Abstract
Rif1 protein is present in eukaryotic cells from yeast to human. In yeast, Rif1 is important for telomere homeostasis. Despite conservation in its domain organization, human Rif1 is not part of the telomere complex but was recently reported to work at DNA double-strand breaks (DSBs) with 53BP1 to inhibit 5′ strand degradation (resection) and stimulate a subset of nonhomologous end-joining (NHEJ) reactions. Martina et al 1 report in this issue of EMBO reports that yeast Rif1 is also recruited to DSBs, but in contrast to its human counterpart, it promotes resection. The authors propose that Rif1 stimulates resection by limiting the access of Rad9, an ortholog of 53BP1, to DSBs.
Most proteins involved in DNA repair and recombination are well conserved throughout evolution. A notable exception is Rif1 and Rad9, which while sharing a high degree of structural conservation with their mammalian orthologs, Rif1 and 53BP1 have acquired new functions in the DNA damage response in higher eukaryotes. Yeast Rif1, together with Rap1 and Rif2, assemble a capped structure at telomeres (Fig 1A), which negatively regulates telomerase activity. In addition, Rif1 prevents G2/M checkpoint activation and degradation of uncapped or short telomeres. This function is not conserved in higher eukaryotes, as Rif1 in mammals is not part of the telomere complex. Rif1 also plays a major role in the regulation of replication, which appears to be a conserved function. Rif1 was shown to control the firing of replication origins by negatively regulating the replicative helicase through association with the PP1 phosphatase Glc7 (Fig 1B) 2.
Figure 1. Multiple functions of Rif1 in yeast and human.
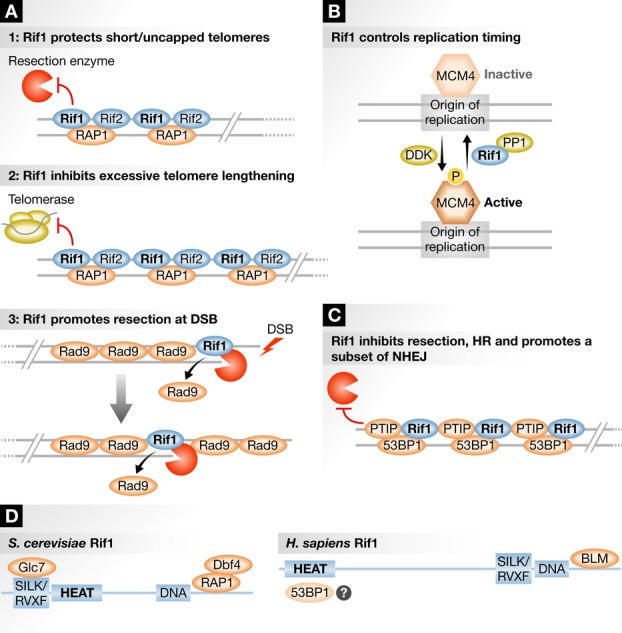
(A) Yeast-specific functions of Rif1. (B) Evolutionary-conserved function of Rif1 in the regulation of replication timing. (C) Human-specific functions of Rif1 in NHEJ. (D) Yeast and human Rif1 proteins’ conserved domains. HEAT repeats, responsible for interaction with proteins; DNA, putative DNA-binding domain; SILK/RVXF, Protein Phosphatase 1 (PP1)-interacting domain; BLM, BLM helicase interaction domain.
In mammals, Rif1 has acquired new functions in the DNA damage response. Rif1 functions at DSBs in a complex with 53BP1 to inhibit homologous recombination (HR) and direct DNA repair toward NHEJ (Fig 1C). Unlike core NHEJ proteins, 53BP1-Rif1 promote only a subset of NHEJ reactions that involve distal joining of DNA ends. For example, they facilitate end-joining during physiological V(D)J recombination and class switch recombination (CSR) in lymphocytes when the two ends are greater than 100 kb apart. In pathological contexts, they generate telomere fusions when telomeric DNA ends are uncapped, and long-range radial chromosomal fusions in BRCA1-deficient cells. Two activities of 53BP1 contribute to its function in these specialized NHEJ reactions. First, 53BP1 inhibits resection at DSB ends, and resection is known to decrease NHEJ in favor of homologous recombination (HR). Second, 53BP1 promotes the synapsis or mobility of DNA ends by as yet unknown mechanisms in long-range end-joining reactions.
The mechanism by which 53BP1 inhibits resection remains unclear. 53BP1 is recruited to DSBs but its mere physical presence at breaks does not inhibit HR or promote NHEJ unless it is phosphorylated at its N-terminus by ATM 3–7. Rif1 interacts with chromatin-bound and phosphorylated 53BP1 at DSBs, where it facilitates some of 53BP1’s functions. Like 53BP1, Rif1 is required for CSR, and in its absence, the immunoglobulin switch regions undergo extensive resection 3,4. However, analysis of mutants deficient in Rif1 and 53BP1 revealed that many of the phenotypes observed in Rif1−/− cells are actually very mild when compared to 53BP1−/−. For example, Rif1 deficiency does not fully restore HR in BRCA1 deficient cells 5–7 and only has a very mild impact on the fusion of dysfunctional telomeres 7. These observations suggested that additional effector proteins are required downstream of 53BP1. This scenario turned out to be correct as an additional 53BP1 N-terminal binding protein, PTIP (also known as PAXIP), was shown to be necessary for 53BP1-mediated inhibition of HR in BRCA1-deficient cells and the fusion of dysfunctional telomeres 8.
Yeast do not have BRCA1 or PTIP, nor do they require distal joining of DNA ends; therefore, one might expect that the 53BP1-Rif1-PTIP regulatory module of recombination would be absent in yeast. Nevertheless, yeast Rad9, similarly to 53BP1, blocks extensive resection, both at DSBs and at uncapped telomeres, although so far this activity has not been correlated with increased NHEJ or telomere fusions. Moreover, the domain structure of Rif1 is partly conserved (Fig 1D). Yeast and human Rif1 carry HEAT repeats, which presumably are responsible for most of Rif1’s protein–protein interactions, and a PP1 phosphatase-binding domain, which is critical for its role in replication 2. In yeast, the C-terminal domain serves as a Rap1 binding region, whereas in mammals, the C-terminal domain is required for interaction with BLM, a RecQ DNA helicase that is critical for HR.
Given the role of mammalian Rif1 in blocking resection downstream of 53BP1, a logical question is to inquire about the role (if any) of yeast Rif1 in resection. Martina et al 1 demonstrated that Rif1 is recruited to DSBs also in yeast. Rif1 was found to be epistatic with Rad51 with respect to DNA damage sensitivity, suggesting that Rif1 functions in HR. In stark opposition to its role in mammalian cells, the authors discovered that Rif1 does not inhibit, but rather promotes resection. This function was specific for Rif1 as Rif2 had no role in resection at DSBs.
Resection in yeast and human is initiated by the MRX complex (hMRN), with Sae2 (hCTIP) forming short ssDNA and recruiting enzymes responsible for extensive resection, Exo1 or Sgs1/Dna2 (hEXO1, BLM, DNA2). Deletion of RIF1 did not have an impact on initial resection; however, extensive resection was significantly decreased 1. Consistent with a defect in resection, Rif1 was found to be important in single strand annealing, a DSB repair pathway that requires extensive resection of sequences between direct repeats. Further, Rif1 was shown to facilitate the Sgs-mediated resection pathway while its role in the Exo1 pathway could not be tested due to the growth defect of rif1∆sgs1∆.
How does yeast Rif1 promote resection? In rif1∆ cells, increased amounts of Rad9 accumulate at DSBs. It was therefore proposed that Rif1 facilitates access of resection enzymes to chromatin by antagonizing Rad9 1. This would be analogous to the antagonism between 53BP1 and BRCA1 in the regulation of DNA repair choice in mammals. One interesting possibility is that Rif1 may facilitate resection via its PP1-binding domain. As previously noted, the SILK-RVXF domain in Rif1 interacts with Gcl7 phosphatase 2 (Fig 1D). Longhese’s group demonstrated that Glc7 is recruited to DNA damage sites where it dephosphorylates H2AX, an important step for timely recovery from the DNA damage checkpoint 9. Since H2AX phosphorylation is needed for efficient Rad9 binding, Rif1 may antagonize Rad9 through Glc7-mediated dephosphorylation of H2AX. It is tempting to speculate that the regulatory functions of Rif1 in recombination in yeast and mammals might be similarly executed by association with the respective PP1 phosphatase.
Taken together, Rad9 blocks extensive resection in yeast by opposing the action of Rif1, whereas mammalian 53BP1 blocks resection in conjunction with Rif1. At first glance, these data point to a major functional diversification of Rif1 in eukaryotes. However, evidence suggests that Rif1 both inhibits and promotes resection in a context-dependent manner in both organisms. Yeast Rif1 prevents nucleolytic degradation of uncapped telomeres and inhibits one type of telomere recombination in telomerase negative cells. In mammals, in addition to inhibiting resection during CSR and at uncapped telomeres, Rif1 was proposed to promote repair of DSBs at stalled replication forks by HR 10. Yet another study showed that recruitment of the resection helicase BLM to DSBs was dependent on Rif1, suggesting that Rif1 has a role in HR repair that is independent of 53BP1 6. Thus, like in yeast, mammalian Rif1 may have both pro- and anti-HR functions. Such opposing functions are not unprecedented for recombination proteins. For example, depending on the context, yeast Sgs1 and Srs2 DNA helicases can either promote or inhibit HR, and in mammals, BRCA1, ATM and H2AX can have pro- and anti-HR functions. As if research in this area were not turbulent enough, the finding that Rif1 can ‘have it both ways’ heightens the need for clearer mechanistic understanding of how cells ‘make up their mind’ to repair either by HR or NHEJ. Only then might we be able figure out how Rif1 manages to eat its cake and have it too.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Martina M, Bonetti D, Villa M, et al. EMBO Rep. 2014 doi: 10.1002/embr.201338338. DOI 10.1002/embr.201338338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Alvino GM, Chang F, et al. Genes Dev. 2014;28:372–383. doi: 10.1101/gad.231258.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, et al. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, et al. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, et al. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, et al. J Biol Chem. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, et al. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, et al. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi M, Mantiero D, Trovesi C, et al. Mol Cell Biol. 2010;30:131–145. doi: 10.1128/MCB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Wu Y, Ferguson D, et al. J Cell Biol. 2009;187:385–398. doi: 10.1083/jcb.200902039. [DOI] [PMC free article] [PubMed] [Google Scholar]


