Abstract
Upon invasion of the erythrocyte cell, the malaria parasite remodels its environment; in particular, it establishes a complex membrane network, which connects the parasitophorous vacuole to the host plasma membrane and is involved in protein transport and trafficking. We have identified a novel subtelomeric gene family in Plasmodium falciparum that encodes 11 transmembrane proteins localized to the Maurer's clefts. Using coimmunoprecipitation and shotgun proteomics, we were able to enrich specifically for these proteins and detect distinct peptides, allowing us to conclude that four to 10 products were present at a given time. Nearly all of the Pfmc-2tm genes are transcribed during the trophozoite stage; this narrow time frame of transcription overlaps with the specific stevor and rif genes that are differentially expressed during the erythrocyte cycle. The description of the structural properties of the proteins led us to manually reannotate published sequences, and to detect potentially homologous gene families in both P. falciparum and Plasmodium yoelii yoelii, where no orthologs were predicted uniquely based on sequence similarity. These basic proteins with two transmembrane domains belong to a larger superfamily, which includes STEVORs and RIFINs.
Infection of human red blood cells by P. falciparum, the most dangerous species of parasites causing human malaria, results in extensive modifications to the host cell that are required for persistent infection. These modifications allow the parasite to mediate import and export of nutrients and waste products, facilitate host cell lysis, and affect the adherence properties (cytoadherence) of infected cells (Udeinya et al. 1981). The latter of these functions is particularly significant to the pathogenesis of severe malaria disease. Cytoadherence is widely believed to aid circumvention of the immune system by sequestering infected cells within internal organ capillaries, thus avoiding circulation through the spleen and detection by major components of the immune system (Miller et al. 2002). High levels of sequestration within capillaries and microvasculatures in the brain result in a cerebral malaria syndrome, a condition that is the cause of most malaria-related deaths (Beeson and Brown 2002). Rosetting, the process by which infected erythrocytes bind uninfected cells, has also been strongly correlated with severe disease by histological examination of postmortem tissues (Heddini et al. 2001).
The parasite contains itself within a self-constructed parasitophorous vacuole (PV), and forms a tubovesicular membrane (TVM) network that extends throughout the erythrocyte cytoplasm and associates with flattened vesicular structures beneath the red-cell membrane, called the Maurer's clefts (MCs), which are believed to translocate proteins secreted from the parasite to the erythrocyte surface (Barnwell 1990; Hinterberg et al. 1994). A more recent model describes the MCs as part of a continuous membrane network extending from the PV (Wickert et al. 2003b). This complex membrane network is believed to be a secretory organelle established by the parasite outside of its own cytoplasm and involved in both import and export (Przyborski et al. 2003). Specifically, the MCs have been implicated in transporting and possibly assembling proteins destined for the knob-like protrusions that extend from the erythrocyte surface (Przyborski et al. 2003). The knobs are dramatic features characteristic of infection and comprise the major sites of cytoadherence between infected erythrocytes and endothelial cells. The main class of proteins localized on the surface of the knobs and involved in cytoadhesion is the PfEMP1 family, which encodes the immunovariant erythrocyte membrane proteins 1 (Craig and Scherf 2001). A number of proteins have been shown to localize to the MCs. In particular, homologs to the COPII proteins Sar1 (PFD0810w), Sec31 (PFB0640c), and Sec23 (PF08_0036) have been detected in MCs (Albano et al. 1999; Adisa et al. 2001; Wickert et al. 2003a), indicating that the MCs participate in hostcell remodeling by transporting parasite-derived proteins to the erythrocyte surface via COPII-coated vesicles. By determining the protein components involved in the transport of cytoadhesion factors to the erythrocyte surface and those that are essential for host-cell remodeling, new drug and vaccine targets could be deduced and applied to fight severe disease.
To identify resident proteins of the parasite-derived membrane network extending through the erythrocyte cytoplasm, a library of monoclonal antibodies (mAb) against P. falciparum asexual stages was prepared (Sam-Yellowe et al. 2001). The isolated SP1C1 mAb identified a 20-kD protein resistant to sodium carbonate extraction (i.e., integral to the membrane). Indirect immunofluorescence assays using SP1C1 mAb clones localized the protein to patchy structures, corresponding to large vesicles in the cytoplasm of trophozoite- and schizont-infected erythrocytes (Fig. 1). By indirect immunofluorescence and confocal microscopy, SP1C1 mAb colocalized with SP1A6 mAb, which is specific for a 130-kD membrane-associated protein (Pf130) that has been detected in both Maurer's clefts and knobs (Sam-Yellowe et al. 2001; Fig. 1). To identify the protein target(s) of the SP1C1 antibody, multidimensional protein identification technology (MudPIT) was applied. MudPIT combines inline high-resolution liquid chromatography with tandem mass spectrometry to separate and identify peptides obtained from proteolytically digested protein samples (Washburn et al. 2001). This strategy has been previously applied to analyze the proteome isolated from whole-cell lysates of different stages of P. falciparum in order to identify components unique or common to particular life cycle stages (Florens et al. 2002). In this communication, we characterize the proteins coimmunoprecipitated by SP1C1 mAb. We describe a new family of transmembrane proteins encoded by genes located within the subtelomeric regions of P. falciparum chromosomes, expressed during the erythrocytic cycle and localized at the Maurer's clefts, a protein family potentially involved in trafficking of antigenic variants.
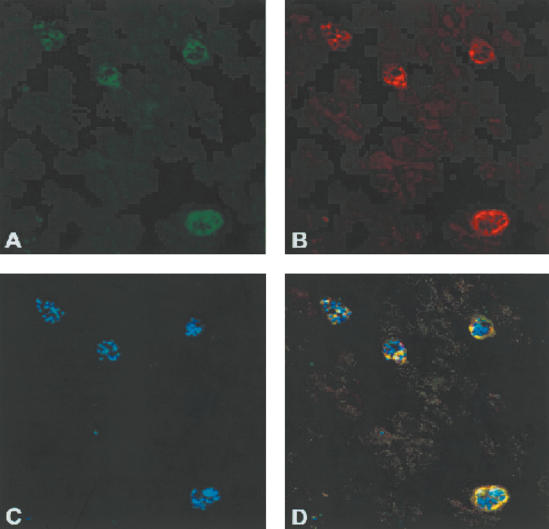
Figure 1.
Colocalization immunofluorescence of PfMC-2TM with Pf130. Trophozoite and schizont-infected erythrocytes were fixed with ice-cold acetone and incubated sequentially with the primary antibodies SP1C1 and SP1A6 specific for PfMC-2TM and Pf130, respectively. The monoclonal antibodies were contained in spent hybridoma culture supernatant and were used undiluted. The anti-mouse secondary antibodies conjugated to Alexa 488 and Alexa 568 (Molecular Probes) were diluted 1: 1000 in 1× PBS and added separately. Following incubation for 1 h at 37°C, slides were washed three times in 1× PBS and once in distilled water. The smears were mounted in Vectashield (Vector) containing 4′, 6-diamidino-2-phenylindole (DAPI) to stain parasite DNA. Parasites were examined by a Nikon epifluorescence microscope. (A) P. falciparum-infected erythrocytes incubated with mAb SP1A6 followed by anti-mouse 488 Alexa conjugate (green). (B) P. falciparum schizont-infected erythrocytes incubated with mAb SP1C1, followed by anti-mouse 568 Alexa conjugate (red). (C) Parasite nuclei counterstained by DAPI. (D) Overlay of SP1A6 and SP1C1 antibodies showing colocalization (yellow) of PfMC-2TM and Pf130 Maurer's cleft proteins.
RESULTS AND DISCUSSION
Identification of the Proteins Coimmunoprecipitated by the SP1C1 Monoclonal Antibody
Monoclonal antibodies from SP1C1 clones were pooled and complexed with goat anti-mouse sepharose beads. After several washes, P. falciparum schizont extracts, prepared as described (Sam-Yellowe et al. 2001), were added to the beads and incubated overnight. Bound immune complexes were eluted twice with 0.1 M glycine (pH 2.8), and a final step was carried out with 10% 1,4-dioxane to elute the more hydrophobic proteins. Two independent coimmunoprecipiations were carried out along with a control, one without SP1C1 mAb bound to the sepharose beads. After TCA precipitation, proteins were denatured, reduced, S-alkylated, digested with endoproteinase Lys-C followed by trypsin, and analyzed by MudPIT. Tandem mass (MS/MS) spectra obtained from digests of the immunecomplexed proteins were interpreted by a modified version of the SEQUEST algorithm (Eng et al. 1994), PEP_PROBE (Sadygov and Yates III 2003), that provides a statistical confidence for each peptide match. MS/MS data sets were searched against a database combining mammalian host proteins (human, mouse, and rat sequences from NCBI RefSeq) with the latest release of the P. falciparum genome (Gardner et al. 2002).
The SP1C1 antibody consistently pulled down 10 hypothetical proteins detected by a large number of peptides (Supplemental Table S1). These proteins shared stretches of sequences, as most of the identified peptides were common between them. A multiple sequence alignment revealed that these 10 proteins were highly homologous (Fig. 2). Intriguingly, whereas five proteins (PFB0985c, PFC1080c, PF11_0014, MAL7P1.5, and PF11_0025) were encoded by two-exon genes and had a putative N-terminal signal sequence and two putative C-terminal transmembrane segments, four others (PFA0680c, MAL6P1.15, PF10_0390, and PFA0065w) were lacking the signal sequence. PFB0960c was the most divergent, as it did not have a signal peptide and was much shorter, missing the C-terminal transmembrane domains. The complete sequences of the chromosomes bearing these genes were downloaded from PlasmoDB (Bahl et al. 2002) and investigated using the Artemis genome sequence viewer (Rutherford et al. 2000). We were able to extend the sequences of these five genes beyond the boundaries preliminarily predicted by the gene-modeling algorithm (Fig. 2, underlined sequences). In all five cases, a 5′ ORF corresponding to the short exon 1 could be detected, whereas a frameshift in exon 2 was responsible for the premature ending of PFB0960c. Because the sequences of these genes were modified from their primary annotation, we now refer to them with their published locus name and an asterisk (Fig. 2). A BLASTP search against the complete P. falciparum database detected MAL7P1.58, an eleventh member of the gene family, from which no peptides were detected. We also searched a P. falciparum database containing all possible ORFs to potentially detect additional genes, but no other sequence was identified.
Figure 2.
Multiple sequence alignments. The sequences of the Maurer's cleft 2-transmembrane domain proteins were aligned using CLUSTAL W (1.82) (Thompson et al. 1994). (*) Conserved residues. Bold, underlined sequences indicate regions of the proteins that were extended from the originally published gene models (Gardner et al. 2002), on the basis of the complete chromosome sequences that were obtained from PlasmoDB (Bahl et al. 2002). The red-dotted vertical line indicates the boundary between exon 1 and exon 2. The regions covered by peptides identified during the MS/MS analyses are highlighted. The green, magenta, cyan, and yellow color coding stands for peptides unique to a protein, and peptides common to two, three, and more than three proteins, respectively. The green boxes indicate the position of signal peptide and transmembrane domains as predicted by SignalP (Nielsen et al. 1997) and TMHMM (Krogh et al. 2001), respectively. The arrow points at the potential processing site. The red and blue boxes define potential bipartite vacuolar translocation signals (VTSs) as described by Lopez-Estrano et al. (2003).
Peptides unique to PFC1080c and PFB0960c* were detected (highlighted in green in Fig. 2), indicating that both proteins were present in the analyzed samples. PFB0985c and PFA0680c* share 94% sequence identity, consequently no unique peptides were recovered from either of them. However, a peptide matching only these two proteins (highlighted in magenta in Fig. 2) confirmed that at least one of them was present. The same holds true for the MAL7P1.5/PF11_0014 pair, sharing 75% sequence identity. On the basis of these unique peptide combinations, we concluded that between four and 10 distinct members of the protein family were present in our samples. This gene family encodes basic membrane proteins of around 230 amino acid residues, ca 27 kD (Table 1). Hence, these sizes are in agreement with the approximate 20 kD measurement obtained by Western blotting and immunoprecipitation of metabolically labeled proteins by the SP1C1 mAb (Sam-Yellowe et al. 2001). Although the alignments of some P. falciparum chromosomes are still not complete, which makes the localization of the genes along chromosomes tentative, with the exception of MAL7P1.58, all of the Pfmc-2tm genes are located within subtelomeric regions of chromosomes (Table 1). On the basis of their known subcellular localization and predicted membrane topology, we propose to name this gene family Pfmc-2tm, for Maurer's cleft two transmembrane proteins.
Table 1.
Plasmodium Subtelomeric Multigene Family Encoding 2-Transmembrane Proteins: Structural Properties
| Namea | Locusb | Chrc | Locd | Exons | SubCe | Length | MW | pI | SPf | TMg | S1g | E1g | S2g | SE2g | Looph |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PfMC-2TM_1.2 | PFA0680c* | 1 | ST | 2 | MC | 229 | 26899.8 | 9.69 | 1 | 2 | 157 | 179 | 184 | 203 | 4 |
| PfMC-2TM_1.1 | PFA0065w* | 1 | ST | 2 | MC | 234 | 27446.4 | 9.66 | 1 | 2 | 160 | 182 | 186 | 208 | 3 |
| PfMC-2TM_2.2 | PFB0985c | 2 | ST | 2 | MC | 229 | 27063.0 | 9.57 | 1 | 2 | 156 | 178 | 182 | 204 | 3 |
| PfMC-2TM_2.1 | PFB0960c* | 2 | ST | 2 | MC | 225 | 26752.6 | 9.52 | 1 | 2 | 157 | 179 | 184 | 203 | 4 |
| PfMC-2TM_3 | PFC1080c | 3 | ST | 2 | MC | 231 | 27004.6 | 9.47 | 1 | 2 | 162 | 184 | 189 | 208 | 4 |
| PfMC-2TM_6 | MAL6P1.15* | 6 | ST | 2 | MC | 231 | 27183.1 | 9.60 | 1 | 2 | 157 | 179 | 183 | 205 | 3 |
| PfMC-2TM_7.1 | MAL7P1.5 | 7 | ST | 2 | MC | 235 | 27557.2 | 9.57 | 1 | 2 | 162 | 184 | 188 | 210 | 3 |
| PfMC-2TM_7.2 | MAL7P1.58 | 7 | 2 | 231 | 27329.0 | 9.55 | 1 | 2 | 159 | 181 | 186 | 208 | 4 | ||
| PfMC-2TM_10 | PF10_0390* | 10 | ST | 2 | MC | 230 | 27217.9 | 9.55 | 1 | 2 | 153 | 175 | 185 | 204 | 9 |
| PfMC-2TM_11.2 | PF11_0025 | 11 | ST | 2 | MC | 231 | 27194.1 | 9.57 | 1 | 2 | 157 | 179 | 184 | 206 | 4 |
| PfMC-2TM_11.1 | PF11_0014* | 11 | ST | 2 | MC | 231 | 27179.8 | 9.29 | 1 | 2 | 162 | 184 | 189 | 208 | 4 |
Protein names a are proposed based on known subcellular localization e. Genes b, which were modified from their published annotation (Gardner et al. 2002), are marked with an asterisk. The gene locations within chromosomes c are reported in d. The presence of a signal peptide (SP)f, as predicted by SignalP (Nielsen et al. 1997), and the number of transmembrane domains (TM)g, as predicted by TMHMM (Krogh et al. 2001) are reported, as well as the position of these TM segments within the protein sequenceg and the number of amino acid residues separating the two TMsh.
Analysis of Pfmc-2tm Gene Expression
We next asked whether the mRNA abundance pattern for the Pfmc-2tm genes within the erythrocytic cycle mirrored their protein patterns by examining expression data obtained using a custom-made high-density oligonucleotide microarray (Le Roch et al. 2003). As the array bears probes to both predicted coding regions as well as intergenic regions of the P. falciparum genome, we were able to examine the expression pattern of probes outside of the original gene models in order to obtain additional empirical evidence for the new annotations. We thus followed 78 probes uniquely mapped to the Pfmc-2tm family, but not necessarily unique to a gene (Supplemental Table S2). The background subtraction was performed using a probabilistic model (Le Roch et al. 2003), so that all probe intensities are positively defined. The global noise level was estimated to be ∼10–20 expression units; however, the estimated noise does not serve as the presence/absence threshold for probes, due to the considerable variations of their hybridization properties. Only 41 of 78 probes with a minimal standard deviation of 20 across their life-cycle expression profile were considered to contain true signals and were analyzed further. The probe expression levels were floored by the noise level of 20.0 and log scaled. Pairwise Pearson correlation coefficients were calculated between the life-cycle expression profiles of different probes, which were then hierarchically clustered. The clustering results are reported in Supplemental Table S2. The subtree containing 33 probes sharing an average correlation coefficient >0.75 between themselves are highlighted in red in the “Corr>0.75” column. Computer simulation demonstrated that two unrelated probes would have less than a 2% probability to generate a correlation >0.75 by chance.
The 33 probes showed a highly coordinated expression profile within a narrow 12-h time frame of the erythrocytic life cycle, obtained under two independent synchronization experiments (Supplemental Table S2). With the exception of MAL7P1.5, probes unique to every Pfmc-2tm gene were expressed during late ring to late trophozoite stages. This pattern is very similar to the differentially expressed erythrocytic stevors (PF10_0395 and PF11_0516) and rifins (PFD1240w, PF10_006) (Le Roch et al. 2003). Furthermore, three of the correlated probes cover the regions corresponding to the short first exon; one covers multiple genes starting at position 47, another hits PF11_0014 and PFA0065w* at position 47, and the other is specific to PFB0960c* at position 44. Altogether, these data support our manual reannotation of PFB0960c* as a 2-exon gene.
Detection of Structurally Homologous Proteins
BLASTP searches against the nonredundant database did not return any homologous proteins and a search against the P. y. yoelii genome (Carlton et al. 2002) showed that there were no orthologs in P. y. yoelii. This suggested that the Pfmc-2tm gene family was unique to P. falciparum. However, we decided to query the P. y. yoelii and P. falciparum sequences for the structural features characterizing the PfMC-2TM proteins as follows: (1) a length of 200 to 300 amino acid residues, (2) an N-terminal hydrophobic sequence, and especially (3) two C-terminal transmembrane domains separated by a very short stretch of residues (<10).
Three additional P. falciparum hypothetical proteins were uncovered through this structure-based query, PFA0715c, and two adjacent genes on chromosome 8, MAL8P1.160 and MAL8P1.161. All three were encoded by subtelomeric genes (Supplemental Table S3). These proteins were clearly related to one another (Supplemental Figure S1), yet distinct from the PfMC-2TM family. The published amino terminus of MAL8P1.160 did not seem to align properly with the other two proteins, which prompted us to investigate alternate choices as first exon. We found an ORF upstream of MAL8P1.160*, whose sequence aligned with the other two proteins (Supplemental Figure S1). We propose to name this small protein family Pf ST-2TM.
Eighteen P. y. yoelii hypothetical proteins were identified via the structure-based query (Supplemental Figure S1), all of which belong to the pyst-b gene family (Carlton et al. 2002). These genes do not have direct orthologs in P. falciparum, as defined by sequence similarities (Carlton et al. 2002). The pyst-b gene family contains 54 genes and is fairly heterogeneous at both the gene and protein levels; the gene structures range from 1 to 6 exons, whereas some of the cognate proteins are predicted to contain up to 6 transmembrane domains. The 18 pyst-b genes identified by the structure-based query are all predicted to have two transmembrane regions separated by a maximum of four residues; hence, we propose to name this subset pyst-b-2tm. Although the genome of P. y. yoelii has been sequenced using a whole-genome shotgun approach (Carlton et al. 2002), some of the contigs were assigned to subtelomeric regions (http://www.tigr.org/tdb/e2k1/pya1/pya1-telo.shtml). At least 12 pyst-b-2tm genes are located within subtelomeric regions on P. y. yoelii chromosomes (Supplemental Table S3). After multiple sequence alignment, the start sites of several genes (denoted by *) were manually reannotated. These reannotations ranged from removing two N-terminal residues to discarding entire 5′ exons. Specifically, two exons 5′ of PY04210* were discarded (40 N-terminal residues), an internal exon was removed from PY02142*, and the published 5′ exon for PY01904, which clearly did not align with the rest of the gene family, was removed. Searching for alternate ORFs in the P. y. yoelii genome revealed 5′ ORFs on contig 522 (1736–1805) and contig 1303 (8079–8147) likely to correspond to PY01904* and PY04315* first exons, respectively. The sequence of contig 2074 did not extend far enough upstream of PY06190 to find a first exon for this gene (Supplemental Figure S1). The pyst-b-2tm genes have a structure similar to that of Pfmc-2tm and Pfst-2tm, that is, a short first exon of 69 bases and a long second one. All full-length PyST-B-2TM proteins have a weakly hydrophobic N-terminal region, predicted to be a cleavable signal sequence by SignalP (Nielsen et al. 1997; Supplemental Figure S1). However, the PyST-B-2TM family shows more heterogeneity than PfMC-2TM and PfST-2TM at the amino acid level. In particular, whereas PfMC-2TM and PfST-2TM proteins are all very basic, the isoelectric points predicted for the PyST-B-2TM proteins covers a wide range of pH from 5.3 to 9.3 (Supplemental Table S3).
A Superfamily of Two-Transmembrane Proteins
Multicopy gene families present exclusively in one Plasmodium species have been described for the var, cir, yir, bir, and vir genes (Janssen et al. 2001). These genes encode large polymorphic erythrocyte-surface proteins, which are highly immunogenic (Janssen et al. 2001). However, the high-sequence variation of these antigens limits their potential use as vaccine candidates. In P. falciparum, other multicopy gene families have been characterized and include the rif, stevor, and sep/etramps genes, which comprise 147, 27, and 13 genes in the P. falciparum genome, respectively. Like Pfmc-2tm and Pfst-2tm, stevor, sep/etramps, and rif genes are located predominantly within subtelomeric regions of chromosomes and encode basic proteins. RIFINs have been detected at the erythrocyte surface (Kyes et al. 1999), STEVORs (Kaviratne et al. 2002) are Maurer's cleft transmembrane proteins, and SEP/ETRAMPs have been localized to the parasitophorous vacuole membrane (PVM; Spielmann et al. 2003) and/or the MCs (Birago et al. 2003).
Although no direct homology can be inferred on the basis of the sequence comparisons, structural alignments highlight a clear relationship between RIFINs, STEVORs, PfMC-2TMs, PfST-2TMs, and PyST-B-2TMs (Fig. 3). All display the following consensus structural characteristics. An N-terminal weakly hydrophobic region potentially corresponds to a secretory signal sequence. Whether the N-terminal leader sequence is cleaved off remains to be ascertained, as the SignalP algorithm could not predict cleavage sites for all of the proteins investigated. No peptide covering the signal-sequence region was detected in our proteomic analysis of the PfMC-2TM family (Fig. 2), hinting toward a processed amino terminus in the mature protein. As reported for STEVORs and RIFINs proteins (Cheng et al. 1998), conserved pairs of cysteine residues can be found along the sequences of the PfMC-2TM, PfST-2TM, and PyST-B-2TM (Fig. 2; Supplemental Fig. S1). When a cysteine position was not conserved across all family members, it was most often replaced by tyrosine or tryptophan residues (one nucleotide difference from a cysteine codon), which might be indicative of sequencing errors. These cysteine residues could be involved in the folding of the large N-terminal intracellular soluble domain via disulfide bridges. The third feature of the superfamily is a lysine-rich C-terminal tail after the second transmembrane segment.
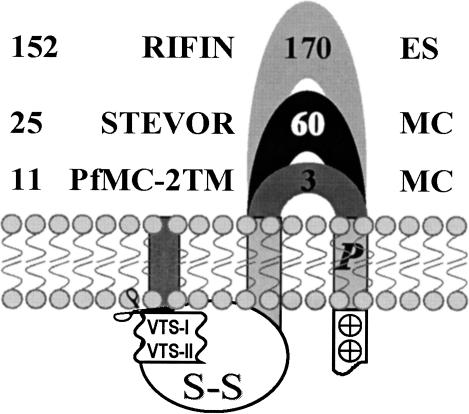
Figure 3.
Schematic representation of the Plasmodium falciparum 2-transmembrane proteins encoded by subtelomeric multicopy gene families. The number of genes and known subcellular localizations (ES, erythrocyte surface and MC, Maurer's Cleft), as well as the average number of amino acid residues in the extracellular loop are reported for the RIFIN, STEVOR, and PfMC-2TM families.
However, two major structural differences between these protein families are to be highlighted (Fig. 3). First, whereas the two transmembrane domains are separated by up to 60 and 170 residues in STEVORs and RIFINs, respectively, only three residues, on average, link the two helices in PfMC-2TM, PfST-2TM, and PyST-B-2TM. In all cases, this loop is predicted to be the only extracellular domain of the proteins and where most of the sequence polymorphism occurs. Such an extremely short extracellular loop should exclude the PfMC-2TM, PfST-2TM, and PyST-B-2TM proteins from being involved in antigenic variation, especially as we have shown that almost all the members of PfMC-2TM were expressed at the same time. Second, a notable feature of the PfMC-2TM, PfST-2TM, and PyST-B-2TM proteins is the presence of conserved proline residues in at least one of their predicted transmembrane domains (Figs. 2 and 3; Supplemental Fig. S1). Proline residues internal to helices are commonly found in many transporters, channels, and receptors, and are often conserved between homologous proteins (Sansom 1992). The resulting hinged/kinked molecular conformations (Sansom and Weinstein 2000) have been shown to be involved in the gating mechanism of ion channels (Bright et al. 2002), in the conformational flexibility of transporters (Tamori et al. 1994), and in the signal transduction within G protein-coupled receptors (Fernandez and Puett 1996).
Recent analyses of two parasite histidine-rich proteins (HRPs) has highlighted the existence of cooperative domains defining a malarial membrane transport/translocation signal (Lopez-Estrano et al. 2003). In addition to a canonical N-terminal signal sequence, which is sufficient to deliver these proteins to the PV, PfHRPI and PfHRPII contain a bipartite vacuolar translocation signal (VTS) necessary to transport them to the cytoplasmic face of MCs. A first domain of ∼30 amino acid residues immediately follows the signal sequence and is particularly rich in asparagines, whereas the second domain is embedded within the histidine-rich region of these proteins. Minimal histidine- or arginine-containing sequences can substitute for domain II, and are sufficient for export to the MCs, arguing the need for positively charged residues in the VTS. Interestingly, asparagine-rich domains following the signal peptide and stretches of consecutive basic residues (mostly lysines) can be found within the sequences of PfSTEVOR, PfMC-2TM, (Fig. 2), PfST-2TM, and PyST-B-2TM (Supplemental Figure S1). The length and position of each domain varies depending on the protein family considered. The presence of such VTS domains in PfRIFINS and PfSEP/ETRAMPs is less clear; in particular, domain I seems to be missing in some variants or could be comprised of other residues (glutamine, aspartate, glutamate). The VTS defined by Lopez-Estrano et al. (2003) could therefore be involved in targeting of soluble proteins such as the HRPs, as well as integral membrane proteins, suggesting shared transport machinery.
A Role in Protein Trafficking?
The definite biological function of the PfMC-2TM proteins remains to be determined, and our data does not show whether the SP1C1 mAb exerts any influence on the assembly or expression of malaria antigens on the surface of infected erythrocytes or of secreted proteins. However, the observations from IFA staining and Western blots confirm the tight window of expression observed in the microarray analysis, and suggest that the proteins may have a role in this time frame relevant for translocation/trafficking of parasite proteins. The most intense IFA staining using the SP1C1 mAb was observed in trophozoites/early schizonts, and intense staining could be detected through mature trophozoites/schizonts (1–8 nuclei; ∼12–36 h post infection). As full segmentation occurred (>8 nuclei), antibody staining was highly reduced (some showed no staining), and colocalization with lipid staining was lost (Sam-Yellowe et al. 2001). The intramembranous network appears to be fully functional, beginning in the ring stage and continuing to the mature schizont stages (∼36 h post infection) when most of the macromolecular transport in the parasite occurs. This transport appears to peak at schizogony with the initial formation of the merozoites, due to a sharp increase in the demand for membrane formation (Kilejian 1980; Pouvelle et al. 1994; Haldar 1998). We are currently investigating the distribution of the PfMC-2TM proteins along with other Maurer's cleft-specific proteins to determine their localization and/or pattern of translocation through the intracellular membranous network.
METHODS
Coimmunoprecipitation
Monoclonal antibodies from SP1C1 clones were pooled and complexed with goat anti-mouse sepharose beads (ICN Cappel) and the bead-antibody complex incubated overnight. The beads were centrifuged for 5 min at 18,000 g. Unbound antibodies were removed and the beads were washed by centrifugation 3× in buffer A (1× PBS, 1% NP40, 1 mM EDTA, 1% BSA). Following the last wash, P. falciparum schizont extracts prepared as described (Sam-Yellowe et al. 2001) were added to the beads. The beads were resuspended gently and incubated overnight at 4°C. Following incubation, centrifugation, and removal of unbound proteins, the beads were washed twice in buffer A, twice in buffer B (Buffer A + 0.5 M NaCl), twice in buffer C (1× PBS, 1 mM EDTA, 1% NP40), and once in buffer D (1× PBS, 1 mM EDTA). Bound immune complexes were eluted with 0.3–0.4 mL of 0.1 M glycine (pH 2.8). After centrifugation, the eluate was collected and transferred to a fresh tube and immediately neutralized with 2 M Tris-HCl (pH 8.0). The elution was performed twice and the eluates pooled. The beads were washed twice with buffer D and the wash supernatant removed. A differential elution was carried out by adding 0.1 mL of 10% 1,4-dioxane to beads. The beads were centrifuged and supernatants collected. The Dioxane supernatants were placed in a Speed Vac evaporator (Savant) to dry the samples. The glycine eluates were used to resuspend the Dioxaneeluted proteins. A final centrifugation was performed to pellet any remaining beads and the supernatant transferred to a fresh tube. For each mAb, two coimmunoprecipiation experiments were analyzed independently.
Protein Digestion
The method follows that of Washburn et al (2001) with modifications. Proteins were concentrated using a TCA (trichloroacetic acid) precipitation. Solutions were brought to 400 μL with Tris-HCl (pH 8.5), and TCA was added to 20%. Mixtures were incubated overnight on ice, then centrifuged for 30 min, and supernatants removed. Pellets were washed two times with acetone, placed in an evaporator for 5 min, and resuspended in 30 μL of 100 mM Tris-HCl (pH 8.5). TCA-precipitated proteins were resuspended in 30 μL 0.1 M Tris-HCl (pH 8.5). Solid urea was added to 8 M. After reduction [5 mM Tris(2-carboxyethyl)phosphine hydrochloride, TCEP, Roche] and alkylation (20 mM iodoacetamide, IAM, Sigma), Endoproteinase Lys-C (Roche) was added to a 1: 100 enzyme:substrate ratio, overnight at 37°C. After 4× dilution with 100 mM Tris-HCl (pH 8.5) and addition of CaCl2 to 2 mM, proteins were further digested using modified Trypsin (Roche), 1: 100 enzyme:substrate ratio, at 37°C, overnight.
Multidimensional Chromatography
Peptide mixtures were pressure loaded onto a 100-μm inner diameter, fused-silica column packed first with from 8 to 9 cm of 5-μm C18 reverse phase particles (Polaris 2000; Metachem Technologies), followed by 4–5 cm of 5μm-strong cation exchange material (Partisphere SCX, Whatman). Loaded microcapillary columns were installed inline with a Quaternary Agilent 1100 series HPLC pump. An overflow tubing was used to decrease the flow rate to ∼200–300 nL/min. The application of a 2.4-V distal voltage electrosprayed the eluting peptides directly into an LCQ-Deca ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (ThermoFinnigan). Three different elution buffers were used as follows: 5% ACN, 0.1% formic acid (Buffer A), 80% ACN, 0.1% formic acid (Buffer B), and 500 mM ammonium acetate, 5% ACN, 0.1% formic acid (Buffer C). Fully automated 6-step chromatography runs were carried out. In such sequences of chromatographic events, peptides were sequentially eluted from the SCX resin to the RP resin by increasing salt steps (increase in Buffer C concentration), followed by organic gradients (increase in Buffer B concentration). The last chromatography step consists of a high-salt wash with 100% Buffer C, followed by the acetonitrile gradient. Full MS spectra were recorded on the peptides over a 400–1600 m/z range, followed by three tandem mass (MS/MS) events sequentially generated in a data-dependent manner on the first, second, and third most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan).
Interpretation of MS/MS Data Sets
PEP_PROBE (Sadygov and Yates III 2003) was used to match MS/MS spectra to peptides in a database containing 5370 P. falciparum sequences (Gardner et al. 2002), combined with 31,853 human, mouse, and rat protein sequences (from RefSeq NCBI). PEP_ PROBE is a modified version of SEQUEST (Eng et al. 1994) and uses a hypergeometric probability model to calculate the confidence for a match to be nonrandom. The validity of peptide/spectrum matches was therefore assessed using the SEQUEST-defined parameters, cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCn), as well as the PEP_PROBE-defined parameters, probability, and confidence for a match to be nonrandom. As specified in the table, spectra/peptide matches were only retained if they had a DeltaCn of at least 0.08 and minimum XCorr of 1.8 for +1, 2.5 for +2, and 3.5 for +3 spectra. An 85% confidence for the peptide/spectrum matches not to be random (as defined by PEP_PROBE) was used as cut-off. In addition, the minimum sequence length was 7 amino acid residues. DTASelect (Tabb et al. 2002) was used to select and sort peptide/spectrum matches passing this criteria set. Peptide hits from multiple runs were compared using CONTRAST (Tabb et al. 2002). Proteins were considered detected in the immune complexes if they were identified by at least two peptides passing all of the selection criteria (or one peptide appearing in at least two independent runs) and were not detected in the control run (no specific mAb added to the beads).
Acknowledgments
We thank LaShonda Everett for excellent technical assistance. This study was funded by NIH grant AI36470 and Cleveland State University's EFFRD award to T.S.Y. J.R.Y. acknowledges the support of the Office of the Naval Research (Cooperative Agreement N00014-01-2-0003), the US Army Medical Research and Material Command, and the National Institutes of Health. E.A.W. is funded by the Ellison Medical Research Foundation. The opinions expressed are those of the authors and do not reflect the official policy of the Department of the Navy, Department of Defense, or the U.S. government.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2126104. Article published online before print in May 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adisa, A., Albano, F.R., Reeder, J., Foley, M., and Tilley, L. 2001. Evidence for a role for a Plasmodium falciparum homologue of Sec31p in the export of proteins to the surface of malaria parasite-infected erythrocytes. J. Cell. Sci. 114: 3377–3386. [DOI] [PubMed] [Google Scholar]
- Albano, F.R., Berman, A., La Greca, N., Hibbs, A.R., Wickham, M., Foley, M., and Tilley, L. 1999. A homologue of Sar1p localises to a novel trafficking pathway in malaria-infected erythrocytes. Eur. J. Cell. Biol. 78: 453–462. [DOI] [PubMed] [Google Scholar]
- Bahl, A., Brunk, B., Coppel, R.L., Crabtree, J., Diskin, S.J., Fraunholz, M.J., Grant, G.R., Gupta, D., Huestis, R.L., Kissinger, J.C., et al. 2002. PlasmoDB: The Plasmodium genome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished). Nucleic Acids Res. 30: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell, J.W. 1990. Vesicle-mediated transport of membrane and proteins in malaria-infected erythrocytes. Blood Cells 16: 379–395. [PubMed] [Google Scholar]
- Beeson, J.G. and Brown, G.V. 2002. Pathogenesis of Plasmodium falciparum malaria: The roles of parasite adhesion and antigenic variation. Cell. Mol. Life Sci. 59: 258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birago, C., Albanesi, V., Silvestrini, F., Picci, L., Pizzi, E., Alano, P., Pace, T., and Ponzi, M. 2003. A gene-family encoding small exported proteins is conserved across Plasmodium genus. Mol. Biochem. Parasitol. 126: 209–218. [DOI] [PubMed] [Google Scholar]
- Bright, J.N., Shrivastava, I.H., Cordes, F.S., and Sansom, M.S. 2002. Conformational dynamics of helix S6 from Shaker potassium channel: Simulation studies. Biopolymers 64: 303–313. [DOI] [PubMed] [Google Scholar]
- Carlton, J.M., Angiuoli, S.V., Suh, B.B., Kooij, T.W., Pertea, M., Silva, J.C., Ermolaeva, M.D., Allen, J.E., Selengut, J.D., Koo, H.L., et al. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419: 512–519. [DOI] [PubMed] [Google Scholar]
- Cheng, Q., Cloonan, N., Fischer, K., Thompson, J., Waine, G., Lanzer, M., and Saul, A. 1998. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 97: 161–176. [DOI] [PubMed] [Google Scholar]
- Craig, A. and Scherf, A. 2001. Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion. Mol. Biochem. Parasitol. 115: 129–143. [DOI] [PubMed] [Google Scholar]
- Eng, J.K., McCormack, A.L., and Yates III, J.R. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Fernandez, L.M. and Puett, D. 1996. Identification of amino acid residues in transmembrane helices VI and VII of the lutropin/choriogonadotropin receptor involved in signaling. Biochemistry 35: 3986–3993. [DOI] [PubMed] [Google Scholar]
- Florens, L., Washburn, M.P., Raine, J.D., Anthony, R.M., Grainger, M., Haynes, J.D., Moch, J.K., Muster, N., Sacci, J.B., Tabb, D.L., et al. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419: 520–526. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R.W., Carlton, J.M., Pain, A., Nelson, K.E., Bowman, S., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar, K. 1998. Intracellular trafficking in Plasmodium-infected erythrocytes. Curr. Opin. Microbiol. 1: 466–471. [DOI] [PubMed] [Google Scholar]
- Heddini, A., Pettersson, F., Kai, O., Shafi, J., Obiero, J., Chen, Q., Barragan, A., Wahlgren, M., and Marsh, K. 2001. Fresh isolates from children with severe Plasmodium falciparum malaria bind to multiple receptors. Infect. Immun. 69: 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterberg, K., Scherf, A., Gysin, J., Toyoshima, T., Aikawa, M., Mazie, J.C., da Silva, L.P., and Mattei, D. 1994. Plasmodium falciparum: The Pf332 antigen is secreted from the parasite by a brefeldin A-dependent pathway and is translocated to the erythrocyte membrane via the Maurer's clefts. Exp. Parasitol. 79: 279–291. [DOI] [PubMed] [Google Scholar]
- Janssen, C.S., Barrett, M.P., Turner, C.M., and Phillips, R.S. 2002. A large gene family for putative variant antigens shared by human and rodent malaria parasites. Proc. R. Soc. London B Biol. Sci. 269: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaviratne, M., Khan, S.M., Jarra, W., and Preiser, P.R. 2002. Small variant STEVOR antigen is uniquely located within Maurer's clefts in Plasmodium falciparum-infected red blood cells. Eukaryot. Cell 1: 926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilejian, A. 1980. Stage-specific proteins and glycoproteins of Plasmodium falciparum: Identification of antigens unique to schizonts and merozoites. Proc. Natl. Acad. Sci. 77: 3695–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kyes, S.A., Rowe, J.A., Kriek, N., and Newbold, C.I. 1999. Rifins: A second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. 96: 9333–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch, K.G., Zhou, Y., Blair, P.L., Grainger, M., Moch, J.K., Haynes, J.D., De la Vega, P., Holder, A.A., Batalov, S., Carucci, D.J., et al. 2003a. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 108: 7025. [DOI] [PubMed] [Google Scholar]
- Lopez-Estrano, C., Bhattacharjee, S., Harrison, T., and Haldar, K. 2003. Cooperative domains define a unique host cell-targeting signal in Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. 100: 12402–12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, L.H., Baruch, D.I., Marsh, K., and Doumbo, O.K. 2002. The pathogenic basis of malaria. Nature 415: 673–679. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Pouvelle, B., Gormley, J.A., and Taraschi, T.F. 1994. Characterization of trafficking pathways and membrane genesis in malaria-infected erythrocytes. Mol. Biochem. Parasitol. 66: 83–96. [DOI] [PubMed] [Google Scholar]
- Przyborski, J.M., Wickert, H., Krohne, G., and Lanzer, M. 2003. Maurer's clefts—A novel secretory organelle? Mol. Biochem. Parasitol. 132: 17–26. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.A., and Barrell, B. 2000. Artemis: Sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Sadygov, R.G. and Yates III, J.R. 2003. A hypergeometric probability model for protein identification and validation using tandem mass spectral data and protein sequence. Analyt. Chem. 75: 3792–3798. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe, T.Y., Fujioka, H., Aikawa, M., Hall, T., and Drazba, J.A. 2001. A Plasmodium falciparum protein located in Maurer's clefts underneath knobs and protein localization in association with Rhop-3 and SERA in the intracellular network of infected erythrocytes. Parasitol. Res. 87: 173–185. [DOI] [PubMed] [Google Scholar]
- Sansom, M.S. 1992. Proline residues in transmembrane helices of channel and transport proteins: A molecular modelling study. Protein Eng. 5: 53–60. [DOI] [PubMed] [Google Scholar]
- Sansom, M.S. and Weinstein, H. 2000. Hinges, swivels and switches: The role of prolines in signalling via transmembrane α-helices. Trends Pharmacol. Sci. 21: 445–451. [DOI] [PubMed] [Google Scholar]
- Spielmann, T., Fergusen, D.J., and Beck, H.P. 2003. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol. Biol. Cell 14: 1529–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb, D.L., McDonald, W.H., and Yates III, J.R. 2002. DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1: 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamori, Y., Hashiramoto, M., Clark, A.E., Mori, H., Muraoka, A., Kadowaki, T., Holman, G.D., and Kasuga, M. 1994. Substitution at Pro385 of GLUT1 perturbs the glucose transport function by reducing conformational flexibility. J. Biol. Chem. 269: 2982–2986. [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeinya, I.J., Schmidt, J.A., Aikawa, M., Miller, L.H., and Green, I. 1981. Falciparum malaria-infected erythrocytes specifically bind to cultured human endothelial cells. Science 213: 555–557. [DOI] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates III, J.R. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19: 242–247. [DOI] [PubMed] [Google Scholar]
- Wickert, H., Rohrbach, P., Scherer, S.J., Krohne, G., and Lanzer, M. 2003a. A putative Sec23 homologue of Plasmodium falciparum is located in Maurer's clefts. Mol. Biochem. Parasitol. 129: 209–213. [DOI] [PubMed] [Google Scholar]
- Wickert, H., Wissing, F., Andrews, K.T., Stich, A., Krohne, G., and Lanzer, M. 2003b. Evidence for trafficking of PfEMP1 to the surface of P. falciparum-infected erythrocytes via a complex membrane network. Eur. J. Cell. Biol. 82: 271–284. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.tigr.org/tdb/e2k1/pya1/pya1-telo.shtml; The TIGR database listing Plasmodium yoelii Telometric and Subtelometric Contigs.