Abstract
Recombination systems based on λ and Cre/loxP have been described to facilitate gene transfer from one vector to another in a high-throughput fashion, avoiding the bottlenecks associated with traditional cloning. However, no system described to date is suitable for the cloning of affinity reagents selected from display libraries, which requires that the recombination signals flanking the affinity reagent are translated with a minimum impact on functionality. As affinity reagents will be essential tools in the functional characterization of proteomes, and display technologies represent the most effective means to generate such affinity reagents on a genomic scale, we developed a Cre/loxP-based system, using mutually exclusive heterologous loxP sites placed 5′ (Lox 2372) and 3′ (Lox WT) of an affinity reagent (scFv). The translated lox sites have minimal impact on scFv expression or functionality, and, in association with a conditionally lethal gene (SacB) permit efficient, high-fidelity transfer to destination vectors. This approach will considerably facilitate the high-throughput downstream use of affinity reagents selected by display technologies, as well as being widely applicable to general recombinatorial cloning for genomic purposes.
Biological processes are increasingly being investigated at the organismal level. Whereas genomic sequences are an all important first step in this endeavor, ultimately, a detailed mechanistic understanding requires information acquired at the protein level. The most extensive functional genomic studies have been carried out in yeast, with individual gene knock outs (Ross-Macdonald et al. 1999), overexpression and proteome chips (Zhu et al. 2001), intracellular localization by tagging (Kumar et al. 2002), protein–protein interaction studies by phage display (Tong et al. 2002), yeast two-hybrid (Schwikowski et al. 2000; Uetz et al. 2000), and widespread mass spectrometric (MS) analysis of purified complexes (Gavin et al. 2002; Ho et al. 2002) having provided large amounts of information. One reason yeast has been used so extensively is the availability of homologous recombination, permitting the replacement of endogenous genes by modified copies. In fact, most of the studies cited above would not have been possible without exploiting this technique, which often involves the genetic fusion of a tag—either a detection peptide recognized by a monoclonal antibody (e.g., myc; Evan et al. 1985), or a “tandem affinity purification tag” (Rigaut et al. 1999), which can be used for purification and subsequent mass spectrometry (MS) of complexes. Homologous recombination has also been used to transfer selected antibodies from yeast display vectors to secretion vectors (Feldhaus et al. 2003). As homologous recombination is not available for most genomes, the only alternative to the fusion of a general tag (using a single detection reagent) is the derivation of specific antibodies, or binding ligands, for all gene products that can be used in standard immunological techniques (Western blotting, immunoprecipitation, immunofluorescence, immunohistochemistry, and purification), as well as new proteomic techniques still under development (antibody chips, MS), and potentially in applications such as biosensors. However, research at a genomic scale requires both a high-throughput capacity, and an ability to derive antibodies against well-conserved proteins, neither of which traditional immunization is capable of achieving. In particular, the generation of antibodies against conserved proteins is difficult, due to clonal deletion of B (Burnet 1959; Talmage 1959) or T (Werlen et al. 2003) cells, as well as receptor selection (Nemazee 2000) at the B cell level. Although antibodies against conserved antigens can be generated, and tolerance overcome by chemical coupling to adjuvants, genetic fusion of T cell epitopes (Dalum et al. 1996, 1997) or prolonged immunization strategies (Cattaneo et al. 1988), these procedures are not suitable for high-throughput antibody generation.
Antibody fragments, such as Fabs or single-chain Fvs (scFv), in which the antigen-specific immunoglobulin variable domains from both the heavy (VH) and light (VL) chains are linked into a single DNA-coding sequence (Bird et al. 1988; Huston et al. 1988), have been proposed as alternate recognition ligands with high affinity and specificity for use in the previously mentioned functional genomic applications. Functional scFvs, displayed on the surface of bacteriophage particles (McCafferty et al. 1990), can be rapidly isolated against any target from libraries typically >109 in complexity (Vaughan et al. 1996; Sheets et al. 1998; de Haard et al. 1999; Sblattero and Bradbury 2000), without the need for complex antigenic preparations to overcome tolerance problems related to immunological tolerance or toxicity, and with the benefit that the gene encoding the selected antibody is simultaneously cloned for downstream genetic manipulations. This latter point has allowed scFvs originally selected by phage display to be easily recloned for expression in different cellular compartments (Persic et al. 1997a), as full-length immunoglobulins (Persic et al. 1997b) or as fusion proteins containing different functional domains (Griep et al. 1999; Muller et al. 1999; Hink et al. 2000). The ability to manipulate selected scFv antibody genes in a potentially high-throughput format greatly enhances the impact of this technology in functional genomic applications.
The need to transfer scFvs from a selection vector to a downstream vector has been traditionally carried out by ensuring that downstream vectors have restriction sites that are compatible with those found in the selection vector (e.g., Krebber et al. 1997; Persic et al. 1997a). However, this procedure is time consuming, requiring DNA production, cleavage, purification, ligation, transformation, and screening. Recombination, which avoids the need for such manipulation, has been proposed as an alternative to rapidly transfer genes from source vectors to destination vectors, with the commercially available Gateway kit (Invitrogen; Hartley et al. 2000) based on λ bacteriophage being most widely used to create large libraries of individual gene-specific ORFs (e.g., Caenorhabditis elegans; Reboul et al. 2001). Unfortunately, this system (see Discussion) is not appropriate for selection from phage display libraries, as the length of some of the recombination signals, resulting from the asymmetric nature of the recombination process, would prevent the functional expression of scFvs in all vectors, requiring multiple recombination rounds for transfer.
Another bacteriophage, P1, also uses a site-specific recombination system that is responsible for partitioning newly synthesized genomic copies during replication (Abremski and Hoess 1984; Hoess and Abremski 1984). This system is composed of a 38-kD phage-encoded Cre recombinase that mediates symmetrical recombination between two 34-bp loxP sites (Abremski and Hoess 1984), which are recreated after recombination. Recombination between two compatible loxP sites will excise or invert the intervening DNA in the case of an intramolecular reaction or transfer suitably flanked loxP DNA in an intermolecular double cross-over recombination event. The Cre/loxP system does not require accessory factors to carry out recombination in vivo or in vitro, and studies have identified several hetero-specific loxP sequences that exclusively recombine with themselves, but not with wild-type lox (Hoess et al. 1986; Sauer 1992; Lee and Saito 1998; Siegel et al. 2001). The combination of WT loxP and 511 loxP has been successfully used for a double reciprocal cross-over reaction to transfer a DNA segment or gene to a predetermined target site (Bethke and Sauer 1997; Feng et al. 1999; Trinh and Morrison 2000), and this has proved to be an important tool for the in vivo manipulation of eukaryotic genomes, including gene activation and deactivation studies in both mammalian cells and transgenic mice. Recent reports, however, have suggested recombination can occur between wild-type and 511 loxP sequences (Lee and Saito 1998; Feng et al. 1999), the levels depending upon the orientation of the sites with respect to one another (Siegel et al. 2001). A translated loxP recombination site has also been used as a peptide spacer between VH and VL chains in order to create large phagemid antibody libraries (Griffiths et al. 1994; Sblattero and Bradbury 2000), suggesting that the amino acid sequence derived can be relatively innocuous.
We recently published work describing the use of an in vivo system to identify noncompatible heterologous loxP sequences (Siegel et al. 2001). On the basis of that study, we have examined the possibility of using the Cre/lox system for transferring genes of interest from one vector to another. We modified our standard filamentous phagemid display vector flanking the scFv gene by 2372 and wild-type loxP sequences. The extraneous amino acids encoded by each loxP site did not substantially lower the ability of the antibody to be functionally incorporated into the phage particle for display or alter the ability to recognize its target antigen. A model destination vector was constructed containing the appropriate loxP sequences flanking a negative selection marker and accurate and efficient transfer of an scFv gene from the display to the destination vector was demonstrated, with the scFv maintaining functionality in the new vector. This will considerably facilitate the implementation of affinity reagents selected using phage display in functional genomics and proteomics.
RESULTS
Functionality of scFv-lox (pDAN11) Vectors
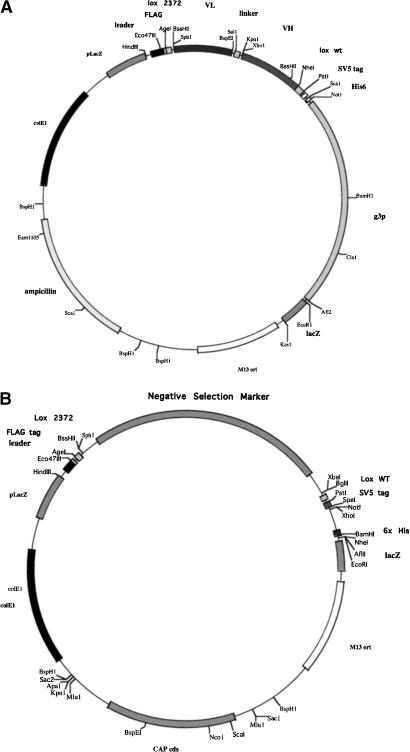
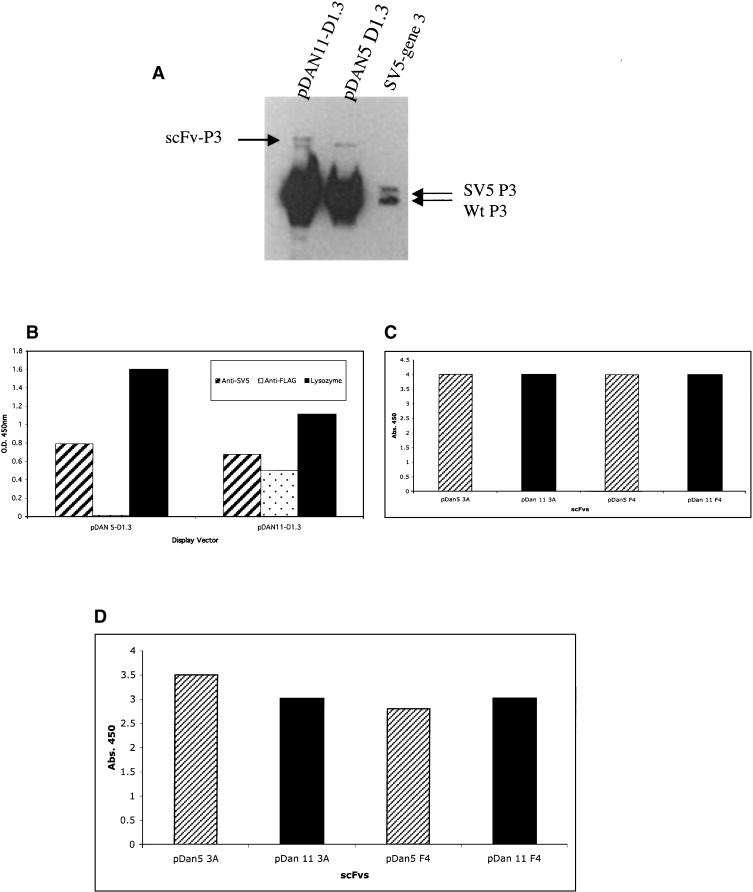
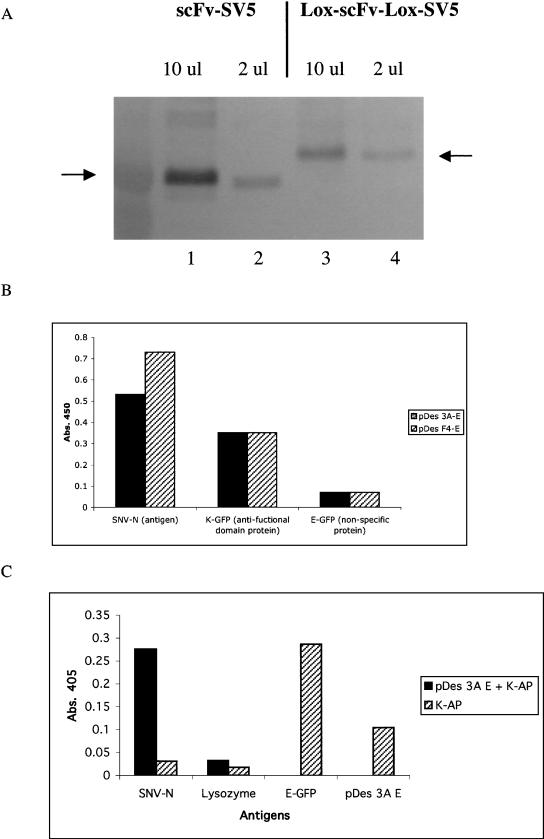
To use a recombinatorial method to transfer selected scFv genes from one vector to another in an expedient manner, a display vector containing an scFv gene flanked by mutually exclusive loxP recombination sites was constructed. Our standard phagemid display vector (pDAN5; Sblattero and Bradbury 2000) was modified to include at the amino terminus of the scFv the FLAG epitope tag and the Lox 2372 recombination sequence; and between the carboxyl terminus of the scFv and gene 3, the Lox WT recombination sequence and the SV5 epitope tag (Fig. 1A). The lox sites are DNA recombination signals that are translated as fusion tags appended to each end of the scFv, in the same frame previously shown to be functional in an scFv linker (Sblattero and Bradbury 2000). Although the lox site was effective in this context, we wanted to ensure that the addition of the loxP sites had no deleterious effect on the ability of scFv antibodies to be displayed on the phage particle or interfered with their ability to recognize a target antigen. Equivalent numbers of phagemid particles from the pDAN11 vector containing the D1.3 scFv gene (pDAN11-D1.3) flanked by translated lox sites, or a control vector that does not (pDAN5-D1.3) were subjected to Western blot analysis using an anti-gene 3 antibody (Fig. 2A). The level of scFv display obtained with the the pDAN11-D1.3 vector (Fig. 2A, lane 1) was comparable to that observed from the control vector (Fig. 2A, lane 2). Similar display levels were also observed using an antibody to the SV5 epitope located between the carboxy-terminal portion of the scFv antibody and the gene 3 coat protein in both pDAN5 and pDAN11 constructs (data not shown). Two gene 3 fusion proteins appear to be present in the pDAN11-D1.3 phagemid particles, and this may represent a certain level of proteolytic cleavage, probably within either the FLAG epitope or the Lox 2372 sequence located at the amino terminus. The control lane (Fig. 2A, lane 3) consists of a phagemid containing only the SV5 tag and no antibody, rescued with helper phage. The two bands observed correspond to (bottom) helper phage p3 and (top) p3 fused to the SV5 tag, which is slightly bigger.
Figure 1.
(A) Map of the lox phagemid display vector pDAN11. The Flag affinity tag and Lox 2372 site (ATAACTTCGTATAGGATACCTTATACGAAGTTAT; 13-bp inverted repeats are underlined) are located upstream of the scFv antibody gene, whereas the Lox WT (ATAACTTCGTATAGCATACATTATACGAAGTTAT) and SV5 affinity tags are located downstream of the scFv gene and upstream of the bacteriophage gene 3-coding sequence, resulting in display of the fusion protein. (B) Generic Lox Destination Vector. A negative selection marker (either Tetr or sacB) is flanked by Lox 2372 and Lox WT, which, after exchange with the scFv gene, creates an in-frame fusion with the periplasmic leader sequence and the SV5 epitope and 6x His tags.
Figure 2.
(A) scFv phage display levels. (Lane 1) pDAN11-D1.3; (lane 2) pDAN5-D1.3; (lane 3) a control vector containing only the SV5 tag fused to gene 3 to mark the size of unfused gene 3 protein. Lower arrows denote native gene 3, and upper arrow shows the scFv-gene3 fusion protein. The 2 × 109 phage particles were loaded in lanes 1 and 2, and both the native and fusion forms of gene 3 were visualized using anti-gene3 mAb (1:2000). (B) Phage ELISA signals observed for either pDAN5 D1.3 or pDAN11 D1.3 against anti-SV5 and anti-Flag mAbs, and lysozyme adsorbed to a 96-well plate. Bound phage were determined using anti-M13 HRP (1:5000) conjugate with the background subtracted from the reported values. (C) Phage ELISA. (D) scFv ELISA signals observed for either pDAN5 or pDAN11 3A and F4 scFv against SNV-N adsorbed to a 96-well plate. The bound phage were detected by anti-M13 horseradish peroxidase conjugate (Pharmacia), and the bound scFvs by SV5 anti-tag antibody (Hanke et al. 1992) followed by anti-mouse antibody conjugated to horseradish peroxidase.
These phage were also tested for their ability to bind SV5 mAb (which recognizes both pDAN11 and pDAN5), anti-FLAG mAb (which only recognizes pDAN11 and lysozyme [the antigen recognized by D1.3, the scFv cloned]). As seen in Figure 2B, both vectors give comparable display levels, with only pDAN11 being recognized (as expected) by FLAG. Phage from both vectors displaying the D1.3 scFv also bind lysozyme at similar levels (Fig. 2B), indicating that the presence of 5′ and 3′ lox sites in pDAN11 do not appear to interfere with scFv function when displayed on phage.
As a further test of functionality, two scFvs (3A, F4) selected against the Sin Nombre Virus Nucleocapsid protein (SNV-N) were also cloned into pDAN11. The anti SNV-N scFv were analyzed for their ability to recognize antigen in both phage-displayed and soluble scFv formats. Both gave similar signals by phage and scFv ELISAs (Fig. 2C,D) when compared with versions that lacked the translated lox sites (pDAN5). These results showed that the lox-scFv-lox format does not appear to interfere with the functionality of the single chains when used as soluble proteins or displayed on phage.
Functionality of Tetr/SacB Lox-Destination Vectors
Having shown that the incorporation of translated heterologous loxP sites flanking the scFv antibody gene does not significantly perturb functionality of the phagemid display vector, we constructed a vector capable of receiving an scFv gene from the display vector in a Cre-mediated double cross-over recombination event (lox-destination vector, Fig. 1B). To facilitate the propagation of only desired clones (destination vectors containing the scFv gene) after transduction of the recombination products into Escherichia coli cells, we incorporated two selection gates. First, the pDAN11 phagemid display vector (ampicillin) and the lox-destination vector (chloramphenicol) contain different antibiotic resistance genes. Second, the lox-destination vector contains a negative selection marker that is toxic under appropriate induction conditions. Previous studies have used the ccdB gene in this capacity (Bernard 1996); however, cells carrying the F pilus (required for phagemid infection) are not adversely affected by the ccdB gene product, as the antidote gene (ccdA) is encoded on the F plasmid. Furthermore, this system is not inducible, and requires different bacteria for selection and propagation, necessitating the search for an alternative marker.
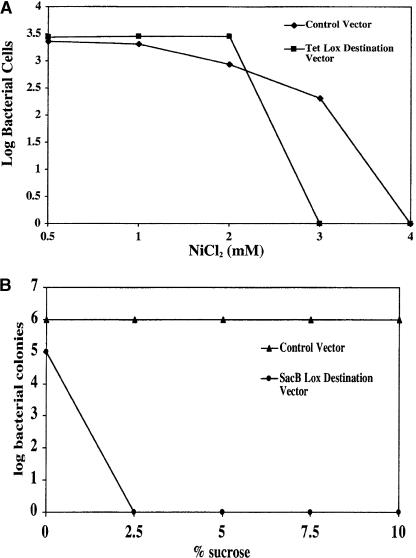
Both the tetracycline resistance gene (Tetr) and the Bacillus subtilis levansucrase-encoding gene (sacB) have been shown to inhibit E. coli growth in the presence of either NiCl2 or sucrose, respectively (Gay et al. 1985; Podolsky et al. 1996). Therefore, we constructed two different lox-destination vectors containing either the Tetr or sacB genes flanked by the Lox 2372 and Lox WT recombination sequences (Fig. 1B). E. coli cells were separately transduced with either of these vectors and tested for the ability to grow on chloramphenicol plates with increasing amounts of NiCl2 or sucrose as compared with a control vector that did not contain a negative selection marker. Cells containing the Tet destination vector were able to grow on plates that contained 1 or 2 mM NiCl2, but no growth was observed on plates with a NiCl2 concentration of 3 mM or above. Control cells lacking the Tetr negative selection marker were selectively enriched greater than two logs at 3 mM NiCl2, but, like the cells containing the Tet vector, were unable to grow in the presence of 4 mM NiCl2 (Fig. 3A), leading to a relatively narrow selection window. Results with the SacB destination vector (Fig. 3B) were more permissive. Cells containing the sacB gene were unable to survive on CAP plates containing 2.5% sucrose or higher. However, unlike NiCl2, the control cells were unaffected at all sucrose concentrations tested, resulting in a rigorous six-log enrichment without any apparent toxicity.
Figure 3.
(A) NiCl2 titration for Tetr negative selection marker. Bacterial cells with vectors that either contained the CAP resistance gene (control vector) or both the CAPr and Tetr (Tet lox-destination vector) were plated on CAP containing the indicated concentration of NiCl2. The log units of surviving cells are shown on the y-axis. (B) Sucrose titration for sacB negative selection marker. Bacterial cells with vectors that contained either the SacB lox-destination vector (CAPr and sacB) or a control vector (CAPr only) were plated on CAP containing the indicated percentage of sucrose. The log units of surviving cells are shown on the y-axis.
Recombinatorial Cloning and Efficiency
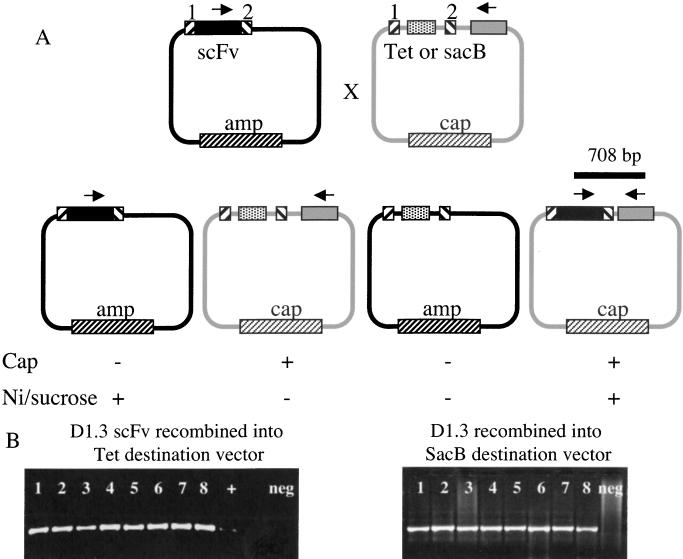
A scheme of the Cre-mediated double cross-over transfer of an scFv gene from the pDAN11 phagemid display vector to the lox-destination vector with all possible recombination products is shown in Figure 4A. From the previous experiment, we anticipated that the incorporation of a negative selection marker would ensure that only those destination vectors that have replaced this negative selection marker with the desired scFv gene during recombination would lead to colony formation under appropriate conditions. As an initial test, we cotransduced the pDAN11 phagemid display vector containing the D1.3 scFv gene with either the Tetr or the SacB lox-destination vector into BS1365 cells that constitutively express Cre recombinase. Phage particles containing the recombination products were isolated and transduced into DH5αF' cells. Infected cells were plated on chloramphenicol plates containing either 3 mM NiCl2 for the Tet destination vector or 8% sucrose for the SacB destination vector.
Figure 4.
(A) scFv recombinatorial subcloning scheme. Recombination between the loxP sites (hatched boxes) mediates exchange of the scFv gene (black box) and the negative selection marker (white box) between the ampicillin (AMP) and chloramphenicol (CAP) resistant vectors. Arrows indicate diagnostic PCR primer binding sites. (B) PCR confirmation of scFv recombination into destination vector. Colonies from the double-selection plate after recombination were analyzed by PCR using a set of diagnostic primers. One primer is internally located in the D1.3 scFv gene with the other external to the Lox WT site (SacB vector; bottom). Negative control lanes are marked.
As expected, only those colonies that had been infected with phage containing the D1.3 scFv in the destination vector backbone were able to survive the double selection gate. Random colonies able to survive the double selection were analyzed by PCR to confirm the presence of the D1.3 scFv gene in the destination vector following recombination. The primers used hybridized to the D1.3 light-chain CDR3 and a region of the destination vector external to the Lox WT recombination sequence, and so were only able to produce an amplification product when these two sequences were brought together on the same plasmid (see Fig. 4A). This product (708 bp) was consistently generated in eight of eight colonies analyzed from chloramphenicol/8% sucrose plates, and on many (but not all) occasions on colonies on the chloramphenicol/3 mM NiCl2 plates. Examples of a representative experiment are shown in Figure 4B. On the basis of the increased robustness of the sacB selection, this was chosen as the negative selectable marker. The appearance of undesired plasmids after transduction of the recombination products obtained with the Tet destination vector is probably explained by the narrow selection window observed in Figure 3A.
Functionality of scFvs Recombined Into Lox-Destination (pDes) Vector
Effects on scFv Expression
Although we have demonstrated that the addition of lox sites does not affect scFv functionality or display, this was within the context of an scFv cloned by traditional means into a phage display vector, and not one that had been transferred to a destination vector by recombination. Therefore, the level of scFv expression after recombination into the SacB-destination (expression) vector was compared with a control vector that was identical, except that it did not contain the loxP sequences. DH5αF' cells were induced for soluble scFv expression (no longer fused to the bacteriophage coat protein), and the resulting culture supernatants subjected to SDS-PAGE and analyzed by Western analysis using SV5, a monoclonal antibody recognizing a carboxy-terminal tag present in both scFv antibody constructs. Figure 5A demonstrates that scFvs are expressed at comparable levels in the two vectors, with the scFv flanked by lox sites being expressed only slightly less than the corresponding control. The figure also shows the relatively modest increase in size imparted by the two lox sequences and the FLAG epitope tag.
Figure 5.
(A) Soluble scFv Western. (Lanes 1,2) D1.3 scFv expressed from a control vector that produces scFv with attached SV5 and 6x His tags at the carboxyl terminus; (lanes 3,4) D1.3 scFv expressed from the pD1.3 destination vector that produces the D1.3 scFv with FLAG and translated Lox 2372 at the amino terminus and Lox WT, SV5, and 6x His tags at the carboxyl terminus. A total of 10 μL of supernatant was used in lanes 1 and 3, and 2 μL in lanes 2 and 4 (see Methods). Arrows denote correctly sized scFv visualized with anti-SV5 and goat anti-mouse-AP conjugate antibodies. (B) scFv ELISA was performed with pDes 3A and F4 against SNV-N, E-coil GFP, and K-coil GFP coated on a 96-well plate. Antigen reactive scFv were detected by anti-SV5 tag antibody followed by anti-mouse antibody conjugated to horse radish peroxidase. (C) scFv ELISA was performed with pDes 3A E-coil scFv incubated with K-coil alkaline phosphatase (K-AP) against SNV-N and lysozyme. Control reactions measured K-coil reactivity toward SNV-N, lysozyme, E-coil GFP, and pDes 3A-E scFv.
Functional Analyses of scFvs Transferred by Recombination
On the basis of the demonstration that Cre-mediated recombination efficiently transfers selected scFvs from the phage display vector, we constructed destination vectors (pDES) that fuse functional domains to the expressed scFv. One such pDES vector adds an E coil to the carboxyl terminus of the scFv, which is able to form a coiled-coil with its partner K coil with an affinity of 60 pM (De Crescenzo et al. 2003). As a result, after recombination, each scFv should be covalently linked to an E coil that is able to bind both to its specific antigen and a K coil. These analyses were performed with the anti-SNV-N scFvs (3A and F4). The ability of the scFvs to bind their antigen was tested by ELISA with SNV-N coated on the ELISA plate (Fig. 5B). The presence of the E coil fused to these scFvs was tested by ELISA using GFP-K coil fusion as the antigen. The specificity of the E–K coil interaction was tested using GFP–E coil fusion as a negative control (Fig 5B). The results show that the scFvs are still able to bind to their antigens and are now also recognized by the K coil. Although this demonstrates function of both scFv and E coil, it does not formally show that functional scFvs are necessarily connected to functional E coils. To demonstrate this point, an ELISA was performed with E coil–scFvs bound to K coil–alkaline phosphatase. In these experiments, only scFvs with both antigen and K coil-binding ability were able to produce a positive signal. Our results showed that scFv 3A-E coil produced positive signal with its specific antigen SNV-N, and not with an irrelevent antigen (lysozyme; Fig. 5C).
DISCUSSION
We have developed a symmetrical recombination system that is capable of transferring an isolated scFv antibody gene from a selection vector into any number of designated downstream vectors using site-specific recombination. Mutually exclusive heterologous loxP sites were placed immediately upstream (Lox 2372) and downstream (Lox WT) of the DNA encoding the scFv antibody (Fig. 1A). Incorporation of these sequences did not alter the degree of scFv display on the phagemid particle (Fig. 2A,B), nor the ability to recognize the target antigen (Fig. 2B,C,D). A recombination protocol was devised to expedite the transfer of the scFv gene contained within the phagemid display vector to another destination vector devoid of the bacteriophage coat protein-coding sequence (Fig. 1A). The correct recombinant was obtained by imposing antibiotic resistance selection for the desired construct and a secondary negative selection against the starting molecules, and the individual components (antibody, E coil, and detection tags) of the destination vector were all shown to be functional. Two different negative selection markers were examined. The efflux pump responsible for tetracycline resistance also allows Ni2+ to permeate the cell wall at lower concentrations than cells lacking this protein, thereby decreasing a bacterial cell's susceptibility to Ni2+ toxicity (Podolsky et al. 1996; Fig. 3A). The levansucrase protein encoded by the Bacillus subtilis sacB gene synthesizes a substrate, in response to sucrose, that is toxic to gram-negative bacteria when uncoupled from the remaining regulatory enzymes (Gay et al. 1985; Fig. 3B). Unlike NiCl2, which tended to kill cells lacking the tetracycline resistance gene relatively easily, the elevated concentrations of sucrose used to select against sacB containing bacteria had no effect, making it the more robust negative selectable marker. When used in model experiments, it was able to allow the growth of only those clones in which the destination vector contained scFvs transferred by recombination (Fig. 3A,B).
To fully exploit scFvs selected using in vitro methods often requires moving the scFv gene from the phagemid display vector into a variety of more specialized vectors, creating fusion proteins better suited for a particular functional use of the isolated scFv antibody. Site-specific recombination offers many advantages over traditional cloning approaches. The elimination of the need to use restriction endonucleases allows any scFv gene to be cloned into any vector of choice (both containing appropriate recombination signals) without having to address problems related to the presence of compatible or cryptic restriction enzyme sites contained within the gene to be subcloned. This issue is particularly troublesome when the exact sequence of the scFv gene has not yet been determined, even when restriction enzymes that are rare in V genes are used (Persic et al. 1997a; Sblattero and Bradbury 1998). In addition, because transfer by site-specific recombination does not rely on PCR-based amplification, sequence mutations to the subcloned gene are not expected, minimizing the need for sequence validation of each destination clone created. Furthermore, the site-specific nature of this subcloning prevents reading frame changes, allowing the faithful transfer of scFv genes into destination vectors providing amino-terminal and/or carboxy-terminal fusions. The speed and ability to subclone scFv genes into any number of destination vectors in a parallel fashion further enhance the potential for the use of scFv antibodies in functional genomic applications.
We have chosen to use the Cre/loxP recombination system using mutually exclusive heterologous loxP sequences flanking the scFv gene to mediate transfer from the phagemid display vector rather than a Cre/loxP system using only one loxP site (Liu et al. 1998) or the λ bacteriophage site-specific recombination system (Hartley et al. 2000). The univector plasmid-fusion system (Liu et al. 1998) uses Cre/loxP recombination to mediate fusion between the plasmid containing the gene of interest (donor vector) with a host vector containing regulatory information. This system utilizes a single loxP WT sequence on each plasmid, and recombination events fusing the donor and acceptor plasmids are genetically selected by antibiotic resistance conferred by a combination of the host-strain genotype and the origin of replication present in the fused recombinant expression construct. Another available Cre/loxP system (Creator System, Clontech) places the gene of interest along with an antibiotic resistance marker between two identical loxP WT sequences. This defined segment of the donor plasmid is then recombined into an acceptor vector containing a single copy of the same loxP WT sequence. Recombined clones are positively selected using the antibiotic marker contained within the loxP sites on the donor plasmid in conjunction with a negative selection marker on the donor vector backbone. However, neither of these systems is feasible for an scFv antibody system that requires the ability to transfer the gene of interest in a directional manner from one plasmid to another without the presence of additional (regulatory) sequences flanking the scFv.
Site-specific recombination based on the λ phage recombination system is available in a commercial format (Hartley et al. 2000; Gateway Technology, Invitrogen) and allows user-defined DNA to be transferred between plasmids directionally. This requires the phage-encoded integrase (Int) and host-derived integration host factor (IHF) to catalyze recombination between a 24-bp attB site (originally in the bacterial genome) and a 243-bp attP site (originally in the phage genome) to generate the 100-bp attL and 168-bp attR sites (that originally flank the newly integrated λ DNA), and Int, IHF, and exisionase (Xis) to carry out the reverse reaction (for review, see Landy 1989). Whereas this system has been extensively and successfully used in cloning collections of individual genes into a nonfunctional source vector followed by recombination into functional destination vectors (Walhout et al. 2000; Reboul et al. 2001), it is not feasible for systems that require expression of the scFv gene, and therefore translation of each recombination site, with each vector used. For λ recombination to be used, either the display vector or the destination expression vector would (at some stage) have to contain the attL sites that contain stop codons in all three frames. Futhermore, the presence of different flanking sequences (based on attB or attL) in different vectors complicate the system. The λ system could be used if antibodies in the original phage display library were flanked by attB sites and recombined first into an attR-containing vector, and subsequently into attL-destination final vectors. Although this would maintain the 24-bp attB sites in initial and final vectors, it would involve a complex double-recombination procedure of unknown efficiency. The λ recombination system also requires a number of different enzymes to be present (Int, IHF, and Xis) for recombination to occur, whereas the Cre system requires a single recombinase easing in vivo manipulations as shown in the protocol we have used in this work.
The Cre/loxP system has been primarily used to facilitate the generation of transgenic animals expressing novel gene products or null mutants after precise alteration of endogenous genes. A double reciprocal cross-over replacement event between the targeting vector and the chromosomal target using heterospecific loxP sites has been shown to greatly increase the recovery of integrants, while eliminating the unnecessary incorporation of vector DNA into the genome (Bethke and Sauer 1997). Building upon this principle, we have created a recombinatorial cloning system using Cre/loxP to transfer scFv antibody genes between various vectors. In this study, we examined expression and functionality of the scFv antibody both in the source vector, which contained gene 3, as well in the destination vector, in which gene 3 was removed and an additional functionality was introduced (an E coil) (Figs, 4A and 5A). The fact that the scFvs were functional in both vectors suggests that flanking translated lox sites do not appear to be inhibitory to antibody function, and will allow us to create a number of different destination vectors fulfilling a variety of applications. For example, the occasional scFv gene that is not expressed at high yields in bacteria could be recombined into a shuttle vector allowing expression in Pichia (Freyre et al. 2000; Powers et al. 2001) or as a fusion protein to enhance expression, solubility or detection (Berdichevsky et al. 1999; Bach et al. 2001). Bivalent scFv antibodies can be created by fusion to complimentary amphipathic coiled coils (Pack and Pluckthun 1992; Pelletier et al. 1999; Arndt et al. 2000), whereas fusion to fluorescent proteins could create intrinsically fluorescent primary-binding ligands (Hink et al. 2000; Morino et al. 2001) and fusions to metal-binding domains allow electron spectroscopic imaging after saturation with nickel or cobalt (Malecki et al. 2002). The ability to create these various constructs in such a robust and parallel manner using the Cre/loxP site-specific recombination system greatly expands the functionality of any given scFv antibody and enhances the applicability to functional genomics. Furthermore, although we have developed this technology for use with scFvs, it is likely that libraries of other affinity reagents, on the basis of alternative scaffolds (for review, see Skerra 2000) could just as easily be created within the context of the lox sites described here. The potential applications could also extend beyond affinity reagents to collections of genes derived either individually or en masse, in a fashion similar to the λ recombination system.
METHODS
Bacterial Strains
DH5αF': (GIBCO BRL): F'/endA1 hsdR17 (rK– mK+) supE44 thi-1 recA1 gyrA (Nair) relAI Δ (lacZYA-argF) U169 deoR (ø80dlacΔ)lacZ) M15)
BS1365: BS591 F' kan (Bs591: recA1 endA1 gyrA96 thi-1 Δ lacU169 supE44 hsdR17 [λ imm434 nin5 X1-cre] (Sauer and Henderson 1988).
JL-1 expresses cre recombinase constitutively and also expresses LacIQ. It was created (by Jianlong Lou, UCSF) in two steps. In the first step, TOP10 F' was mated with Stbl2, to create ST-1, by selecting for growth on minimal medium without leucine and with proline and tetracycline. In the second step, ST-1 was mated with BS591, to create JL-1 by selecting on minimal medium and tetracycline. JL-1 was tested by infecting with pWT/WT-lox-GFP phagemid as previously described (Siegel et al. 2001).
ST-1: F'{lac Iq, Tn 10(TetR)} mcr A Δ(mcrBC-hsdRMS-mrr) recA1 endA1 lon gyrA96 thi supE44 relA1 λ – Δ (lac-proAB).
JL-1: F'{lac Iq, Tn 10(TetR)} recA1 endA1 gyrA96 thi-1D lacU169 supE44 hsdR17[λ imm 434 nin5 X1-cre].
Construction of pDAN11 D1.3, 3A, F4
pDAN5 D1.3 (Sblattero and Bradbury 2000) was partially digested with SphI and to completion with EcoRI, and then filled in and cut back with T4 DNA polymerase. The full-length vector fragment (5273 bp) was purified and religated. The correct construct recreated the EcoRI site and resulted in the plasmid pDAN5 D1.3-SphI. This differed from pDAN5 by removing the SphI and lox site found at the 3′ end of gene 3 in pDAN5. The leader sequence from pDAN5 was PCR amplified with two primers as follows: pDAN9.1 for (GAAGTTATGGTCGACCCGCTAGCGGCCATG GCGGCCGATAACTTCGTATAATGTATGCTATACGAAGTTATAC CGGTCTCATCTTTATAATCGGCAAGAGCGCTGCTTGCTGC GAG) and M13 rev (AGCGGATAACAATTTCACACA). The 244-bp product was cloned into pDAN5 D1.3-SphI using HindIII and SalI to create pDAN8. This replaced the leader-VL gene with a polylinker with the following structure: leader in which BssHII was changed to Eco47III, FLAG tag, AgeI, Lox WT site, SfiI/NcoI, SalI.
The D1.3 scFv gene was amplified with pDAN11BH5 for (GATGAGACCGGTATAACTTCGTATAGGATACCTTATACGAA GTTATCGGGCGCGCATGCCGACATT) and pDAN11Pst1 rev (ATGGTGAGTACTATCCAGGCCCAGCAGTGGGTTTGGGATTG GTTTGCCTGCAGATAACTTCGTATAATGTATG) primers. These primers added an Age1 restriction site, the Lox 2372 recombination site (ATAACTTCGTATAGGATACCTTATACGAAGTTAT; 13-bp inverted repeats are underlined) plus BssH2 and Sph1 sites upstream of the D1.3 scFv gene and the Lox WT recombination site (ATAACTTCGTATAGCATACATTATACGAAGTTAT) and Pst1, SV5 epitope tag and Sca1 site downstream of the gene. The amplified DNA was digested with Age1 and Sca1 and ligated into similarly digested pDAN8 vector DNA to create pDAN11. This flanks the scFv with the same restriction sites used in our original display vector, pDAN5. The final vector structure is illustrated in Figure 1A.
The D1.3 scFv gene in pDAN11 D1.3 was replaced with 3A or F4 scFv (recognizing the SNV-N protein from the Sin Nombre Hanta Virus) genes from pDAN5 vectors using BssHIII and NheI restriction digestion, to create pDAN11 3A and pDAN11 F4.
Construction of Tetr and SacB Lox-Destination Vectors
The ampicillin resistance gene from pDAN11 was replaced with the chloramphenicol (CAPR) resistance gene from pBSL121 (Alexeyev et al. 1995), to create pDAN11-CAP, by amplifying the CAPR gene with primers that appended BspH I sites at either end, and using the two BspHI flanking the β lactamase gene as cloning sites. This intermediate vector was then digested with Sph1 and Nhe1 to remove the scFv gene contained between the 5′ Lox site (2372) and 3′ Lox site (WT). The gene encoding tetracycline resistance (Tetr) was amplified from pBSL190 (Alexeyev et al. 1995) with primers that added a Sph1 restriction site at the 5′ end (5′ TET Sph1: ACATGCATGCCAATTCTCATGTTTGACAGC) and a Spe1 site on the 3′ end (3′ TET Spe1: GGACTAGTAGATCTAGAC AGCTTGATCAAGGGTTGGTTTGC) of the amplified DNA. The PCR product was digested with SphI and SpeI (NEB) and ligated into pDAN11-CAP cleaved with SphI and NheI to create pDAN11-CAP/Tet, which contains the Tetr gene in place of a scFv gene between the 2372 and wild-type loxP sites. pDAN11-CAP/SacB was constructed by recloning the SacB gene from pUCD800 (Gay et al. 1985) into pBluescript SK (Stratagene) using BamHI and PstI. After testing in pBluescript SK, the tetracycline resistance gene in pDAN11-CAP/Tet was replaced with the SacB gene using SphI and PstI. The DNA encoding the bacteriophage minor coat protein gene 3 was removed from pDAN11-CAP/Tet after digestion with Pst1 and Afl2 restriction enzymes and ligated to an appropriately digested adapter fragment containing the SV5 epitope tag, 6X His tag, an E coil gene (Chao et al. 1996, 1998) and NotI, XhoI, BamHI, and NheI (NEB), to create pDES-EC-tet, the Tet lox-destination vector, (see Fig. 1B for arrangement). The SacB lox-destination vector (pDES-EC-SacB), was created by recombining pDAN11-CAP/SacB with pDES-EC-tet, and testing the recombination products by PCR, using primers specific for SacB and the E coil.
Phagemid ELISA
Nunc Maxisorp ELISA plates (Fisher Scientific) were coated with 100 μL of antigen or anti-tag antibody overnight at 4°C (anti-FLAG 10 μg/mL, anti-SV5 monoclonal antibodies 10 μg/mL, lysozyme 50 μg/mL, and SNV-N 5 μg/mL). Plates were rinsed and blocked with either 2% BSA or 4% fish gelatin (Sigma) for 1 h. Phagemid particles were added to each well and incubated at room temperature for 1.5 h. Bound phagemid were rinsed three times with PBST (PBS containing 0.1% Tween 20) and three times with PBS, and detected using 1:5000 dilution of anti-M13 horseradish peroxidase conjugate (Pharmacia).
scFv Expression
Single colonies of scFv expressing plasmids were inoculated into 1 mL of 2xTY/Ampicillin 100 μg/mL, 1% glucose, and grown overnight at 37°C. A total of 10 μL of this pre-culture was diluted into 1 mL of the same medium and grown to OD600 0.5 at 37°C. Cells were harvested and resuspended into 1 mL of 2xTY/Amp/250 μM IPTG, and grown overnight at 30°C. Cells were harvested, and the supernatants were collected. scFvs were also isolated from the periplasm by osmotic shock; the cell pellet was resuspended in 20% sucrose (1/40 culture volume), centrifuged for 6 min 6000g, and the supernatant retained and mixed with the supernatant obtained after resuspending the cell pellet in 5 mM MgSO4 (1/100 culture volume) and recentrifuging. If needed, scFvs were purified using the His tag and Nickel-NTA agarose beads (Qiagen) according to manufacturer's instructions.
scFv ELISA
Nunc Maxisorp ELISA plates (Fisher Scientific) were coated with 100 μL of antigen overnight at 4°C. The antigens used in this study were SNV-N, 5 μg/mL, E-coil GFP, 10 μg/mL, and K-coil GFP, 10 μg/mL. E- and K-coil GFP are GFP that have been tagged with E or K coils (Chao et al. 1996). Plates were rinsed and blocked with either 2% BSA or 4% fish gelatin (Sigma) for 1 h. Supernatant containing scFv were added to each well and incubated at room temperature for 1.5 h. Washing was carried out three times with PBST and three times with PBS, and scFvs were detected using anti-SV5 tag antibody followed by anti-mouse horseradish peroxidase conjugate (DAKO Corp.). When coiled coil ELISAs were carried out (to demonstrate functionality of both SV5 tag and coiled coil), 100 μL of 10-μg/mL concentration antigens were coated on Nucn Maxisorp plates for 1 h at 37°C. The plates were blocked with 200 μL of 4.5% fish gelatin for 1 h. Meanwhile, an equal amount of K coil-Alkaline phosphatase was incubated with either E-coil-scFvs or PBS and allowed to form the E/K coil interaction for 15 min. The mixture was then allowed to bind to antigens for 1.5 h in the presence of 1% fish gelatin. The plate was washed three times with PBST, three times with PBS, and the AP Elisa substrate (phosphatase substrate kit – Pierce) was added. The ELISA signals were read at 405 nm.
Negative Selection Marker Titration
Cells transformed with either destination vector (Tet or SacB) or a control vector encoding only resistance to chloramphenicol (pDAN11-CAP) were grown to OD600 = 1 and 37°C and plated onto selective medium. The Tet destination vector cells were plated on chloamphenicol (100 μg/mL) 2XTY plates containing 0.5%, 1%, 2%, 3%, or 4% NiCl2. The SacB destination vector cells were plated on chloramphenicol (100 μg/mL) 2XTY plates containing 0%, 2.5%, 5%, 7.5%, or 10% sucrose. After overnight growth at 37°C, surviving colonies were counted and compared with the number of colonies obtained on chloramphenicol plates without the negative selector.
Cre Recombination
BS1365 cells (constitutively expressing Cre recombinase) were grown to an OD600 of 0.5 at 37°C and cotransduced with pDAN11-D1.3 and either lox-destination vectors at a multiplicity of infection (MOI) of 10:1 for each. Infected cells were diluted fivefold into 2xYT/chloramphenicol 100 μg/mL/Amp 100 μg/mL/3% glucose, and allowed to recombine at 37°C until an OD600 of 0.5 was again achieved (typically 2–4 h). Cells were then infected with M13K07 helper phage for 30 min at 37°C and grown overnight at 30°C in 2xYT/chloramphenicol 100 μg/mL/Amp 100 μg/mL/Kanamycin 100 μg/mL to produce phagemid particles containing the various recombination products. Recombination could also be carried out overnight, and the culture was diluted 20-fold and grown to an OD600 of 0.5 prior to helper phage infection. Lox-destination vectors containing the D1.3, 3A, F4 scFv gene were specifically obtained by infecting DH5aF' cells at an OD600 of 0.5 with the recombined phagemid at an MOI of 1:1 for 30 min at 37°C and plating on 2xYT agar containing 100 μg/mL chloramphenicol with the appropriate negative selection reagent (3 mM NiCl2 or 8% sucrose). The accuracy of recombination was determined by carrying out PCR using primers annealing separately in the two vectors prior to recombination, and together in the correct product of recombination. For transfer of D1.3 from pDAN11 to pDES-EC, VL D1.3 CDR3 5′ (TTTGG AGTACTCCTCGG) and RS 1 rev (CGGAATTCGGATCCTTAGT GATGGTGATGGTGATGCTTCTCAAGAGCGCTCACTTCCTTT) were used, for transfer of 3A and F4 from pDAN11 to pDES-EC, pDpH-5′ (GCAGCCGCTGGATTGTTATTA) and RS 1 rev were used.
scFv Western Analysis
scFv supernatants were denatured in 2-mercapthoethanol containing sample loading buffer for 3 min at 100°C and loaded onto NuPAGE gradient 4%–12% Bis-Tris SDS gel (Invitrogen). Proteins were then transferred to nitrocellulose membrane (Schleicher and Schuell), blocked with 4% fish gelatin (Sigma), incubated with anti-SV5 mouse monoclonal antibody and goat anti-mouse IgG-AP conjugate (Dako Corp.), following by incubation in 1 Step NBT/BCIP (Pierce Inc.).
Acknowledgments
We thank Dr. Clarence I. Kado, University of California, Davis, for the kind donation of SacB plasmid, Dr. Brian Sauer for BS1365 for the cre expression strain, Drs. Jim Marks and Jianlong Lou for the JL-1 Cre-Tet bacterial strain, Dr. Roberto Marzari and Dr. Fiorella Florian for the pDAN8 vector, Dr. Geoff Waldo for enlightened discussions, and Ms. Priya Dighe for technical support. This work was supported by DOE grants DE-FG02-98ER62647 and 06SCP1002, and LANL LDRD-DR grant X1MF, both awarded to A.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1821804.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C.I. Kado, B. Sauer, J. Marks, J. Lou, R. Marzari, and F. Florian.]
References
- Abremski, K. and Hoess, R. 1984. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J. Biol. Chem. 259: 1509–1514. [PubMed] [Google Scholar]
- Alexeyev, M., Shokolenko, I., and Croughan, T. 1995. Improved antibiotic-resistance gene cassettes and ω elements for Eschericia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160: 63–67. [DOI] [PubMed] [Google Scholar]
- Arndt, K.M., Pelletier, J.N., Muller, K.M., Alber, T., Michnick, S.W., and Pluckthun, A. 2000. A heterodimeric coiled-coil peptide pair selected in vivo from a designed library-versus-library ensemble. J. Mol. Biol. 295: 627–639. [DOI] [PubMed] [Google Scholar]
- Bach, H., Mazor, Y., Shaky, S., Shoham-Lev, A., Berdichevsky, Y., Gutnick, D.L., and Benhar, I. 2001. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J. Mol. Biol. 312: 79–93. [DOI] [PubMed] [Google Scholar]
- Berdichevsky, Y., Ben-Zeev, E., Lamed, R., and Benhar, I. 1999. Phage display of a cellulose binding domain from Clostridium thermocellum and its application as a tool for antibody engineering. J. Immunol. Methods 228: 151–162. [DOI] [PubMed] [Google Scholar]
- Bernard, P. 1996. Positive selection of recombinant DNA by CcdB. Biotechniques 21: 320–323. [DOI] [PubMed] [Google Scholar]
- Bethke, B. and Sauer, B. 1997. Segmental genomic replacement by Cre-mediated recombination: Genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res. 25: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, R.E., Hardman, K.D., Jacobson, J.W., Johnson, S., Kaufman, B.M., Lee, S.M., Lee, T., Pope, S.H., Riordan, G.S., and Whitlow, M. 1988. Single-chain antigen-binding proteins [published erratum appears in Science 1989 Apr. 28;244(4903):409]. Science 242: 423–426. [DOI] [PubMed] [Google Scholar]
- Burnet, F.M. 1959. The clonal selection theory of acquired immunity. The University Press, Cambridge, UK.
- Cattaneo, A., Rapposelli, B., and Calissano, P. 1988. Three distinct types of monoclonal antibodies after long term immunization of rats with mouse NGF. J. Neurochem. 50: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Chao, H., Houston Jr., M.E., Grothe, S., Kay, C.M., O'Connor-McCourt, M., Irvin, R.T., and Hodges, R.S. 1996. Kinetic study on the formation of a de novo designed heterodimeric coiled-coil: Use of surface plasmon resonance to monitor the association and dissociation of polypeptide chains. Biochemistry 35: 12175–12185. [DOI] [PubMed] [Google Scholar]
- Chao, H., Bautista, D.L., Litowski, J., Irvin, R.T., and Hodges, R.S. 1998. Use of a heterodimeric coiled-coil system for biosensor application and affinity purification. J. Chromatogr. B Biomed. Sci. Appl. 715: 307–329. [DOI] [PubMed] [Google Scholar]
- Dalum, I., Jensen, M.R., Hindersson, P., Elsner, H.I., and Mouritsen, S. 1996. Breaking of B cell tolerance toward a highly conserved self protein. J. Immunol. 157: 4796–4804. [PubMed] [Google Scholar]
- Dalum, I., Jensen, M.R., Gregorius, K., Thomasen, C.M., Elsner, H.I., and Mouritsen, S. 1997. Induction of cross-reactive antibodies against a self protein by immunization with a modified self protein containing a foreign T helper epitope. Mol. Immunol. 34: 1113–1120. [DOI] [PubMed] [Google Scholar]
- De Crescenzo, G., Litowski, J.R., Hodges, R.S., and O'Connor-McCourt, M.D. 2003. Real-time monitoring of the interactions of two-stranded de novo designed coiled-coils: Effect of chain length on the kinetic and thermodynamic constants of binding. Biochemistry 42: 1754–1763. [DOI] [PubMed] [Google Scholar]
- de Haard, H.J., van Neer, N., Reurs, A., Hufton, S.E., Roovers, R.C., Henderikx, P., de Bruine, A.P., Arends, J.W., and Hoogenboom, H.R. 1999. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 274: 18218–18230. [DOI] [PubMed] [Google Scholar]
- Evan, G.I., Lewis, G.K., Ramsay, G., and Bishop, J.M. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell Biol. 5: 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaus, M.J., Siegel, R.W., Opresko, L.K., Coleman, J.R., Feldhaus, J.M., Yeung, Y.A., Cochran, J.R., Heinzelman, P., Colby, D., Swers, J., et al. 2003. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat. Biotechnol. 21: 163–170. [DOI] [PubMed] [Google Scholar]
- Feng, Y.Q., Seibler, J., Alami, R., Eisen, A., Westerman, K.A., Leboulch, P., Fiering, S., and Bouhassira, E.E. 1999. Site-specific chromosomal integration in mammalian cells: Highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 292: 779–785. [DOI] [PubMed] [Google Scholar]
- Freyre, F.M., Vazquez, J.E., Ayala, M., Canaan-Haden, L., Bell, H., Rodriguez, I., Gonzalez, A., Cintado, A., and Gavilondo, J.V. 2000. Very high expression of an anti-carcinoembryonic antigen single chain Fv antibody fragment in the yeast Pichia pastoris. J. Biotechnol. 76: 157–163. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gay, P., Le Coq, D., Steinmetz, M., Berkelman, T., and Kado, C.I. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164: 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep, R.A., van Twisk, C., Kerschbaumer, R.J., Harper, K., Torrance, L., Himmler, G., van der Wolf, J.M., and Schots, A. 1999. pSKAP/S: An expression vector for the production of single-chain Fv alkaline phosphatase fusion proteins. Protein Expr. Purif. 16: 63–69. [DOI] [PubMed] [Google Scholar]
- Griffiths, A.D., Williams, S.C., Hartley, O., Tomlinson, I.M., Waterhouse, P., Crosby, W.L., Kontermann, R.E., Jones, P.T., Low, N.M., Alison, T.J., et al. 1994. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 13: 3245–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke, T., Szawlowski, P., and Randall, R.E. 1992. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J. Gen. Virol. 73: 653–660. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink, M.A., Griep, R.A., Borst, J.W., van Hoek, A., Eppink, M.H., Schots, A., and Visser, A.J. 2000. Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. J. Biol. Chem. 275: 17556–17560. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Hoess, R.H. and Abremski, K. 1984. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. 81: 1026–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess, R., Wierzbicki, A., and Abremski, K. 1986. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 14: 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston, J.S., Levinson, D., Mudgett, H.M., Tai, M.S., Novotny, J., Margolies, M.N., Ridge, R.J., Bruccoleri, R.E., Haber, E., Crea, R., et al. 1988. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. 85: 5879–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber, A., Bornhauser, S., Burmester, J., Honeggar, A., Willuda, J., Bosshard, H.R., and Pluckthun, A. 1997. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods 201: 35–55. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Agarwal, S., Heyman, J.A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., et al. 2002. Subcellular localization of the yeast proteome. Genes & Dev. 16: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy, A. 1989. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu. Rev. Biochem. 58: 913–949. [DOI] [PubMed] [Google Scholar]
- Lee, G. and Saito, I. 1998. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216: 55–65. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8: 1300–1309. [DOI] [PubMed] [Google Scholar]
- Malecki, M., Hsu, A., Truong, L., and Sanchez, S. 2002. Molecular immunolabeling with recombinant single-chain variable fragment (scFv) antibodies designed with metal-binding domains. Proc. Natl. Acad. Sci. 99: 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty, J., Griffiths, A.D., Winter, G., and Chiswell, D.J. 1990. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 348: 552–554. [DOI] [PubMed] [Google Scholar]
- Morino, K., Katsumi, H., Akahori, Y., Iba, Y., Shinohara, M., Ukai, Y., Kohara, Y., and Kurosawa, Y. 2001. Antibody fusions with fluorescent proteins: A versatile reagent for profiling protein expression. J. Immunol. Methods 257: 175–184. [DOI] [PubMed] [Google Scholar]
- Muller, B.H., Chevrier, D., Boulain, J.C., and Guesdon, J.L. 1999. Recombinant single-chain Fv antibody fragment-alkaline phosphatase conjugate for one-step immunodetection in molecular hybridization. J. Immunol. Methods 227: 177–185. [DOI] [PubMed] [Google Scholar]
- Nemazee, D. 2000. Receptor selection in B and T lymphocytes. Annu. Rev. Immunol. 18: 19–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack, P. and Pluckthun, A. 1992. Miniantibodies: Use of amphipathic helices to produce functional, flexibly linked dimeric FV fragments with high avidity in Escherichia coli. Biochemistry 31: 1579–1584. [DOI] [PubMed] [Google Scholar]
- Pelletier, J.N., Arndt, K.M., Pluckthun, A., and Michnick, S.W. 1999. An in vivo library-versus-library selection of optimized protein–protein interactions. Nat. Biotechnol. 17: 683–690. [DOI] [PubMed] [Google Scholar]
- Persic, L., Righi, M., Roberts, A., Hoogenboom, H.R., Cattaneo, A., and Bradbury, A. 1997a. Targeting vectors for intracellular immunization. Gene 187: 1–8. [DOI] [PubMed] [Google Scholar]
- Persic, L., Roberts, A., Wilton, J., Cattaneo, A., Bradbury, A., and Hoogenboom, H. 1997b. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene 187: 9–18. [DOI] [PubMed] [Google Scholar]
- Podolsky, T., Fong, S.T., and Lee, B.T. 1996. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid 36: 112–115. [DOI] [PubMed] [Google Scholar]
- Powers, D.B., Amersdorfer, P., Poul, M., Nielsen, U.B., Shalaby, M.R., Adams, G.P., Weiner, L.M., and Marks, J.D. 2001. Expression of single-chain Fv-Fc fusions in Pichia pastoris. J. Immunol. Methods 251: 123–135. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332–336. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald, P., Coelho, P.S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.H., Sheehan, A., Symoniatis, D., Umansky, L., et al. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413–418. [DOI] [PubMed] [Google Scholar]
- Sauer, B. 1992. Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. J. Mol. Biol. 223: 911–928. [DOI] [PubMed] [Google Scholar]
- Sauer, B. and Henderson, N. 1988. The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene 70: 331–341. [DOI] [PubMed] [Google Scholar]
- Sblattero, D. and Bradbury, A. 1998. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology 3: 271–278. [DOI] [PubMed] [Google Scholar]
- Sblattero, D. and Bradbury, A. 2000. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 18: 75–80. [DOI] [PubMed] [Google Scholar]
- Schwikowski, B., Uetz, P., and Fields, S. 2000. A network of protein–protein interactions in yeast. Nat. Biotechnol. 18: 1257–1261. [DOI] [PubMed] [Google Scholar]
- Sheets, M.D., Amersdorfer, P., Finnern, R., Sargent, P., Lindqvist, E., Schier, R., Hemmingsen, G., Wong, C., Gerhart, J.C., and Marks, J.D. 1998. Efficient construction of a large nonimmune phage antibody library; the production of panels of high affinity human single-chain antibodies to protein antigens. Proc. Natl. Acad. Sci. 95: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.W., Jain, R., and Bradbury, A. 2001. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 505: 467–473. [DOI] [PubMed] [Google Scholar]
- Skerra, A. 2000. Engineered protein scaffolds for molecular recognition. J. Mol. Recognit. 13: 167–187. [DOI] [PubMed] [Google Scholar]
- Talmage, D.W. 1959. Immunological specificity, unique combinations of selected natural globulins provide an alternative to the classical concept. Science 129: 1643–1648. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Drees, B., Nardelli, G., Bader, G.D., Brannetti, B., Castagnoli, L., Evangelista, M., Ferracuti, S., Nelson, B., Paoluzi, S., et al. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295: 321–324. [DOI] [PubMed] [Google Scholar]
- Trinh, K.R. and Morrison, S.L. 2000. Site-specific and directional gene replacement mediated by cre recombinase. J. Immunol. Methods 244: 185–193. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Vaughan, T.J., Williams, A.J., Pritchard, K., Osbourn, J.K., Pope, A.R., Earnshaw, J.C., McCafferty, J., Hodits, R.A., Wilton, J., and Johnson, K.S. 1996. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 14: 309–314. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575–592. [DOI] [PubMed] [Google Scholar]
- Werlen, G., Hausmann, B., Naeher, D., and Palmer, E. 2003. Signaling life and death in the thymus: Timing is everything. Science 299: 1859–1863. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293: 2101–2105. [DOI] [PubMed] [Google Scholar]