Abstract
The membrane topology of Om45 in the yeast mitochondrial outer membrane (OM) is under debate. Here, we confirm that Om45 is anchored to the OM from the intermembrane space (IMS) by its N-terminal hydrophobic segment. We show that import of Om45 requires the presequence receptors, Tom20 and Tom22, and the import channel of Tom40. Unlike any of the known OM proteins, Om45 import requires the TIM23 complex in the inner membrane, a translocator for presequence-containing proteins, and the membrane potential (ΔΨ). Therefore, Om45 is anchored to the OM via the IMS by a novel import pathway involving the TIM23 complex.
Keywords: mitochondria, Om45, translocator, yeast
Introduction
Most mitochondrial proteins are encoded by the nuclear genome and synthesized in the cytosol. They are subsequently imported into mitochondria and sorted to intramitochondrial compartments such as the outer membrane (OM), intermembrane space (IMS), inner membrane (IM), and matrix, with the aid of translocator complexes in the OM and IM 1–3. Most matrix proteins and some IM proteins are synthesized as a precursor protein with an N-terminal cleavable presequence, which contains a mitochondrial targeting signal. Presequence-containing precursor proteins use the TOM40 complex in the OM to cross the OM and the TIM23 complex in the IM to cross or get inserted into the IM. The TOM40 complex consists of multiple subunits including presequence receptors, Tom20 and Tom22, and a channel-forming protein, Tom40. Tim50 of the TIM23 complex receives a presequence from the Tom40 channel and transfers it to the Tim23 channel, depending on the membrane potential (ΔΨ) across the IM 4–6. Matrix-targeted proteins subsequently use an import motor mitochondrial Hsp70 (mtHsp70) and its partner proteins for translocation and unfolding, while IM-sorted proteins with a hydrophobic sorting signal downstream of the matrix-targeting signal in the presequence require translocation arrest at the TIM23 channel followed by lateral release into the IM.
Mitochondrial OM proteins can be categorized into two classes: β-barrel proteins and proteins with α-helical transmembrane (TM) segment(s). Newly synthesized β-barrel proteins cross the OM through the Tom40 channel and are transferred via small Tim proteins to the TOB/SAM complex, which facilitates the formation of a β-barrel structure and insertion into the OM 7. Insertion of OM proteins with multiple α-helical TM segments is facilitated by Tom13/Mim1 together with Mim2. On the other hand, there is apparently no common pathway for the insertion of OM proteins with a single TM segment.
Om45 is one of the most abundant proteins in the OM of yeast mitochondria 8,9. The protein level of Om45 increases under nonfermentable conditions 10, and the role of Om45 in the regulation of porin channels was proposed 11. Om45 has a stretch of hydrophobic residues near the N-terminus (residues 5–22) like Tom20 and Tom70, both of which are anchored to the OM by their N-terminal TM segments and expose the C-terminal domains to the cytosol (Nin-Cout topology). Om45 has thus been assumed to take a similar Nin-Cout topology. Consistent with this model, Tom20 and Tom70 variants whose N-terminal TM segments are replaced with residues 1–32 of Om45 are functional in vivo 12. Besides, anchoring of Om45 to the OM was reported to require no proteinaceous component, but to take place spontaneously 13. However recently, the Nin-Cout topology of Om45 was challenged by the observation that Om45 in isolated mitochondria is not digested by externally added protease 11, although conflicting results of protease digestion were reported previously 8,14. OM proteins inserted into the OM from the IMS are unusual, and the import pathway for such proteins and involvement of possible proteinaceous components should be of great interest; only Taz1, a yeast ortholog of human tafazzin, was reported to be inserted into both the OM and IM from the IMS 15.
In this report, we re-examined the submitochondrial localization of Om45 in yeast mitochondria and found that Om45 is indeed integrated into the OM from the IMS. We found that Om45 travels to the IMS for insertion into the OM via a novel import pathway mediated by the TIM23 complex and driven by ∆Ψ.
Results and Discussion
Om45 is an outer membrane protein exposed to the IMS
To re-examine the membrane orientation of Om45, we treated isolated mitochondria with a relatively high concentration (125 μg/ml) of proteinase K (PK) or trypsin. OM proteins with a cytosolic domain, Tom70 and Tom22, were digested by both proteases, while Tim50, an IM protein exposed to the IMS, and Kgd1, a matrix protein, were not (Fig 1A). Om45 was protected from PK and trypsin, while it was completely digested when mitochondrial membranes were solubilized with Triton X-100 (Fig 1A). Although the expression level of Om45 was markedly enhanced under nonfermentable conditions (Fig 1A, Lactate) as compared with fermentable conditions (Fig 1A, Glucose), protease resistance of Om45 was not affected by the medium (Fig 1A, Glucose, Om45 (Dark)). When the OM was ruptured by osmotic swelling and the resultant mitoplasts were treated with PK, Om45 was digested like the IM proteins exposed to the IMS, Tim50 and Tim23, but not like the matrix protein Kgd1 (Fig 1B). These results clearly show the localization of Om45 in the IMS, not on the mitochondrial surface.
Figure 1. Om45 is an OM protein exposed to the IMS.
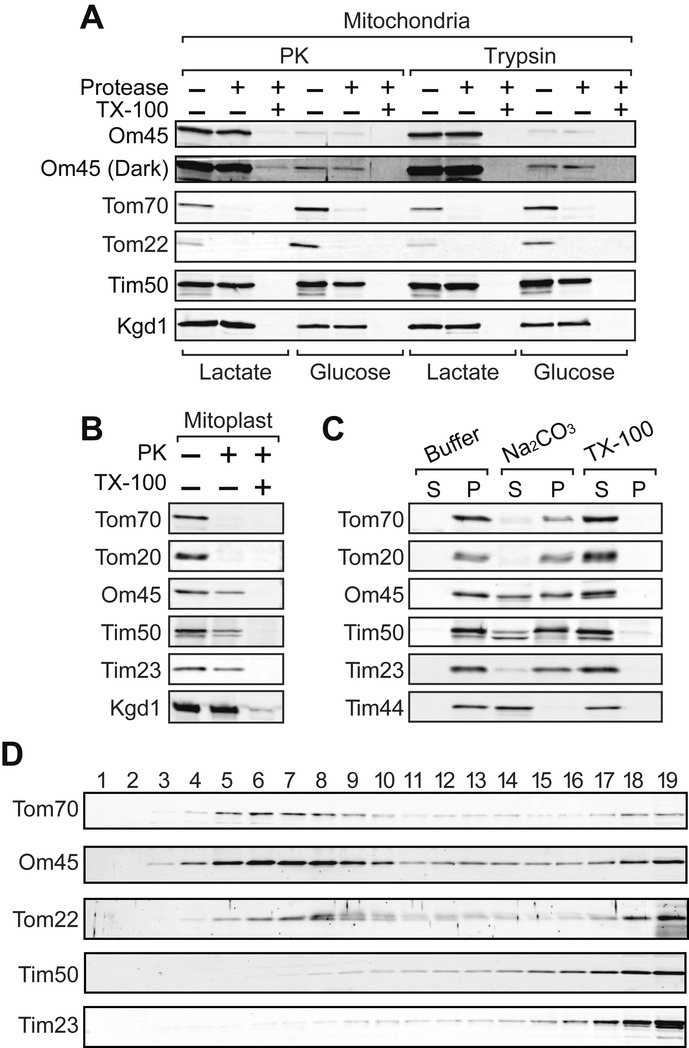
A Mitochondria isolated from the cells cultured in YPD or lactate medium were treated with 125 μg/ml of PK or trypsin in the presence or absence of 0.2% Triton X-100 for 20 min on ice. Proteins were analyzed by SDS–PAGE and immunoblotting with the indicated antibodies. In Om45 (Dark), contrast of the image was enhanced.
B Mitoplasts were PK-treated, and proteins were analyzed as in (A).
C Mitochondria were incubated in SEM buffer (Buffer), with 0.1 M Na2CO3 or with 1% Triton X-100 (TX-100), on ice for 20 min and then subjected to ultracentrifugation. The resulting supernatants and pellets were analyzed as in (A).
D OM and IM vesicles generated from mitochondria were subjected to sucrose density-gradient centrifugation. Each fraction was analyzed as in (A).
Then, we assessed association of Om45 with membranes by alkaline treatment. While integral membrane proteins Tom20, Tom70, Tim23, and Tim50 were resistant against extraction with Na2CO3 and recovered mainly (Tom20, Tom70, and Tim23) or partly (Tim50) in the pellet, Tim44, a peripheral IM protein, was extracted to the supernatant (Fig 1C). Om45 was recovered in both the supernatant and pellet, suggesting that Om45 is at least partly integrated in the membrane. Then, we asked which of the membranes, the OM and IM, Om45 is associated with. We prepared OM and IM vesicles from mitochondria by sonication and separated them by sucrose density-gradient centrifugation. Immunoblotting of the fractions collected from the sucrose gradient showed that Om45 as well as OM proteins, Tom70 and Tom22, was mainly recovered in the lighter fractions (Fig 1D, lanes 4–10), while IM proteins, Tim50 and Tim23, were recovered in the heavier fractions (Fig 1D, lanes 15–19), indicating that Om45 is associated with the OM vesicles. Taken together, we conclude that Om45 is an OM protein associated with the OM from the IMS side.
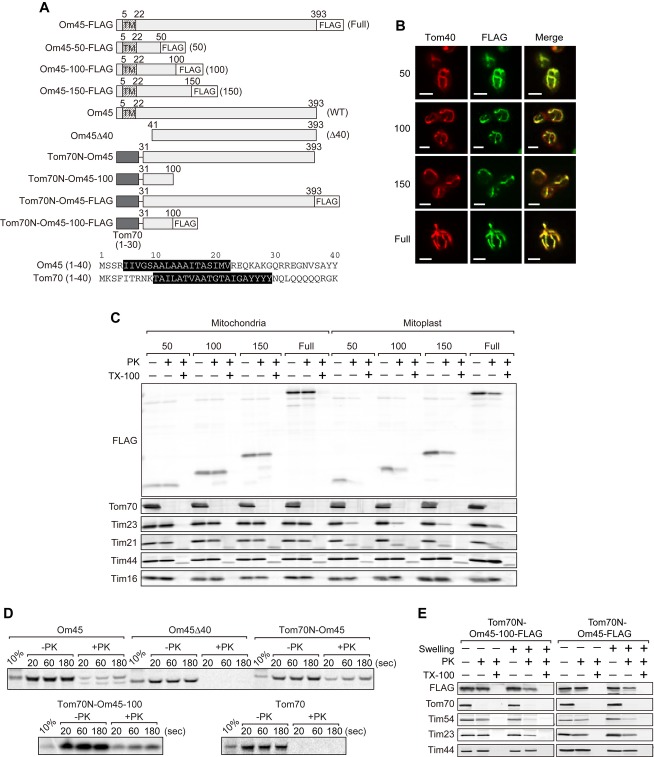
N-terminal segments of Om45 and Tom70 can direct Om45 to the IMS
Since the revealed membrane topology of Om45 is unusual, we reasoned that Om45 has a novel targeting signal and follows a novel import pathway. We thus expressed full-length Om45 or its C-terminally truncated versions fused to the FLAG epitope tag (Fig 2A) in yeast cells. By immunofluorescence with the anti-FLAG antibody, truncated Om45 variants as short as 50 residues were localized to mitochondria, which were stained with anti-Tom40 antibodies (Fig 2B). Then, we performed protease treatment of mitochondria with these Om45 variants. The C-terminal FLAG tag of the Om45 variants as well as full-length Om45 was mostly resistant against PK, while they became protease-sensitive after disrupting the OM by osmotic swelling, showing that they are localized in the IMS (Fig 2C).
Figure 2. The N-terminal 50 residues direct Om45 to the IMS.
A Top: A schematic diagram of Om45 variants (with the FLAG tag). Bottom: The predicted TM segments in residues 1–40 of Om45 and Tom70 are indicated by white letters on black background.
B Localization of the Om45 variants was assessed by immunofluorescence with the anti-FLAG antibody (FLAG). The mitochondria were visualized with anti-Tom40 antibodies (Tom40). Scale bar, 3 μm.
C Mitochondria or mitoplasts with the indicated Om45 variants were incubated with 125 μg/ml PK with or without 0.2% Triton X-100 (TX-100) for 20 min on ice. Proteins were analyzed by SDS–PAGE and immunoblotting with the indicated antibodies.
D The indicated 35S-labeled proteins were imported into mitochondria in vitro for the indicated times and PK-treated. Imported proteins were analyzed by SDS–PAGE and radioimaging.
E Mitochondria (−Swelling) or mitoplasts (+Swelling) with the indicated Om45 variants were analyzed as in (C).
We next tested in vitro import of Om45 lacking the N-terminal 10–40 residues (Om45Δ10, Om45Δ20, and Om45Δ40) and fusion proteins consisting of the N-terminal 30 residues of Tom70 followed by residues 31-393 of Om45 (Tom70N-Om45) or residues 31–100 of Om45 (Tom70N-Om45-100) (Fig 2A and Supplementary Fig S1A). Om45 lacking N-terminal 10–40 residues was imported into mitochondria only inefficiently in vitro (Fig 2D and Supplementary Fig S1B), suggesting that Om45 is guided to the mitochondrial IMS by its N-terminal 40 residues. Surprisingly, Tom70N-Om45 and Tom70N-Om45-100 became resistant against PK like Om45, while Tom70 remained sensitive to PK (Fig 2D). Tom70N-Om45 and Tom70N-Om45-100 expressed in yeast cells were resistant against PK in mitochondria, while they were sensitive to PK in mitoplasts (Fig 2E). These results indicate that the N-terminal 30 residues including the TM segment of both Om45 and Tom70 can direct the C-terminal domain of Om45 to the IMS, but not the one of Tom70. Inability of the N-terminal 30 residues of Tom70 to direct the Tom70 C-terminal domain may be due to the tight folding of Tom70 as compared with Om45 (Supplementary Fig S2).
The TOM40 complex facilitates Om45 import
How is the mitochondrial targeting signal of Om45 recognized and how does Om45 cross the OM? We tested involvement of the presequence-receptor subunits of the TOM40 complex, Tom20 and Tom22, in Om45 import. For this purpose, we took advantage of the yeast mutant strains expressing Tom20 and Tom22 variants, whose receptor domains can be selectively deleted by a tobacco etch virus (TEV) protease in vitro (Fig 3A) 16. Import of Om45 and Tom70N-Om45 was, like that of the presequence-containing Hsp60 precursor, retarded in mitochondria lacking the receptor domain of Tom20 or Tom22 (Fig 3B–D). The targeting signals of Om45 and Tom70 that direct Om45 to the IMS are thus recognized by Tom20 and Tom22 despite the dissimilarity of the signals to typical mitochondrial presequences. On the other hand, import of Om45 does not depend on Tom70 and Tom71, receptors for presequence-less proteins (Supplementary Fig S3).
Figure 3. The TOM40 complex facilitates import of Om45.
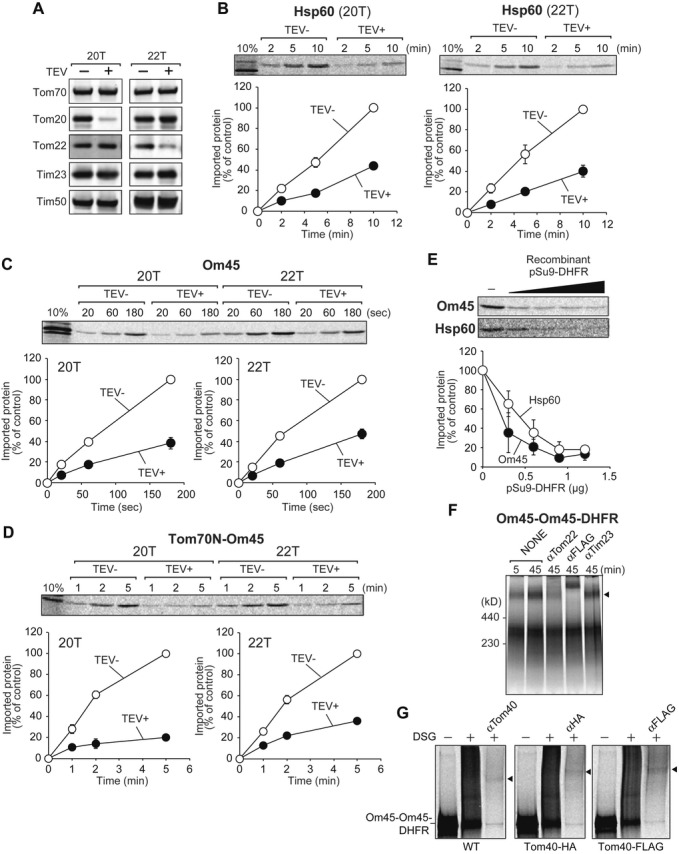
A Mitochondria with Tom20-TEV (20T) or Tom22-TEV (22T) were treated with TEV protease and subjected to SDS–PAGE followed by immunoblotting using the indicated antibodies.
B–D 35S-labeled Hsp60 precursor (B), Om45 (C), and Tom70N-Om45 (D) were incubated with the indicated TEV protease-treated mitochondria for the indicated times at 25°C. After PK digestion, imported proteins were analyzed by SDS–PAGE and radioimaging. Amounts of the imported proteins at the longest incubation in control mitochondria were set to 100%. Values are mean ± SEM (n = 3).
E The indicated 35S-labeled proteins were premixed with increasing amounts of pSu9-DHFR, then imported into mitochondria at 25°C for 2 min and 5 min, respectively, and analyzed as in (B–D). Amounts of the imported proteins in the absence of pSu9-DHFR were set to 100%.
F 35S-labeled Om45-Om45-DHFR was imported to Tom40-FLAG mitochondria for the indicated time. After import, mitochondria were washed with SEM buffer and solubilized in 1% digitonin. Then, a clarifying spin followed, and the supernatant was incubated with the indicated antibodies on ice for 50 min, and then analyzed by 4–10% BN PAGE and radioimaging. Om45-Om45-DHFR stuck at the TOM40 complex is indicated with arrowheads.
G 35S-labeled Om45-Om45-DHFR was incubated with mitochondria containing Tom40 (WT), Tom40-HA, or Tom40-FLAG for 10 min at 25°C. Mitochondria were then washed with SEM buffer and subjected to crosslinking with 200 μM DSG. The samples were solubilized and subjected to immunoprecipitation using the indicated antibodies, and immunoprecipitated proteins were detected by radioimaging. Crosslinked products are indicated with arrowheads.
We next asked whether Om45 uses the Tom40 channel to cross the OM. We tested kinetic competition in import between Om45 and excess amounts of recombinant pSu9-mouse dihydrofolate reductase (DHFR), a fusion protein between the presequence of Fo-ATPase subunit 9 and DHFR. pSu9-DHFR uses the TOM40 complex and TIM23 complex for translocation across the OM and IM, respectively. Import of Om45 was, like the matrix-targeted Hsp60 precursor, significantly suppressed by the presence of increasing concentrations of pSu9-DHFR, suggesting that import of Om45 and pSu9-DHFR competes with each other at the receptor and/or channel level (Fig 3E). We then probed direct physical interactions between translocating Om45 and the TOM40 complex. When we incubated Om45-DHFR or Om45-Om45-DHFR, a fusion protein consisting of Om45 or a tandem repeat of Om45 followed by DHFR, respectively, with isolated mitochondria and then treated the mitochondria with PK, Om45-Om45-DHFR generated fragments that were partially protected against degradation, while Om45-DHFR was digested completely (data not shown). When we incubated Om45-Om45-DHFR with mitochondria with Tom40-FLAG and then analyzed the digitonin-solubilized mitochondria by blue-native (BN) PAGE, Om45-Om45-DHFR was migrated as a large complex (Fig 3F, arrowhead), whose band was gone or shifted to a higher MW region after incubation with anti-Tom22 antibodies or the anti-FLAG antibody. Therefore, Om45-Om45-DHFR likely represents a stable translocation intermediate for the Om45 pathway at the TOM40 complex level. Incubation of mitochondria accumulating stuck Om45-Om45-DHFR with a bifunctional crosslinker, disuccinimidyl glutarate (DSG), generated a DSG-dependent higher MW band, which shifted upward when we replaced endogenous Tom40 with Tom40-HA or Tom40-FLAG (Fig 3G). This band hence arose from the crosslinked product between Om45-Om45-DHFR and Tom40, indicating the physical contact between the translocation intermediate of Om45-Om45-DHFR with Tom40. Therefore, Om45 most likely uses the Tom40 channel to cross the OM to reach the IMS. Inability of Om45 to direct a passenger protein like tightly folded DHFR to the IMS may explain the previous results that chimeric proteins with the Om45 targeting signal are often localized on the mitochondrial surface 12.
The TIM23 complex mediates Om45 import
How is the translocation of Om45 across the OM driven? Since Om45 is recognized by Tom20 and Tom22, which mainly recognize the presequence of the TIM23 pathway substrates, we examined whether the TIM23 complex is involved in Om45 import. To this end, we utilized tim23-71,78 and tim50-279,282,286 strains, which exhibit defects in protein import via the TIM23 pathway 6. We isolated mitochondria from these mutant cells and corresponding wild-type cells and confirmed that mutant mitochondria contain normal levels of the translocator components (Supplementary Fig S4A). We then performed in vitro import of 35S-labeled substrate proteins into these mitochondria. Interestingly, import of Om45 and Tom70N-Om45 was impaired in tim23-71,78 and tim50-279,282,286 mitochondria (Fig 4A and B), although Om45 or Tom70N-Om45 does not contain a presequence-like signal. As a control, import of pSu9-DHFR, a substrate for the TIM23 pathway, was strongly impaired in both tim23-71,78 and tim50-279,282,286 mitochondria (Fig 4C). Import of a presequence-less IM protein Tim23, a substrate for the TIM22 pathway, not the TIM23 pathway, was efficient or even enhanced in these mutant mitochondria as compared with the wild-type mitochondria (Fig 4D). Om45 is thus the first OM protein that uses the TIM23 complex in the IM for its import.
Figure 4. The TIM23 complex facilitates import of Om45.
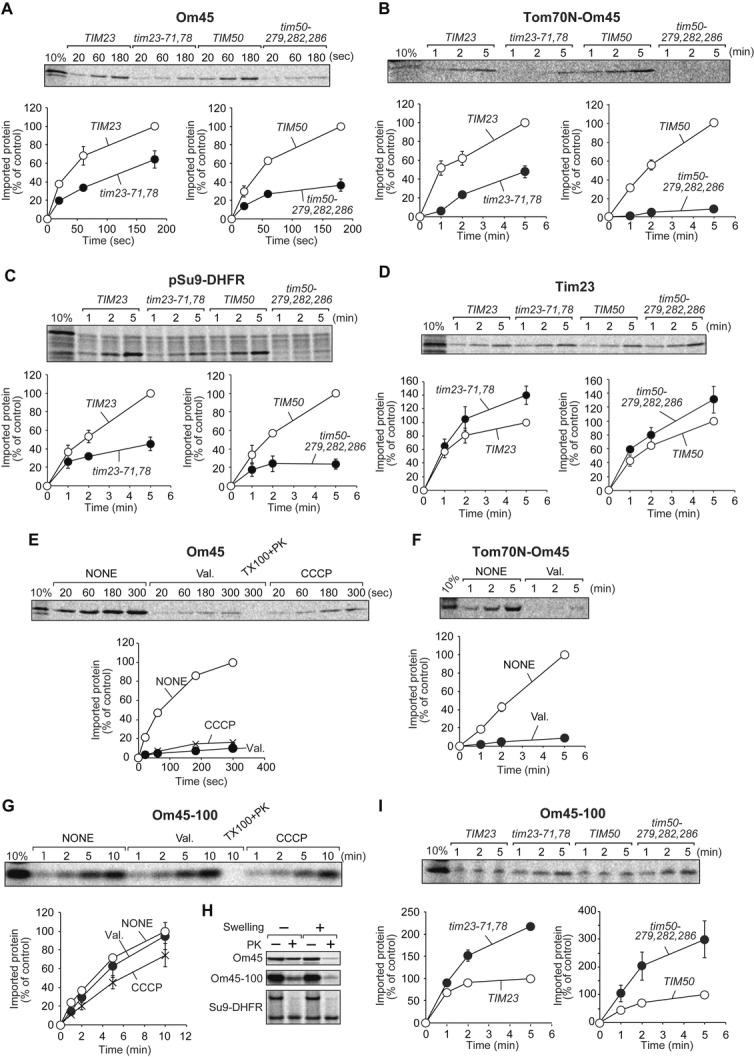
A–D The indicated 35S-labeled proteins were imported into tim23-71,78, tim50-279,282,286, or corresponding wild-type mitochondria and analyzed as in Fig 3B–D.
E–G The indicated 35S-labeled proteins were imported into mitochondria with (NONE) or without (Val. or CCCP) ∆Ψ at 25°C and analyzed as in (A–D).
H The indicated 35S-labeled proteins were imported into mitochondria for 10 min at 25°C. Then, mitochondria (−Swelling) were converted to mitoplasts (+Swelling) and treated with 50 μg/ml PK. Proteins were analyzed as in (A–D).
I 35S-labeled Om45-100 was imported into mitochondria isolated from tim23-71,78, tim50-279,282,286, and their corresponding wild-type cells (TIM23 and TIM50) at 25°C and analyzed as in (A–D).
We then asked whether Om45 import depends on the components involved in the TIM22 pathway for IM proteins or the Tim40/Mia40 pathway for IMS proteins (Supplementary Fig S4B–D). However, Om45 import did not strongly depend on these components as compared with the controls, ruling out the involvement of the TIM22 or Tim40/Mia40 pathway for sorting of Om45 to the IMS.
Membrane potential drives Om45 import
ΔΨ is required for the early step of protein translocation across or insertion into the IM via the TIM23 complex. We thus asked whether Om45 and Tom70N-OM45 import requires ∆Ψ in vitro. Isolated mitochondria were pretreated with valinomycin or carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to dissipate ∆Ψ, and then incubated with 35S-labeled substrates. Import of Om45 and Tom70N-Om45 was strongly impaired for the de-energized mitochondria (Fig 4E and F), indicating that Om45 import crucially requires ∆Ψ. We then tested whether import of the C-terminally truncated Om45 variant, Om45-100, into the IMS also depends on ∆Ψ. Interestingly, Om45-100 was efficiently imported into de-energized mitochondria without ∆Ψ as compared with energized mitochondria (Fig 4G) and reached the IMS (Fig 4H). Besides, Om45-100 was imported into tim23-71,78 and tim50-279,282,286 mitochondria even more efficiently than wild-type mitochondria (Fig 4I). These results suggest that ∆Ψ and the TIM23 complex are required for driving translocation of full-length Om45, but not its truncated version, across the OM.
Protein translocation across the IM and/or protein unfolding by the TIM23 complex requires the action of mtHsp70 powered by ATP hydrolysis in the matrix. We thus analyzed in vitro import of Om45 into ssc1-2 and ssc1-3 mitochondria with temperature-sensitive mtHsp70 mutants 17. Although import of matrix-targeted pSu9-DHFR was impaired with ssc1-3 mitochondria, import of Om45 was not affected (Supplementary Fig S5A), suggesting that mtHsp70 import motor is not required for Om45 import. However interestingly, depletion of Pam17, another subunit of the TIM23 complex that may link the TIM23 core part and mtHsp70 import motor 18, impaired import of Om45 like pSu9-DHFR (Supplementary Fig S5B).
Conclusions
Om45 is an OM protein that is inserted into the OM from the IMS. In contrast to the previously proposed spontaneous-insertion model, Om45 is recognized by the presequence receptors, Tom20 and Tom22, on the surface of mitochondria and crosses the OM through the TOM40 complex. Then, the targeting signal of Om45 is transferred to the TIM23 complex like presequence-containing precursors. Although the precise roles of the TIM23 complex and ΔΨ in the Om45 import are not clear, it is likely that the TIM23 complex uses ΔΨ to provide a driving force for translocation of the remaining domain of Om45 across the OM. Both the N-terminal segments of Om45 and Tom70 can guide the C-terminal domain of Om45 to the IMS via this pathway. Finally, Om45 is re-inserted into the OM from the IMS side and assembled into a complex with, for example, Om14 11. Although a Mim1 dependence of the final step of Om45 assembly is reported in a parallel study 19, involvement of other component(s), if any, in this new pathway remains to be investigated in future studies.
Materials and Methods
Plasmids, strains, and growth conditions
Construction of plasmids and strains and growth conditions are described in Supplementary Methods online.
Protease digestion and in vitro protein import
Protease digestion and TEV protease treatment 16 of mitochondria and in vitro protein import are described in Supplementary Methods online.
Competition experiments
Recombinant pSu9-DHFR was purified from E. coli cells as described previously 20. Different amounts of purified recombinant pSu9-DHFR were first mixed with 35S-labeled Om45 or the Hsp60 precursor on ice. Om45 or the Hsp60 precursor was then incubated with energized mitochondria at 25°C. Import reaction was stopped by adding valinomycin for the Hsp60 precursor or ice-cold SEM buffer (250 mM sucrose, 10 mM MOPS-KOH, pH 7.2, 1 mM EDTA) for Om45, and unimported proteins were digested with 50 μg/ml PK. Mitochondria were washed with SEM buffer, and proteins were analyzed by SDS–PAGE and radioimaging.
Crosslinking
35S-labeled Om45-Om45-DHFR was incubated with mitochondria for 10 min at 25°C. Mitochondria were collected by centrifugation, washed with SEM buffer, and then suspended in SEM buffer for crosslinking with DSG. Details of the crosslinking procedures are described in Supplementary Methods online.
Acknowledgments
We thank Dr. Elizabeth A. Craig for the ssc1-2 and ssc1-3 strains, Dr. Janet M. Shaw for the tom70∆tom71∆ strain, and Dr. Kaori Yunoki-Esaki for recombinant pSu9-DHFR. We are grateful to the members of the Endo lab for valuable discussions. We acknowledge support of this work by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and a CREST Grant from the Japan Science and Technology Corporation (JST).
Author contributions
JS, YT, TY, and TE designed experiments. JS and TY performed experiments and analyzed the data. JS, YT, TY, and TE wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]
- Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. Tim23–Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol. 2009;184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- Riezman H, Hay R, Gasser S, Daum G, Schneider G, Witte C, Schatz G. The outer membrane of yeast mitochondria. Isolation of outside-out sealed vesicles. EMBO J. 1983;2:1105–1111. doi: 10.1002/j.1460-2075.1983.tb01553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Jensen RE, Guido EC. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989;264:21091–21096. [PubMed] [Google Scholar]
- Bruckmann A, Hensbergen PJ, Balog CIA, Deelder AM, Brandt R, Snoek ISI, Steensma HY, van Heusden GPH. Proteome analysis of aerobically and anaerobically grown Saccharomyces cerevisiae cells. J Proteomics. 2009;71:662–669. doi: 10.1016/j.jprot.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Lauffer S, Mäbert K, Czupalla C, Pursche T, Hoflack B, Rödel G, Krause-Buchholz U. Saccharomyces cerevisiae porin pore forms complexes with mitochondrial outer membrane proteins Om14p and Om45p. J Biol Chem. 2012;287:17447–17458. doi: 10.1074/jbc.M111.328328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T, Stan T, Neupert W, Rapaport D. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J Biol Chem. 2003;278:42064–42071. doi: 10.1074/jbc.M305736200. [DOI] [PubMed] [Google Scholar]
- Merklinger E, Gofman Y, Kedrov A, Driessen AJM, Ben-Tal N, Shai Y, Rapaport D. Membrane integration of a mitochondrial signal-anchored protein does not require additional proteinaceous factors. Biochem J. 2012;442:381–389. doi: 10.1042/BJ20111363. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol. 2006;174:379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Yatsukawa Y, Esaki M, Aiken-Hobbs AE, Jensen RE, Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko O, Melin J, Schulz C, Kilisch M, Hutu DP, Rehling P. Signal recognition initiated reorganization of the presequence translocase during protein import. EMBO J. 2013;32:886–898. doi: 10.1038/emboj.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz LS, et al. The presequence pathway is involved in protein sorting to the mitochondrial outer membrane. EMBO Rep. 2014 doi: 10.1002/embr.201338144. DOI 10.1002/embr.201338144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki M, Kanamori T, Nishikawa S, Shin I, Schultz PG, Endo T. Tom40 protein import channel binds to non-native proteins and prevents their aggregation. Nat Struc Biol. 2003;10:988–994. doi: 10.1038/nsb1008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.