Abstract
Scope
We re-evaluated previously reported associations between variants in pathways of one-carbon (folate) transfer genes and ovarian carcinoma (OC) risk, and in related pathways of purine and pyrimidine metabolism, and assessed interactions with folate intake.
Methods and Results
Odds ratios (OR) for 446 genetic variants were estimated among 13,410 OC cases and 22,635 controls and among 2,281 cases and 3,444 controls with folate information. Following multiple testing correction, the most significant main effect associations were for DPYD variants rs11587873 (OR=0.92, P=6x10−5) and rs828054 (OR=1.06, P=1x10−4). Thirteen variants in the pyrimidine metabolism genes, DPYD, DPYS, PPAT and TYMS, also interacted significantly with folate in a multi-variant analysis (corrected P=9.9x10−6) but collectively explained only 0.2% of OC risk. Although no other associations were significant after multiple testing correction, variants in SHMT1 in one-carbon transfer, previously reported with OC, suggested lower risk at higher folate (Pinteraction=0.03-0.006).
Conclusions
Variation in pyrimidine metabolism genes, particularly DPYD, which was previously reported to be associated with OC, may influence risk; however, stratification by folate intake is unlikely to modify disease risk appreciably in these women. SHMT1 SNP-byfolate interactions are plausible but require further validation. Polymorphisms in selected genes in purine metabolism were not associated with OC.
Keywords: case-control, DPYD, folate, polymorphism, SHMT1
Introduction
Global statistics estimated that ovarian carcinoma (OC) afflicted 225,000 women and resulted in 140,000 deaths in 2008 [1]. There are no specific screening methods or unique symptoms to detect OC in early stages [2]. Risk stratification strategies may have appreciable impact in reducing the incidence and suffering from OC by identifying those women at greatest risk of developing the disease who would benefit from preventive measures [3]. Germline mutations in high-risk genes (e.g., BRCA1 and BRCA2) remain the best-defined genetic risk factors [4], but explain just 10-15% of all OC [5-7]. About 4% of the polygenic risk is explained by common, but poorly understood, low-risk polymorphisms [8-11] and most non-genetic risk factors (oral contraceptive use [12-14], parity [15, 16], breast-feeding [15, 17], tubal ligation [18], endometriosis [19] and smoking [20]) are not conducive to public health recommendations for risk modification.
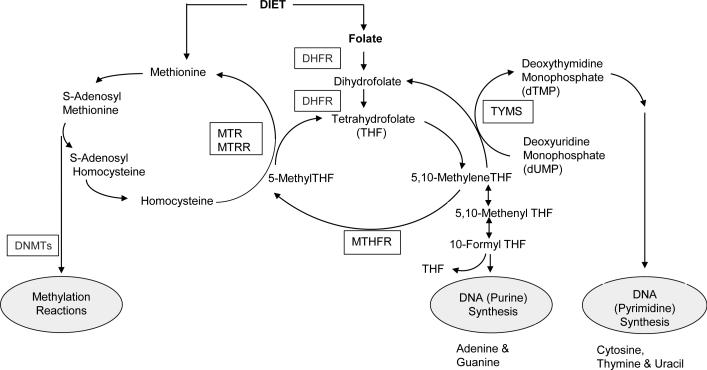
Stratification of genetic risk by dietary factors may prevent some cancers. For example, folates participate in one-carbon (1-C) transfer reactions that are essential for the biosynthesis of purines (adenine and guanine) and pyrimidines (cytosine, thymine and uracil), which are incorporated into DNA and RNA, as well as for the biosynthesis of methyl groups for DNA methylation [21] (Figure 1). Perturbation of the coenzymatic role of folate, or of key enzymes in 1-C transfer or purine or pyrimidine metabolism, can have broad consequences that lead to tumor initiation [22] and progression [23] and thus could alter risk among a substantial proportion of individuals. Numerous genetic disorders of purine and pyrimidine metabolism have been characterized in humans [24]. Although these are rare and inherited in Mendelian fashion, genes encoding enzymes in these pathways have been associated with various cancers [25-30]. We previously examined associations between 180 tagging common single nucleotide polymorphisms (tagSNPs) in 21 genes involved in 1-C transfer and risk of OC in 1,770 participants [31] and also reported risk modification by multivitamin intake, a proxy for folate intake [31, 32]. Ten SNPs in eight genes (AHCYL1, DNMT3A, DPYD, MTHFD1, MTHFS, SHMT1, SLC19A1 and TYMS) were associated with OC at P≤0.05 in either ordinal or co-dominant genetic risk models [31] and eight SNPs in five genes (DNMT3A, DNMT1, MTHFR, MTHFD1 and ATIC) were associated with OC at P≤0.05 in interaction analyses with multivitamin use [31, 32]. In those studies, the strongest evidence for association was for a haplotype and a single SNP (rs9909104) in SHMT1 for which we calculated a false positive report probability of 9% to 16%, respectively [31], and for two SNPs in ATIC interacting with multivitamin use at a false discovery rate <0.25 and P<0.05 [32]. SNPs in DNMT3A, DPYD, MTHFD1 and MTHFS did not replicate in a subsequent genotyping effort among 16,000 participants [33].
Figure 1.
Overview of the role of folate and key enzymes involved in one-carbon transfer for DNA synthesis and methylation reactions. DHFR, dihydrofolate reductase; DNMTs, DNA methyltransferases; MTHFR, methylenetetrahydrofolate reductase; MTR, 5-methyltetrahydrofolate-homocysteine methyltransferase; MTRR, 5-methyltetrahydrofolatehomocysteine methyltransferase reductase; TYMS, thymidylate synthase.
Our objectives in the current investigation were to re-evaluate previously reported genetic associations in a larger sample size, evaluate additional variants in the related pathways of purine and pyrimidine metabolism and to assess effect modification by dietary folate intake. We performed these analyses in over 36,000 women contributing DNA in the Ovarian Cancer Association Consortium (OCAC).
Materials and Methods
Gene and SNP selection
Genes were selected according to two categories. The first category consisted of six genes (ATIC, DNMT3B, DPYD, MTR, SHMT1 and TYMS) with previously observed SNP associations with OC [31, 32] and were included for replication. These SNPs were selected with high gene coverage using minor allele frequency (MAF) ≥0.01 and pair-wise linkage disequilibrium (LD) threshold of r2<0.8 (ATIC, DNMT3B, DPYD and MTR) or <0.9 (SHMT1 and TYMS). The second category consisted of nine genes related to purine metabolism (ADSL, ADSS, DCK, GART, GMPS, IMPDH1, IMPDH2, PAICS and PFAS) and 11 genes involved in pyrimidine metabolism (AK3, CAD, CMPK1, CTPS, DHODH, DPYS, NME6, PPAT, PRPS2, RRM2B and UMPS) (Supplementary Figure 1). These SNPs were selected with MAF≥0.05 and LD threshold of r2<0.8. All SNPs within 5kb up- and downstream of the largest cDNA isoform (Human Genome build 36) of each gene was selected using information from 60 unrelated individuals of European ancestry sequenced in the pilot phase of the 1000 Genomes Project [34] and binned using the Haploview program [35]. We prioritized tagSNPs for genotyping that were coding SNPs, had the highest MAF in each bin and, if available, met criteria for predicted likelihood of successful genotyping based on Illumina quality score metrics. In January 2010, 803 tagSNPs were submitted for genotyping: 31% of these SNPs were unique to the 1,000 Genomes Project and not found in dbSNP.
Study Subjects
Subjects (n=47,630) from 43 individual studies participating in OCAC were grouped into 34 geographically similar study strata [11]. Of 44,308 subjects whose DNA passed genotyping quality control criteria (see below), we further excluded subjects with borderline tumors, subjects of non-European ancestry and those with prior history of cancer other than non-melanoma skin cancer, leaving 36,045 eligible subjects (13,410 cases and 22,635 controls) for analysis. Informed consent was obtained in each of the individual studies and local human research investigations committees approved each study.
Genotyping and Quality Control (QC)
Details of the genotyping have been described elsewhere [11]. In brief, we used an Illumina Infinium custom iSelect BeadChip developed for the international Collaborative Oncology Gene-environment Study (iCOGS). Centralized genotyping calls and QC were performed at the University of Cambridge. Quality control for samples has been detailed previously [11]. We excluded SNPs that failed genotyping, had call rates <95% and MAF >0.05 or call rates <99% and MAF <0.05, departed from Hardy-Weinberg equilibrium (P value <10−7), had discordant genotypes >2% between duplicates and monomorphic SNPs. Of 803 tagSNPs submitted for genotyping, 203 SNPs failed genotyping, 127 were monomorphic and 27 had MAF <0.01 leaving 446 SNPs that passed QC. Genotyping failures and monomorphic SNPs reflected the large number of polymorphisms that were subsequently found to be falsely positive in the pilot phase sequencing data of the 1000 Genomes Project.
Covariate and Dietary Data
Key clinical, demographic and questionnaire data were harmonized across study centers and merged into a common dataset. Dietary intakes of folate and total energy were estimated with validated food frequency questionnaires (FFQs) in six studies (AUS [36], DOV [37], HAW and STA [38], NEC [39] and NJO [40]) pertaining to the year preceding recruitment or for the time period approximately four years before the reference date (DOV). Data on the use of multivitamins and single vitamin and mineral supplements were also available and total folate intake was estimated by summing intakes from both food sources and from supplements. Nutrient and genotype data were available for 2,281 cases and 3,444 controls of European ancestry.
Statistical Analysis
Genotypes were used to estimate allele frequencies and pair-wise LD between SNPs was estimated with r2 values using Haploview [35]. Data from the 34 study strata were combined into a single dataset following confirmation of no statistical heterogeneity in SNP associations across study sites. We estimated odds ratios (OR) and 95% confidence intervals (CI) for each SNP using unconditional logistic regression treating the number of variant alleles carried as an ordinal (log-additive) variable. Secondary analyses also considered co-dominant (non-additive) risk models. Interactions between each SNP and total folate intake were evaluated with the Wald test in models that also included a one degree-of-freedom product term for the ordinal coding for genotype and total folate intake group (below/above the energy-adjusted median intake: ≤484 μg/d vs >484 μg/d ≈ approximately the dietary reference intake of 400 μg/d for folate, which is also the folic acid content of a typical multivitamin supplement). Risk models were adjusted for age (continuous), study stratum and the first five eigenvalues from principal components analysis to account for sub-strata of European ancestry across the 34 international studies (see ref [11]). Additional adjustment for non-genetic risk factors did not change estimates and these variables were excluded from the models (data not shown).
Multi-variant analysis
Because some of the genes selected for replication belonged to either the purine (ATIC) or pyrimidine (DPYD and TYMS) metabolism pathways, while DNMT3B, MTR and SHMT1 belonged to 1-C transfer, we evaluated associations according to these three pathways. However, we considered evidence for replication if SNPs in these six genes reached statistical significance according to the criteria described below.
To assess the likelihood of false-positive findings, we performed a multi-variant analysis that accounted for the potential correlations between SNPs within genes in a pathway. Since our primary interest was to evaluate SNP-by-folate interactions, we prioritized these associations for evaluation of multiple testing as follows. A likelihood ratio test (LRT) statistic was calculated by comparing a regression model with and without significant SNP-by-folate interaction terms. Permutation-based tests were then used to compute P-values from a null distribution of the LRT statistic generated by permuting case status 10,000 times. The generation of a null distribution was performed five times, each time with a different seed. For evaluation of individual genes that showed SNP associations at P<0.05, we applied a conservative Bonferroni correction of the Type I error using the number of SNPs tested in that gene’s pathway (44 SNPs in 1-C transfer, 100 SNPs in purine metabolism and 302 SNPs in pyrimidine metabolism). The corresponding thresholds were P=0.001 for 1-C transfer, P=5x10-4 for purine metabolism and P=1.6x10−4 for pyrimidine metabolism.
We also estimated haplotype frequencies of >1% for selected genes with and without stratification by total folate intake using an expectation-maximization algorithm [41] as described in detail elsewhere [32]. The generation of haplotypes using 129 DPYD tagSNPs resulted in an infinite recursion so we selected 29 tagSNPs, one for each haplotype block constructed according to the Gabriel criteria [42] in Haploview [35] and two located outside of a haplotype block. These tagSNPs were selected based on significant P values in main effect or interaction analyses, highest MAF or highest D' values with other tagSNPs in the haplotype block. Individual haplotype associations were interpreted carefully in the absence of global haplotype significance.
To assess the population importance of the SNP-by-folate interactions, we used the Genome-wide Complex Trait Analysis (GCTA) program to estimate the percent variance in risk of OC explained by the SNP-by-folate interaction terms [43]. In principle, the GCTA program can be used to evaluate a subset of SNPs or SNP-by-environment interactions and has been used by the developers in this context (J Yang, personal communication, December 2013). Briefly, we first estimated the pairwise genetic relationship matrix (GRM) of the subjects using the SNPs of interest and then fitted the GRM in a regression model that also included age, study stratum, five eigenvalues, total folate intake group and SNP-by-folate interaction terms. Restricted maximum likelihood was applied to deconstruct the phenotypic variance into the percentages explained by the SNPs, the SNP-by-folate interaction terms and residual environmental component.
Statistical tests were two-sided and, unless stated otherwise, were implemented with SAS version 9 (SAS Institute, NC), R [44] and Plink v1.07 [45] software.
Results
The distribution of cases and controls stratified by study is shown in Supplementary Table 1. Descriptive information on the 446 SNPs is provided in Supplementary Table 2.
Twenty-three SNPs were associated with risk of OC at P<0.05, including two SNPs in SHMT1 (1-C transfer) and 12 SNPs in DPYD (pyrimidine metabolism) (Table 1). We reported associations with SNPs in both of these genes previously, although with different variants [31]. In the current study, the two SNPs with the smallest P value were in DPYD in pyrimidine metabolism: rs11587873 (OR, 0.92; 95% CI, 0.89-0.96; P=6x10−5) and rs828054 (OR, 1.06; 95% CI, 1.03-1.10; P=1x10−4). These two SNPs remained statistically significant at the corrected P=1.6x10−4. The other 10 DPYD SNPs were correlated with either rs11587873 or rs828054. There was no statistical heterogeneity in ORs across study strata. Associations were similar when restricted to high-grade serous OC histology (Table 1). Associations for the remaining SNPs are shown in Supplementary Table 3.
Table 1.
Per-allele (log-additive) associationsa) between variants in genes in 1-C transfer and purine and pyrimidine metabolism pathways and risk of ovarian carcinoma: 13,410 cases (5,813 high-grade serous-only) and 22,635 controls of European ancestry in OCAC.
| All cases | High-Grade Serous cases | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | OR | 95% CI lower | 95% CI upper | P-value | P-value (SNP*site interaction) | OR | 95% CI lower | 95% CI upper | P-value | P-hetb) |
| 1-C transfer | |||||||||||
| DNMT3B | rs4911256 | 1.04 | 1.00 | 1.07 | 0.025 | 0.88 | 1.04 | 0.99 | 1.09 | 0.082 | 0.26 |
| SHMT1 | rs669340 | 0.96 | 0.93 | 0.99 | 0.021 | 0.58 | 0.94 | 0.90 | 0.98 | 0.006 | 0.09 |
| SHMT1 | rs4925179 | 0.97 | 0.93 | 1.00 | 0.040 | 0.69 | 0.95 | 0.91 | 1.00 | 0.037 | 0.21 |
| Purine metabolism | |||||||||||
| ATIC | pos215884573 | 1.05 | 1.00 | 1.10 | 0.036 | 0.34 | 1.08 | 1.01 | 1.14 | 0.016 | 0.08 |
| IMPD2 | rs4974081 | 0.96 | 0.93 | 1.00 | 0.043 | 0.68 | 0.96 | 0.92 | 1.01 | 0.169 | 0.66 |
| Pyrimidine metabolism | |||||||||||
| AK3c) | rs691941 | 0.95 | 0.91 | 0.99 | 0.020 | 0.13 | 0.94 | 0.89 | 1.00 | 0.061 | 0.11 |
| DPYD | rs828054 | 1.06 | 1.03 | 1.10 | 0.0001 | 0.80 | 1.07 | 1.03 | 1.12 | 0.002 | 0.02 |
| DPYD | rs11587873 | 0.92 | 0.89 | 0.96 | 0.00006 | 0.08 | 0.93 | 0.88 | 0.98 | 0.008 | 0.01 |
| DPYD | rs12120388 | 0.94 | 0.91 | 0.97 | 0.0004 | 0.83 | 0.94 | 0.90 | 0.98 | 0.004 | 0.02 |
| DPYD | rs676686 | 1.06 | 1.02 | 1.09 | 0.001 | 0.85 | 1.07 | 1.02 | 1.12 | 0.003 | 0.01 |
| DPYD | rs914959 | 1.05 | 1.01 | 1.08 | 0.007 | 0.65 | 1.05 | 1.00 | 1.10 | 0.028 | 0.08 |
| DPYD | rs7537668 | 1.04 | 1.01 | 1.08 | 0.011 | 0.61 | 1.05 | 1.01 | 1.10 | 0.026 | 0.03 |
| DPYD | rs4128474 | 0.95 | 0.90 | 1.00 | 0.031 | 0.99 | 0.96 | 0.90 | 1.02 | 0.178 | 0.34 |
| DPYD | rs494271 | 1.04 | 1.00 | 1.07 | 0.034 | 0.80 | 1.04 | 1.00 | 1.10 | 0.052 | 0.02 |
| DPYD | rs6678858 | 0.95 | 0.91 | 1.00 | 0.034 | 1.00 | 0.97 | 0.91 | 1.03 | 0.321 | 0.70 |
| DPYD | rs7555294 | 0.96 | 0.92 | 1.00 | 0.041 | 0.90 | 0.97 | 0.91 | 1.02 | 0.229 | 0.73 |
| DPYD | rs4434871 | 0.95 | 0.91 | 1.00 | 0.041 | 0.97 | 0.98 | 0.92 | 1.04 | 0.491 | 0.65 |
| DPYD | rs7522938 | 1.03 | 1.00 | 1.07 | 0.048 | 0.72 | 1.02 | 0.98 | 1.07 | 0.330 | 0.26 |
| DHODHd) | rs3213423 | 1.05 | 1.01 | 1.09 | 0.018 | 0.10 | 1.01 | 0.96 | 1.06 | 0.757 | 0.77 |
| DHODH | rs11864453 | 0.97 | 0.93 | 1.00 | 0.037 | 0.19 | 0.97 | 0.93 | 1.01 | 0.170 | 0.54 |
| NME6 | rs7651161 | 0.97 | 0.94 | 1.00 | 0.048 | 0.73 | 0.96 | 0.92 | 1.01 | 0.089 | 0.49 |
| RRM2B | pos103299244 | 2.67 | 1.15 | 6.19 | 0.022 | 0.92 | 2.42 | 0.78 | 7.48 | 0.126 | 0.27 |
| TYMS | pos656897 | 1.27 | 1.09 | 1.48 | 0.002 | 0.27 | 1.31 | 1.07 | 1.62 | 0.010 | 0.008 |
Adjusted for age (continuous), study stratum and the first five eigenvalues from principal components analysis; P < 0.05.
P-value for tumor heterogeneity comparing odds ratios between all controls and each of high-grade serous OC, low-grade serous OC, mucinous OC, endometrioid OC and clear cell OC.
SNP is located 3′ upstream of gene.
SNP is located in flanking 5′ region of gene; all other SNPs are located in introns; ‘pos’ SNPs are novel and identified by chromosomal position.
When SNPs were examined by interactions with total folate intake (Table 2), 22 SNPs showed interactions at P<0.05, including two SNPs that were associated with OC risk overall (SHMT1 rs4925179 and DPYD rs7522938). Three of four SHMT1 SNPs in 1-C transfer (rs56001517, rs7216214 and rs2273026) were associated with a 23% to 30% decreased risk of OC at higher total folate intake (smallest P=0.006) and these three SNPs were correlated with each other (r2 = 0.61 to 0.92) but did not pass the multiple testing significance threshold of P=0.001. Fifteen of the 22 SNPs (60%) were in pyrimidine metabolism genes and a large proportion of these were in four genes (DPYS, DPYD, PPAT and TYMS) that encode enzymes in the sub-pathway of fluoropyrimidine metabolism, which is an important pharmacogenomics pathway targeted by anti-folate chemotherapy. We, therefore, evaluated the 13 SNP-by-folate interactions in these four genes collectively in a multi-variant analysis. Despite generating five null distributions, none achieved a LRT statistic that included the observed LRT statistic: permuted maximum χ2 = 37.95 to 45.74 with 13 degrees-of-freedom (df; smallest P=1.6x10−5) compared to observed χ2 = 46.92 with 13 df (P=9.9x10−6). This suggested the observed value was more extreme than would be expected. Associations for the remaining SNP-by-folate interactions are shown in Supplementary Table 4. We estimated the percentage of variance in risk of OC explained by the 13 SNP-by-folate interaction terms in pyrimidine metabolism to be 0.1994 % (95% CI, 0.1991 to 0.1997) compared to 1x10-4 % (95% CI, 9.9x10−5 to 1x10−4) explained by the 13 SNPs alone.
Table 2.
Associationsa) between variants in genes in 1-C transfer and purine and pyrimidine metabolism pathways and risk of ovarian carcinoma stratified by total folate intake: 2,281 cases and 3,444 controls of European ancestry participants in OCAC.
| Total folate Intake ≤ 484 μg/d | Total folate intake > 484 μg/d | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | |||||||||||||||||||
| Gene | SNP | AAb) | AB | BB | AA | AB | BB | OR | 95% CI lower |
95% CI upper |
P value |
AA | AB | BB | AA | AB | BB | OR | 95% CI lower |
95% CI upper |
P value |
P intc) |
| 1-C transfer | ||||||||||||||||||||||
| SHMT1d) | rs56001517 | 1506 | 232 | 8 | 991 | 162 | 5 | 1.05 | 0.85 | 1.29 | 0.656 | 1475 | 215 | 6 | 1011 | 108 | 2 | 0.70 | 0.55 | 0.89 | 0.004 | 0.012 |
| SHMT1 | rs7216214 | 1394 | 335 | 18 | 907 | 237 | 15 | 1.08 | 0.91 | 1.28 | 0.396 | 1341 | 340 | 13 | 925 | 182 | 12 | 0.76 | 0.63 | 0.92 | 0.004 | 0.006 |
| SHMT1 | rs4925179 | 711 | 786 | 250 | 501 | 526 | 131 | 0.88 | 0.79 | 0.99 | 0.03 | 703 | 775 | 217 | 469 | 509 | 144 | 1.03 | 0.92 | 1.16 | 0.606 | 0.048 |
| SHMT1 | rs2273026 | 1409 | 323 | 15 | 931 | 213 | 15 | 1.03 | 0.86 | 1.22 | 0.767 | 1363 | 322 | 11 | 938 | 172 | 11 | 0.77 | 0.64 | 0.94 | 0.008 | 0.027 |
| Purine metabolism | ||||||||||||||||||||||
| IMPD1d) | rs6948333 | 991 | 653 | 101 | 624 | 453 | 77 | 1.09 | 0.96 | 1.23 | 0.171 | 950 | 637 | 105 | 638 | 435 | 46 | 0.91 | 0.80 | 1.03 | 0.139 | 0.039 |
| PFAS | rs11649742 | 1049 | 606 | 92 | 721 | 378 | 60 | 0.93 | 0.82 | 1.06 | 0.257 | 1020 | 592 | 84 | 624 | 443 | 55 | 1.14 | 1.00 | 1.30 | 0.045 | 0.029 |
| PFAS | pos8110739 | 1659 | 85 | 0 | 1122 | 37 | 0 | 0.64 | 0.43 | 0.96 | 0.03 | 1630 | 64 | 2 | 1068 | 51 | 1 | 1.18 | 0.81 | 1.71 | 0.384 | 0.034 |
| Pyrimidine metabolism | ||||||||||||||||||||||
| CTPS | rs6675122 | 1030 | 611 | 106 | 615 | 475 | 69 | 1.18 | 1.04 | 1.33 | 0.009 | 960 | 619 | 118 | 624 | 442 | 56 | 0.98 | 0.86 | 1.11 | 0.706 | 0.034 |
| CTPS | rs41268101 | 1121 | 521 | 92 | 677 | 402 | 61 | 1.16 | 1.02 | 1.32 | 0.022 | 1026 | 568 | 88 | 670 | 387 | 45 | 0.95 | 0.83 | 1.09 | 0.441 | 0.034 |
| DPYD | rs667565 | 671 | 810 | 266 | 378 | 573 | 207 | 1.18 | 1.06 | 1.31 | 0.003 | 614 | 814 | 268 | 439 | 508 | 175 | 0.92 | 0.82 | 1.03 | 0.127 | 0.001 |
| DPYD | rs4520446 | 501 | 879 | 363 | 318 | 566 | 274 | 1.09 | 0.98 | 1.21 | 0.135 | 464 | 824 | 397 | 331 | 555 | 234 | 0.90 | 0.81 | 1.00 | 0.057 | 0.016 |
| DPYD | rs7522938 | 752 | 745 | 216 | 440 | 540 | 164 | 1.15 | 1.03 | 1.29 | 0.011 | 712 | 760 | 208 | 498 | 469 | 145 | 0.96 | 0.86 | 1.08 | 0.482 | 0.019 |
| DPYD | rs2811182 | 562 | 887 | 297 | 413 | 538 | 208 | 0.96 | 0.86 | 1.07 | 0.43 | 617 | 784 | 296 | 360 | 553 | 209 | 1.12 | 1.00 | 1.25 | 0.041 | 0.042 |
| DPYS | rs1962267 | 550 | 872 | 325 | 417 | 555 | 187 | 0.87 | 0.78 | 0.97 | 0.014 | 586 | 802 | 309 | 378 | 535 | 209 | 1.02 | 0.92 | 1.14 | 0.689 | 0.033 |
| DPYS | rs2853160 | 489 | 889 | 369 | 303 | 580 | 276 | 1.08 | 0.97 | 1.20 | 0.158 | 460 | 839 | 398 | 317 | 569 | 236 | 0.93 | 0.83 | 1.03 | 0.169 | 0.036 |
| DPYS | rs17834440 | 1295 | 427 | 25 | 908 | 239 | 12 | 0.82 | 0.69 | 0.97 | 0.018 | 1318 | 359 | 20 | 853 | 250 | 19 | 1.11 | 0.94 | 1.31 | 0.223 | 0.008 |
| DPYS | rs2853178 | 859 | 750 | 138 | 601 | 475 | 83 | 0.93 | 0.83 | 1.05 | 0.242 | 889 | 680 | 128 | 571 | 448 | 103 | 1.10 | 0.97 | 1.24 | 0.142 | 0.049 |
| DPYS | rs13263121 | 751 | 782 | 212 | 530 | 492 | 136 | 0.93 | 0.84 | 1.04 | 0.227 | 764 | 751 | 181 | 480 | 496 | 145 | 1.13 | 1.01 | 1.27 | 0.034 | 0.02 |
| DPYS | rs1319371 | 1053 | 613 | 81 | 672 | 422 | 65 | 1.10 | 0.97 | 1.25 | 0.157 | 1011 | 592 | 94 | 683 | 387 | 52 | 0.90 | 0.79 | 1.02 | 0.106 | 0.03 |
| DPYS | rs35450967 | 1034 | 626 | 87 | 654 | 435 | 70 | 1.11 | 0.98 | 1.26 | 0.1 | 982 | 616 | 99 | 671 | 397 | 54 | 0.88 | 0.77 | 1.01 | 0.061 | 0.012 |
| PPAT | rs13135046 | 516 | 857 | 374 | 362 | 579 | 216 | 0.91 | 0.82 | 1.01 | 0.079 | 519 | 837 | 338 | 314 | 569 | 239 | 1.08 | 0.96 | 1.20 | 0.198 | 0.023 |
| TYMS | rs2298582 | 1336 | 393 | 18 | 914 | 234 | 11 | 0.86 | 0.73 | 1.02 | 0.09 | 1356 | 327 | 14 | 864 | 235 | 22 | 1.21 | 1.02 | 1.44 | 0.027 | 0.006 |
Adjusted for age (continuous), study stratum and the first five eigenvalues from principal components analysis. SNP effect was determined using a log-additive logistic regression model.
Allele counts: AA=homozygous wildtype allele carriers; AB=heterozygous allele carriers; BB=homozygous variant allele carriers.
P-value for interaction.
SNP is located 5′ downstream of gene; all other SNPs are located in introns; ‘pos’ SNPs are novel and identified by chromosomal position.
There were no significant associations of haplotypes at the global (gene) level with risk of OC for DPYD, DPYS or SHMT1 (Supplementary Table 5). Interestingly, SHMT1 haplotype #6 comprised minor alleles of the three correlated SHMT1 SNPs (rs56001517, rs7216214 and rs2273026) mentioned above and showed a decreased risk with OC at higher total folate intake (haplotype OR=0.68, 95% CI=0.53-0.87, P=0.002) that mirrored those of the individual SNP findings in Table 2. The selection of 29 haplotype block tagSNPs produced a single haplotype that was not significant, while several individual haplotypes of low frequency in DPYS were observed at P<0.05. Folate intake was not independently associated with risk of OC in multivariable-adjusted models (OR for >484 μg/d vs ≤484 μg/d = 1.04, 95% CI=0.91-1.18), nor when using different total folate intake cutpoints (OR for >400 μg/d vs ≤400 μg/d = 1.00, 95% CI=0.88-1.13 and OR for >683 μg/d [>75% percentile] vs ≤683 μg/d = 0.98, 95% CI=0.84-1.14).
Discussion
The results of the current study suggested a potential role for inherited variation in DPYD in pyrimidine metabolism with risk of OC. Folate intake may modify genetic risk of OC in the pyrimidine metabolism pathway, but the population effect is likely to be small. A possible role for SHMT1 SNP-by-folate interactions in the one-carbon transfer pathway may exist, but requires further validation. Selected genes in purine metabolism were not associated with risk of OC.
In the current study, 12 SNPs with additive effects in DPYD were found to be associated with OC and the strongest association (rs11587873) suggested a modest 8% decreased risk. We previously reported that the main effect of another DPYD SNP, rs1801265 (Arg29Cys), was associated with increased risk of OC among homozygous rare allele carriers [31], although that association was not replicated elsewhere [33] or in the current study. DPYD was represented by five SNPs in our earlier study of 1,770 participants [31] and these participants were also included in the current investigation of 129 SNPs. A haplotype analysis did not support an association of a single DPYD haplotype with risk of OC; however, this may be due, in part, from selecting 29 haplotype block tagSNPs to overcome the infinite recursion when using all 129 tagSNPs. We, therefore, cannot rule out a role for DPYD in risk of OC.
During pyrimidine metabolism, uracil and thymine concentrations are determined, in part, by the availability of 1-C units from folates and are catabolized to β-alanine and to valine/leucine/isoleucine, respectively (Supplementary Figure 1). DPYD encodes dihydropyrimidine dehydrogenase, the initial and rate-limiting step, whereas DPYS encodes dihydropyrimidinase, which catalyzes the secondary step. DPYD and DPYS enzyme deficiency or inhibition can cause decreased production of β-alanine or accumulation of the pyrimidines, uracil/dihydrouracil and thymine/dihydrothymine, and has shown considerable phenotypic variation ranging from severe neurological and developmental disorders associated with inborn errors to milder symptoms of lethargy, dizziness [46-48] and gastrointestinal abnormalities (gastroesophageal reflux, malabsorption) [49]. The pyrimidine metabolism pathway is identical for the degradation of fluoropyrimidines including 5-fluorouracil (5-FU), one of the most commonly prescribed chemotherapeutic agents in cancers [50, 51]. DPYD or DPYS enzyme deficiency results in toxicity among cancer patients from the inability to metabolize 5-FU [52, 53]. Screening programs for inborn errors of pyrimidine degradation have also identified individuals without symptoms, indicating an incomplete knowledge of the full spectrum of genetic, gene-environment, biochemical and clinical manifestations of DPYD and DPYS impairment [46, 49]. It will, therefore, be important to also investigate these associations with survival outcomes.
We had also previously reported increased risk with the main effect of SHMT1 variant rs9909104 [31], but could not replicate the main effect association here. The expanded analysis of SHMT1 SNPs and haplotypes in the present study suggested that risk may be modified by higher folate intake, although these associations did not meet the criteria of significance following multiple testing correction. SHMT1 encodes the serine hydroxymethyltransferase 1 (soluble) enzyme that catalyzes the reversible conversion of glycine and tetrahydrofolate to serine and 5,10 methylenetetrahydrofolate in the cytoplasm for the synthesis of methionine, pyrimidines (e.g., thymidylate) and purines [54]. Serine synthesis, nucleotide synthesis and the pentose phosphate pathway, which generates ribose-5-P (see Supplementary Figure 1), are implicated as important mechanisms of metabolic reprogramming in cancer cells [55]. Polymorphisms in SHMT1 have also been associated with carcinomas of the lung [56] and head and neck [57] and have been shown to interact with dietary folate to alter risk of non-Hodgkin lymphoma [58].
The strengths of this investigation include the gene-environment risk analysis in a targeted pathway using a large assembly of women with OC and the rigorous centralized genotyping and quality control standards. We improved upon our previous work [31, 32] by refining the associations using total folate intake instead of multivitamin supplement use. The median cutpoint for total folate intake approximated the dietary reference intake of 400 μg/d; therefore, the SNP-by-folate interactions can be interpreted as comparing women who meet or exceed recommendations to those who do not. There are also limitations to the current study. Folate intake was assessed at time of diagnosis using a FFQ that asked about average intake over the last year that may not represent habitual intake and may be affected by recall bias. Another potential limitation of the SNP-by-folate interactions is the analysis of all tumor types without stratification by tumor histology. This was decided a priori to maximize statistical power, although the evaluation of SNP main effects suggested no significant differences in risk estimates across the histological types for most SNPs. Another limitation is the inability to distinguish potentially causal SNPs at this time. We genotyped tagSNPs and some genes, such as DPYD, were large and were represented by several correlated tagSNPs that suggested either decreased or increased risk. The evaluation of a DPYD haplotype did not satisfactorily overcome this challenge and will need further clarification.
Conclusions
SNPs in DPYD may have modest effects on risk of OC and will require further evaluation in order to disentangle putative causal variants. Our findings suggest that exceeding the recommendations for folate intake does not negatively modify susceptibility in selected genes in pyrimidine metabolism to influence risk of OC. A possible role for SHMT1 SNP-by-folate interactions in the one-carbon transfer pathway may exist, but will require further validation. Polymorphisms in selected genes in purine metabolism do not appear to be associated with OC.
Supplementary Material
Acknowledgments
This study would not have been possible without the contributions of the following: P. Hall, (COGS); D. F. Easton, A. M. Dunning and A. Lee (Cambridge); J. Benitez, A. Gonzalez-Neira and the staff of the CNIO genotyping unit; D. C. Tessier, F. Bacot, D. Vincent, S. LaBoissière and F. Robidoux and the staff of the Genome Quebec genotyping unit; S. E. Bojesen, S. F. Nielsen, B. G. Nordestgaard, and the staff of the Copenhagen DNA laboratory; and S. A. Windebank, C. A. Hilker, J. Meyer and the staff of Mayo Clinic Genotyping Core Facility. We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, and D. Whiteman (AUS); G. Peuteman, T. Van Brussel, and D. Smeets (BEL); U. Eilber (GER); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T.W. Park-Simon, K. Beer-Grondke, and D. Schmidt (HJO); J. Vollenweider (MAY); the MD Anderson Center for Translational and Public Health Genomics (MDA); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao, and M. Stagner (POL); C. Luccarini, P. Harrington, the SEARCH team and ECRIC (SEA); the Scottish Gynaecological Clinical Trails group and SCOTROC1 investigators (SRO); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan, and J. Ford (UKO); and Carole Pye (UKR). We thank Jian Yang for assistance with the GCTA program.
Funding
The COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 - HEALTH-F2-2009-223175). The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The scientific development and funding for this project were supported by the Canadian Institutes of Health Research (MOP-86727) and the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112). Funding of the constituent studies was provided by the Canadian Institutes of Health Research (MOP-84340), WorkSafeBC 14, and OvCaRe: BC’s Ovarian Cancer Research Team; the American Cancer Society (CRTG-00-196-01-CCE); the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124, C536/A13086, C536/A6689); the Celma Mastry Ovarian Cancer Foundation ; the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal (Oak Foundation); the Fred C. and Katherine B. Andersen Foundation; the German Cancer Research Center; the German Federal Ministry of Education and Research of Germany, Program of Clinical Biomedical Research (01GB 9401); the Helsinki University Central Hospital Research Fund; Helse Vest; Imperial Experimental Cancer Research Centre (C1312/A15589); the L & S Milken Foundation; the Lon V. Smith Foundation (LVS-39420); the Mayo Foundation; the Mermaid I project; the Minnesota Ovarian Cancer Alliance; the National Health and Medical Research Council (NHMRC) of Australia (199600, 209057, 251533, 396414, 400281, and 504715); Nationaal Kankerplan of Belgium; the Norwegian Cancer Society; the Norwegian Research Council; the OHSU Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); Pomeranian Medical University; Radboud University Medical Center; the Roswell Park Cancer Institute Alliance Foundation; the Royal Marsden Hospital; the Rudolf-Bartling Foundation; the Sigrid Juselius Foundation; the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685); the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge and the University College London Hospitals; the US Army Medical Research and Material Command (DAMD17-98-1- 8659, DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-10-1-0280); the US National Cancer Institute (K07-CA095666, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA15083, P50-CA105009, P50- CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01- CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063682, R01-CA064277, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA076016, R01-CA080742, R01-CA080978, R01-CA083918, R01-CA087538, R01- CA092044, R01-095023, R01-CA106414, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01- CA136924, R01-CA149429, R03-CA113148, R03-CA115195, R37-CA070867, R37-CA70867, U01-CA069417, U01- CA071966 and Intramural research funds); the US National Institutes of Health/National Center for Research Resources/General Clinical Research Center (MO1-RR000056); and the US Public Health Service (PSA-042205).
L.E.K. was supported by a Canadian Institutes of Health Research Investigator award (MSH-87734). P.M.W. is supported by the NHMRC of Australia. B.Y.K. holds an American Cancer Society Early Detection Professorship (SIOP-06-258-01- COUN). F.M. is supported by a K-award from the National Cancer Institute (K07-CA080668).
List of Abbreviations
- 1-C
one-carbon
- ADSL
adenylosuccinate lyase
- ADSS
adenylosuccinate synthase
- AK3
adenylate kinase 3
- ATIC
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
- CAD
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
- CMPK1
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
- CTPS
CTP synthase 1
- DCK
deoxycytidine kinase
- DHODH
dihydroorotate dehydrogenase (quinone)
- DNMT3B
DNA (cytosine-5-)-methyltransferase 3 beta
- DPYD
dihydropyrimidine dehydrogenase
- DPYS
dihydropyrimidinase
- GART
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
- GCTA
Genome-wide Complex Trait Analysis
- GMPS
guanine monphosphate synthase
- iCOGS
international Collaborative Oncology Gene-environment Study
- IMPDH1
IMP (inosine 5'-monophosphate) dehydrogenase 1
- IMPDH2
IMP (inosine 5'-monophosphate) dehydrogenase 2
- LD
linkage disequilibrium
- LRT
likelihood ratio test
- MAF
minor allele frequency
- NME6
NME/NM23 nucleoside diphosphate kinase 6
- OC
Ovarian carcinoma
- OCAC
Ovarian Cancer Association Consortium
- OR
odds ratio
- PAICS
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase
- PFAS
phosphoribosylformylglycinamidine synthase
- PPAT
phosphoribosyl pyrophosphate amidotransferase
- PRPS2
phosphoribosyl pyrophosphate synthetase 2
- QC
quality control
- RRM2B
ribonucleotide reductase M2 B (TP53 inducible)
- SHMT1
serine hydroxymethyltransferase 1 (soluble)
- SNP
single nuctleotide polymorphism
- TYMS
thymidylate synthase
- UMPS
uridine monophosphate synthetase
Footnotes
Conflict of Interest Statement
The authors declare that there are no financial or commercial conflicts of interest.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Gentry-Maharaj A, Menon U. Screening for ovarian cancer in the general population. Best Pract. Res. Clin. Obstet. Gynaecol. 2012;26:243–256. doi: 10.1016/j.bpobgyn.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan S, Coward JI, Bast RC, Jr., Berchuck A, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall JM, Lee MK, Newman B, Morrow JE, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 5.Boyd J, Rubin SC. Hereditary ovarian cancer: molecular genetics and clinical implications. Gynecol. Oncol. 1997;64:196–206. doi: 10.1006/gyno.1996.4572. [DOI] [PubMed] [Google Scholar]
- 6.Narod SA, Madlensky L, Bradley L, Cole D, et al. Hereditary and familial ovarian cancer in southern Ontario. Cancer. 1994;74:2341–2346. doi: 10.1002/1097-0142(19941015)74:8<2341::aid-cncr2820740819>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 7.Risch HA, McLaughlin JR, Cole DE, Rosen B, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am. J. Hum. Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Ramus SJ, Tyrer J, Bolton KL, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolton KL, Tyrer J, Song H, Ramus SJ, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 2013;45:362–370. 370e361–362. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hankinson SE, Colditz GA, Hunter DJ, Spencer TL, et al. A quantitative assessment of oral contraceptive use and risk of ovarian cancer. Obstet. Gynecol. 1992;80:708–714. [PubMed] [Google Scholar]
- 13.The reduction in risk of ovarian cancer associated with oral-contraceptive use. The Cancer and Steroid Hormone Study of the Centers for Disease Control and the National Institute of Child Health and Human Development. N. Engl. J. Med. 1987;316:650–655. doi: 10.1056/NEJM198703123161102. [DOI] [PubMed] [Google Scholar]
- 14.Beral V, Doll R, Hermon C, Peto R, Reeves G. Ovarian cancer and oral contraceptives: collaborative reanalysis of data from 45 epidemiological studies including 23,257 women with ovarian cancer and 87,303 controls. Lancet. 2008;371:303–314. doi: 10.1016/S0140-6736(08)60167-1. [DOI] [PubMed] [Google Scholar]
- 15.Whittemore AS, Harris R, Itnyre J. Characteristics relating to ovarian cancer risk: collaborative analysis of 12 US case-control studies. II. Invasive epithelial ovarian cancers in white women. Collaborative Ovarian Cancer Group. Am. J. Epidemiol. 1992;136:1184–1203. doi: 10.1093/oxfordjournals.aje.a116427. [DOI] [PubMed] [Google Scholar]
- 16.Risch HA, Marrett LD, Howe GR. Parity, contraception, infertility, and the risk of epithelial ovarian cancer. Am. J. Epidemiol. 1994;140:585–597. doi: 10.1093/oxfordjournals.aje.a117296. [DOI] [PubMed] [Google Scholar]
- 17.Luan NN, Wu QJ, Gong TT, Vogtmann E, et al. Breastfeeding and ovarian cancer risk: a meta-analysis of epidemiologic studies. Am. J. Clin. Nutr. 2013;98:1020–1031. doi: 10.3945/ajcn.113.062794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sieh W, Salvador S, McGuire V, Weber RP, et al. Tubal ligation and risk of ovarian cancer subtypes: a pooled analysis of case-control studies. Int. J. Epidemiol. 2013;42:579–589. doi: 10.1093/ije/dyt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearce CL, Templeman C, Rossing MA, Lee A, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol. 2012;13:385–394. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber MT, Kjaer SK, Dehlendorff C, Chang-Claude J, et al. Cigarette smoking and risk of ovarian cancer: a pooled analysis of 21 case-control studies. Cancer Causes Control. 2013;24:989–1004. doi: 10.1007/s10552-013-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J. Nutr. 2000;130:129–132. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 22.Blount BC, Mack MM, Wehr CM, MacGregor JT, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YI. Role of folate in colon cancer development and progression. J. Nutr. 2003;133:3731S–3739S. doi: 10.1093/jn/133.11.3731S. [DOI] [PubMed] [Google Scholar]
- 24.Rodwell VW. In: Harper's Biochemistry. Murray RK, Granner DK, Mayes PA, Rodwell VW, editors. McGraw-Hill; New York: 2000. pp. 386–401. [Google Scholar]
- 25.Penuelas S, Noe V, Ciudad CJ. Modulation of IMPDH2, survivin, topoisomerase I and vimentin increases sensitivity to methotrexate in HT29 human colon cancer cells. Febs J. 2005;272:696–710. doi: 10.1111/j.1742-4658.2004.04504.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi Y, Uemura H, Ishiguro H, Kitamura H, et al. Expression of thymidylate synthase, dihydropyrimidine dehydrogenase, thymidine phosphorylase, and orotate phosphoribosyl transferase in prostate cancer. Prostate Cancer Prostatic Dis. 2005;8:260–265. doi: 10.1038/sj.pcan.4500817. [DOI] [PubMed] [Google Scholar]
- 27.Khan S, Abdelrahim M, Samudio I, Safe S. Estrogen receptor/Sp1 complexes are required for induction of cad gene expression by 17beta-estradiol in breast cancer cells. Endocrinology. 2003;144:2325–2335. doi: 10.1210/en.2002-0149. [DOI] [PubMed] [Google Scholar]
- 28.Spurr IB, Birts CN, Cuda F, Benkovic SJ, et al. Targeting tumour proliferation with a small-molecule inhibitor of AICAR transformylase homodimerization. Chembiochem. 2012;13:1628–1634. doi: 10.1002/cbic.201200279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Beaumais TA, Jacqz-Aigrain E. Intracellular disposition of methotrexate in acute lymphoblastic leukemia in children. Curr Drug Metab. 2012;13:822–834. doi: 10.2174/138920012800840400. [DOI] [PubMed] [Google Scholar]
- 30.Yanamoto S, Kawasaki G, Yoshitomi I, Mizuno A. Expression of p53R2, newly p53 target in oral normal epithelium, epithelial dysplasia and squamous cell carcinoma. Cancer Lett. 2003;190:233–243. doi: 10.1016/s0304-3835(02)00588-8. [DOI] [PubMed] [Google Scholar]
- 31.Kelemen LE, Sellers TA, Schildkraut JM, Cunningham JM, et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 2008;68:2498–2506. doi: 10.1158/0008-5472.CAN-07-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelemen LE, Wang Q, Dinu I, Vierkant RA, et al. Regular Multivitamin Supplement Use, Single Nucleotide Polymorphisms in ATIC, SHMT2, and SLC46A1, and Risk of Ovarian Carcinoma. Front Genet. 2012;3:33. doi: 10.3389/fgene.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelemen LE, Goodman MT, McGuire V, Rossing MA, et al. Genetic variation in TYMS in the one-carbon transfer pathway is associated with ovarian carcinoma types in the Ovarian Cancer Association Consortium. Cancer Epidemiol. Biomarkers Prev. 2010;19:1822–1830. doi: 10.1158/1055-9965.EPI-09-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abecasis GR, Altshuler D, Auton A, Brooks LD, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Ibiebele TI, Parekh S, Mallitt KA, Hughes MC, et al. Reproducibility of food and nutrient intake estimates using a semi-quantitative FFQ in Australian adults. Public Health Nutr. 2009;12:2359–2365. doi: 10.1017/S1368980009005023. [DOI] [PubMed] [Google Scholar]
- 37.Patterson RE, Kristal AR, Tinker LF, Carter RA, et al. Measurement characteristics of the Women's Health Initiative food frequency questionnaire. Ann. Epidemiol. 1999;9:178–187. doi: 10.1016/s1047-2797(98)00055-6. [DOI] [PubMed] [Google Scholar]
- 38.Hankin JH, Wilkens LR, Kolonel LN, Yoshizawa CN. Validation of a quantitative diet history method in Hawaii. Am. J. Epidemiol. 1991;133:616–628. doi: 10.1093/oxfordjournals.aje.a115934. [DOI] [PubMed] [Google Scholar]
- 39.Willett W, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am. J. Epidemiol. 1985;122:51–65. doi: 10.1093/oxfordjournals.aje.a114086. [DOI] [PubMed] [Google Scholar]
- 40.Bandera EV, King M, Chandran U, Paddock LE, et al. Phytoestrogen consumption from foods and supplements and epithelial ovarian cancer risk: a population-based case control study. BMC Womens Health. 2011;11:40. doi: 10.1186/1472-6874-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nature reviews. Genetics. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Development Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2008. http://www.R-project.org. [Google Scholar]
- 45.Purcell S, Neale B, Todd-Brown K, Thomas L, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. http://pngu.mgh.harvard.edu/purcell/plink/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Kuilenburg AB, Vreken P, Abeling NG, Bakker HD, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum. Genet. 1999;104:1–9. doi: 10.1007/pl00008711. [DOI] [PubMed] [Google Scholar]
- 47.van Gennip AH, Abeling NG, Vreken P, van Kuilenburg AB. Inborn errors of pyrimidine degradation: clinical, biochemical and molecular aspects. J. Inherit. Metab. Dis. 1997;20:203–213. doi: 10.1023/a:1005356806329. [DOI] [PubMed] [Google Scholar]
- 48.Connolly GP, Simmonds HA, Duley JA. Pyrimidines and CNS regulation. Trends Pharmacol. Sci. 1996;17:106–107. doi: 10.1016/0165-6147(96)20001-x. [DOI] [PubMed] [Google Scholar]
- 49.van Kuilenburg AB, Dobritzsch D, Meijer J, Meinsma R, et al. Dihydropyrimidinase deficiency: Phenotype, genotype and structural consequences in 17 patients. Biochim. Biophys. Acta. 2010;1802:639–648. doi: 10.1016/j.bbadis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Hertz R, Li MC, Spencer DB. Effect of methotrexate therapy upon choriocarcinoma and chorioadenoma. Proc. Soc. Exp. Biol. Med. 1956;93:361–366. doi: 10.3181/00379727-93-22757. [DOI] [PubMed] [Google Scholar]
- 51.Farber S. Some observations on the effect of folic acid antagonists on acute leukemia and other forms of incurable cancer. Blood. 1949;4:160–167. [PubMed] [Google Scholar]
- 52.Johnson MR, Diasio RB. Importance of dihydropyrimidine dehydrogenase (DPD) deficiency in patients exhibiting toxicity following treatment with 5-fluorouracil. Adv. Enzyme Regul. 2001;41:151–157. doi: 10.1016/s0065-2571(00)00011-x. [DOI] [PubMed] [Google Scholar]
- 53.Thomas HR, Ezzeldin HH, Guarcello V, Mattison LK, et al. Genetic regulation of dihydropyrimidinase and its possible implication in altered uracil catabolism. Pharmacogenet. Genomics. 2007;17:973–987. doi: 10.1097/FPC.0b013e3282f01788. [DOI] [PubMed] [Google Scholar]
- 54.Schirch L. Serine hydroxymethyltransferase. Adv. Enzymol. Relat. Areas Mol. Biol. 1982;53:83–112. doi: 10.1002/9780470122983.ch3. [DOI] [PubMed] [Google Scholar]
- 55.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 56.Wang L, Lu J, An J, Shi Q, et al. Polymorphisms of cytosolic serine hydroxymethyltransferase and risk of lung cancer: A case-control analysis. Lung Cancer. 2007;57:143–151. doi: 10.1016/j.lungcan.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z, Shi Q, Sturgis EM, Spitz MR, Wei Q. Polymorphisms and haplotypes of serine hydroxymethyltransferase and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Pharmacogenet. Genomics. 2005;15:557–564. doi: 10.1097/01.fpc.0000170915.19522.b2. [DOI] [PubMed] [Google Scholar]
- 58.Li Q, Lan Q, Zhang Y, Bassig BA, et al. Role of one-carbon metabolizing pathway genes and gene-nutrient interaction in the risk of non-Hodgkin lymphoma. Cancer Causes Control. 2013;24:1875–1884. doi: 10.1007/s10552-013-0264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.