Summary
A hallmark of retroviral replication is integration of the viral genome in the host cell DNA. This characteristic makes retrovirus-based vectors attractive delivery vehicles for gene therapy. However, adverse events in gene therapeutic trials, caused by activation of proto-oncogenes due to Murine Leukemia Virus (MLV)-derived vector integration, hamper their application. Here we show that bromodomain and extraterminal (BET) proteins (BRD2, BRD3 and BRD4) and MLV integrase specifically interact and co-localize within the nucleus of the cell. Inhibition of the BET proteins chromatin interaction via specific bromodomain inhibitors blocks MLV virus replication at the integration step. MLV integration site distribution parallels the chromatin binding profile of BET proteins, and expression of an artificial fusion protein of the BET integrase binding domain with the chromatin interaction domain of the lentiviral targeting factor LEDGF/p75, retargets MLV integration away from TSS and into the body of actively transcribed genes, conform to the Human Immunodeficiency Virus (HIV) integration pattern. Together these data validate BET proteins as MLV integration targeting factors.
Introduction
Integration of a DNA copy of the retroviral RNA genome into the host chromatin is a pivotal step in retroviral replication and links the fate of the invading virus with that of the infected cell. This characteristic makes retrovirus-based vectors suitable to deliver therapeutic genes into cells to correct genetic diseases. MLV-derived vectors have been used successfully to correct primary immunodeficiency disorders like X-linked severe combined immunodeficiency (SCID-X1) (Cavazzana-Calvo et al., 2000; Gaspar et al., 2004; Hacein-Bey-Abina et al., 2002). However, their use led to adverse events in a subset of patients due to LTR driven activation of proto-oncogenes (i.e. insertional mutagenesis) resulting in uncontrolled clonal cell proliferation and leukemia. Retroviral integration site distribution is not random. Whereas the host protein lens epithelium-derived growth factor (LEDGF/p75) targets lentiviral (e.g. HIV) integration towards the body of active transcription units (Ciuffi et al., 2005), gammaretroviral (e.g. MLV) integration is independent of LEDGF/p75 and preferentially occurs near transcription start sites (TSSs), CpG islands and DNaseI hypersensitive sites (DHS) (Cattoglio et al., 2010; Felice et al., 2009; Mitchell et al., 2004; Wu et al., 2003). In addition, retroviral integration is favored on the outward-facing major groove of nucleosome-wrapped DNA (Roth et al., 2011; Wang et al., 2007). It is generally accepted that cellular proteins, co-factors, dictate target site selection. In this study, we identified the cellular determinants that target MLV integration. In earlier work with hybrid HIV viruses it was shown that transferring the MLV integrase (IN) coding region into HIV caused the chimeras to integrate with a specificity close to that of MLV, revealing IN as the principal viral determinant of integration specificity (Lewinski et al., 2006). Therefore we screened for cellular MLV IN interaction partners that could act as MLV-specific tether. While an earlier study picked up BRD2 as a MLV IN-interacting protein (Studamire and Goff, 2008), we show here that members of the bromodomain and extraterminal domain containing (BET) family of proteins (BRD2, BRD3 and BRD4) interact with MLV IN and orchestrate gammaretroviral integration, in agreement with a recent report by Sharma et al. (Sharma et al., 2013) and that engineered BET proteins can retarget MLV replication.
Results
BET proteins bind MLV integrase
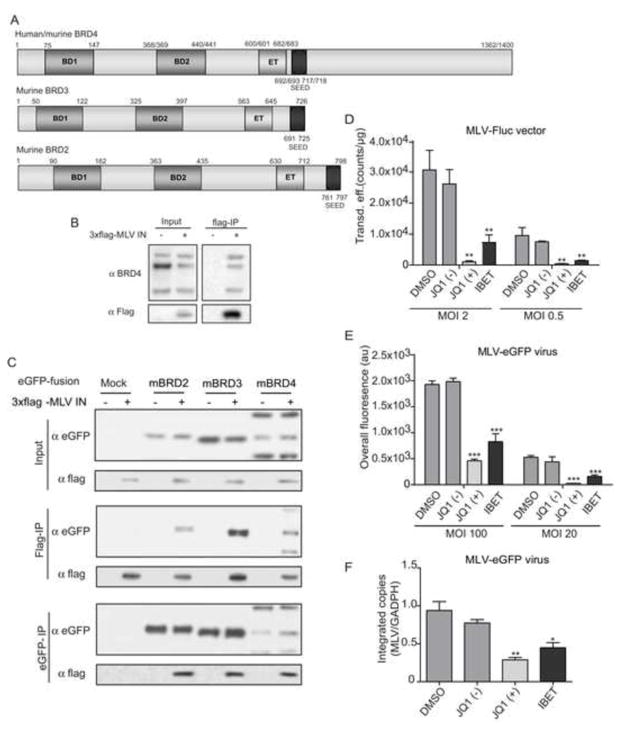
We singled out MLV integrase (IN)-interacting proteins from 293T cell extracts expressing triple flag-tagged MLV IN via co-immunoprecipitation (co-IP) of IN using flag-affinity matrix. Eluted proteins were identified by mass spectrometry (MS). Wild-type 293T cells were analyzed in parallel as control. Bromodomain containing protein 4 (BRD4) was represented with the largest set of peptides (data not shown), but we also identified BRD3. Of note, an earlier study picked up BRD2 as MLV IN-interacting protein (Studamire and Goff, 2008). All these proteins are members of the bromodomain and extraterminal (BET) protein family (Figure 1A) and share two chromatin-interacting bromodomains, recognizing acetylated histone tails, and a protein-interacting extraterminal (ET) domain (for a review see (Devaiah and Singer, 2013)). To confirm the interaction of endogenous BRD4 with MLV IN, transiently expressed flag-tagged MLV IN was immunoprecipitated from 293T nuclear extracts. BRD4 was readily detected in the pull-down fraction by Western blot (Figure 1B). In addition, 3xflag MLV IN interacted with both human and mouse BRD4 (hBRD4 and mBRD4) (Co-IP, data not shown). A similar co-IP experiment in cells transiently expressing eGFP-tagged BET proteins and flag-tagged MLV IN showed that next to eGFP-BRD4, MLV IN interacts with eGFP-tagged mBRD2 and mBRD3 as well (Figure 1C). When expressed alone, eGFP-tagged BET proteins located to the nucleus of NIH3T3 cells, while MLV IN fused to the monomeric red fluorescent protein (mRFP-MLV IN) predominantly located to the cytoplasm with only trace amounts in the nucleus (Figure S1A). However, co-expression of either BRD2, 3 or 4 with mRFP-MLV IN relocated MLV IN to the nucleus of the cell, co-localizing with the respective BET proteins (Figure S1A and B). Similar data were obtained in HeLa cells (data not shown).
Figure 1. BET proteins interact with MLV IN and are important for viral integration.
A) Schematic representation of human/murine BET proteins. Numbers correspond to aa positions. BD = bromodomain; ET = extraterminal domain; SEED = serine, glutamic acid, aspartic acid-rich domain. B) Co-immunoprecipitation of endogenous BRD4 with flag-tagged MLV IN analyzed by Western blot. C) 293T cells were cotransfected with flag-tagged MLV IN and eGFP, eGFP-mBRD2, eGFP-mBRD3 or eGFP-mBRD4 expression constructs. MLV IN (Flag-IP) or mBRD proteins (eGFP-IP) were precipitated and analyzed by Western blot. D-E) NIH3T3 cells were transduced with MLV-derived vectors (MLV-Fluc) (D) or infected with an eGFP expressing viral clone (MLV-eGFP virus) (E) in the presence of 200 nM JQ1(−) or JQ1(+), 500 nM I-BET or an equivalent amount of DMSO as a negative control. Two multiplicities of infection (MOI) are presented. 24 hours post transduction/infection, cells were washed and in case of the viral clone, raltegravir (1 μM) was added to prevent multiple-round replication. 24 hours later transduction or infection efficiency was determined. Data are plotted as average ± standard deviation of triplicate measurements. F) NIH3T3 cells were infected with MLV-eGFP as in panel E. Subsequently, cells were expanded and split until 10 days post infection. The number of integrated copies was determined via qPCR and normalized to GADPH. Average values and standard deviations of a triplicate measurement are shown. In all panels, differences were determined using a student’s T-test. * p < 0.05, ** p < 0.01, *** p < 0.001.
Bromodomain inhibitors reduce MLV replication
To evaluate the role of BET proteins in MLV replication, we exploited the recently identified BET protein bromodomain inhibitors JQ1 (+) and I-BET (Filippakopoulos et al., 2010; Nicodeme et al., 2010). JQ1 (−) (the inactive R-enantiomer) and DMSO served as negative controls. Based on 50% cytotoxicity (CC50) and inhibitory concentrations (IC50) (Supplementary Table 1) we used JQ1 (+) and I-BET concentrations of 200 nM and 500 nM, respectively. NIH3T3 cells were either transduced with a retroviral vector encoding firefly luciferase (MLV-Fluc vector) (Figure 1D), or infected with a viral clone expressing eGFP (MLV-eGFP virus) (Figure 1E). Both vector transduction and virus infection were inhibited 5- to 10-fold compared to JQ1 (−) or DMSO control, respectively, whereas transduction with an HIV-derived vector (HIV-Fluc) was not inhibited (Figure. S1C). Similar results were obtained in HeLa cells using the MLV-Fluc vector (data not shown). Reporter gene expression following transfection of the MLV-eGFP molecular clone was not affected by the presence of JQ1 (+) or I-BET excluding a transcriptional effects (Figure S1D). To determine the step in the viral replication cycle where bromodomain inhibitors inhibit MLV replication, viral DNA intermediates were quantified via Q-PCR. Quantification of the integrated proviral copies at 10 days post transduction/infection in the presence of BET inhibitors revealed that the integrated MLV (virus or vector) copies were reduced 2- to 3-fold, while HIV-Fluc integration was not inhibited (Figure 1F and Figures S1E, S1F, for MLV-eGFP virus, MLV-Fluc vector and HIV-Fluc, respectively). Since the amount of total DNA at early time points after infection, which is a measure of reverse transcription, was not reduced in the presence of BET inhibitors (Figure S1G), we conclude that BET proteins act at a step between reverse transcription and integration.
The MLV IN C-terminus binds the BET extraterminal domain
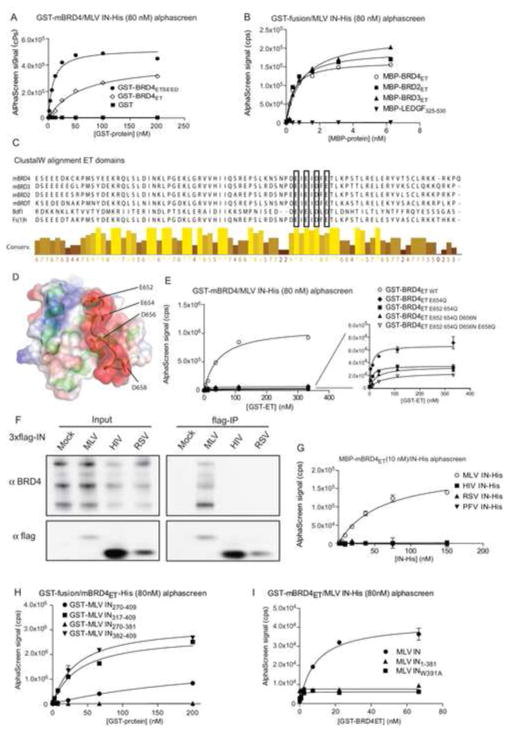
Pull-down experiments using a panel of eGFP-tagged mBRD4 truncation mutants pinpointed the ET domain (BRD4ET, aa 601–685) as the minimal IN binding domain (Figures S2A,B). Confocal microscopy experiments corroborated that BRD4ET is the minimal domain required for co-localization with mRFP-MLV IN (Figure S2C and data not shown). To confirm a direct interaction, recombinant Glutathione S-transferase (GST)-tagged BRD4ET and BRD4ETSEED (aa 601–721) and His6-tagged MLV IN were shown to interact in an AlphaScreen protein-protein interaction assay (apparent Kd of 8.56±1.55 nM and 58.70±8.05 nM, respectively) (Figure 2A). As expected recombinant BRD2ET and BRD3ET interacted with MLV IN as well, with Kd values in the same range as BRD4ET (Figure 2B). Considering the conservation in the ET domain among BET proteins (Figure 2C) and the BRD4ET NMR structure (Lin et al., 2008), we introduced E652Q, E654Q, D656N and E658Q mutations in the BRD4 ET-domain (Figure 2D). Maximal loss of binding was obtained with the quadruple mutant (referred to as BRD4ETmut in further experiments) (Figure 2E). The observed interaction with BET-proteins was specific for MLV IN, as IN proteins from other retroviral families did not interact with BET proteins as revealed by co-IP of lysates of 293T cell transiently expressing flag-tagged MLV, HIV or RSV (Rous Sarcoma Virus) IN (Figure 2F). Similar data were obtained in an AlphaScreen assay measuring the interaction of MBP-tagged BRD4ET with His-tagged MLV, HIV, RSV or PFV (Prototype Foamy Virus) IN (Figure 2G). To pinpoint the MLV IN-BET interacting domain different flag-tagged MLV IN deletion constructs were generated, transiently expressed in 293T cells, and tested in Co-IP (Figure S2D). Only MLV IN fragments containing the C-terminal domain (MLV IN207-409) pulled down endogenous BRD4 from 293T cell lysates. Specific interaction in cells was corroborated by co-localization of the mRFP-MLV IN207-409 and eGFP-tagged mBRD4 using confocal microscopy (Figure S2E). AlphaScreen analysis using smaller truncation mutants of the MLV IN C-terminal domain revealed that the last 27 residues of MLV IN (aa 382–409) were sufficient to interact with BRD4ET (Figure 2H). Finally, alanine scanning of the latter domain revealed W391 to be critically important for the interaction (Figure 2I and data not shown). Similar results were obtained for BRD2ET and BRD3ET (data not shown). Taking into account that BET proteins are known to associate with promoter regions through their bromodomains (Leroy et al., 2012) and interact with MLV IN through their ET-domain in a gammaretrovirus-specific manner, we considered BET proteins as good candidate MLV targeting factors.
Figure 2. The BET protein ET domain interacts with the MLV-IN C-terminus.
A) Direct interaction of His6-tagged MLV IN (80 nM) with an increasing amount of GST-tagged mBRD4ET, GST-tagged mBRD4ETSEED or GST alone as measured by AlphaScreen. Apparent Kd’s of 8.56 ± 1.50 nM (ETSEED) and 58.70 ± 8.05 nM (ET) were determined using a non-linear regression curve fit for specific binding. B) Comparable affinity of different BET proteins for MLV IN. 40 nM His6-tagged MLV IN was titrated against the indicated MBP-fused ET domains. Apparent Kd’s of 0.60 ± 0.17, 0.97 ± 0.18 and 0.41 ± 0.03 nM were determined for MBP-BRD2, -3 and -4, respectively. MBP-LEDGF325-530 was used as a negative control. C) Sequence alignment (ClustalW) of the ET domain of mBRD4 (Q9ESU6, aa 601–683), mBRD3 (Q8K2F0, aa 563–645), mBRD2 (Q7JJ13, aa 630–712), mBRDT (Q91Y44, aa 496–578), Saccharomyces cerevisiae (Sc) Bromodomain factor (Bdf1) (P35817, aa 518–598) and Drosophila melanogaster (Dm) Female Sterile Homeotic (Fs(1)h) (P13709, aa942–1024). D) NMR structure of the ET domain of BRD4 (Lin et al., 2008). E) Interaction of His6-tagged MLV IN (80 nM) with GST-tagged mBRD4601-685 mutants as measured by AlphaScreen. F) Co-immunoprecipitation of endogenous BRD4 with flag-tagged MLV, HIV or RSV IN analyzed by Western blot. G) Interaction of 100 nM GST-tagged mBRD4ET with His6-tagged MLV, HIV, RSV or PFV IN measured by AlphaScreen. The Kd for MLV IN (56.7 ± 23.08 nM) was determined using a non-linear regression curve fit for specific binding. The other INs showed no binding to BRD4601-721. H) Interaction of 80 nM His-tagged mBRD4ET with increasing amounts of the GST-tagged MLV IN C-terminal domain (aa 270–409) and truncation mutants thereof as measured by AlphaScreen. I) Interaction of 80 nM His-tagged mBRD4ET with GST-tagged MLV IN, the indicated C-terminal MLV-IN deletion mutant (aa 1–381) or MLV IN with a W391A point mutation as measured by AlphaScreen. All AlphaScreen experiments were performed three times. Representative experiments are shown. Error bars indicate the standard deviations of triplicate data points.
MLV vectors integrate near BET protein hot spots
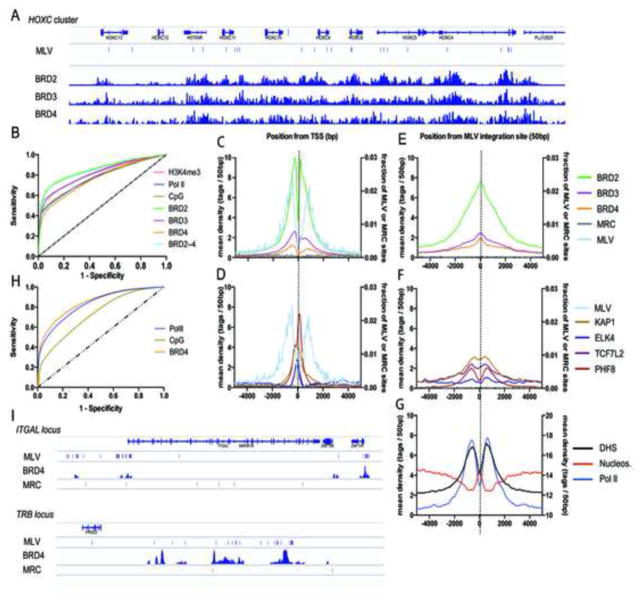
To correlate MLV integration with BET-chromatin binding sites, we determined 10,514 unique MLV-vector integration sites in 293T cells and computationally generated matched random control (MRC) sites. Integration sites were compared with the BRD2, -3 and -4 chromatin binding profile (Supplementary Table 2) (Leroy et al., 2012). HOX gene clusters are enriched for BRD2–4 bound nucleosomes (Leroy et al., 2012). Indeed, we detected 11, 10 and 23 MLV integration sites in the HOXA, HOXB (data not shown) and HOXC (Figure 3A) clusters, respectively, versus 1, 0 and 0 MRC sites. Genome-wide MLV integration was significantly (p<0.0001, Mann-Whitney U test) enriched in regions bound by either of the three BET proteins: 42.7, 23.5 and 15% of integration sites were situated in BRD2-, 3- or 4-islets, respectively, compared to 1.9, 0.9 and 0.6% of MRC (Supplementary Table 3). In addition, we observed stronger correlation with BET protein binding than with previously described markers associated with MLV integration such as Pol II binding, H3K4me3 or CpG islands (9.1, 6.9, 4.4 %, respectively) (Supplementary Table 3) (Cavazza et al., 2013; Santoni et al., 2010). BRD2 proved to be the best predictor for MLV integration (Figure 3B), with over 50% of MLV integration sites locating within 149 bp of a BRD2 binding site (Supplementary Table 3). Moreover, both MLV integration sites and BRD2–4 ChIP-Seq tags concentrate around RefGene TSSs with a similar bimodal distribution (Figure 3C) (Cattoglio et al., 2010), which differs from the pattern of other transcription factors defined in 293T cells (KAP1, ELK4, TCF7L2 and PHF8; Supplementary Table 2 and Figure 3D). Analysis of the distribution of BRD2–4 around MLV integration sites revealed that the BRD2–4 occupancy is highest at the integration site itself (Figure 3E), while the maximal tag density for other transcription factors is adjacent to the integration site (Figure 3F). In addition, we observed a clear peak of nucleosome occupancy at the site of integration (Figure 3G), supporting the notion that MLV preferentially integrates into nucleosomal DNA in vivo (Roth et al., 2011). Indeed, open chromatin (Kundaje et al., 2012; Natarajan et al., 2012) and RNA Pol II are favored at either side of the integration site, in accordance with the preference for nucleosomal targeting (Figure 3G). Similar results were obtained when MLV integration sites were compared to ChIP-Seq data for BRD4 in human primary CD4+ T-cells (Roth et al., 2011; Zhang et al., 2012). 18.8% of MLV integrations were located to BRD4 islands compared to 0.7% of MRCs, 17.0% of Pol II peaks and 2.3% CpG islands (Supplementary Table 3). Receiver-operator characteristic (ROC) analysis confirmed that MLV integration sites correlate best with the BRD4 chromatin binding profile (Figure 3H and Supplementary Table 3). Analysis of CD4+ T-cell specific loci that are active and bound by BRD4, such as the ITGAL and the TRB locus, underscored the link between BET proteins (BRD4) and MLV integration (Figure 3I). The ITGAL locus (16p11.2) for instance encodes integrin alpha-L (CD11A), contains 4 BRD4 islands all of which are associated with MLV integration sites (MLV n=52, MRC n=6). A similar pattern was observed at the 3′ end of the T-cell receptor beta locus (TRB, location 7q34) in the joining and constant segment coding region (MLV n=33, MRC n=2).
Figure 3. MLV vectors integrate in BET protein hot spots.
A) Schematic representation of the BET chromatin binding profile (Leroy et al., 2012) and MLV integration sites (MLV) in the HOXC cluster in 293T cells. Matched random control sites (MRC) were absent in this region. B) Receiver-operator characteristic (ROC) analysis of BRD2–4, Pol II, H3K4me3 and CpG islands (Berry et al., 2006). The area under the curve (AUC) is calculated for the different markers and shown in Supplementary Table 3. (C–G) Mean background-subtracted sequencing read density in 50 bp bins in a 10 kb window around (C, D) transcription start sites (TSSs) or (E–G) MLV integration sites for (C, E) BRD2–4 (ChIP-Seq), (D, F) the unrelated transcription factors PHF8, ELK4, KAP1 and TCF7L2 (ChIP-Seq) or (G) open chromatin (DNase I Hypersensitivity, DHS), Pol II (ChIP-Seq) and nucleosome positions (Micrococcal Nuclease-Seq, MN-Seq) plotted on the right y-axis. In (C–F) the number of MLV and MRC sites is plotted on the right y-axis as a fraction of the total number of respective sites. (H) ROC analysis of BRD4, Pol II and CpG islands in human primary CD4+ T-cells. Corresponding AUC values are given in Supplementary Table 3. (I) Schematic representation of the ITGAL (Integrin alpha-L) locus and the 3′ end of the TRB (T-cell receptor beta) locus highlighting MLV integration sites, BRD4 peaks and MRC sites.
BRD4 hybrids retarget MLV integration
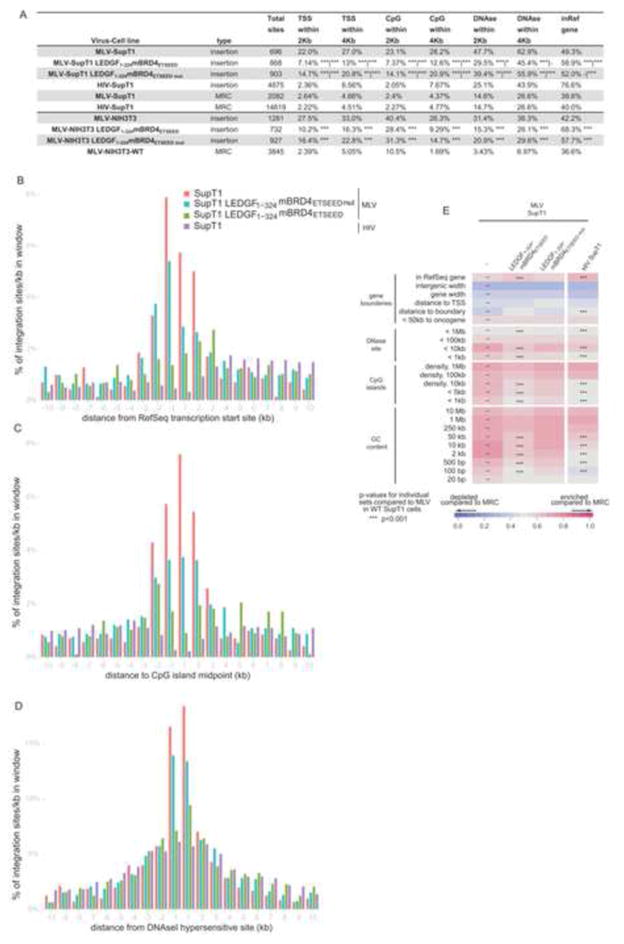
Taken together, our data are consistent with a role of BET proteins in gammaretroviral integration site targeting. If this is the case, fusions of the MLV IN-interacting domain with another chromatin binding domain should redirect integration away from the MLV-like pattern. To unambiguously prove this hypothesis we generated NIH3T3 and SupT1 cell lines stably expressing a chimeric fusion protein linking mBRD4ETSEED with the chromatin binding domain of the lentiviral targeting factor, LEDGF/p75 (aa 1–324) (LEDGF1-324mBRD4ETSEED). As a control, we also generated cell lines expressing the quadruple interaction-defective mutant (LEDGF1-324mBRD4ETSEEDmut). Protein expression, verified by Western blotting, did not affect cell growth (data not shown). Following transduction with a MLV-based vector, we amplified MLV integration sites and analyzed their distribution. In line with an earlier report, MLV integration in wild-type SupT1 cells was enriched within a 2 kb window near TSSs (22.0%), CpG islands (23.1%) and DNAse hypersensitive sites (DHS) (47.7%) (p<0.001 compared to MRC) (Figure 4A). Expression of LEDGF1-324mBRD4ETSEED shifted integration away from these features and towards RefSeq genes (from 49.3% to 58.9%) while an intermediate phenotype was observed for the interaction-deficient mutant. Even more pronounced results were obtained in NIH3T3 cells (Figure 4A). When integration sites were binned based on their distance to TSSs and CpG or DHS island midpoints (Figure 4B,C,D, respectively), LEDGF1-324mBRD4ETSEED overexpression targeted integration away from the TSS, CpG island and DHS midpoints (compare red and green bars), a pattern reminiscent of that of lentiviral vector integration (purple bars) (Wu et al., 2003). In addition, when analyzing integration preferences relative to a wide range of genomic and epigenetic features (Figure 4E and S3), expression of the LEDGF1-324mBRD4ETSEED shifted integration from a MLV to an HIV-like phenotype for all investigated markers, in contrast to overexpression of the ETSEED quadruple mutant protein. Together these data show that overexpression of a LEDGF1-324mBRD4ETSEED fusion protein efficiently shifts the MLV integration profile, corroborating that BET proteins function as integration targeting factors for MLV.
Figure 4. A LEDGF-BRD4 chimeric protein retargets MLV integration towards an HIV-like pattern.
A) MLV or HIV Integration sites obtained from SupT1 or NIH3T3 cells and their genomic distribution. The percentage of integrations in RefSeq genes and around TSS, CpG islands and DNAseI hypersensitive sites (2 kb and 4 kb window) is shown. Matched random control sites (MRC) for MLV and HIV integration sites in wild type cells are shown. TSS; transcription start site, CpG; CpG rich island, DNase; DNase I hypersensitive site. Asterisks depict pairwise Fisher’s test compared to MLV-SupT1|HIV-SupT1 *, p<0.05; **, p<0.01; ***, p<0.001. All data reach significance, p<0.001, compared to MRC. B–D) Integration frequencies surrounding RefSeq TSSs, CpG islands and DNAseI hypersensitive sites (DHS) in SupT1 cells. E) Heat map of integration frequency relative to genomic features in SupT1 cells, summarizing the relation between proviral integration sites and genomic features. Integration data sets are indicated above the columns. Genomic features analyzed are shown to the left of the corresponding row of the heat map. Tile color indicates whether a particular feature is favored (red) or disfavored (blue) for integration for the respective data sets relative to their MRCs, as detailed in the colored ROC area scale at the bottom of the panel. p values (asterisks) show significance of departures from the MLV integration sites in WT SupT1 cells (*, p < 0.05; **, p < 0.01; ***, p < 0.001, Wald statistics referred to χ2 distribution). The naming of the genomic features is described in Brady et al. (Brady et al., 2009); TSS, transcription start site.
Discussion
While HIV integration site targeting is mediated by LEDGF/p75 (Cherepanov et al., 2003; Ciuffi et al., 2005; Gijsbers et al., 2010; Llano et al., 2006; Schrijvers et al., 2012; Shun et al., 2007) the cellular co-factor driving MLV integration site targeting remained unknown. Here we describe BET proteins as the MLV targeting factors. In agreement with Sharma and co-workers (Sharma et al., 2013), we show that BRD2, BRD3 and BRD4 specifically interact with MLV IN and that bromodomain inhibitors can block MLV replication at the integration step. Further, it was shown that a recombinant BRD4 deletion mutant containing the bromodomains and the ET domain stimulated MLV concerted integration in vitro (Sharma et al., 2013). BET protein knockdown or treatment with JQ1(+) decreases integration around TSSs and CpG islands (Sharma et al., 2013). In addition, we demonstrate that, even in the presence of endogenous BET proteins, MLV integration efficiently shifts towards an HIV phenotype upon expression of a LEDGF1-324mBRD4ETSEED fusion, underscoring the role of BET proteins in MLV targeting. Retroviruses tend to direct integration into outward-facing major grooves on nucleosome-wrapped DNA (Roth et al., 2011). TSSs of expressed genes are nucleosome depleted, while the TSSs of the same genes when not expressed, are nucleosome bound (Struhl and Segal, 2013). Since MLV integrates at TSSs of actively transcribed genes, the bimodal MLV integration pattern naturally follows. However, the exact role of BET proteins, known to bind polyacetylated histone tails found around TSSs, remains to be investigated.
The specificity of the interaction of BET proteins for gammaretroviral IN, explains MLV integration site distribution. Lentiviral (HIV), alpharetroviral (RSV) nor spumaviral (PFV) IN interact with BET proteins. When and why different retroviral families evolved to interact with distinct targeting factors and how this relates to replication kinetics and pathogenesis remains poorly understood.
Our in vitro analysis revealed that the evolutionary conserved BRD4 ET-domain amino acids E652, E654, D656 and E658, are pivotal for interaction with MLV IN. Still, no interaction was detected between MLV IN and the yeast Bdf1 ET domain, suggesting the existence of other important interaction points (data not shown). While LEDGF/p75 binds HIV IN across the catalytic core domain dimer interface, BET proteins interact with the 27 C-terminal aa of MLV IN. A single point mutant (W391A) is sufficient to abolish this interaction. Future research will show whether it is possible to replace the BET binding region by alternative chromatin interaction domains, possibly resulting in a safer retroviral integration site profile.
In conclusion, we propose a model for MLV integration targeting incorporating previous insights on the function of MLV p12 and the newly described MLV IN-BET interaction (Figure S4) (Elis et al., 2012; Schneider et al., 2013).
Experimental Procedures
Extended experimental procedures
Compounds
The BET compounds JQ1 (Filippakopoulos et al., 2010) (the active, positive (JQ1 (+)) and inactive, negative (JQ1 (−)) enantiomer) and I-BET (Nicodeme et al., 2010) were kindly provided by J. Bradner (Harvard University, Boston, USA) and dissolved in DMSO.
Retroviral vector transduction
NIH3T3 (2*104) or SupT1 (8*104) cells were seeded in 96-well plates. After 24 h, cells were transduced with MLV- or HIV-derived vector particles. After 48h, cells were washed and cultured for another 24 h in normal growth medium. Subsequently, cells were split and 50% was reseeded for luciferase assays or fluorescence-activated cell sorting (FACS) analysis, while the remaining 50% was kept in culture to determine integrated copies and/or to perform integration site analysis.
Virus infection
To monitor early MLV-eGFP replication in the presence/absence of BET inhibitors, 2*106 NIH3T3 cells were seeded in a 6-well plate. Twenty-four hours later, cells were infected with MLV-eGFP at an MOI of 5000 RTU per well with or without the indicated compounds. Four hours after infection, cells were washed twice with phosphate-buffered saline (PBS) and fresh medium containing the indicated compounds. Cells were trypsinized and pelleted at 4, 8, 10, 12 and 24 h post transfection. To measure integrated copies, cells were passaged over 10 days in the presence of 1 μM raltegravir to block viral replication.
Analysis of next-generation sequencing data
Data were obtained from the Gene Expression Omnibus (GEO) (Edgar et al., 2002) or the Sequence Read Archive (SRA) (Leinonen et al., 2011) as detailed in Supplementary Table 2. Raw sequencing reads were mapped to the GRCh37/hg19 human genome assembly using the Bowtie2 short read aligner (Langmead and Salzberg, 2012). To delineate regions significantly enriched in BRD2–4, we used a shape-based peak calling approach (Hower et al., 2011) considering an average fragment length of 150 bp, the size of nucleosomal DNA and a p-value cut-off of 0.001. Distances from MLV integration or MRC sites to BRD2, 3, 4, Pol II, H3K4me3 or CpG islands were determined and analyzed (Berry et al., 2006) using Prism 5.0 (GraphPad software).
Sequence read densities were determined in 10 kb windows around MLV integration, MRC or refGene TSSs by counting fragment length-extended sequence tags in 50 bp bins for the sample and (when available) control libraries. The signal density was calculated as the difference between these two with negative values set to 0, after normalization by total sequencing depth. MLV and MRC sites were counted according to the same procedure in 50 bp bins in the studied regions and normalized to the total number of sites.
Supplementary Material
Highlights.
The BET ET domain specifically interacts with the MLV integrase C-terminus
BET chromatin binding inhibitors block MLV replication at the integration step
The MLV integration site distribution correlates with the BET chromatin binding profile
MLV vector integration can be retargeted by engineered BET proteins
Acknowledgments
We thank Nam Joo Van der Veken, Martine Michiels, and Paulien Van de Velde for excellent technical assistance. We acknowledge SyBioMa (formerly known as Prometa, KU Leuven) for technical assistance with LC-MS analysis and S. Aerts and A. Verfaillie for help with the BRD ChIP-Seq analysis. Work at KU Leuven was supported by grants and fellowships from the FWO, IWT and the DOC research council (OT, IDO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady T, Lee YN, Ronen K, Malani N, Berry CC, Bieniasz PD, Bushman FD. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 2009;23:633–642. doi: 10.1101/gad.1762309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, Schmidt M, von Kalle C, Howe S, Thrasher AJ, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- Cattoglio C, Pellin D, Rizzi E, Maruggi G, Corti G, Miselli F, Sartori D, Guffanti A, Di Serio C, Ambrosi A, et al. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- Cavazza A, Moiani A, Mavilio F. Mechanisms of retroviral integration and mutagenesis. Hum Gene Ther. 2013;24:119–131. doi: 10.1089/hum.2012.203. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, Glimm H, Gyapay G, Prum B, Fraser CC, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Singer DS. Two faces of brd4: mitotic bookmark and transcriptional lynchpin. Transcription. 2013;4:13–17. doi: 10.4161/trns.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elis E, Ehrlich M, Prizan-Ravid A, Laham-Karam N, Bacharach E. p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog. 2012;8:e1003103. doi: 10.1371/journal.ppat.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice B, Cattoglio C, Cittaro D, Testa A, Miccio A, Ferrari G, Luzi L, Recchia A, Mavilio F. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS One. 2009;4:e4571. doi: 10.1371/journal.pone.0004571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J, Brouns G, Schmidt M, Von Kalle C, Barington T, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, Thrasher AJ, Wulffraat N, Sorensen R, Dupuis-Girod S, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Hower V, Evans SN, Pachter L. Shape-based peak identification for ChIP-Seq. BMC Bioinformatics. 2011;12:15. doi: 10.1186/1471-2105-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, Batzoglou S, Sidow A. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 2012;22:1735–1747. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy G, Chepelev I, Dimaggio PA, Blanco MA, Zee BM, Zhao K, Garcia BA. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YJ, Umehara T, Inoue M, Saito K, Kigawa T, Jang MK, Ozato K, Yokoyama S, Padmanabhan B, Güntert P. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 2008;17:2174–2179. doi: 10.1110/ps.037580.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder ARW, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SL, Malani N, Bushman FD. Gammaretroviral integration into nucleosomal target DNA in vivo. J Virol. 2011;85:7393–7401. doi: 10.1128/JVI.00635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni FA, Hartley O, Luban J. Deciphering the code for retroviral integration target site selection. PLoS Comput Biol. 2010;6:e1001008. doi: 10.1371/journal.pcbi.1001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Brzezinski JD, Aiyer S, Malani N, Gyuricza M, Bushman FD, Roth MJ. Viral DNA tethering domains complement replication-defective mutations in the p12 protein of MuLV Gag. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1221736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijvers R, De Rijck J, Demeulemeester J, Adachi N, Vets S, Ronen K, Christ F, Bushman FD, Debyser Z, Gijsbers R. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012;8:e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B, Goff SP. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology. 2008;5:48. doi: 10.1186/1742-4690-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G, Tebas P, June CH, Bushman FD. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther. 2009;17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJT, Thiessen N, Pettersson S, Jones SJM, Knapp S, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.