Abstract
Early endosomes are essential for regulating cell signalling and controlling the amount of cell surface molecules during neuronal morphogenesis. Early endosomes undergo retrograde transport (clustering) before their homotypic fusion. Small GTPase Rab5 is known to promote early endosomal fusion, but the mechanism linking the transport/clustering with Rab5 activity is unclear. Here we show that Drosophila Strip is a key regulator for neuronal morphogenesis. strip knockdown disturbs the early endosome clustering and Rab5-positive early endosomes become smaller and scattered. Strip genetically and biochemically interacts with both Glued (the regulator of dynein-dependent transport) and Sprint (the guanine nucleotide exchange factor for Rab5), suggesting that Strip is a molecular linker between retrograde transport and Rab5 activation. Overexpression of an active form of Rab5 in strip mutant neurons suppresses the axon elongation defects. Thus, Strip acts as a molecular platform for the early endosome organization that plays important roles in neuronal morphogenesis.
Neurons are highly polarized cells, possessing two morphologically and functionally distinct domains, axons and dendrites1,2. In developing neurons, membrane trafficking is essential for delivering diverse organelles and molecules that are required for elongation and guidance of the growing axon or dendrite processes3,4,5. Especially bi-directional endosomal trafficking between cell body and neurite termini is crucial to respond to continuously changing surrounding environments; endosomal trafficking regulates the number and activity of cell-surface molecules at the neurite termini in time and space6. For example, brain derived neurotrophic factor (BDNF) driven Trk receptor endocytosis at growth cone affects axon growth7,8. Asymmetric clathrin-mediated endocytosis of adhesion receptors such as integrins is required for repulsive growth cone guidance9.
Among vesicular structures in endocytic pathway, early endosomes are essential for cell signalling, serving both as signalling centres and as sorting centres to degrade or recycle cargoes10,11. The organization of early endosomes consists of three steps; endocytosis, clustering, and fusion. Internalized small early endosomes are clustered and then undergo fusion to become matured early endosomes. Rab5 is a key regulator for ligand sequestration at plasma membrane and repetitive fusion of internalized early endosomes by recruiting its effectors such as VPS34/p150, EEA1, Rabaptin-5, and Rabenosyn-5 (Rbsn-5)12-17. In addition, the dynein-dependent transport of small early endosomes from the periphery to the cell centre is important for clustering them18,19. Recently, the essential roles of endocytosis in axon elongation of mouse sympathetic neurons7 and endocytosis and endosomal trafficking in the dendrite arborization of Drosophila dendritic arborization (da) sensory neurons20,21 were reported. However, the physiological significance of clustering of early endosomes and the molecular link between the transport/clustering and Rab5 activity are largely unknown.
We here performed a forward genetic mosaic screen to identify genes for their cell-autonomous functions in dendrite and axonal development, and identified an evolutionarily conserved protein, Striatin-interacting protein (Strip), that works as a molecular linker between retrograde transport and Rab5 activity in early endosome organization. Strip and its orthologs were reported to be a component of Striatin-interacting phosphatase and kinase (STRIPAK) complex22-24, however, the function of Strip has not been reported yet, to our knowledge. Interestingly, we found that Strip forms the protein complex with both Glued, the ortholog of mammalian p150Glued, and Sprint, the ortholog of RIN-1. Glued is an essential component of the dynactin complex and regulates the initiation of retrograde transport on microtubule25-29 and Sprint is supposed to be a guanine nucleotide exchange factor (GEF) for Rab530, 31. We found that Strip affects the transport of early endosomes by forming complex with Glued in developing neurons. Furthermore, clustering of early endosomes is defective in strip knockdown Drosophila S2 cells and HeLa cells; early endosomes become smaller and scattered as reported in dynein-inhibited HeLa cells18. Moreover, similar with other GEFs, Sprint and Strip have a higher affinity for guanosine diphosphate (GDP)-bound forms of Rab5 than for guanosine triphosphate (GTP)-bound form. In addition, Strip seems to stabilize protein level of Sprint. Finally, the expression of constitutively active form of Rab5 in strip mutant neurons suppresses the axon elongation defect. These data demonstrate that Strip coordinates dynein-dependent transport and Rab5 activation at the clustering and fusion of early endosomes, which are required for axon elongation.
Results
stripdogi PNs show defects in neuronal morphogenesis
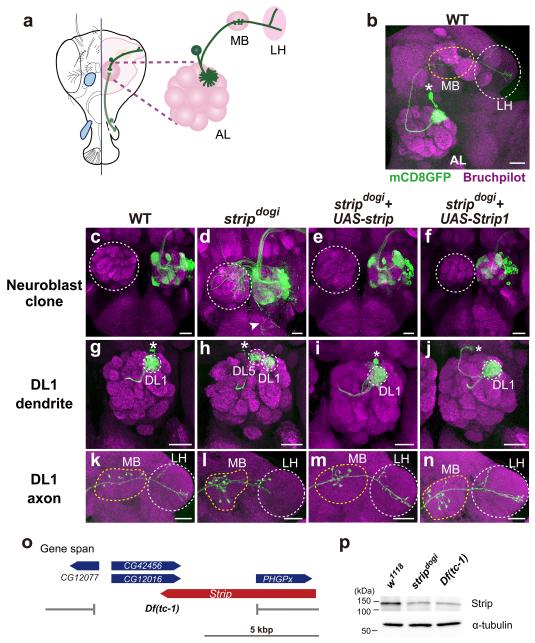
Drosophila olfactory projection neurons (PNs) (Fig. 1a), which are analogous to vertebrate mitral/tufted cells, provide an excellent model system for studying the molecular mechanisms of neuronal morphogenesis. Neuronal targeting and branching patterns of PNs are genetically defined32-34, and the mosaic analysis with a repressible cell marker (MARCM) method allows us to manipulate the genotype of a single PN (single-cell clone) or of a group of PNs that share the same lineage (neuroblast clone) and are positively labelled in vivo35. We performed a MARCM-based forward genetic screen of PNs to identify the genes essential for neuronal morphogenesis36, and isolated a mutant doubled glomeruli (dogi). As we will mention below, the causal gene of dogi mutant is strip gene, thus we name this mutant allele stripdogi hereafter. Wild-type PN axons target two higher olfactory centres, the mushroom body and the lateral horn, while dendrites target specific glomeruli in the antennal lobe, the first olfactory centre consisting of approximately 50 glomeruli32, 37, 38 (Fig. 1a,b). In neuroblast clones homozygous for stripdogi, neuronal guidance and targeting were severely defective, including ectopic innervation to the contralateral antennal lobe (Fig 1d, dotted circle) or the subesophageal ganglion (SOG) (Fig. 1d, arrowhead), which was never observed in wild-type neuroblast clones (Fig. 1c).
Figure 1.
Branching and elongation defects of stripdogi homozygous projection neurons.
(a) Schematic view of projection neurons (PNs). Olfactory information is transmitted from the olfactory receptor neurons (light green) to PNs (dark green) at the antennal lobe (AL, pink circle). The dendrite of a single PN targets a specific glomerulus out of 50 glomeruli in the AL, while the axon elongates and branches at the mushroom body (MB) and the lateral horn (LH). (b) Representative image of wild-type (WT) DL1 mosaic analysis with a repressible cell marker (MARCM) clone, DL1 PN (green). Magenta: Bruchpilot staining. Asterisk: cell body. Yellow and white dotted circles indicate MB and the LH, respectively. (c–f) Representative images of WT (c), stripdogi neuroblast clones (d), stripdogi neuroblast clones expressing cDNA of strip (e) or cDNA of mouse homolog of strip, Strip1 (f). Contralateral ALs are marked by dotted circles and neurite innervation to subesophageal ganglion (SOG) is indicated by an arrowhead (d). (g–n) Representative images of DL1 MARCM single-cell clone of each genotype. (g–j) Dendrites of DL1 PNs. DL1 and DL5 glomeruli are indicated by dotted circles and cell bodies are labelled with asterisks. (k–n) Axons of DL1 PNs. Yellow and white dotted circles indicate the mushroom body (MB) and the lateral horn (LH), respectively. Scale bar: 25 μm. (o) Genomic structure of strip. The small deletion allele Df(tc-1) lacks 7 kb of the genomic region containing strip, CG42456, CG12016, and CG12077. Scale bar: 5 kbp. (p) Strip expression in WT, the zygotic mutant homozygous for stripdogi or Df(tc-1). Embryo lysates from 18–21 h after egg-laying (AEL) were used for immunoblotting; Strip protein was detected by the anti-Strip antibody. The Strip protein is about 130 kDa.
To examine PN morphologies at a higher resolution, we generated single-cell clones homozygous for stripdogi (hereafter we call this single neuron as stripdogi PN). PNs that target a single glomerulus are categorized into roughly 50 classes, each with stereotyped dendrite and axon branching patterns32,37,38. In the wild-type DL1 PN, dendrites targeted the posterior, dorsolateral glomerulus DL1 and arborized within the DL1 glomerulus (Fig. 1b,g), while the axon exhibited stereotypical L-shape branching in the lateral horn (Fig. 1b,k). In the stripdogi DL1 PN, both axons and dendrites were defective. Unlike wild-type PNs, in which the dendrite shaft is generated by one branch that forms off the main branch and never branches further, stripdogi PN dendrites had an additional branch appearing from the proximal side of the dendrite shaft (magnified image in Fig. 2f arrowhead, penetrance of additional branch: 35.3%, n = 24/68), resulting in dendrite targeting of two glomeruli rather than a single glomerulus (Fig. 1h). The axons of stripdogi DL1 PNs did not elongate or form L-shaped branches within the lateral horn (Fig. 1l, penetrance of short axon: 98.2%, n = 56/57).
Figure 2.
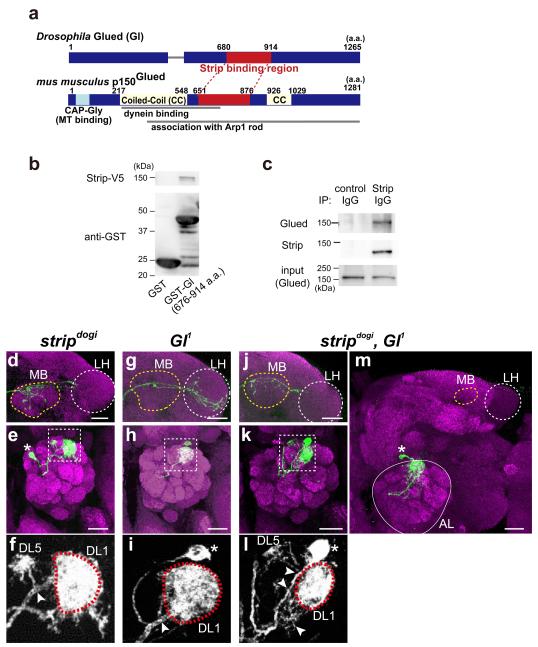
Glued (Gl) physically and genetically interacts with Strip
(a) Comparison between Drosophila Gl and its mouse orthologue, p150Glued. The 680–914 amino-acid region of Gl bound strongly to Strip in a yeast two-hybrid screen. This is analogous to the 651–876 amino acid region of p150Glued (68.9% similarity and 23.0% identity); this region interacts with the dynein motor protein and the Arp1 rod. (b) Lysates of 0–22 h after egg-laying (AEL) embryos that express Strip-V5 were pulled down with GST-fused Gl domain that binds to Strip in the yeast two-hybrid screen. (c) Immunoprecipitation of S2 lysates. Endogenous Gl was co-immunoprecipitated when endogenous Strip was precipitated by the anti-Strip antibody. (d–m) Genetic interactions between strip and Gl in projection neurons (PNs). MARCM DL1 single-cell clones for stripdogi (d–f), Gl1(g–i), or both stripdogi and Gl1 (j–m) were generated at 0–24 h ALH (green). Magenta: Bruchpilot staining. Scale bar: 25 μm. (d,g,j) Axons. Yellow and white dotted circles indicate the mushroom body (MB) and the lateral horn (LH), respectively. (e,f,h,i,k,l) Dendrites. Cell bodies are indicated by asterisks. (f), (i), and (l) are magnified images of dotted rectangles of (e), (h), and (k), respectively. Only green channels are shown in grey scale. DL1 glomeruli are labelled with dotted red circles. Additional dendrite branches are indicated by arrowheads. (m) Representative image of stripdogi and Gl1 DL1 PN exhibiting the “extremely short axon” phenotype; the axon did not exit the antennal lobe (AL, white circle) and did not enter the mushroom body (MB, yellow dotted circle) or the lateral horn (LH, white dotted circle).
Strip is required for PN morphogenesis
To identify the causal gene for these stripdogi PN phenotypes, we performed meiotic recombination mapping using single nucleotide polymorphisms (SNPs)39 and P-elements40; this placed the stripdogi mutation in an approximately 28-kb region of 63D1 (see Methods for details). Within this region, we generated a small deletion allele, Df(tc-1), which lacked 7 kb of the genomic region containing strip (CG11526), CG42456, CG12016, and CG12077 (Fig. 1o). The MARCM Df(tc-1) clones exhibited defects in axon elongation, similar to those seen in stripdogi PNs (Supplementary Fig. 1a), and also in dendrite branching (Supplementary Fig. 1b,c arrowheads), which was observed in some stripdogi PNs (Fig. 2e,f arrowhead). Next, we generated a short hairpin RNA against strip (shRNA-strip) to knock down strip expression; the expression of shRNA-strip in wild-type DL1 PN recapitulated the dendrite branching and axon elongation defects seen in stripdogi PNs (Supplementary Fig. 1d–f; see Supplementary Fig. 1g–j for the evaluation of shRNA-strip). Lastly, the expression of strip cDNA in stripdogi PN clones rescued all the stripdogi phenotypes (Fig. 1e,i,m). These findings confirmed that strip is responsible for the stripdogi PN phenotypes, and is cell-autonomously required for neuronal morphogenesis, specifically for dendrite branching and axon elongation.
Strip protein is highly conserved from yeast to humans (Supplementary Fig. 1k), with 45.9% amino-acid identity and 81.4% similarity between Drosophila and human STRIP1, and known as a component of Striatin-interacting phosphatase and kinase (STRIPAK) complex22-24. However, Strip has no characterized functional domains and its function has not been studied yet. When the mouse orthologue of strip, Strip141,42 (MGI: 2443884) was expressed only in stripdogi PNs using the MARCM strategy, all the stripdogi phenotypes were rescued (Fig. 1f,j,n), suggesting that the function of Strip is conserved between Drosophila and mammals. To characterize the Strip protein, we generated an antibody against Strip and confirmed its reliability by staining PNs homozygous for Df(tc-1) at 0 h after puparium formation (APF) (Supplementary Fig. 1l–o); immunostaining was not observed in Df(tc1)-homozygous cell bodies (Supplementary Fig. 1o). This result indicates that Strip is expressed in PNs during dendrite branching and axon elongation. Then, we used the Strip antibody for western blotting of embryo lysates from zygotic stripdogi and Df(tc-1) mutants, and found that the amount of Strip protein was significantly and similarly reduced in both lysates (Fig. 1p). The remaining expression of Strip in zygotic stripdogi or Df(tc-1) mutants is probably due to the maternally-derived strip gene product. Thus, the stripdogi allele used for phenotypic analysis is a strong loss-of-function or null allele.
Strip interacts with Glued and Sprint
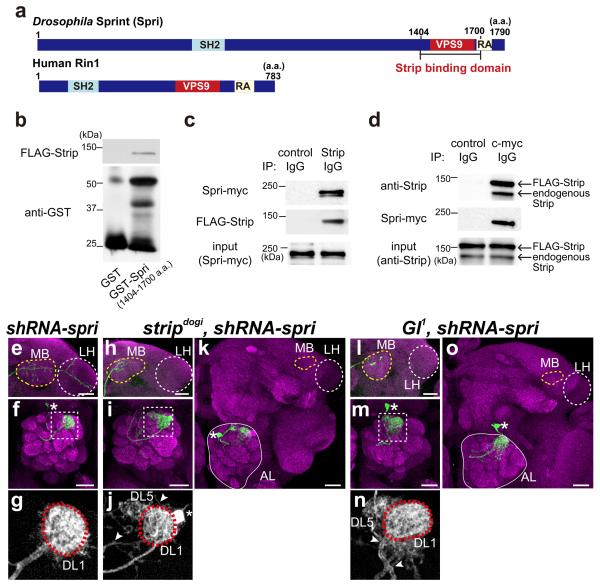
To examine how Strip regulates dendrite branching and axon elongation, we performed a yeast two-hybrid screen using full-length Strip as bait and the Drosophila whole-embryo cDNA library as prey, and identified Glued (Gl) and Sprint (Spri) as the binding partners of Strip (Fig. 2a,3a). Gl is a major component of the dynactin complex, which is a positive regulator of the microtubule (MT)-dependent retrograde motor, dynein, and regulates the initiation of retrograde transport25-29. Gl and its mouse ortholog, p150Glued, have an MT-binding domain, a dynein intermediate chain-binding domain, and an Arp1-associated domain (Fig. 2a). The Gl cDNA fragment isolated from the yeast two-hybrid screen encoded 235 amino acids, from position 680 to 914, corresponding to the dynein- or Arp1-binding domain (Fig. 2a). We confirmed this physical interaction by performing glutathione S-transferase (GST) pull-down and co-immunoprecipitation using Drosophila embryos and Drosophila S2 cell lysates (Fig. 2b,c). The other binding partner of Strip, Spri is an ortholog of human RIN-1 and has a vacuolar protein sorting 9 (VPS9) domain that is crucial for its function as a GEF for the small GTPase protein, Rab543 (Fig. 3a). The spri cDNA fragment isolated from the yeast two-hybrid screen encoded 397 amino acids, from position 1404 to 1700, corresponding to the VPS9 domain (Fig. 3a). We also confirmed this interaction by performing GST pull-down and co-immunoprecipitation using Drosophila embryos and Drosophila S2 cell lysates (Fig. 3b–d).
Figure 3.
Sprint (Spri) physically and genetically interacts with Strip
(a) Comparison between Drosophila Spri and its human orthologue, RIN-1. The 1404–1700 amino-acid region of Spri bound strongly to Strip in a yeast two-hybrid screen. This is analogous to the vacuolar protein sorting homology domain 9 (VPS9) that is required for guanine nucleotide exchange activity. SH2: Src homology domain 2, RA: Ras association domain. (b) Lysates of 0–22 h after egg-laying (AEL) embryos expressing FLAG-tagged Strip (FLAG-Strip) was pulled down with GST-fused Spri (Strip binding domain). (c, d) Immunoprecipitation of S2 lysates expressing c-myc tagged Spri (Spri-myc) and FLAG-Strip. Spri-myc was co-immunoprecipitated when FLAG-Strip was precipitated by the anti-Strip antibody (c). FLAG-Strip was co-immunoprecipitated when Spri-myc was precipitated by the anti-c-myc antibody. Since FLAG-Strip was detected with the anti-Strip antibody, the lower band represents endogenous Strip (d). (e–o) Genetic interactions between spri and strip or spri and Gl in projection neurons (PNs). MARCM DL1 single-cell clones expressing shRNA-spri (e–g), stripdogi DL1 single-cell clones expressing shRNA-spri (h–k), or Gl1 DL1 single-cell clones expressing shRNA-spri (l–o) were induced (green) at 0–24 h ALH. Magenta: Bruchpilot staining. Scale bar: 25 μm. (e,h,l) Axons. Yellow and white dotted circles indicate the mushroom body (MB) and the lateral horn (LH), respectively. (f,g,i,j,m,n) Dendrites. Cell bodies are indicated by asterisks. (g), (j), and (n) are magnified images of dotted rectangles of (f), (i), and (m), respectively. Only green channels are shown in grey scale. DL1 glomeruli are labelled with dotted red circles. Additional dendrite branches are indicated by arrowheads. (k.o) representative images of stripdogi DL1 PNs expressing the shRNA-spri (k), and Gl1 DL1 PNs expressing the shRNA-spri (o) exhibiting the “extremely short axon” phenotype, i.e., the axon did not exit the antennal lobe (white circle) and did not enter the mushroom body (MB, yellow dotted circle) or the lateral horn (LH, white dotted circle).
We next examined the genetic interactions between strip, Gl, and spri. Gl1 (dominant negative form of Gl) DL1 PNs (Fig. 2g–i) or PNs expressing the shRNA-spri (Fig. 3e–g) did not exhibit obvious defects in axon elongation (Fig. 2g and 3e) or dendrite branching (Fig. 2h,i arrowhead, 3f,g). Interestingly, axon elongation or dendrite branching phenotypes of stripdogi DL1 PNs (Fig. 2d–f) became more severe by adding Gl1 (double mutant clones for stripdogi and Gl1, Fig. 2j–m) or expressing the shRNA-spri (Fig. 3h–k). In both cases, we observed an extremely short axon phenotype, i.e., the axon did not exit the antennal lobe (Fig. 2m (penetrance of extremely short axon: 15.6%, n = 5/32) and 3k (penetrance of extremely short axon: 13.0%, n = 3/23)), which was never observed in stripdogi DL1 PNs (penetrance of extremely short axon: 0%, n= 0 /68). We also found an obvious genetic interaction between strip and Gl in the rough-eye phenotype, in which Gl is known to be involved44 (Supplementary Fig. 2a–d). These biochemical and genetic data suggest that Strip functions with Gl and Spri in dendrite branching and axon elongation. Since stripdogi, Gl1 PNs (double mutant clones) and stripdogi PNs expressing shRNA-spri exhibit the similar “extremely short axon” phenotype, we wondered whether Gl and spri genetically interact. As we expected, axon elongation and dendrite branching defects were observed Gl1 PNs expressing shRNA-spri (Fig. 3l–o, penetrance of extremely short axon in o: 38.9%, n= 7/18), suggesting that strip, Gl and spri cooperatively regulate axon elongation and dendrite branching.
Strip is involved in transporting early endosomes
Since the main function of Gl is to regulate the initiation of dynein-based retrograde transport28, 29, it is possible that stripdogi mutation affects intracellular transport. Thus, to examine the function of Strip, we first analysed the subcellular localization of synaptotagmin, a representative axonal transport cargo regulated by the dynein-dynactin complex45. We expressed hemagglutinin (HA)-tagged synaptotagmin (syt-HA)46 in single-cell DL1 PN clones. Most of the syt-HA was delivered properly to the presynaptic sites in stripdogi PNs, but some of it accumulated abnormally in the axon stalks (Supplementary Fig. 2h–j, quantification in k). This accumulation was not observed in wild-type PNs (Supplementary Fig. 2e–g, quantification in k); thus, abnormal accumulation of syt-HA in stripdogi PN axon did not result from syt-HA overexpression. These data indicate that Strip is required for the intracellular transport of synaptic proteins in PN axons.
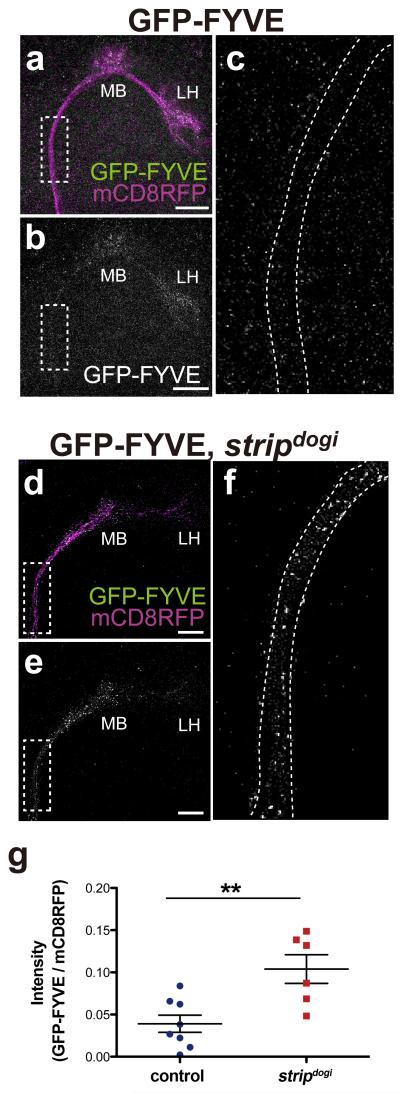
We also found defective subcellular localization of early endosomes in stripdogi PNs. Since the proper regulation of endosomal trafficking is required for neuronal morphogenesis6, we hypothesized that Strip affects the transport of early endosomes during axon elongation. When the early endosome marker, the green florescent protein (GFP)-fused FYVE domain (GFP-FYVE)47 was expressed in developing wild-type PNs, it was mainly localised at the termini but not in the stalk of axons at 21 hours after puparium formation (Fig. 4a–c, quantification in g). Interestingly, GFP-FYVE showed significantly higher levels in the axon stalk of stripdogi PNs (Fig. 4d–f, quantification in g) compared to that in the control (Fig. 4a–c, quantification in g). Thus, Strip affects the subcellular localization of early endosomes in PN axons when axon elongation is taking place.
Figure 4.
Localization of early endosomes was altered in the axons of developing strip projection neurons (PNs)
(a–f) A GFP-fused FYVE domain (GFP-FYVE) was expressed in PNs, and its localization was investigated at 21 h after puparium formation (APF). Axons of wild-type (WT, a–c) and stripdogi PNs (d–f). (a, d) GFP-FYVE and MARCM neuroblast clones labelled by mCD8-RFP are shown in green and magenta, respectively. (b,e) Only green channels are shown in grey scale. (c) and (f) are magnified images of dotted rectangles of (b) and (e), respectively. Scale bar: 25 μm. (g) The intensity of GFP-FYVE in the axon stalk (200 μm2) was normalized to the intensity of mCD8RFP and then plotted. Error bars are defined as SEM. **p = 0.0047, t-test.
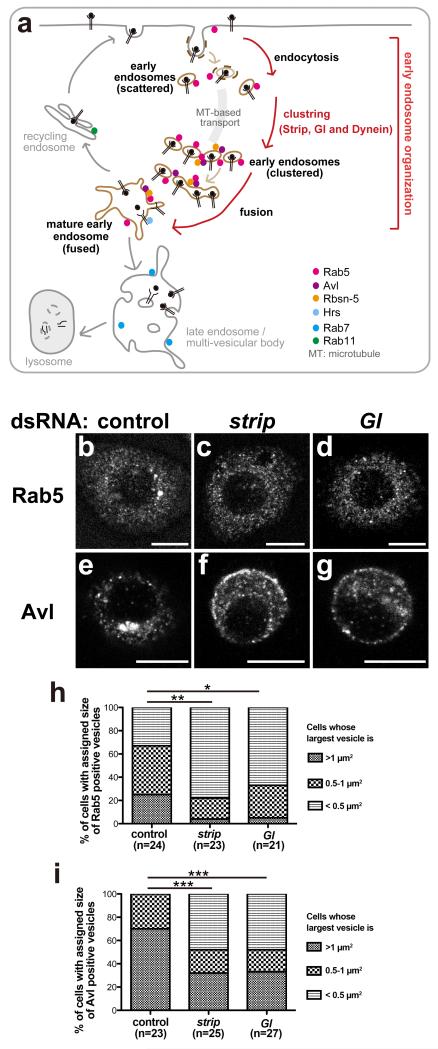
Strip and Gl are involved in clustering of early endosomes
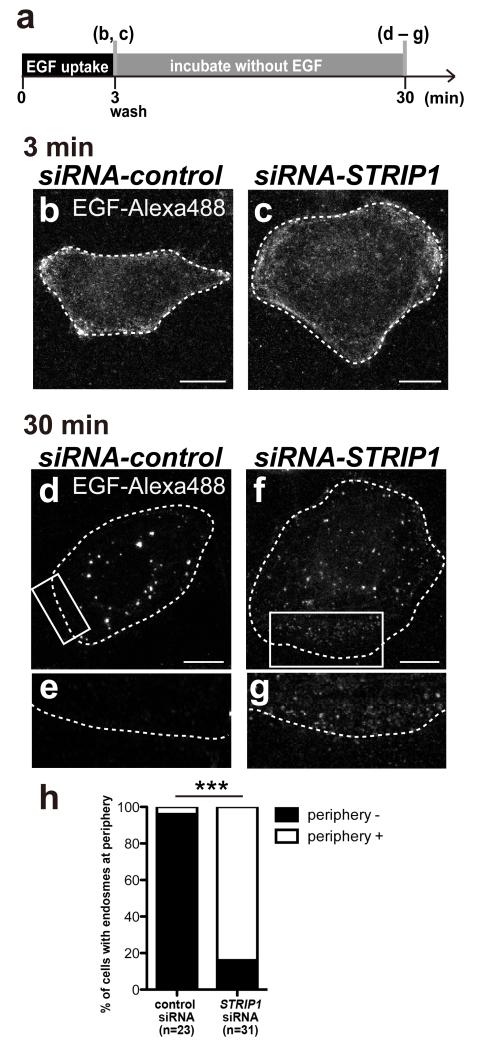
To examine the effect of Strip on endosomal trafficking, the localization of Rab5- and Avalanche (Avl; Drosophila ortholog of Syntaxin 7, 12)48-positive early endosomes was examined in Drosophila S2 cells. The organisation of early endosomes consists of three steps; endocytosis, clustering, and fusion (Fig. 5a). Retrograde transport is required for clustering of internalized vesicles (small early endosomes) that are formed by endocytosis18,19. After clustering, small early endosomes undergo homotypic fusion to form mature early endosomes12,13,15. Interestingly, the size of Rab5- and Avl-positive early endosomes was significantly affected in S2 cells treated with strip dsRNA. Enlarged Rab5- and Avl-positive early endosomes were frequent in cells treated with control dsRNA (Fig. 5b,e, Rab5 is quantified in 5h, and Avl is quantified in 5i), but they were smaller in cells treated with strip dsRNA (Fig. 5c,f,h,i). Next, we tried to elucidate which step of early endosome orgnization is defective in strip knockdown. We examined the endocytosis ability of cells treated with strip dsRNA using the FM1-43 styryl dye and found that strip knockdown did not inhibit endocytosis (Supplementary Fig. 3, see Methods for details). Furthermore, the same “smaller Rab5- and Avl-positive early endosomes” phenotype was observed when the cells were treated with Gl dsRNA (Fig. 5d,g–i). These results suggest that Strip affects, at least, the clustering of early endosomes by forming a complex with Gl, which is consistent with the previous report that dynein-dependent early endosome transport from the periphery to the centre of a cell is required for the organization of early endosomes18,19. We also revealed that Strip function in early endosome clustering is evolutionarily conserved. We treated HeLa cells with control or human STRIP1 siRNA and stimulated them with fluorescent-labelled-EGF (EGF-Alexa488) for three minutes followed by washing to remove labelled EGF from the medium (Fig. 6a). Like FM1-43 dye uptake assay performed in Drosophila S2 cells, endocytosis of EGFR bound to EGF-Alexa488 was not defective in STRIP1 siRNA-treated HeLa cells, suggesting that STRIP1 knockdown does not inhibit endocytosis. EGF-Alexa488-positive vesicles were localized at cell periphery both in control or STRIP1 siRNA-treated HeLa cells just after washing out the EGF-Alexa488 (Fig. 6b,c). After thirty minutes incubation, the localization of EGF-Alexa488 was different between control and STRIP1 siRNA-treated HeLa cells (Fig. 6d–g, Supplementary Fig. 4). In control cells, EGF-Alexa488-positive vesicles disappeared from cell periphery and mainly localized at peri-nuclear region (Fig. 6d,e, quantified in h). On the other hand, in STRIP1 siRNA-treated HeLa cells, most of cells possessed small and scattered EGF-Alexa488-positive vesicles, which are mainly at cell periphery (Fig. 6f,g, quantified in h). In addition, those small EGF-Alexa488-positive vesicles are EEA1-negative (Supplementary Fig. 4d–f arrowheads), suggesting they are immature early endosomes. Thus, we concluded that Drosophila strip and human STRIP1 are involved in clustering of early endosome organization.
Figure 5.
Localization defects of early endosomes in strip knockdown Drosophila S2 cells
(a) Schematic view of endocytic pathways. Early endosome organization consists of three steps, endocytosis, clustering and fusion of small early endosomes. (b–g) Localization of early endosome markers (Rab5 and Avalanche, Avl) in the control (b,e), strip (c,f), and Gl (d,g) dsRNA-treated S2 cells. (b–d) Rab5-positive early endosomes. (e–g) Avl-positive early endosomes. Scale bar: 7.5 μm. (h,i) Quantification of the size of Rab5-positive early endosomes (h) and Avl-positive early endosomes (i). The area of each vesicle was measured with the Particle Analyzer of the ImageJ software. The percentage of cells whose largest vesicle with sizes >1 μm2, >0.5 μm2 but <1 μm2, and <0.5 μm2 are shown. *p = 0.049, **p = 0.0068, ***p < 0.0001, chi-square test.
Figure 6.
Localization defects of EGF-Alexa488 in STRIP1 knockdown HeLa cells
(a) Summary of EGF-Alexa488 uptake assay. (b–g) Localization of EGF-Alexa488 in the control (b,d,e) and STRIP1 (c,f,g) siRNA-treated HeLa cells. HeLa cells were transfected with control or STRIP1 siRNA for 3 days, and endocytosis uptake was assayed with EGF-Alexa488. After a 3 min uptake of EGF-Alexa488, cells were fixed (b,c) or washed with DMEM and cultured for 27 min and fixed (d–g). The rectangle regions of (d) and (f) are magnified at (e) and (g), respectively. Scale bar: 10 μm. (h) All cells were classified into having or not having EGF-Alexa488 at cell periphery by a blind test. ***p<0.0001, chi-square test.
Strip regulates axon elongation by controlling Rab5 activity
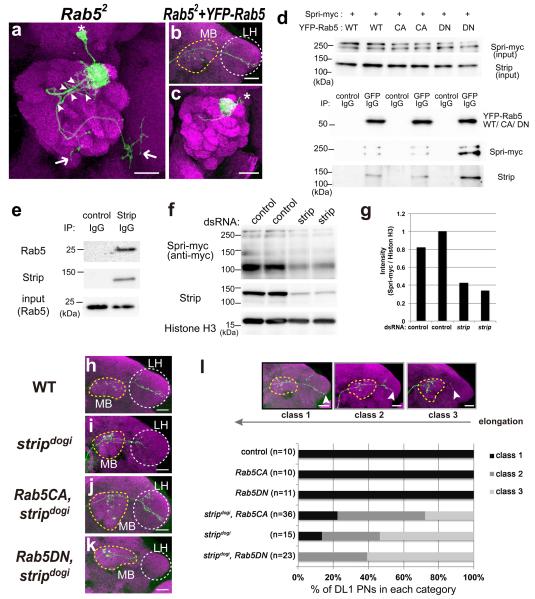
Given that Strip makes complexes with Gl and Spri, we hypothesized that Strip affects the organization of early endosomes by properly clustering them with Gl and promoting early endosome fusion with Spri-Rab5, which are required for axon elongation. To confirm our hypothesis, we first investigated whether Rab5 is required for axon elongation in PNs by generating MARCM single-cell PN clones homozygous for a Rab5 null mutation47 (Fig. 7a). As in the stripdogi DL1 PN clones, the dendrites in Rab52 DL1 PN had additional branches appearing from the proximal side of the dendrite shaft, with a penetrance of 85.7% (Fig. 7a arrowheads, n = 18/21). Some Rab52 DL1 dendrites targeted both DL1 and DL5, the same glomeruli targeted by the stripdogi DL1 PNs (Supplementary Fig. 5). In addition, Rab52 DL1 PNs had an extremely short axon phenotype (47.6%, n = 10/21), i.e., the axons stopped in the vicinity of the antennal lobe and did not extend toward the mushroom body or the lateral horn (Fig. 7a, arrows). These phenotypes were similar to those observed in double mutant clones for stripdogi and Gl1 (Fig. 2m), stripdogi PNs expressing shRNA-spri (Fig. 3k), or Gl1 PNs expressing shRNA-spri (Fig. 3o). The expression of yellow fluorescent protein (YFP)-fused Rab5 (YFP-Rab5)49 rescued Rab52 PNs phenotypes (Fig. 7b,c), indicating that Rab5 is cell-autonomously required for dendrite branching and axon elongation in PNs.
Figure 7.
Strip affects Rab5 activity in axon elongation
(a–c) Representative image of Rab52 DL1 PNs (a, green) and Rab52 DL1 PNs expressing YFP fused Rab5 (b,c, green). Magenta: Bruchpilot staining. Asterisk: Cell body. Yellow and white dotted circles indicate the mushroom body (MB) and the lateral horn (LH), respectively. Scale bar: 25 μm. (d) Immunoprecipitation of S2 lysates expressing c-myc tagged Spri (Spri-myc) and YFP-tagged Rab5 (YFP-Rab5WT), YFP-tagged dominant negative form of Rab5 (YFP-Rab5DN) or YFP-tagged constitutively active form of Rab5 (YFP-Rab5CA). Spri-myc and Strip were co-immunoprecipitated when YFP-Rab5 WT/CA/DN were precipitated by the anti-GFP antibody. (e) Immunoprecipitation of S2 lysates. The anti-Strip antibody co-immunoprecipitated Strip and endogenous Rab5. (f) The expression level of Spri-myc in control and strip dsRNA-treated S2 cells. Knockdown efficiency was confirmed by anti-Strip antibody and anti-Histone H3 was used as a loading control. (g) Densitometric analysis of Spri-myc in (f). The intensity of Spri-myc was measured by using Image J software and normalized against the intensity of Histone H3 and showed in graph. (h–k) Representative image of DL1 wild-type (WT, h), stripdogi PN (i), stripdogi PN expressing constitutively active form of Rab5 (Rab5CA, Rab5Q88L, j), and stripdogi PN expressing dominant-negative form of Rab5 (Rab5DN, Rab5S43N, k) labelled by mCD8RFP (green). Magenta: Bruchpilot staining. Yellow and white dotted circles indicate the mushroom body (MB) and the lateral horn (LH), respectively. Scale bar: 25 μm. (l) The axons of DL1 PNs were categorized into three classes according to the extent of elongation in a blind test. Class 1 axons reached the end of the LH, while class 3 axons terminated near the mushroom body (l, arrowheads). The percentage of PNs in each category is shown in (l).
Next, we examined the physical interactions among Strip, Spri, and Rab5. Since Spri is an ortholog of RIN-1 that exhibits Rab5-specific guanine nucleotide exchange activity43, one would expect that Drosophila Spri could have a higher affinity for GDP-bound forms of Rabs than for GTP-bound forms, which is the common feature of GEFs. As expected, c-myc-tagged Spri (Spri-myc) formed complex preferentially with a dominant-negative form of Rab5 (Rab5DN, Rab5S43N)49, which mimics the GDP-bound or free form of Rab5, and weakly to constitutively active form (Rab5CA, Rab5Q88L)49, which mimics the GTP-bound form (Fig. 7d). In accordance with our hypothesis, Strip also had a higher affinity for GDP-bound forms of Rab5 than for GTP-bound forms (Fig. 7d). In addition, we also confirmed the physical interaction between endogenous Strip and Rab5 (Fig. 7e). Furthermore, we found that the level of Spri-myc was decreased when strip was knocked down in S2 cells (Fig. 7f, quantification in g), indicating that Strip affects the protein level of Spri. Together, these results suggest that Strip is involved in Spri-mediated Rab5 activation by stabilizing Spri.
Since Strip seems to regulate the activity of Rab5 by forming a complex with Spri, we examined whether a constitutively active form of Rab5 (Rab5CA, Rab5Q88L) could compensate for the loss of Strip function in early endosomes during axon elongation. Indeed, the stripdogi short-axon phenotype (Fig. 7i) was partially suppressed in stripdogi DL1 PN clones expressing Rab5CA (Fig. 7j, quantification in l), with some axons even exhibiting L-shaped branching at the lateral horn (Fig. 7j, white dotted circle). In contrast, the short-axon phenotype was enhanced in stripdogi DL1 PNs expressing a dominant-negative form of Rab5 (Rab5DN, Rab5S43N) (Fig. 7k, quantification in l), i.e., most of axons were terminated at the calyx of the mushroom body (Fig. 7l “class 3”). These data support our hypothesis that the Strip regulates axon elongation by controlling the activity of Rab5.
Discussion
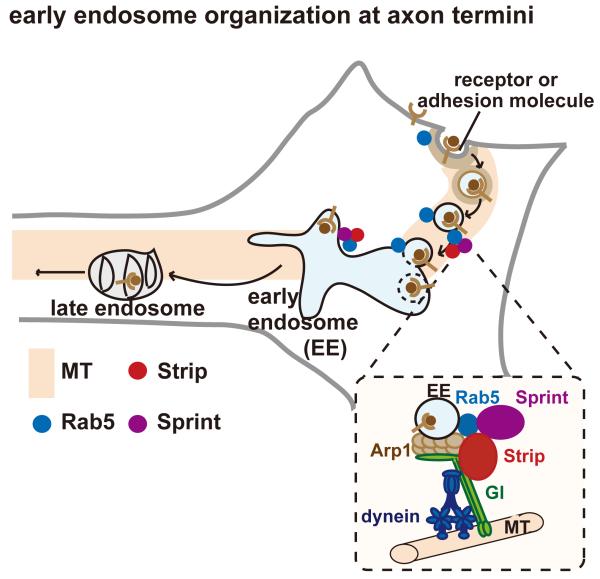
During neural development, axon termini are exposed to a constantly changing surrounding environment. Thus, the spatio-temporal regulation of the amount of adhesion molecules and receptors at the axon growth cone is essential6. In this study, we identified uncharacterized protein Strip that forms complexes with Gl and Spri, and positively regulates clustering (retrograde transport along microtubule) of early endosomes. To form mature early endosomes from internalized small early endosomes, the coordination between clustering and fusion of early endosomes is crucial. For this, Strip serves as a molecular linker by interacting with Gl in clustering, and with Spri, Rab5 and probably with Vps45 in fusion. Vps45 is a Sec-1/Munc18 (SM) protein that targets Avl to promote fusion of early endosomes, which seems to function with Strip in the wing development (genetic interactions among strip, Gl, and regulators of early endosome fusion: Supplementary Fig. 6). Since we also observed genetic interaction between spri and Gl in axon elongation (Fig. 3l,o), Strip seems to be a platform for early endosome organization by forming a dynein-Gl-Strip-Spri-Rab5 complex. Herein, we propose the following model to explain the physiological role of Strip in the organization of early endosomes (Fig. 8, See Fig. 8 legend for details). During axon elongation, the strength of the elongation signal could be properly regulated by the dynein-Gl-Strip-Spri-Rab5 complex by affecting the organization of mature early endosomes. Mature early endosomes are essential vesicular structures for cell signalling, serving both as signalling centres and as sorting centres to degrade or recycle cargoes. In vertebrate neurons, target-derived nerve growth factor (NGF) mediates axon elongation via retrograde axon transport of TrkA-containing signalling endosomes locally or to the cell body to stimulate the expression of downstream target genes essential for long-term axon growth and target innervation7,8. Furthermore, the amount of cell adhesion molecules might also be regulated by Strip-mediated early endosome organization. It has been reported that the regulation of the amount and localization of molecules such as L1 and integrin in the axon growth cone is essential for growth cone motility9.
Figure 8.
Hypothetical model for the function of Strip in axon elongation.
Strip (red) binds to the dynein accessory proteins Glued (green) and Sprint (purple), and forms a complex with the Rab5 (blue)-containing early endosomes, suggesting that Strip seems to be in a large molecular complex, dynein-Gl-Strip-Spri-Rab5 complex (see the dotted rectangle). Thus, Strip seems to serve as a molecular linker of clustering and fusion steps of internalized small early endosomes, which is essential for the organization of mature early endosomes containing a ligand-receptor complex analogous to the vertebrate neurotrophin-TrkA complex that sends signals for axon growth.
Further investigations are still needed to identify which Strip domains are important for the formation of the dynein-Gl-Strip-Spri-Rab5 complex. Because Gl and Spri were identified using a yeast two-hybrid screen, it seems that the binding between Gl and Strip, and that between Spri and Strip are likely to be direct. Furthermore, we think that Gl and Spri bind to Strip simultaneously and form a tertiary complex for two reasons; 1) Spri and Gl showed genetic interaction in PNs and exhibited a phenotype similar to that of stipdogi PNs and 2) Vps45 and Gl exhibited genetic interaction at the wing vein formation (Supplementary Fig. 6). Thus, Strip seems to serve as a molecular linker between early endosome clustering and fusion by simultaneously forming a complex with Gl and Spri. Additionally, Strip seems to stabilize Spri protein level. Unfortunately, we were unable to produce a usable anti-Spri antibody; therefore we could not examine Spri localization. However, we speculate that Spri stabilized by Strip could determine the spatio-temporal localization and the activity of Rab5. Regarding the binding between Strip and Gl, we hypothesize that Strip may act as an adaptor between early endosomes and dynein. Unlike the highly diverse kines in superfamily proteins (KIFs), which encompass 45 genes, cytoplasmic dyneins have only two heavy chain family members in mammals3. Thus, adaptor proteins for the dynein motor are thought to be important to determine the cargo selectivity, and some adaptor proteins have already been reported3,50,51. Although Kif16b links Rab5 and microtubules52, no specific molecule linking the dynein heavy chain with Rab5-positive early endosomes has been reported yet. Therefore, we speculate that Strip could serve as an adaptor protein between Rab5-positive early endosomes and the dynein/dynactin complex by binding to Gl, similar to RILP and ORP1L that serve as adaptors between late endosomes and the dynein/dynactin complex50,51.
We have provided genetic evidences that Strip, Gl, Spri, and Rab5 are required for both axon elongation and dendrite branching in PNs. However, the expression of a constitutively active form of Rab5 partially suppressed axon elongation and did not suppress dendrite branching defects of stripdogi PNs (Fig. 7h–l, Supplementary Fig. 7). We consider that the level and the spatio-temporal pattern of Rab5 activity are crucial. Axon elongation and dendrite branching may require different levels or patterns of Rab5 activity. Furthermore, the failure to suppress the dendrite phenotype could also be explained by the fact that not only formation, but also localization of mature early endosomes is required for dendrite branching. In Drosophila dendritic arborization (da) neurons, the proximal branching phenotype has been observed in the dendrite of Rab52, dynein light intermediate chain (dlic), or the kinesin heavy chain (khc) mutant20. Consistent with this report, we also observed dendrite proximal branching phenotype in Rab52 or stripdogi PNs (Fig. 2e,f,7a). The additional dendrite branched out from the dendrite stalk at a more proximal side in Rab52 or stripdogi PNs than in wild-type PNs. Therefore, we hypothesized that the constitutively active form of Rab5 could not suppress the dendrite phenotype in stripdogi PNs because the expression of the constitutively active form of Rab5 in stripdogi PNs could promote early endosome maturation in axon and dendrites, but could not adjust early endosome subcellular localization, which is crucial for dendrite branching.
Strip is structurally and functionally conserved. Branching and elongation defects in stripdogi PNs were rescued by the expression of the mouse homolog, Strip1 (Fig. 1f,j,n). In addition, we also found that Strip1 is involved in neuronal migration in the mouse developing cerebral cortex (Supplementary Fig. 8). In the mouse developing cerebral cortex, the Rab5-dependent endosomal trafficking pathway is required for neuronal migration53, rather than for neurite extension, probably because it occurs prior to dendrite formation and axon elongation in vivo. Electroporation of Strip1 shRNA into the cerebral cortex at E14 caused migration defects (Supplementary Fig. 8), similar to those observed when Rab5 shRNAs were used53. Thus, strip/Strip1 is involved in Rab5-dependent neural development including neuronal morphogenesis and migration events in both Drosophila and the mouse.
During our study of Strip function, Strip was reported as a member of the STRIPAK complex23,24,42. STRIPAK complex contains the PP2A phosphatase complex that is widely used as a phosphatase22. The core component of the PP2A phosphatase complex shows phosphatase activity, while the regulatory subunit determines the substrate specificity. Thus PP2A phosphatase targets specific molecules by changing the regulatory subunit. When Strip is included in the PP2A complex, Cka serves as a regulatory subunit and targets Hippo and probably RAF which belongs to the RAS-MAPK pathway23,24. Since early endosome organization is important for many signal transduction pathways, it might be possible that axon elongation and dendrite targeting require both the dynein-Gl-Strip-Spri-Rab5 complex and the Strip-STRIPAK (Cka)-Hippo or the Strip-STRIPAK (Cka)-RAS-MAPK complex, but this does not seem possible, at least, in neuronal morphogenesis of PNs. When we generated PNs homozygous for Cka, we did not find any defects in axon elongation or dendrite branching (Supplementary Fig. 9). Thus, we think that the Strip-STRIPAK complex might not play a major role in neuronal morphogenesis.
The disruption of dynein or dynactin function causes neurodegeneration in many species, and this has been suggested to reflect impaired transport of signalling endosomes54. In humans, G59S and G59A missense mutations of p150Glued have been discovered in motor neuropathy 7B55 and Perry syndrome56, respectively. Both mutations are located within the p150Glued cytoskeleton-associated protein glycine-rich (CAP-Gly) domain, which is an MT-binding domain required for efficient retrograde transport. Progressive motor neuron degeneration is observed in p150Glued transgenic mice, with pathological similarities to amyotrophic lateral sclerosis (ALS), including muscle atrophy, axonal swelling, disrupted intracellular trafficking, and proliferation of degenerative organelles such as lysosomes and autophagosomes54,57. Furthermore, motor neurons in Als2 (juvenile ALS-associated gene, Rab5GEF)-deficient mice show axonal degeneration. Moreover, the primary neurons in this mouse model, show disturbed endosomal trafficking of the insulin-like growth factor 1 (IGF-1) and brain-derived neurotrophic factor (BDNF) receptors54. These observations suggest that retrograde transport and Rab5GEF may play a particularly important role in axonal health by transmitting information about the local environment from the axon to the soma. Since Strip affects the early endosome organization by forming a complex with Gl and Spri, studying the functions of Strip may provide new insights into both developmental and pathological processes.
Methods
Genetic mapping of stripdogi mutants
The causal gene, strip was mapped by single nucleotide polymorphism (SNP)-based recombination39, which showed that the mutation responsible for the projection neuron (PN) phenotype in the stripdogi MARCM clone was at a cytological location between 63A and 64A. Next, assuming that stripdogi is homozygous lethal, we performed a lethal complementation test with deficiency lines available for this candidate region. Df(3L)HR232 and Df(3L)HR119 failed to complement the stripdogi lethality, suggesting that the stripdogi chromosome maps within the genomic region from 63C02 to 63D02 (~130 kb). We next measured the recombination rate from genetically mapped P-element insertion sites40. While generating recombinant lines, we obtained three independent recombinants that complemented the stripdogi lethality; analysis of their recombination points narrowed the candidate region down to ~28 kb. Next, we generated a small deficiency, Df(tc-1) (~7 kb) (Fig. 1o), using the f05249 and e04482 PiggyBac lines. Df(tc-1) failed to complement the stripdogi lethality. Furthermore, we made shRNA-strip and PNs expressing shRNA-strip exhibited the phenotypes similar to those observed in stripdogi PN clones (Supplementary Fig. 1d–f). In addition, Strip protein expression was significantly reduced in stripdogi homozygous embryos (Fig. 1p). Lastly, we confirmed that UAS-strip could mostly rescue the stripdogi PN clone phenotypes (Fig. 1e,i,m), indicating that strip was the gene responsible for these phenotypes.
Clonal analysis
We used the MARCM method58. Briefly, we crossed MARCM ready flies that contain FLP recombinase, an FRT site, GAL4, tublin 1 promoter-GAL80 and UAS-mCD8GFP/RFP to a line containing the corresponding FRT and mutation of interest for MARCM analysis. To induce MARCM neuroblast clones, we heat-shocked (37 °C, 1h) flies with the appropriate genotype 0–24 h after larval hatching (ALH). To analyse DL1 PNs, we heat-shocked flies (37 °C, 1h) with the appropriate genotypes at 0–30h ALH for Figure 1g, h, k, and l, or at 0–24h ALH for other analyses. The anterodorsal PN single-cell clones generated in this time period innervate the DL1 glomerulus. Dissections were performed on both genders of adults aged 1–10 days, with the exception of those described in Fig. 4a–f (21 h after puparium formation, APF) and Supplemetrary Fig. 1m–o (0 h APF).
Fly strains
The genotypes of Drosophila melanogaster used in each experiment are shown in Supplementary Table 1. Flies were maintained under standard laboratory condition (25 °C). Both genders of adults aged 1–10 days are used for most studies. As for Supplementary Figure 2a–d and Supplementary Figure 6, adults aged 1–2 days were used. Gl1, UAS-YFP-Rab549, UAS-YFP-Rab5S43N49, and UAS-YFP-Rab5Q88L49, daughterless (da)-Gal4 were obtained from the Bloomington Stock Centre. Rab52 and UAS-GFP-myc-2xFYVE were kind gifts from Marcos González-Gaitán47, UAS-syt-HA is a kind gift from Thomas Schwarz46, UAS-Vps45-RNAi, Vps45JJ2, and avl1 were kind gifts from David Bilder15,59,48, and Cka1 is a kind gift from Steven X. Hou60.
DNA construction and generation of transgenic flies
To generate UAS-strip, UAS-strip-V5 and UAS-FLAG-strip flies, strip cDNA was amplified by PCR using the EST clone (AT25231) and was subcloned into pUAST. A V5 epitope tag was added at the strip C-terminus or a FLAG epitope tag was added at the strip N-terminus. The cDNA of Mus musculus Strip1, was amplified by PCR using the BC023952 EST clone. Since the 12bp of N-terminal sequence of this clone is reported not to align with the mouse genome, we corrected the sequence and subcloned it into pUAST. Transgenic flies were generated by BestGene Inc. The generation of shRNA-strip, shRNA-Gl, shRNA-sprint, and UAS-resistant-strip was designed as follows61. The two sites targeted by shRNA-strip were shRNA-strip-6: 5′-ATGGCTTGGAAACAGAAAAATA-3′ and shRNA-strip-9: 5′-CGGCCACAGCTACTGATATACT-3′; the two sites targeted by shRNA-Gl were shRNA-Gl-1: 5′-GCGAACTAATACCGGTATTATC-3′ and shRNA-Gl-7: 5′ -GGACCGACACAGAACTTTAAGA-3′; the two sites targeted by shRNA-spri were shRNA-sprint-2: 5′ -TTCAGCGGATTACTCATTAAGA-3′ and shRNA-sprint-10: 5′ -GCGGCCAGTAATCCCTTTAATG-3′. For UAS-resistant-strip, the 20th nucleotide of the strip-shRNA-9 target site was altered from A to T. These shRNAs were subcloned into pUAST or pUASTattB, and transgenic flies were generated by BestGene Inc. To generate UAS-spri-myc, sprint cDNA was amplified by PCR using the EST clone (SD06366), and was subcloned in pUAST. A c-myc epitope tag was added at the C-terminus of sprint. To generate shRNA-Strip1, we used the Whitehead Institute’s siRNA prediction tool to research target sites (http://sirna.wi.mit.edu/). We inserted oligonucleotides targeting four distinct regions in the Strip1 coding sequence (shRNA-Strip1-1: 5′-GAATTCCTGATGAATCGAA-3′, shRNA-Strip1-2: 5′-CTATAAGGCTGACGACTCT-3′, shRNA-Strip1-3: 5′-GTTGAACCACATCTACCAG-3′, shRNA-Strip1-4: 5′-CGTACGGCAATGATCTTGA-3′) into the pSilencer 3.0-H1 neo vector (Ambion). All of these regions contained a hairpin loop sequence (5′-TTCAAGAGA-3′). To generate pcDNA3-Strip1-V5, we added a V5 epitope tag to the C-terminus of Strip1 cDNA, and subcloned it into pcDNA3.
Detailed DNA constructions are available upon request.
Bioinformatics
For homology searches, we used the In Paranoid: Eukaryotic Ortholog Groups (http://inparanoid.sbc.su.se/cgi-bin/index.cgi). Identity and similarity were calculated by the GENETYX-MAC. Clustal W (http://clustalw.ddbj.nig.ac.jp/top-j.html) was used to generate a phylogenetic tree of the strip homologs (Supplementary Fig. 1k).
Anti-Strip antibody generation
We generated a recombinant protein representing a 91-amino-acid hydrophilic region of Drosophila Strip (amino acid 309 to 399 of PB, 264 to 354 of PA) in E. coli (pCold vector). The recombinant protein was purified and injected into rabbits or rats to generate polyclonal or monoclonal antibodies, respectively. The IgG fraction of the polyclonal antibody was purified. Antibody generation was performed by the Hokudo Co., Ltd.
Immunohistochemistry
The fixation and immunostaining of fly brains were carried out as follows62. Brains were dissected in 0.3% Triton X-100 (vol/vol) in phosphate-buffer (PBT), followed by fixing in 4% paraformaldehyde (PFA) (wt/vol) with 0.3% Triton X-100 (vol/vol) in PBT at RT for 20 min, and subsequently blocked in PBT with 5% normal goat serum (vol/vol) for 1 h at RT. Primary and secondary antibody incubation were carried out in PBST for overnight at 4 °C. For Fig. 4a–f, pupal brains were dissected in cold Schneider’s medium and incubated with primary antibody in Schneider’s medium with 10% normal goat serum (vol/vol) at RT for 1 h, followed by quickly rinsing with cold PBS for 3 times and fixing in 4% PFA (vol/vol) in PBS at RT for 20 min. Subsequent blocking and antibody incubation were similar with adult brains. For Drosophila S2 cells, cells were cultured on Concanavalin A-coated coverslips, fixed, and immunostained as follows62. S2 cells were grown for 1 h at 26 °C on Concanavalin A-coated coverslips, and were fixed for 20 min in 4% PFA (wt/vol) with 0.3% Triton X-100 (vol/vol) in phosphate-buffer saline (PBST) at RT for 10 min, and subsequently blocked in PBST with 5% normal goat serum (vol/vol) for 30 min. Primary and secondary antibody incubation were carried out in blocking solution for 1 h at RT. For HeLa cells, cells were cultured on polyethylene-imine-coated cover slips, fixed, and immunostained as with the S2 cells. We used the following antibodies: nc82 (mouse, 1:40, DSHB), the anti-mCD8 (rat, 1:200, Invitrogen), anti-GFP (mouse, 1:1000, Clontech), the anti-DsRed (rabbit, 1:500, Clontech), anti-HA 12CA5 (mouse, 0.8 μg/ml, Roche), and anti-EEA1 (mouse, 1:500, BD Transduction Laboratories) antibodies. The anti-Rab547 (rabbit, 1:1000) and the anti-Avl48 (rabbit, 1:200) antibodies are kind gifts from Marcos González-Gaitán, David Bilder and Thomas Vaccari. The anti-Strip (rat) was generated as described above (1:50).
Immunoprecipitation and immunoblotting
S2 cell were sonicated in lysis buffer (25 mM Tris-HCl, pH 7.9, 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 10 mM DTT, and 1 × cOmplete (cocktail of protease inhibitors, Roche)) and incubated with anti-Strip antibody (rat, 1:100) or control IgG overnight. Protein G agarose (Roche) was added, and immunoprecipitation was performed according to the manufacturer’s protocol. For the immunoprecipitation of c-myc tagged Sprint, S2 cells transfected with Actin-Gal4 (pAcPa-Actin5c-Gal4), UAS-spri-myc and UAS-FLAG-strip were lysed and incubated with EZ view Red Anti-c-Myc Affinity Gel (Sigma). For the immunoprecipitation of YFP tagged Rab5, Rab5WT, Rab5CA, and Rab5DN were subcloned in pUAST at first; Rab5WT, Rab5CA, and Rab5DN were amplified by PCR using UAS-YFP-Rab5WT49, UAS-YFP-Rab5Q88L49, and UAS-YRP-Rab5S43N49 flies as template. S2 cells transfected with Actin-Gal4, UAS-spri-myc and UAS-YFP-Rab5 WT/CA/DN were lysed and incubated with anti-GFP antibody (rabbit, 1:500, MBL) overnight. Protein G agarose (Roche) was added and immunoprecipitation was performed according to the manufacturer’s protocol. Western blotting was performed according to standard techniques. Briefly, we subjected 2–10 μg of S2 cell lysates or embryo lysates to SDS-PAGE analysis (12.5% for Histon H3 and Rab5 detection; for others, 7.5% or 10%) and immunoblotting. The following antibodies were used for immunoblotting: the anti-Strip antibody (rabbit, 1:200), the anti-Strip antibody (rat, 1:50), the anti-α-tubulin (mouse, 1:2000, Sigma), the anti-Gl C-term antibody27 (rabbit, 1:2000), the anti-GFP antibody (mouse, 1:2000, Roche), anti-myc antibody (mouse, 1:1000, Invitrogen), anti-FLAG antibody (mouse, 1:1000, Sigma), anti-V5 antibody (mouse, 1:200, Invitrogen), anti-GST (mouse, 1:200, Santa Cruz Biotechnology), and the anti-Histone H3 antibody (rabbit, 1:2000, Active motif). The anti-Rab5 antibody47 (rabbit, 1:1000) is a kind gift from M. González-Gaitán. All data are representative of more than three biological replicates. Images of the full blots for all western blots are provided in Supplementary Fig. 10.
GST pull-down
The Gl fragment from 676 to 914 amino acids was amplified from genomic DNA and the Spri fragment from 1404 to1700 amino acids were amplified by PCR using EST clone (SD06366). Both of these fragments were subcloned into pGEX-5X3. GST, GST-Gl (676-914 a.a.), or GST-Spri (1404-1700 a.a.) was expressed in BL21 (DE3) bacterial cells and purified with Glutathione Sepharose 4 Fast Flow (GE). These glutathione sepharose beads were co-incubated with embryo lysates (UAS-strip-V5/+; daughterless (da)-Gal4/+ or da-Gal4/UAS-FLAG-strip) collected at 0-22 h after egg laying.
Yeast two-hybrid screening
For yeast two-hybrid screening, the full length of strip was used as bait and the Drosophila whole-embryo library was used as prey; the screening was performed by the Hybrigenics.
dsRNA generation and treatment
The dsRNA design, production, and treatment were performed as follows63. 700-900 bp gene-specific sequences with T7 RNA polymerase sequence at both ends were amplified by PCR and in vitro transcription was performed using T7 RiboMAX Express RNAi System (Promega) according to the manufacturer’s protocol. For strip dsRNA, templates for in vitro transcription were generated using the primers 5′-TAATACGACTCACTATAGGGCCTGCATAAACCTGCTGCGC-3′ and 5′-TAATACGACTCACTATAGGGCTAGAGGGCGTCCCAGTCGG-3′. For control and Gl dsRNA, primer sequences were used to amplify the sequence from the bacterial cloning plasmid pBluescript SK63 and the Gl C-terminus27. Cells (1.0 × 106) were cultured in a 35-mm dish; 30 μg dsRNA was applied every two days until the cells were used for analysis. For Figure 5, cells were treated with control, strip, or Gl dsRNA for 8 days. For Figure 7f, S2 cells that stably express YFP-Rab5 were transfected with Actin-Gal4 and UAS-sprint-myc for two days and split into half to treat with control or strip dsRNA for 6 days.
Transfection with shRNA
Actin-Gal4 and desired UAS construct (UAS-shRNA-strip, UAS-FLAG-strip, UAS-spri-myc, UAS-YFP-Rab5WT, DN, or CA) were co-transfected using Effectene Transfection Reagent (Qiagen); 2.0×106 Drosophila S2 cells for 35-mm dish, 6.0×106 cells for 60-mm dish were plated and transfection was performed according to the manufacturer’s protocol. Cells were collected for immunoblots 48 h after transfection. pcDNA3-Strip1-V5 and shRNA-Strip1-1, 2, 3 or 4 were co-transfected using Lipofectamin 2000 (Invitrogen); 1.0 × 105 HEK293T cells were plated on a 35-mm dish, and transfection was performed according to the manufacturer’s protocol. Cells were collected for immunoblots 24 h after transfection.
FM1-43 dye assay
Drosophila S2 cells were plated on Concanavalin A-coated cover slips, and treated with 5 μg/ml FM1-43 dye (Molecular Probes)/ HBSS for 1 min, and incubated for 5 min at 25 °C after a quick wash, and fixed immediately with 4% (vol/vol) PFA/HBSS without washing. Fixation was 10 min at RT. Cells were mounted with SlowFade® Gold antifade reagent with DAPI (Invitrogen).
EGF-Alexa488 uptake assay in HeLa cells
Dharmacon siGENOME non-Targeting siRNA Pool #1 (D-00126-13-05) (100 nM) or siGENOME SMARTpoll human STRIP1 (M-02516091-005) (100 nM) was transfected into HeLa cells (1.0 × 105) with Lipofectamin 2000 (Invitrogen) according to manufacturer’s protocol, cultured for 3 days, and plated on polyethylene-imine-coated cover slips for over night. Cells were pulsed with Alexa Fluor 488 EGF complex (Invitrogen) for 3 min and immediately fixed or rinsed and incubated for 27 min at 37 °C and fixed.
Electroporation in utero
Animals were handled according to the guidelines established by the University of Tokyo and Keio University. All the electroporations were performed on E14 embryos. Electroporation in utero and the measurement of EGFP fluorescence intensity in various regions of the cerebral cortex were performed as follows53. Fluorescent images of frozen section of EGFP expressing mouse brains were captured by TCS-SP5 laser scanning confocal microscopy (Leica). Fluorescence intensities inside similar width rectangles in various regions of the cerebral cortex (layers II–IV, V–VI, IZ, and SVZ/VZ) were measured by the Leica SP5 software. Relative intensities to the total fluorescence were calculated and plotted in the graphs with standard errors.
Image acquisition
For all experiments other than HeLa cell imaging, images were obtained using TCS-SP5 laser scanning confocal microscopy (Leica) with a PL APO CS 40×/1.25 Oil lens (Leica) for PN imaging or a PL APO CS 63×/1.40 Oil lens (Leica) for imaging of S2 cells. For imaging of HeLa cells, TCS-SP8 laser scanning confocal microscopy (Leica) with a PL APO CS 63×/1.40 Oil lens was used. Fields of view were randomly selected and all experiments were repeated more than three times. For PN clones other than Figure 4, more than 10 clones were imaged and representative images were shown in Figures. For PN clones of Figure 4, S2 cells and HeLa cells, the exact number of samples imaged was shown in each Figure.
Image analysis
For Figure 4, fluorescence intensities of FYVE-GFP and mCD8RFP inside axon stalks (200 μm2) were measured by the Leica SP5 software. For Figure 5, image analysis was performed using the Particle Analyzer of ImageJ software. Single section that passes the centre of cells was selected for each S2 cell stained with the anti-Rab5 antibody or the anti-Avl antibody, and the size of vesicles was determined by using the particle analyzer. For Figure 6, Figure 7l, and Supplementary Figure 2k, all images were blinded prior to the classification to avoid experimental bias. The investigator who conducted the blind test was totally independent from the investigator who performed experiments.
Statistical analysis
Statistical analysis was performed using Prism software (GraphPad). Unpaired two-tailed t-tests were used to compare the GFP-FYVE intensity between control and strip PNs, and for the quantitative analysis of the extent of cell migration in the mice cerebral cortex. The chi-square test was used to compare data variations among control, strip, or Gl dsRNA-treated S2 cells or between control and STRIP1 siRNA-treated HeLa cells. The chi-squre test was also used to compare data variations of synaptotagmin localization between control or stripdogi PNs. Exact P-values are provided unless p < 0.0001.
Supplementary Material
Acknowledgements
We thank D. Bilder, M. González-Gaitán, T. Schwarz, S. X. Hou, the Bloomington Drosophila Stock Centre (NIH P40OD018537), and the Kyoto Drosophila Genetic Resource Centre for fly stocks. We thank T. Vaccari for anti-Avl antibody, M. González-Gaitán for anti-Rab5 antibody, C.-H. Chen for advising us on shRNA construction, Y. Hiromi for actin-Gal4 and all members of the Miura and Luo laboratories for their comments on this study. L.L. is an investigator in the Howard Hughes Medical Institute. This work was supported by grants from the National Institute of General Medical Science of the National Institutes of Health (R01-GM085232 to V.I.G.), the National Institutes of Health (R01-DC005982 to L.L.), the Japanese Ministry of Education, Science, Sports, Culture, and Technology (MEXT), the Japan Society for the Promotion of Science, and the Japan Science and Technology Agency (to C.S., K.T., Y.Y., M.M., and T.C.).
Footnotes
Author contributions
C.S. and T.C. designed the study with the help of M.M. and L.L. C.S. conducted most of the experiments. S.H. assisted with some experiments. T.C. performed genetic mosaic screen and mapping of strip gene. T.K., M.S. and Y.Y. performed in utero electroporation experiments. V.I.G. supplied reagents and effective RNAi protocol. C.S. and T.C. analysed the data. C.S. and T.C. wrote the manuscript with contributions from all other authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, Doe CQ. Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 4.Horton A, Ehlers M. Neuronal polarity and trafficking. Neuron. 2003;40:277–295. doi: 10.1016/s0896-6273(03)00629-9. [DOI] [PubMed] [Google Scholar]
- 5.Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap C, Winckler B. Harnessing the power of the endosome to regulate neural development. Neuron. 2012;74:440–451. doi: 10.1016/j.neuron.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodmer D, Ascano M, Kuruvilla R. Isoform-specific dephosphorylation of dynamin1 by calcineurin couples neurotrophin receptor endocytosis to axonal growth. Neuron. 2011;70:1085–1099. doi: 10.1016/j.neuron.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ascano M, Bodmer D, Kuruvilla R. Endocytic trafficking of neurotrophins in neural development. Trends Cell Biol. 2012;22:266–273. doi: 10.1016/j.tcb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tojima T, Hines JH, Henley JR, Kamiguchi H. Second messengers and membrane trafficking direct and organize growth cone steering. Nat Rev Neurosci. 2011;12:191–203. doi: 10.1038/nrn2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrowolski R, De Robertis EM. Endocytic control of growth factor signalling: multivesicular bodies as signalling organelles. Nat Rev Mol Cell Biol. 2012;13:53–60. doi: 10.1038/nrm3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jovic M, Sharma M, Rahajeng J, Caplan S. The early endosome: a busy sorting station for proteins at the crossroads. Histol Histopathol. 2010;25:99–112. doi: 10.14670/hh-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 13.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen E, et al. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison HA, et al. Regulation of early endosomal entry by the Drosophila tumor suppressors Rabenosyn and Vps45. Mol Biol Cell. 2008;19:4167–4176. doi: 10.1091/mbc.E08-07-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T, Nakamura A. The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development. 2008;135:1107–1117. doi: 10.1242/dev.017293. [DOI] [PubMed] [Google Scholar]
- 17.Ohya T, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 18.Driskell O, Mironov A, Allan V, Woodman P. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat Cell Biol. 2007;9:113–120. doi: 10.1038/ncb1525. [DOI] [PubMed] [Google Scholar]
- 19.Hunt S, Stephens D. The role of motor proteins in endosomal sorting. Biochem Soc Trans. 2011;39:1179–1184. doi: 10.1042/BST0391179. [DOI] [PubMed] [Google Scholar]
- 20.Satoh D, et al. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- 21.Yang WK, et al. Nak regulates localization of clathrin sites in higher-order dendrites to promote local dendrite growth. Neuron. 2011;72:285–299. doi: 10.1016/j.neuron.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 22.Hwang J, Pallas DC. STRIPAK complexes: structure, biological function, and involvement in human diseases. The international journal of biochemistry & cell biology. 2014;47:118–148. doi: 10.1016/j.biocel.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Molecular cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Ashton-Beaucage D, et al. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS biology. 2014;12:e1001809. doi: 10.1371/journal.pbio.1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroer T. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 26.King S, Schroer T. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, et al. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd T, et al. The p150(Glued) CAP-Gly Domain Regulates Initiation of Retrograde Transport at Synaptic Termini. Neuron. 2012;74:344–360. doi: 10.1016/j.neuron.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moughamian A, Holzbaur E. Dynactin is required for transport initiation from the distal axon. Neuron. 2012;74:331–343. doi: 10.1016/j.neuron.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo K, Jekely G, Rorth P. Cloning and expression of sprint, a Drosophila homologue of RIN1. Mech Dev. 2001;101:259–262. doi: 10.1016/s0925-4773(00)00565-7. [DOI] [PubMed] [Google Scholar]
- 31.Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Jefferis G, Hummel T. Wiring specificity in the olfactory system. Semin Cell Dev Biol. 2006;17:50–65. doi: 10.1016/j.semcdb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Jefferis G, Marin E, Stocker R, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 34.Marin E, Jefferis G, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109:243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- 35.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 36.Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- 37.Sakuma C, Anzo M, Miura M, Chihara T. Development of olfactory projection neuron dendrites that contribute to wiring specificity of the Drosophila olfactory circuit. Genes & genetic systems. 2014;89:17–26. doi: 10.1266/ggs.89.17. [DOI] [PubMed] [Google Scholar]
- 38.Hong W, Luo L. Genetic control of wiring specificity in the fly olfactory system. Genetics. 2014;196:17–29. doi: 10.1534/genetics.113.154336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger J, Suzuki T, Senti K, Stubbs J, Schaffner G, Dickson B. Genetic mapping with SNP markers in Drosophila. Nat Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- 40.Zhai R, et al. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci U S A. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai SW, et al. Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC Biol. 2011;9:54. doi: 10.1186/1741-7007-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goudreault M, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 44.McGrail M, Gepner J, Silvanovich A, Ludmann S, Serr M, Hays TS. Regulation of cytoplasmic dynein function in vivo by the Drosophila Glued complex. J Cell Biol. 1995;131:411–425. doi: 10.1083/jcb.131.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin M, Iyadurai S, Gassman A, Gindhart H, TS S, M W. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol Biol Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson I, Ranjan R, Schwarz T. Synaptotagmins I and IV promote transmitter release independently of Ca(2+) binding in the C(2) A domain. Nature. 2002;418:336–340. doi: 10.1038/nature00915. [DOI] [PubMed] [Google Scholar]
- 47.Wucherpfennig T, Wilsch-Bräuninger M, González-Gaitán M. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J Cell Biol. 2003;161:609–624. doi: 10.1083/jcb.200211087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7:1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, et al. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordens I, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 51.Johansson M, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoepfner S, et al. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 53.Kawauchi T, et al. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 2010;67:588–602. doi: 10.1016/j.neuron.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Cosker KE, Courchesne SL, Segal RA. Action in the axon: generation and transport of signaling endosomes. Curr Opin Neurobiol. 2008;18:270–275. doi: 10.1016/j.conb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puls I, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 56.Farrer M, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Devon RS, et al. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci U S A. 2006;103:9595–9600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- 59.Menut L, Vaccari T, Dionne H, Hill J, Wu G, Bilder D. A mosaic genetic screen for Drosophila neoplastic tumor suppressor genes based on defective pupation. Genetics. 2007;177:1667–1677. doi: 10.1534/genetics.107.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen HW, et al. CKA, a novel multidomain protein, regulates the JUN N-terminal kinase signal transduction pathway in Drosophila. Molecular and cellular biology. 2002;22:1792–1803. doi: 10.1128/MCB.22.6.1792-1803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 62.Wu J, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- 63.Rogers S, Rogers G. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.