose, D.J., Mau, B., Zhou, S., et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184: 4601–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso, T. and Tautz, D. 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13: 2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B. 1996. The yeast genome project: What did we learn? Trends Genet. 12: 263–270. [DOI] [PubMed] [Google Scholar]
- Fischer, D. and Eisenberg, D. 1999. Finding families for genomic ORFans. Bioinformatics 15: 759–762. [DOI] [PubMed] [Google Scholar]
- Fleischmann, R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J.F., Dougherty, B.A., Merrick, J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496–512. [DOI] [PubMed] [Google Scholar]
- Florea, L., McClelland, M., Riemer, C., Schwartz, S., and Miller, W. 2003. EnteriX 2003: Visualization tools for genome alignments of Enterobacteriaceae. Nucleic Acids Res. 31: 3527–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., Han, C.-G., Ohtsubo, E., Nakayama, K., and Murata, T. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8: 11–22. [DOI] [PubMed] [Google Scholar]
- Hendrix, R.W., Lawrence, J.G., Hatfull, G.F., and Casjens, S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8: 504–508. [DOI] [PubMed] [Google Scholar]
- Juhala, R.J., Ford, M.E., Duda, R.L., Youlton, A., Hatfull, G.F., and Hendrix, R.W. 2000. Genomic sequences of bacteriophages HK97 and HK022: Pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299: 27–51. [DOI] [PubMed] [Google Scholar]
- Karlin, S. 1998. Global dinucleotide signatures and analysis of genomic heterogeneity. Curr. Opin. Microbiol. 1: 598–610. [DOI] [PubMed] [Google Scholar]
- Keiper, B.D. and Wormington, W.M. 1990. Nucleotide sequence and 40 S subunit assembly of Xenopus laevis ribosomal protein S22. J. Biol. Chem. 265: 19397–19400. [PubMed] [Google Scholar]
- Lawrence, J.G. and Ochman, H. 1997. Amelioration of bacterial genomes: Rates of change and exchange. J. Mol. Evol. 44: 383–397. [DOI] [PubMed] [Google Scholar]
- Lerat, E., Daubin, V., and Moran, N.A. 2003. From gene trees to organismal phylogeny in prokaryotes: The case of the γ-Proteobacteria. PLoS Biol. 1: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.-H. 1997. Molecular evolution. In Molecular evolution (ed. W.-H. Li). Sinauer Associates, Inc., Sunderland, MA.
- Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., Iijima, Y., Najima, M., Nakano, M., Yamashita, A., et al. 2003. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V. cholerae. Lancet 361: 743–749. [DOI] [PubMed] [Google Scholar]
- May, B.J., Zhang, Q., Li, L.L., Paustian, M.L., Whittam, T.S., and Kapur, V. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. 98: 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland, M., Sanderson, K.E., Spieth, J., Clifton, S.W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852–856. [DOI] [PubMed] [Google Scholar]
- Moran, N.A. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H. 2002. Distinguishing the ORFs from the ELFs: Short bacterial genes and the annotation of genomes. Trends Genet. 18: 335–337. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Dougan, G., James, K., Thomson, N., Pickard, D., Wain, J., Churcher, C., Mungall, K., Bentley, S., and Holden, M. 2001a. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413: 848–852. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B.W., Thomson, N.R., Titball, R.W., Holden, M.T., Prentice, M.B., Sebaihia, M., James, K.D., Churcher, C., Mungall, K.L., et al. 2001b. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413: 523–527. [DOI] [PubMed] [Google Scholar]
- Pedulla, M.L., Ford, M.E., Houtz, J.M., Karthikeyan, T., Wadsworth, C., Lewis, J.A., Jacobs-Sera, D., Falbo, J., Gross, J., Pannunzio, N.R., et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113: 171–182. [DOI] [PubMed] [Google Scholar]
- Perna, N., Plunkett, G., Burland, V., Mau, B., Glasner, J., Rose, D., Mayhew, G., Evans, P., Gregor, J., and Kirkpatrick, H. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529–533. [DOI] [PubMed] [Google Scholar]
- Perriere, G., Bessieres, P., and Labedan, B. 2000. EMGLib: The enhanced microbial genomes library (update 2000). Nucleic Acids Res. 28: 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E.P. and Danchin, A. 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18: 291–294. [DOI] [PubMed] [Google Scholar]
- Schoolnik, G.K. and Yildiz, F.H. 2000. The complete genome sequence of Vibrio cholerae: A tale of two chromosomes and of two lifestyles. Genome Biol. 1: REVIEWS1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger, D.W., Cheung, K.J., Mei, R., Johansson, E.M., Richmond, C.S., Blattner, F.R., Lockhart, D.J., and Church, G.M. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18: 1262–1268. [DOI] [PubMed] [Google Scholar]
- Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y., and Ishikawa, H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407: 81–86. [DOI] [PubMed] [Google Scholar]
- Siew, N. and Fischer, D. 2003a. Analysis of singleton ORFans in fully sequenced microbial genomes. Proteins 53: 241–251. [DOI] [PubMed] [Google Scholar]
- Siew, N. and Fischer, D. 2003b. Twenty thousand ORFan microbial protein families for the biologist? Structure (Camb.) 11: 7–9. [DOI] [PubMed] [Google Scholar]
- Simpson, A.J., Reinach, F.C., Arruda, P., Abreu, F.A., Acencio, M., Alvarenga, R., Alves, L.M., Araya, J.E., Baia, G.S., Baptista, C.S., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature 406: 151–157. [DOI] [PubMed] [Google Scholar]
- Stover, C.K., Pham, X.Q., Erwin, A.L., Mizoguchi, S.D., Warrener, P., Hickey, M.J., Brinkman, F.S., Hufnagle, W.O., Kowalik, D.J., Lagrou, M., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- Sueoka, N. 1988. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. 85: 2653–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas, I., Klasson, L., Canback, B., Naslund, A.K., Eriksson, A.S., Wernegreen, J.J., Sandstrom, J.P., Moran, N.A., and Andersson, S.G. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296: 2376–2379. [DOI] [PubMed] [Google Scholar]
- Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3: 203–208. [DOI] [PubMed] [Google Scholar]
- Wei, J., Goldberg, M.B., Burland, V., Venkatesan, M.M., Deng, W., Fournier, G., Mayhew, G.F., Plunkett III, G., Rose, D.J., Darling, A., et al. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, R.A., Burland, V., Plunkett III, G., Redford, P., Roesch, P., Rasko, D., Buckles, E.L., Liou, S.R., Boutin, A., Hackett, J., et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. 99: 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://globin.cse.psu.edu/enterix; Percent Identity Plots on the EnteriX server.